Abstract
In the past decade Clostridium difficile has become a bacterial pathogen of global significance. Epidemic strains have spread throughout hospitals, while community acquired infections and other sources ensure a constant inoculation of spores into hospitals. In response to the increasing medical burden, a new C. difficile antibiotic, fidaxomicin, was approved in 2011 for the treatment of C. difficile-associated diarrhea. Rudimentary fecal transplants are also being trialed as effective treatments. Despite these advances, therapies that are more effective against C. difficile spores and less damaging to the resident gastrointestinal microbiome and that reduce recurrent disease are still desperately needed. However, bringing a new treatment for C. difficile infection to market involves particular challenges. This review covers the current drug discovery pipeline, including both small molecule and biologic therapies, and highlights the challenges associated with in vitro and in vivo models of C. difficile infection for drug screening and lead optimization.
1. Introduction
1.1. Epidemiology
Clostridium difficile is a Gram-positive spore forming anaerobic bacterium that has become a significant problem in health care settings and in the community in recent years. It was recognized as an urgent threat to human health in a 2013 CDC report on antibiotic resistance.1 Patients are susceptible to infection when there is a disturbance in the healthy gut microbiome, often due to prior oral antibiotic use, which permits C. difficile to colonize and cause disease in the gastrointestinal tract. C. difficile infection (CDI) severity varies from self-limiting mild diarrhea to severe life-threatening pseudomembranous colitis and toxic megacolon (inflamed colon with abdominal distension). The C. difficile glycosylating toxins, toxin A (TcdA) and toxin B (TcdB), are important virulence factors that promote epithelial tissue damage and inflammation in the infected host, resulting in rapid fluid loss into the intestinal epithelium and diarrhea.2 Some strains produce an additional toxin, binary toxin or CDT. CDT is prevalent in strains commonly associated with severe disease, such as BI/NAP1/027 and ribotype 078 isolates, although the role of this toxin in disease remains undefined (review Gerding et al.3).
C. difficile spores are an ideal vehicle for transmission between patients because they persist in the environment for long periods and are resistant to heat and typical disinfectants such as alcohol based hand washes.4,5 Spores are ingested from the environment and germinate in response to bile salts in the small intestine. The resulting vegetative cells colonize the colon and produce toxins that cause disease symptoms.2 An epidemic fluoroquinolone-resistant group of strains, belonging to the BI/NAP1/027 class, is associated with more severe disease and increased death rates6 and has spread rapidly throughout hospitals and community care facilities at a global scale.7−9 Community acquired C. difficile infection (CA-CDI) rates have also increased with 40% of CA-CDI patients requiring hospitalization, providing a recurrent source of spores in hospitals and making complete eradication of the disease in hospitals a challenging task.10
1.2. Current Antibiotic Therapies
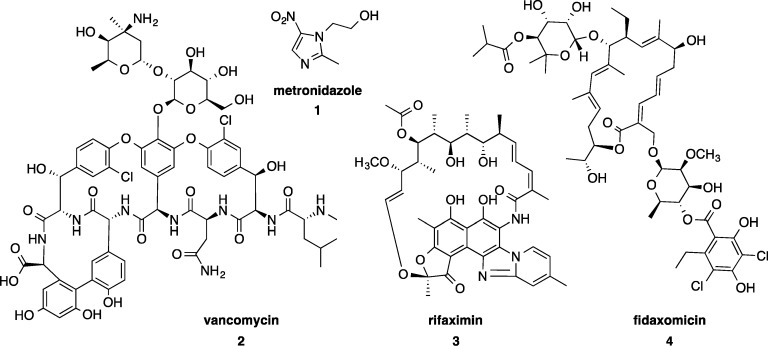
The first line treatment for C. difficile infection is antibiotics, either metronidazole 1 for mild to moderate infection or oral vancomycin 2 for moderate to severe infection (Figure 1). Both of these drugs are generic and have been on the market for over 40 years.11,12 Unfortunately in 14–27% of cases they do not effectively treat the infection or prevent relapsing infection.13 Rifaximin 3 is sometimes used as a “chaser therapy”, following initial treatment.14 Fidaxomicin 4 (Figure 1) is the first new drug on the market specifically designed to treat C. difficile and has been available since 2011. It offers improvements on relapse rates15 by reducing collateral damage to the resident gut microbiota because it is more selective for C. difficile(16,17) over potentially beneficial bacteria, thereby encouraging the suppression of C. difficile colonization and proliferation, as well as purportedly inhibiting spore formation.18
Figure 1.
Current antibiotic treatments primarily used to treat for C. difficile infection: metronidazole 1, vancomycin 2, rifaximin 3 (sometimes used as a chaser therapy), and fidaxomicin 4.
1.2.1. Metronidazole
Metronidazole, a nitroimidazole, is active against a wide spectrum of anaerobic bacteria and parasites. Reduction of the nitro functional group in metronidazole initiates decomposition to toxic radical species. The nitro functional group scavenges electrons from electron carriers, such as reduced ferredoxin, which are at a lower reduction potential than their respective protein homologues in facultative anaerobic bacteria. Reduction forms an unstable nitro radical anion, which in most cases decomposes rapidly to nitrite.19,20 This reduction consumes the compound and drives further uptake into the cell.20 The nitrite and the radical imidazole that form cause damage to bacterial DNA leading to cell death.20 An alternative nitro group reduction pathway via nitroso and hydroxylamine intermediates to the amine is less likely because of the high energy barrier of this process.20 Oral metronidazole is essentially 100% bioavailable, with the systemic absorption resulting in reduced concentrations in the colon that approach the minimal inhibition concentration (MIC) in the colon.21 The relatively low concentration of compound at the site of infection due to systemic absorption is thought to contribute to reduced efficacy in moderate to severe cases of CDI and toward the development of resistance.22
1.2.2. Vancomycin
Vancomycin, a glycopeptide antibiotic, inhibits cell wall synthesis in Gram-positive bacteria by binding to cell wall building blocks. Specifically, vancomycin binds to the C-terminus of polypeptide intermediates terminating in d-alanyl d-alanine (d-Ala-d-Ala), particularly the peptidoglycan precursor lipid II.23 This binding blocks transglycosylase enzymes from transferring the pentapeptide of lipid II to a polyglycan chain. Vancomycin can also bind to the pentapeptide on the polyglycan, inhibiting transpeptidases from linking adjacent pentapeptides to create the cross-linked framework of peptidoglycan.23 Vancomycin is known to dimerize which increases the affinity to the d-Ala-d-Ala terminated pentapeptide.24 Vancomycin is given orally and is minimally systemically absorbed, resulting in high concentrations achieved in the colon.22 The broad-spectrum activity of vancomycin against Gram-positive bacteria contributes to a reduction in microbiome diversity compared to fidaxomicin treatment.16
1.2.3. Rifaximin
