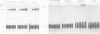Abstract
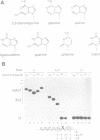
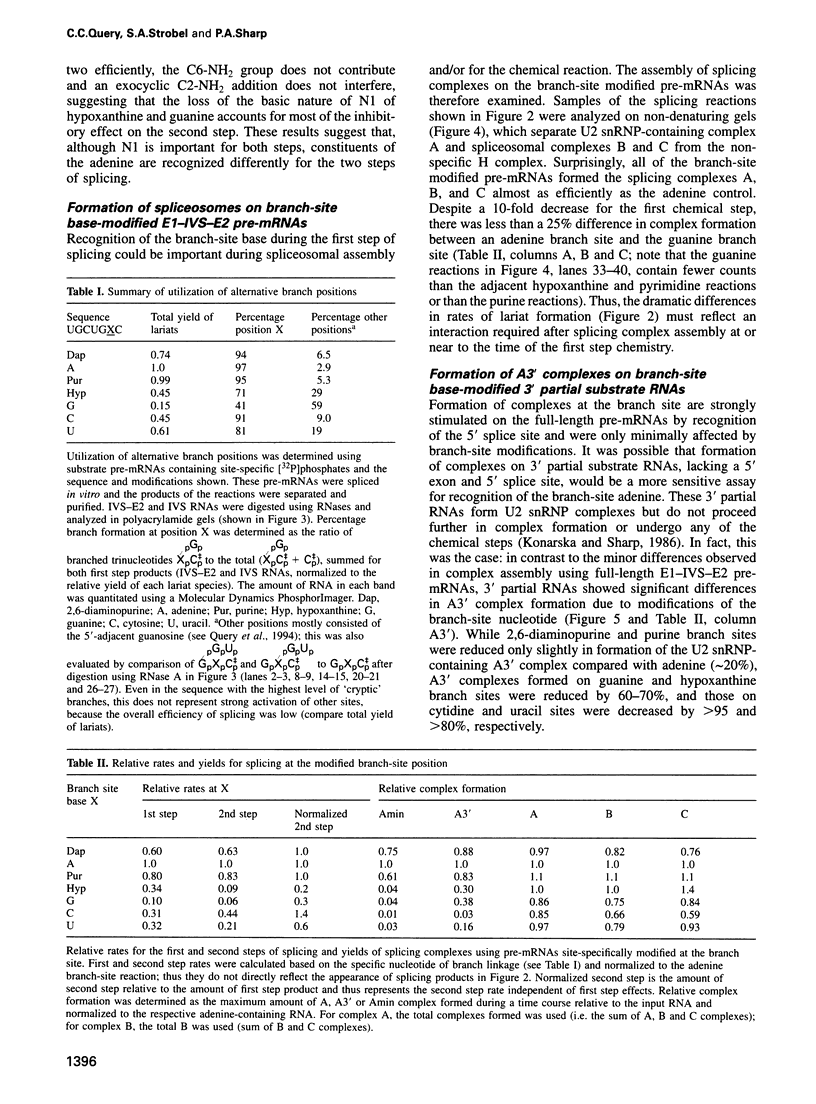
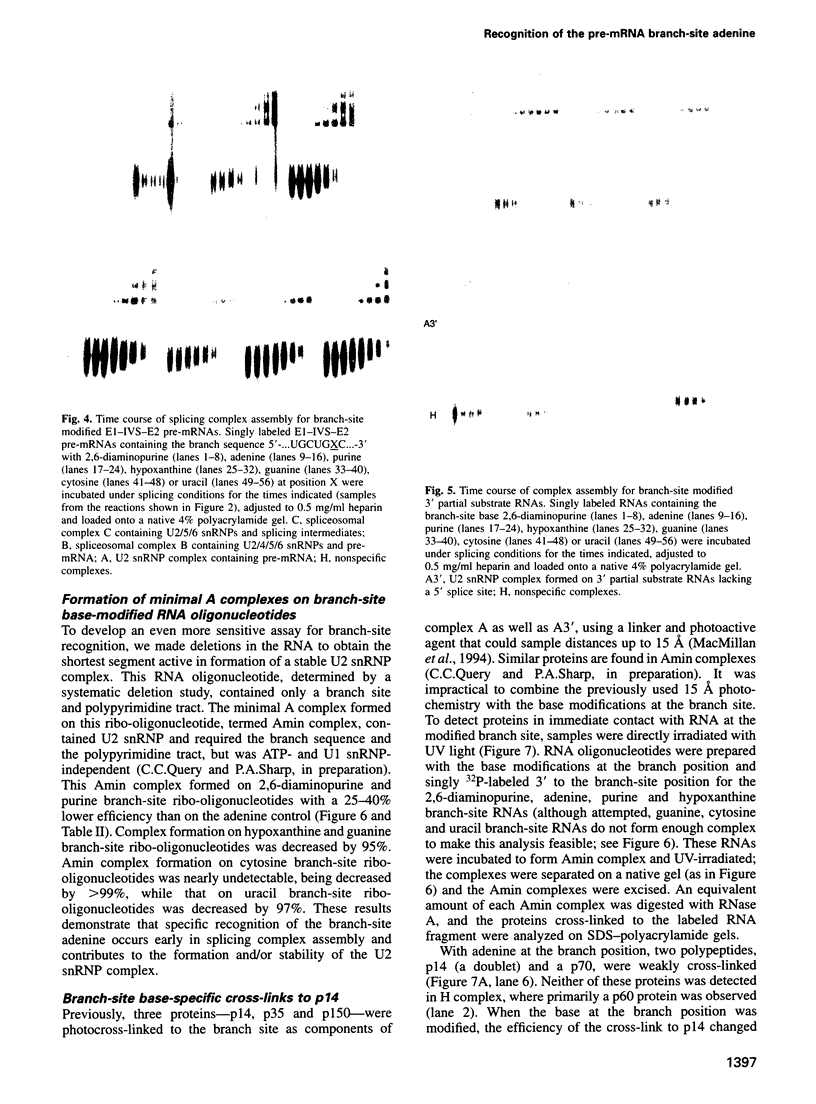
An adenosine at the branch site, the nucleophile for the first transesterification step of splicing, is nearly invariant in mammalian pre-mRNA introns. The chemical groups on the adenine base were varied systematically and assayed for formation of early spliceosome complexes and execution of the first and second steps of splicing. Recognition of constituents of the adenine is critical in formation of a U2 snRNP-containing complex on a minimal branch-site oligonucleotide. Furthermore, the efficiencies of the first and second chemical steps have different dependencies on the functional groups of the adenine. In total, the chemical groups on the adenine base at the branch site are differentially recognized during at least three different processes in the splicing of pre-mRNA. Moreover, a protein, p14, interacts with the adenine in a base-specific fashion and may mediate early recognition of this base.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abovich N., Liao X. C., Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994 Apr 1;8(7):843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- Adema G. J., Bovenberg R. A., Jansz H. S., Baas P. D. Unusual branch point selection involved in splicing of the alternatively processed Calcitonin/CGRP-I pre-mRNA. Nucleic Acids Res. 1988 Oct 25;16(20):9513–9526. doi: 10.1093/nar/16.20.9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adema G. J., van Hulst K. L., Baas P. D. Uridine branch acceptor is a cis-acting element involved in regulation of the alternative processing of calcitonin/CGRP-l pre-mRNA. Nucleic Acids Res. 1990 Sep 25;18(18):5365–5373. doi: 10.1093/nar/18.18.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens S. E., Galisson F., Legrain P., Lührmann R. Evidence that the 60-kDa protein of 17S U2 small nuclear ribonucleoprotein is immunologically and functionally related to the yeast PRP9 splicing factor and is required for the efficient formation of prespliceosomes. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):8229–8233. doi: 10.1073/pnas.90.17.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosi R., Gröning K., Behrens S. E., Lührmann R., Krämer A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993 Oct 1;262(5130):102–105. doi: 10.1126/science.8211112. [DOI] [PubMed] [Google Scholar]
- Brosi R., Hauri H. P., Krämer A. Separation of splicing factor SF3 into two components and purification of SF3a activity. J Biol Chem. 1993 Aug 15;268(23):17640–17646. [PubMed] [Google Scholar]
- Burgess S. M., Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993 Jul 2;73(7):1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- Champion-Arnaud P., Reed R. The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev. 1994 Aug 15;8(16):1974–1983. doi: 10.1101/gad.8.16.1974. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Sharp P. A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989 Dec 1;59(5):789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- Couto J. R., Tamm J., Parker R., Guthrie C. A trans-acting suppressor restores splicing of a yeast intron with a branch point mutation. Genes Dev. 1987 Jul;1(5):445–455. doi: 10.1101/gad.1.5.445. [DOI] [PubMed] [Google Scholar]