The RNA synthesis inhibitor rifaximin is most commonly used to treat CDI as a chaser antimicrobial therapy after an initial dose of vancomycin. It is nonabsorbable and has minimal systemic effects. It is relatively selective on the gastrointestinal microbiota, although the concentrations of administered compound achieved in the gut are likely to cause inhibition of bacteria other than C. difficile.25,26
1.2.4. Fidaxomicin
The macrolide fidaxomicin inhibits RNA synthesis by RNA polymerase. Unlike the broad-spectrum mechanisms of action of metronidazole and vancomycin, fidaxomicin shows a narrower spectrum of activity selective for C. difficile over other gut microbes.27 Fidaxomicin also inhibits spore production,18 with both effects believed to account for reduced relapse rates observed clinically. For example, in comparison to vancomycin, fidaxomicin had 52% fewer second occurrence relapse rates by 28 days after infection in patients with no prior episode of CDI (22.6% of vancomycin treated patients relapsed vs 11.7% of fidaxomicin treated patients).15 A second meta-analysis reported a 40% reduction in persistent diarrhea, recurrence, or death over 40 days.28 These figures demonstrate the improvement in clinical outcomes that can occur with a treatment specifically developed to target C. difficile. Nevertheless, there is still room for further improvement as fidaxomicin fails in 12% of treatments (in contrast to 14–27% treatment failures with vancomycin and metronidazole).13,28 Fidaxomicin is minimally absorbed, which means systemic side effects are avoided despite its cytotoxicity, which is similar to tamoxifen against breast cancer cell lines.29
1.3. Drug Resistance
Metronidazole, vancomycin, and fidaxomicin drug resistance in C. difficile is not widespread at this time. However, resistance has been observed and is of concern given the increased prevalence of C. difficile infections over the past decade and the subsequent requirement for antibiotics now and in the future.30−32 Metronidazole resistance of >32 μg/mL has been reported, but resistance is unstable and is often lost on passaging or freeze–thawing of bacteria.30,33,34 The MIC values observed of >32 μg/mL are likely outside the therapeutic concentrations achieved in the colon, since the concentration of metronidazole in the feces of patients with CDI has been found to be 0.8–24.2 μg/g of stool.21 Reduced susceptibility to vancomycin has been seen, with C. difficile isolates with a MIC of 4 μg/mL reported (compared to sensitive strains with MIC range of ≤0.5–2 μg/mL).31 In clinical trials fidaxomicin resistance up to 16 μg/mL was observed, a significant increase over the normal MIC range of 0.003–1 μg/mL.35 The MIC of fidaxomicin increased to 2 μg/mL against C. difficile in a 13-generation forced resistance study.36 For fidaxomicin, like the rifamycin antibiotics, single amino acid substitutions in the protein targets cause high resistance (MIC > 256 μg/mL), rendering the antibiotic ineffective.37 The clinical relevance of elevated MICs to vancomycin and fidaxomicin is unclear given the high concentrations (generally >1000 μg/g) achieved in the feces.38,39 However, considering the relative ease that C. difficile strains have spread globally, any development of resistance is of concern, particularly if no other new drugs are approved for C. difficile. Therefore, continued investment into the development of new antimicrobials is important to mitigate the potential for development and spread of resistance to the current arsenal of antibiotics.
1.4. Alternative Treatments for Severe CDI: Fecal Bacteriotherapy
Fecal bacteriotherapy is used to treat relapsing or severe CDI that is refractory to treatment with antibiotics and acts to restore balance to the gut microbiota in order to suppress C. difficile outgrowth. Typically patients are given antibiotics to reduce the load of actively growing C. difficile, with antibiotic treatment withheld 2–3 days prior to transplant of homogenized donor feces into the colon through a nasogastric intubation, enema, or colonoscope.40 This therapy is psychologically unappealing and also poses risks in terms of infection from the donor and from the irrigation and colonoscopy procedures performed prior to the transplant.41 Despite these drawbacks, fecal bacteriotherapy is gaining popularity because of the reported success rate of 92% (systematic review by Gough et al.42). In light of the potential benefits, patient acceptance is high with 94% of patients surveyed indicating that they would accept treatment over antibiotics if recommended by a doctor.42−44
Despite evidence of fecal bacteriotherapy being performed since the 4th century during the Dong-Jin dynasty in China,45,46 the first randomized controlled clinical trial for treatment of C. difficile was only reported in 2013.41 This rudimentary treatment is moving toward a refined, standardized practice supported by a hospital based prescreened donor system, with ongoing investigations into frozen fecal preparations and case studies of “designer” synthetic fecal transplants incorporating 33 strains of laboratory cultured bacteria.41,47−49 A standardized laboratory preparation is considerably more palatable, controllable, safer, and marketable, since strains can be sequenced and antibiotic resistant strains or other pathogenic organisms avoided. However, further research is required to ensure longevity of treatment and that laboratory passage does not reduce strain efficacy.50
Medical protocols are reported in the literature,51,52 but do-it-yourself guides53 and YouTube “how to” videos are directly accessible to patients online. In response to this, in April 2013 the FDA moved to regulate fecal bacteriotherapy as a biological therapy and thus require that physicians file an IND before performing fecal transplant.54 However, acknowledgment that this would potentially deny patients a life-saving cure saw an update on this ruling in July 2013, with fecal transplants to treat CDI permitted on compassionate grounds without an IND under an “enforcement discretion” as long as informed consent is obtained with appropriate acknowledgment of risks of the investigational therapy.55 In March 2014 these guidelines were tightened so that the donor must be known to the physician or the provider and that the donor sample be screened under the direction of the provider.56 Fecal bacteriotherapy also faces further regulatory issues, with recent debate over whether the treatment should be classified as an investigational new drug or tissue.56,57
2. Clinical Development
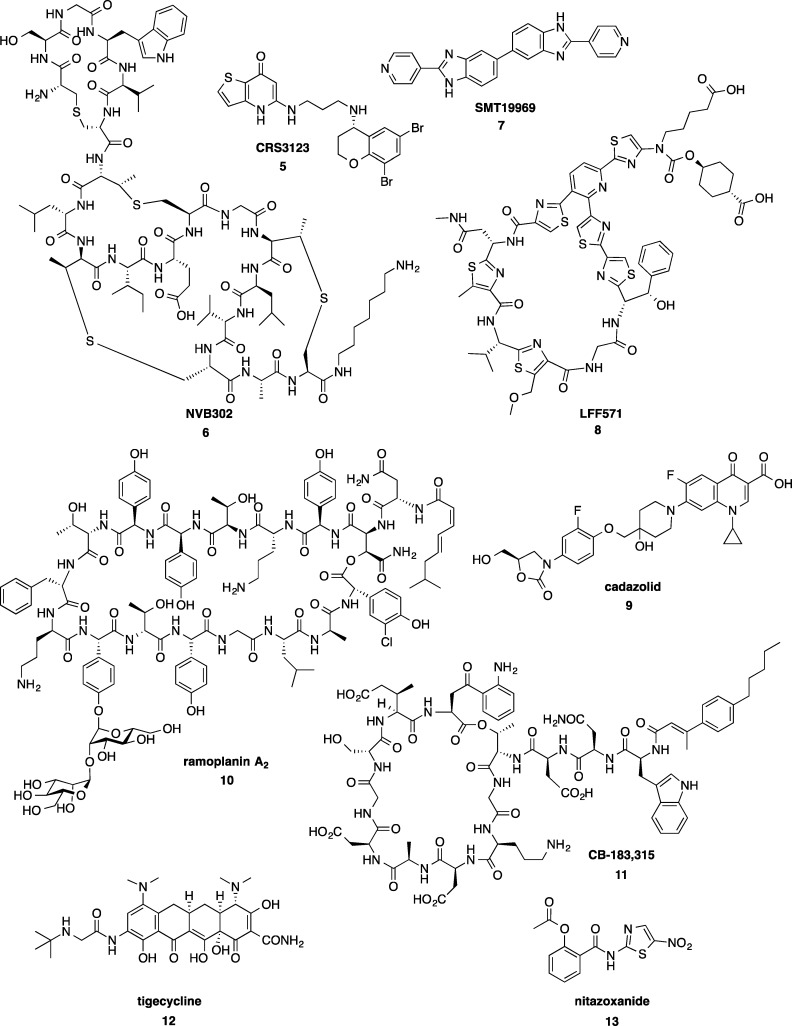
The clinical pipeline for new C. difficile treatments is characterized by traditional antibiotic molecules and nonantibiotic biological therapeutics (recently reviewed by Tsutsumi58) at various stages of development (Figure 2, Table 1).
Figure 2.
Antibiotics in clinical trials to treat CDI.
Table 1. Clinical Pipeline for C. difficile Treatment Includes Antibiotics and Nonantibiotic Therapies at Each Phase of Development.
| compd | phase | developer | class | ref |
|---|---|---|---|---|
| Antibiotic therapies | ||||
| CRS3123, 5 | I | National Institute of Allergy and Infectious Diseases (NIAID) | methionyl-tRNA synthetase inhibitor | (59) |
| NVB302, 6 | I | Novacta Biosystems Limited | lantibiotic | (60) |
| SMT19969, 7 | II | Summit Corporation Plc./Wellcome Trust | bis-benzimidazole | (63) |
| LFF571, 8 | II | Novartis Pharmaceuticals | 4-aminothiazolyl macrocycle | (62) |
| cadazolid, 9 | III | Actelion | quinolonyloxazolidinone | (64) |
| ramoplanin, 10 | II (previously completed phase III) | Nanotherapeutics, Inc. | lipoglycodepsipeptide | (65) |
| rifaximin, 3 | III | Salix Pharmaceuticals | rifamycin | (166) |
| CB-183,315, 11 | III | Cubist Pharmaceuticals | lipopeptide | (67, 68) |
| nitazoxanide, 13 | III | VA Medical Center, Houston/Baylor College of Medicine | nitrothiazolide | (70) |
| Nonantibiotic therapies | ||||
| IC84 | I | Valneva Austria GmbH | vaccine | (72) |
| frozen encapsulated FMT | I | Massachusetts General Hospital | microbiota restoration | (167) |
| frozen FMT | I | Massachusetts General Hospital | microbiota restoration | (168) |
| defined FMT | I | Baylor College of Medicine/Michael Debakey Veterans Affairs Medical Center | microbiota restoration | (169) |
| PF-06425090 | II | Pfizer | vaccine | (170) |
| VP20621 | II | Shire | nontoxigenic C. difficile | (75) |
| GS-CDA1, MDX-1388 | II | University of Massachusetts, Worcester/Medarex | human monoclonal antibodies | (171) |
| FMT | II | NorthShore University HealthSystem | microbiota restoration | (172) |
| FMT in pediatric patients | II | MemorialCare Health System | microbiota restoration | (173) |
| FMT | II | Colleen Kelly, The Miriam Hospital | microbiota restoration | (174) |
| FMT by colonoscopy | II | Catholic University of the Sacred Heart | microbiota restoration | (175) |
| RBX2660 | II | Rebiotix Inc. | microbiota restoration | (80) |
| tolevamer (GT267-004) | III | Genzyme | toxin binder | (73) |
| MK-3415, MK-6072, and MK-3415A | III | Merck | human monoclonal antibodies | (74) |
| ACAM-CDIFF | III | Sanofi-Pasteur | vaccine | (71) |
| Bio-K+ CL-1285 | III | Bio-K Plus International Inc. | microbiota restoration | (79) |
| FMT by capsule versus colonoscopy | II/III | University of Alberta/University of Calgary | microbiota restoration | (176) |
2.1. Traditional Antibiotics in the Clinical Pipeline
Several new antibiotics have entered the first stage of human clinical testing, phase I, including 5 (CRS3123, previously known as REP3123), a methionyl-tRNA synthetase inhibitor being developed by the National Institute of Allergy and Infectious Diseases.59 A type B lantibiotic deoxyactagardine B, 6 (NVB302), under development by Novacta Biosystems has completed phase I.60 Summit Corporation is recruiting for proof of efficacy phase II clinical trials of their bis-benzimidazole 7 (SMT19969),61 while Novartis has completed phase II trials of their 4-aminothiazolyl macrocycle 8 (LFF571).62,63 Acetlion’s cadazolid 9, a quinolonyloxazolidinone chimeric antibiotic, is recruiting for pivotal phase III testing.64 Ramoplanin 10, a lipoglycodepsipeptide antibiotic acquired from Oscient Pharmaceuticals by Nanotherapeutics65 has completed a phase III trial for CDI and will be conducting a phase IIb study for relapse prevention.66 A lipopeptide analogue of daptomycin, 11 (CB-183,315) under development by Cubist, is currently recruiting for phase III.67,68 Two existing drugs are undergoing repurposement trials: the glycycline antibiotic tigecycline 12 (phase unknown)69 and nitazoxanide 13 (phase II),70 a drug currently used to treat protozoan infections.
2.2. Nonantibiotic Therapies in the Clinical Pipeline
In addition to new antibiotic drug candidates for treatment of C. difficile infection, there are numerous biotherapeutic approaches at each phase of the clinical pipeline. Unlike antibiotic-based therapies, several of these approaches target the C. difficile major toxins, TcdA and TcdB, and include active and passive vaccine treatments to boost the immune response. A Sanofi-Pasteur ACAM-CDIFF TcdA- and TcdB-toxoid vaccine in phase III is the most advanced in the clinical pipeline.71 Pfizer and Intercell (IC84) both have vaccine candidates in phase I, both utilizing recombinant toxin-based approaches.72 Another treatment approach tested the ability of polymers to bind the disease-causing toxins secreted during C. difficile vegetative growth, thereby reducing inflammation and severe disease symptoms. Unfortunately, the polymer tolevamer failed to meet efficacy end points in phase III trials and was inferior to metronidazole and vancomycin antibiotic treatment.73 Merck’s human monoclonal antibodies (MK-3415, MK-6072, and MK-3415A), for use in a passive immunotherapeutic approach by inactivating the toxins, are in phase III trials.74
CDI is considered a secondary infection resulting from an initial disruption in the healthy microbial gut microbiota, often caused by antibiotic use. Therefore, another therapeutic approach aims to correct or prevent dysbiosis by manipulating the microbial niche. Treatment with nontoxigenic C. difficile strains, which aims to outcompete toxigenic strains and prevent their colonization, is in phase II trials, by ViroPharma (acquired by Shire in 2013).75 However, the C. difficile toxin-encoding PaLoc region from a toxigenic strain was recently shown to be mobilized to nontoxigenic isolates, supporting the hypothesis that nontoxigenic strains can become toxigenic through horizontal gene transfer events.76 This raises concerns that the use of live nontoxigenic strains in therapeutic approaches is risky, especially since a number of placebo patients were also found to be infected with nontoxigenic C. difficile strains during the clinical trials, apparently because of spore contamination of communal living areas.77 There are also ethical considerations surrounding the use of this approach, since carriers may transmit the strain to nonconsenting individuals. An alternative approach in the future could be the use of Clostridium scindens, since it has recently been shown to promote resistance to C. difficile infection in mouse CDI models in a manner associated with increased production of C. difficile-inhibiting secondary bile metabolites.78
Another option under consideration is the use of probiotic therapies to prevent CDI. Bio-K+ CL-1285 is an encapsulated probiotic propriety mixture of Lactobacillus acidophilus and Lactobacillus casei that has completed phase III clinical trials for prevention of antibiotic-associated diarrhea, including CDI.79 Patients at high-risk of developing CDI (50–70 years age, receiving penicillin, cephalosporin, or clindamycin antibiotic therapy), administered two capsules of Bio-K+ CL-1285 daily, were significantly less likely to develop CDI (23.8% placebo vs 1.2% twice daily capsule treatment, P = 0.002), suggesting that this approach may be efficacious.79
As discussed earlier, microbiota transplantation also aims to restore an imbalanced microbiota using a holistic strategy. Fecal transplant therapies or artificial microbial preparations containing a diverse microbial ecosystem are used in this approach. Clinical trials are currently evaluating issues such as whether frozen and thawed or encapsulated preparations are as effective as the fresh, unprocessed material, since many bacteria, particularly anaerobes, would not survive the treatment and storage phase, perhaps reducing product efficacy compared with the starting material. The aim is to make this therapy easier to distribute, as set donations can be processed, tested for pathogens, prepared ahead of time, and made more palatable by encapsulation of the transplant. A recent 2014 study indicates that frozen capsules are indeed a viable form of administration.49 Rebiotix have focused on developing an off the shelf product for transplant that is consistent and easy to administer. Their product, RBX2660, is a microbial suspension, which has completed enrollment in phase II, and Rebiotix is in discussions with the FDA on design of the phase III trial.80 Monarch Labs in collaboration with BioTherapeutics, Education and Research Foundation is also seeking to develop a fecal transplant therapy, Medical Microbiota, which is a prescreened, cGMP processed product for transplantation. They are also developing a cGMP processing and banking service for autologous transplantation.81
An important factor that may influence the efficacy of microbiota-restoration strategies is the treatment of patients with broad-acting antibiotics such as vancomycin to control CDI, although antibiotics are generally halted shortly prior to administration of microbiota therapy.52 Antibiotic therapy reduces the load of vegetative C. difficile cells in the host prior to infusion of the microbial suspension, which may contribute to the subsequent success of microbiota restoration strategies. However, the necessity of this antibiotic therapy prior to the application of microbial restoration therapies is not known.52 It is clear that the need for antibiotics for the treatment of CDI will continue and that their adjunct use in microbial restoration therapies may be highly beneficial. For this reason, there is a continued market for investment into the development of new antimicrobials against CDI, which will be the focus of the remainder of this review.