- Deirdre A., Scadden J., Smith C. W. Interactions between the terminal bases of mammalian introns are retained in inosine-containing pre-mRNAs. EMBO J. 1995 Jul 3;14(13):3236–3246. doi: 10.1002/j.1460-2075.1995.tb07326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyhr-Mikkelsen H., Kjems J. Inefficient spliceosome assembly and abnormal branch site selection in splicing of an HIV-1 transcript in vitro. J Biol Chem. 1995 Oct 13;270(41):24060–24066. doi: 10.1074/jbc.270.41.24060. [DOI] [PubMed] [Google Scholar]
- Freyer G. A., Arenas J., Perkins K. K., Furneaux H. M., Pick L., Young B., Roberts R. J., Hurwitz J. In vitro formation of a lariat structure containing a G2'-5'G linkage. J Biol Chem. 1987 Mar 25;262(9):4267–4273. [PubMed] [Google Scholar]
- Grabowski P. J., Padgett R. A., Sharp P. A. Messenger RNA splicing in vitro: an excised intervening sequence and a potential intermediate. Cell. 1984 Jun;37(2):415–427. doi: 10.1016/0092-8674(84)90372-6. [DOI] [PubMed] [Google Scholar]
- Hartmuth K., Barta A. Unusual branch point selection in processing of human growth hormone pre-mRNA. Mol Cell Biol. 1988 May;8(5):2011–2020. doi: 10.1128/mcb.8.5.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig H., Aebi M., Weissmann C. Effect of mutations at the lariat branch acceptor site on beta-globin pre-mRNA splicing in vitro. Nature. 1986 Dec 11;324(6097):589–591. doi: 10.1038/324589a0. [DOI] [PubMed] [Google Scholar]
- Jamison S. F., Garcia-Blanco M. A. An ATP-independent U2 small nuclear ribonucleoprotein particle/precursor mRNA complex requires both splice sites and the polypyrimidine tract. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5482–5486. doi: 10.1073/pnas.89.12.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987 Jun 19;49(6):763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- Lamm G. M., Lamond A. I. Non-snRNP protein splicing factors. Biochim Biophys Acta. 1993 Jun 25;1173(3):247–265. doi: 10.1016/0167-4781(93)90122-t. [DOI] [PubMed] [Google Scholar]
- MacMillan A. M., Query C. C., Allerson C. R., Chen S., Verdine G. L., Sharp P. A. Dynamic association of proteins with the pre-mRNA branch region. Genes Dev. 1994 Dec 15;8(24):3008–3020. doi: 10.1101/gad.8.24.3008. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- Michaud S., Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 1991 Dec;5(12B):2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- Moore M. J., Sharp P. A. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature. 1993 Sep 23;365(6444):364–368. doi: 10.1038/365364a0. [DOI] [PubMed] [Google Scholar]
- Moore M. J., Sharp P. A. Site-specific modification of pre-mRNA: the 2'-hydroxyl groups at the splice sites. Science. 1992 May 15;256(5059):992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- Nilsen T. W. RNA-RNA interactions in the spliceosome: unraveling the ties that bind. Cell. 1994 Jul 15;78(1):1–4. doi: 10.1016/0092-8674(94)90563-0. [DOI] [PubMed] [Google Scholar]