3. Early Drug Discovery Research and Preclinical Development Pipeline
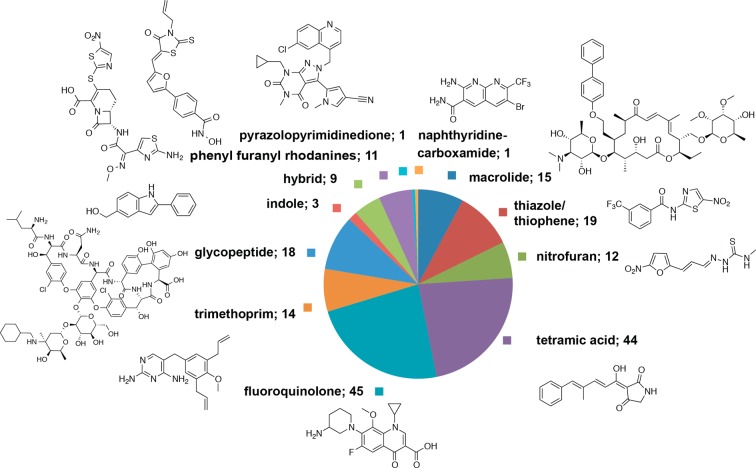
We have undertaken a chemoinformatic approach to understand the historical and recently explored chemical space for compounds with antimicrobial activity against C. difficile at the preclinical stages of drug research and discovery. Compounds with reported antimicrobial activity against C. difficile were abstracted from the ChEMBL82 database. The data were then separated into either known, marketed antibiotics or compounds at the early discovery research phase and preclinical stages of development. The compounds in the early preclinical drug discovery phase were clustered using Pipeline Pilot into antimicrobial classes (Figure 3, methods in Supporting Information). Analysis of the data from ChEMBL showed that the results only contained compounds published in four scientific journals over selected years: Antimicrobial Agents and Chemotherapy (2007–2010), Bioorganic Medicinal Chemistry (one article from 2008), Bioorganic Medicinal Chemistry Letters (1998–2012), and Journal of Medicinal Chemistry (1984–2009). Therefore, the search was widened to include all bioactivities against C. difficile catalogued in the Reaxys Medicinal Chemistry Public Beta as well as manual searches of the literature.
Figure 3.
Compounds in early and preclinical drug discovery phase with antimicrobial activity against C. difficile in the scientific literature from the analysis of compounds with antimicrobial activity against C. difficile curated by ChEMBL. Antibiotics on the market and agents in clinical trials were excluded from analysis.
The majority of studies are 10–20 years old and predate the C. difficile crisis of the past decade. However, the more recent studies that include C. difficile as a greater focus demonstrate the renewed drug discovery efforts for this pathogen. Notably, few of the older studies are focused on medicinal chemistry efforts to specifically optimize compounds for activity against C. difficile. However, the tiacumicin family of antibiotics, including fidaxomicin, was initially discovered in 1975 but the development of this class remained largely unexplored until the late 1990s when Optimer Pharmaceuticals initiated the commercial development of fidaxomicin for CDI.29 Therefore, examining the past can offer direction for future investigations, especially since most of the older research was not conducted in the context of the requirements for a C. difficile-specific antibiotic.
The majority of the reports describe investigations of traditional antibiotic chemotypes with screening against C. difficile performed in parallel to testing against other bacterial pathogens that form the focus of these studies. These studies are summarized in Supporting Information Table 1. Of note is the importance of testing drug candidates against a number of strains, since there is a wide range of MIC results for some drug candidates. The MIC50 and MIC90 values are important to the interpretation of the MIC ranges obtained from testing against multiple strains and thus determining the efficacy against a population of strains. The discussion here focuses on studies where a number of analogs were synthesized and on studies of unique chemotypes tested against C. difficile.
3.1. Nitroheterocycles
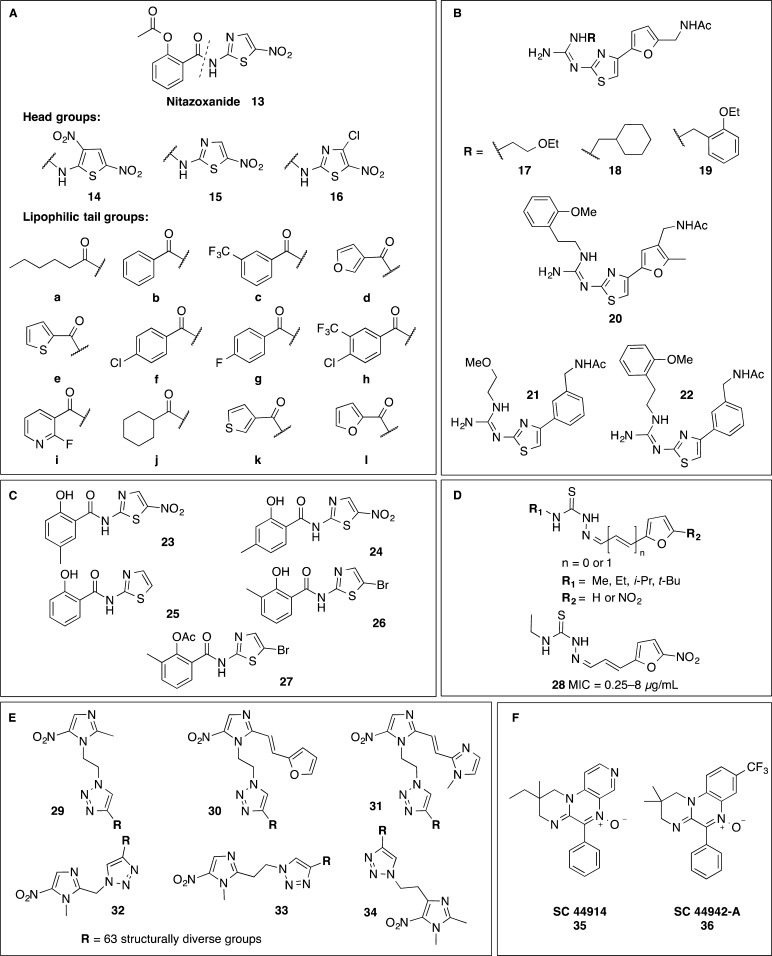
Ballard et al. have explored analogs of the nitrothiazole drug, nitazoxanide 13, for activity against C. difficile as part of a broader study for compounds with activity against Helicobacter pylori (Figure 4A).83 Variations to the thiophene and thiazole “head” groups 14–16 and amide “tail” a–e (Figure 4A)83 were found to influence activity with MICs varying from 0.4 to >28 μM. The headgroup 15 found in nitazoxanide was generally favored, with 15c and 15d the most potent against C. difficile (MICs of 0.4 and 1 μM, respectively). A follow-up study explored tail group f–l and headgroup 15 to give MICs ranging from 0.8 to 5.9 μM (Figure 4A).84 Four furylthiazoles (no nitro group) 17–22 and two phenylthiazoles 21–22 also developed for H. pylori by Fujisawa Pharmaceutical Co. were tested for activity against C. difficile but were not active at 100 μg/mL (Figure 4B).85−87 In contrast, two of five thiazolides, 23 and 24, only those with a nitrothiazole ring similar to nitazoxanide, displayed potent activity (0.06–0.5 and 0.06–0.25 μg/mL, respectively) against 10 strains of C. difficile.88 The three inactive compounds 25–27 (MIC > 32 μg/mL) either contained no nitro group (denitrotizoxanide) 25 or a bromine in place of the nitro group (26 and 28) (Figure 4C).88
Figure 4.
Nitroheterocycles: (A) analogs of nitazoxanide 14–16;83,84 (B) furanyl thiazoles 17–20 and phenylthiazoles 21 and 22 (inactive against C. difficile, MIC > 100 μg/mL);85−87 (C) thiazolides with a nitrothiazole ring, similar to nitazoxanide;88 (D) nitrofuranylsemicarbazone 28;89 (E) the six 5-nitroimidazole scaffolds 29–34 in the library of metronidazole triazole conjugates (crude, >85% purity);90 (F) quinoxalines 35–36.92
A series of nitrofuranylsemicarbazones were investigated for MIC activity against C. difficile, Staphylococcus epidermis, Staphylococcus aureus, methicillin resistant S. aureus, and Propionibacterium acnes (Figure 4D).89 Only compound 28 had reasonable activity against four strains of C. difficile, in the range of 0.25–8 μg/mL, with the other 11 compounds in the range of 32 to >256 μg/mL. While the mechanism of action of nitrofuryls is typically attributed to toxicity caused by reduction of the nitro group,89 the role of this mechanism in this case is unclear as several matched pairs with R2 = H or NO2 did not show large differences in activity. The R1 groups (methyl, isopropyl, and tert-butyl) are likely too far from the nitrofuranyl to affect the reduction process electronically, so they should not cause changes in activity based on a reduction mechanism. Additionally the active compound shows similar activity against the other bacteria assayed, presumably under aerobic conditions, further supporting an alternative mechanism of action for this compound.
The chemical space for nitroheterocyclic compounds with a reduction-based mode of action similar to metronidazole remained relatively unexplored for C. difficile until a series of metronidazole triazole conjugates was published in 2013 (Figure 4E).90 This group synthesized six 5-nitroimidazole azido cores and reacted each core with a library of 63 structurally diverse alkynes to make 378 5-nitroimidazole triazole conjugates 29–34.90 These compounds were tested as crude mixtures (purity of >85% by LCMS) against C. difficile, H. pylori, Trichomonas vaginalis, Giardia lamblia, and Bacteroides fragilis. A second follow-on series of 47 different alkynes reacted with the six cores gave a further 281 compounds, but these were not tested against C. difficile.90 Interestingly, a high proportion of the compounds in the initial series were active against metronidazole resistant G. lamblia (100% of compounds tested) and T. vaginalis (47% of compounds tested) parasite strains. However, the compounds generally lost activity against the microaerophilic bacterium H. pylori containing mutations in both the frxA and rdxA genes that encode reductases involved in the clinical resistance to metronidazole (only 1.4% of compounds active).90,91 This suggests that some metronidazole triazole conjugates may be active against metronidazole resistant C. difficile. However, the effectiveness of these compounds at inhibiting spore formation, germination, and spore outgrowth is not currently known.
The quinoxalines 35 (SC44914) and 36 (SC-44942-A) have a similar spectrum of action as metronidazole, with the mode of action postulated to be similar because of similar reduction potentials (Figure 4F).92 Both quinoxalines were potent against C. difficile with MICs against 20 isolates of ≤0.06 μg/mL for 35 and ≤0.06 to 0.5 μg/mL for 36 (the metronidazole control was ≤0.06 to 2 μg/mL).92
3.2. Glycopeptides
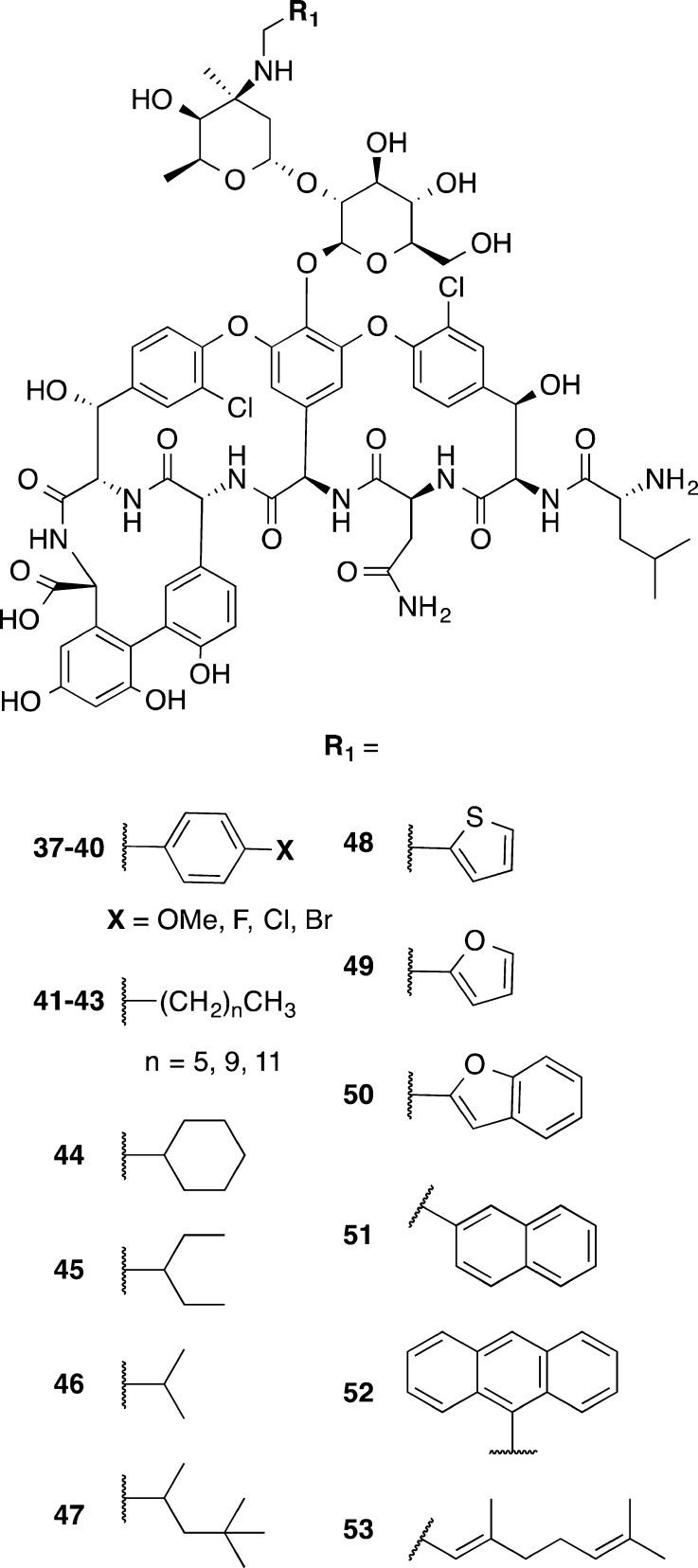
Zhang et al.93 made a series of desmethyl vancomycin analogs 37–53 inspired by telavancin and oritavancin, glycopeptides approved for treatment of complicated skin infections (Figure 5). This series of compounds contained desmethyl vancomycin in which the methyl group on the amino group of the N-terminal residue of vancomycin was absent. Various hydrophobic groups were appended via the amine on the vancosamine sugar (R1 of Figure 5), in a fashion similar to the hydrophobic chains on oritavancin and telavancin. When tested for potency against Enterococcus faecium and C. difficile, the compounds were generally within one dilution of the MICs for the controls (vancomycin and desmethyl vancomycin) for four strains of C. difficile, indicating that variations at these positions do not play a key role in determining activity for C. difficile. This was in contrast to variation observed in the structure–activity relationships for E. faecium.