- Noble J. C., Prives C., Manley J. L. Alternative splicing of SV40 early pre-mRNA is determined by branch site selection. Genes Dev. 1988 Nov;2(11):1460–1475. doi: 10.1101/gad.2.11.1460. [DOI] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G. Evidence for an essential non-Watson-Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature. 1993 Feb 18;361(6413):660–662. doi: 10.1038/361660a0. [DOI] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G., Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987 Apr 24;49(2):229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Query C. C., Moore M. J., Sharp P. A. Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev. 1994 Mar 1;8(5):587–597. doi: 10.1101/gad.8.5.587. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988 Oct;2(10):1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science. 1988 Nov 18;242(4881):1028–1035. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Zamore P. D., Green M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988 Jan 29;52(2):207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Seraphin B., Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989 Oct 20;59(2):349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Staknis D., Reed R. Direct interactions between pre-mRNA and six U2 small nuclear ribonucleoproteins during spliceosome assembly. Mol Cell Biol. 1994 May;14(5):2994–3005. doi: 10.1128/mcb.14.5.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel S. A., Cech T. R., Usman N., Beigelman L. The 2,6-diaminopurine riboside.5-methylisocytidine wobble base pair: an isoenergetic substitution for the study of G.U pairs in RNA. Biochemistry. 1994 Nov 22;33(46):13824–13835. doi: 10.1021/bi00250a037. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Rosbash M. The yeast branchpoint sequence is not required for the formation of a stable U1 snRNA-pre-mRNA complex and is recognized in the absence of U2 snRNA. EMBO J. 1991 May;10(5):1209–1216. doi: 10.1002/j.1460-2075.1991.tb08062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace J. C., Edmonds M. Polyadenylylated nuclear RNA contains branches. Proc Natl Acad Sci U S A. 1983 Feb;80(4):950–954. doi: 10.1073/pnas.80.4.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Zamore P. D., Patton J. G., Green M. R. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992 Feb 13;355(6361):609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- Zhuang Y. A., Goldstein A. M., Weiner A. M. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989 Oct;3(10):1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]
- van Oers C. C., Adema G. J., Zandberg H., Moen T. C., Baas P. D. Two different sequence elements within exon 4 are necessary for calcitonin-specific splicing of the human calcitonin/calcitonin gene-related peptide I pre-mRNA. Mol Cell Biol. 1994 Feb;14(2):951–960. doi: 10.1128/mcb.14.2.951. [DOI] [PMC free article] [PubMed] [Google Scholar]