Figure 5.
Desmethyl vancomycin derivatives 37–53 with lipophilic variations at R1 on the vancosamine sugar.93
3.3. Macrolides
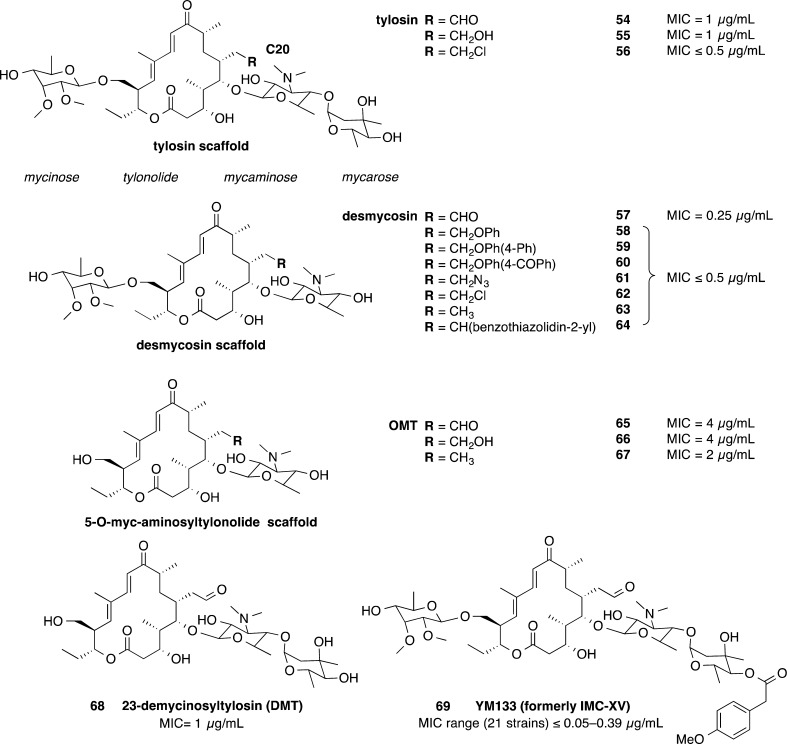
Kirst et al. reported on macrolide antibiotics related to tylosin natural products and synthetically modified derivatives from Eli Lilly Research Laboratories (Figure 6).94 The compounds were tested against a wide range of aerobic and anaerobic bacteria. Fifteen of these compounds 54–68 were tested for activity against C. difficile.94 Six compounds of the desmyocosin scaffold 58–64 showed a MIC of ≤0.5 μg/mL compared with the desmycosin 57 MIC of 0.25 μg/mL. These compounds were also active against a range of anaerobic bacterial strains including B. fragilis, Fusobacterium symbiosum, Peptococcus prevoti, and Peptostreptococcus anaerobius, as well as aerobic bacterial strains including S. aureus and Streptococcus group B, C, and D strains. Three tylosin derived macrocycles also showed good activity at ≤0.5 μg/mL compared to 1 μg/mL for tylosin 54.
Figure 6.
Tylosin derived macrolides 54–69.94
The 23-demycinosyltylosin (DMT) derivative 68 also showed good activity against C. difficile (1 μg/mL) with the 5-O-myc-aminosyltylonolide (OMT) 65 and OMT derivatives 66 and 67 showing weaker activity at 2–4 μg/mL (Figure 6).94 The majority of these compounds were tested using in vivo models, against either Streptococcus pyogenes or S. aureus. The goal of this study was to improve the oral availability of these compounds by chemical modifications. The compounds were dosed subcutaneously and orally. Several compounds lost activity when dosed orally compared to subcutaneous dosing. Peripheral plasma compound levels measured in mice and rats supported the varied oral bioavailability. This study is an example where drug candidates that failed in the past because of properties such as poor oral availability may now be useful in the treatment of C. difficile, where compounds with poor oral availability are desirable for C. difficile treatment because a high concentration of compound remains at the site of infection, with low systemic exposure.
A more recently developed tylosin macrolide 69 (YM133) exhibited potent activity against 21 C. difficile strains with MICs ranging from ≤0.05 to 0.39 μg/mL (Figure 6).95 Compound 69 was also active against macrolide-resistant S. aureus but was less active against strains with 14- and 16-membered macrolide resistance compared to 14-membered macrolide resistant or erythromycin resistant strains.95
3.4. Fluoroquinolones
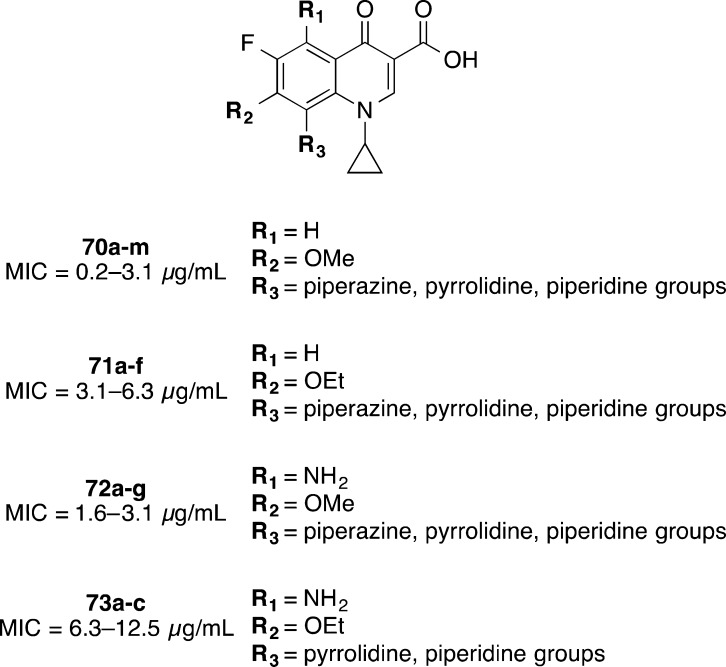
A large number of fluoroquinolones 70a–m, 71a–f, 72a–g, and 73a–c were synthesized and investigated for antibacterial activity by Sanchez et al. from Parke-Davis as a division of Warner-Lambert Pharmaceuticals (now acquired by Pfizer) (Figure 7).96 As a part of this study, 27 compounds were tested against several anaerobic bacteria including C. difficile. The compounds ranged in activity from 0.2 to 12.5 μg/mL, with the most potent series 70a–m containing R1 = H, R2 = OMe, and R3 = variety of piperazine, pyrrolidine, and piperidine groups (Figure 7).96 While C. difficile potency was good, the compounds possessed poor specificity, with broad spectrum activity against Gram-negative and Gram-positive facultative anaerobic bacteria and obligately anaerobic bacteria.96
Figure 7.
Fluoroquinolones 70–73.96
3.5. Tetramic and Tetronic Acids
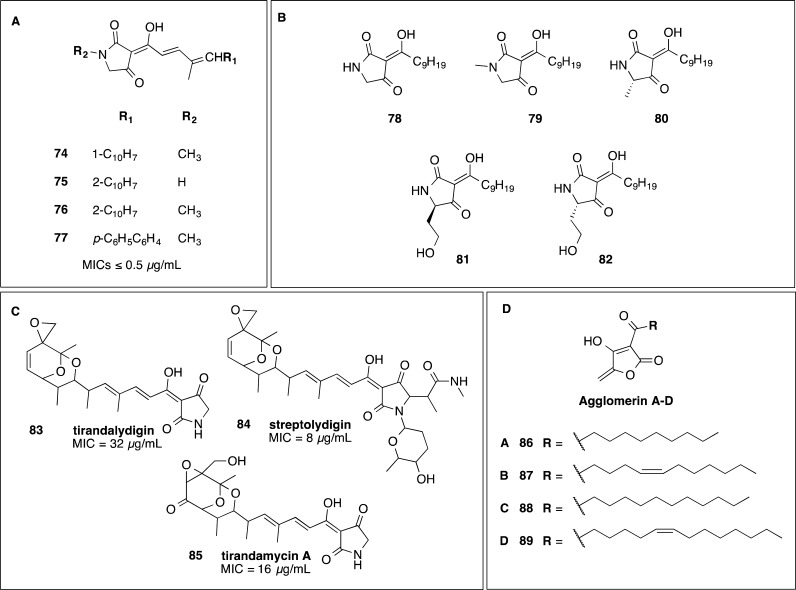
Tetramic acids have been shown to have activity against C. difficile (Figure 8). Aromatic dienoyltetramic acids 74–77 formed the basis of a study by Rosen et al. of Abbott Laboratories.97 The compounds’ activity ranged from 125 to ≤0.5 μg/mL. The derivatives active at ≤0.5 μg/mL generally contained a longer C10H7 lipophilic tail. More recently, Ueda et al. examined tetramic acids, derived from bacterial autoinducers, for their bactericidal activity against C. difficile.98 Compound 78 at 10 μM (2.5 μg/mL) reduced C. difficile numbers to the limit of detection of 2 log CFU/mL after overnight incubation.98 The other tetramic acids tested 79–82 with lower activity (13–15 μg/mL) showed that the keto–enol form with the free hydroxyl group and a longer acyl side chain is important to maintain activity against C. difficile, since tetramic acids lacking these features were inactive at 100 μM (21–31 μg/mL).98 The tetramic acids, tirandalydigin 83 (MIC = 32 μg/mL), streptolydigin 84 (8 μg/mL), and tirandamycin 85 (16 μg/mL), were weakly active against one strain of C. difficile.99 The agglomerins A, B, C, and D 86–89, tetronic acids structurally related to tetramic acids, were reported by Shoji et al. at Shionogi & Co. with activity against C. difficile from 0.78 to 3.13 μg/mL.100 The structures were solved in 1990 by Terui et al.101
Figure 8.
(A–C) Tetramic acids 74–85 investigated for activity against C. difficile(87,97,98) and (D) structurally related tetronic acids 86–89.100
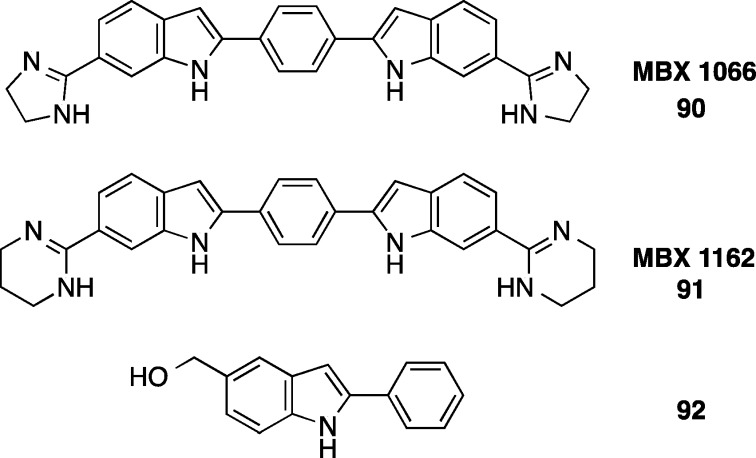
3.6. Bis-Indoles
The bis-indoles 90 (MBX 1066) and analogue 91 (MBX 1162) (Figure 9) investigated by Microbiotix Inc. have broad spectrum activity against Gram-positive and Gram-negative aerobic and anaerobic bacteria including 18 isolates of C. difficile (MIC90 for both compounds was 0.12 μg/mL).102 The compounds inhibit DNA synthesis and are proposed to bind in the minor groove of the DNA duplex because of their similarity to DNA binding agents.103 The bisindole structure is important for activity against C. difficile since a 2-phenyl-1H-indole 92 tested against three C. difficile strains was inactive at the highest concentration tested (100 μg/mL).104
Figure 9.
Bis-indoles 90 and 91 with potent activity against C. difficile compared to the 2-phenyl-1H-indole 92 not active at 100 μg/mL.102,104
3.7. Hybrids
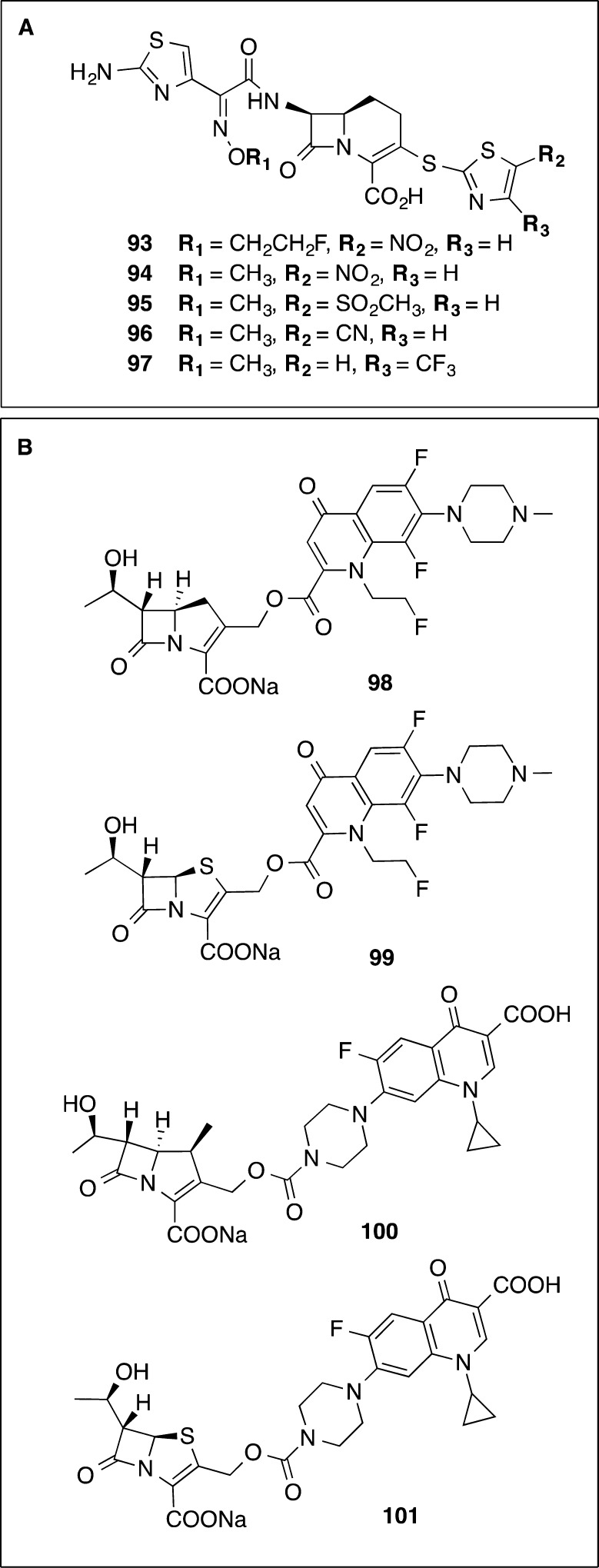
Hybrid-like 1-carba-3-thiathiazole cephalosporins were investigated by Eli Lilly for their activity against anaerobic bacteria (Figure 10A).105 Five compounds 93–97 were reported with thiazoles at the 3-thia positions and either a methyloxime 94–97 or fluoroethyloxime 93 side chain.105 The compound derivatives with a nitrothiazole and a methyloxime 94 or fluoroethyloxime side chain 93 exhibited broad spectrum activity against the panel of anaerobes tested which was generally equivalent to or better than cefoxitin and cefotetan controls. They were active at 2 and 4 μg/mL against C. difficile (cefoxitin, 32 μg/mL; cefotetan, 16 and 32 μg/mL). Thiazole ring substitutions 95–97 were tolerated, and the compounds had similar activity against the aerobic organisms: S. aureus, S. epidermis, Streptococcus pneumoniae, Haemophilus influenza, Escherichia coli, and Enterobacter aerogenes.
Figure 10.
(A) Hybrid 1-carba-3-thiathiazole cephalosporins10593–97 and (B) hybrid penem and carbapenems linked to quinolones 98–101.106
Attempts were made to replace the nitro group on the thiazole because of observed mutagenicity. Methyl sulfoxide 95, nitrile 96, or trifluoromethyl 97 groups increased the activity against C. difficile (MICs of 0.06–0.125 μg/mL). However, loss of the nitro-substituted thiazole resulted in loss of activity against Bacteroides spp. and F. symbiosum, which the researchers deemed as a negative feature in their search for a broad-spectrum agent. Given that C. difficile infection is exacerbated by damage to the gut microbiota, selective agents are now considered desirable. This illustrates the potential value of re-examining drug candidates abandoned in the past because of their lack of broad-spectrum action, as this selectivity is now considered an advantage.
Another example of hybrid compounds is the Hoffmann-La Roche study by Corraz et al. into penems and carbapenems linked at the 2′ position to quinolones (Figure 10B).106 Four hybrid compounds 98–101 were tested against either one or in some instances two strains of C. difficile. The hybrid compounds showed superior MIC activity (0.5–8 μg/mL) against C. difficile when compared to the single antibiotic counterparts (fleroxacin, ciprofloxacin, or imipenem, 4–16 μg/mL). However, in general the hybrid compounds had superior MICs against a broad range of other Gram-negative and Gram-positive bacteria, indicating that these are not good leads to pursue for new selective agents against C. difficile.
3.8. Other DNA Topoisomerase and Gyrase Inhibitors
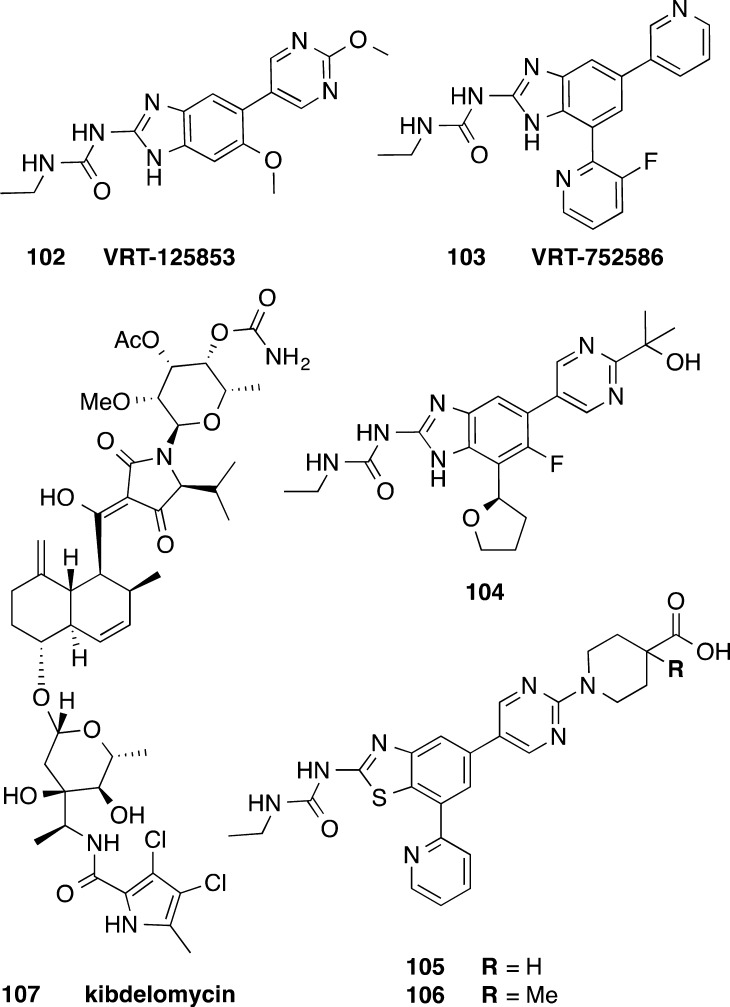
Both synthetic inhibitors and natural products that target topoisomerase II, inhibiting DNA synthesis, have been reported (Figure 11). Dual acting inhibitors of bacterial DNA gyrase and topoisomerase IV, based on an aminobenzimidazole urea core, have been extensively explored by Vertex Pharma as new leads against a range of Gram-positive pathogens (staphylococci, streptococci, and enterococci) and the respiratory Gram-negative pathogen H. influenzae.107 Two lead compounds, 102 (VRT-125853) and 103 (VRT-752586), showed relatively low frequencies of spontaneous resistance.107 Compound 103 was active in S. aureus and S. pneumoniae animal infection models.108 When tested for in vitro activity against C. difficile, 102 displayed a range of potencies against 11 strains of C. difficile (1–16 μg/mL), while 103 was found to be even more potent (0.06–4 μg/mL).107 Further development of the aminobenzimidazole urea compound class by Vertex has alleviated deficiencies identified for 103 such as CYP3A4 inhibition, reactive metabolite formation, short half-life, and poor physicochemical properties, and 104 has been selected as a preclinical candidate.109 However, the activities of 104 and other analogs against C. difficile have not been reported. The Vertex library of aminobenzimidazoles (>330 compounds)110 prepared could be reinvestigated to potentially identify alternative lead compounds with potent, more selective activity against C. difficile and poor oral availability.
Figure 11.
Aminobenzimidazole urea inhibitors of bacterial DNA gyrase and topoisomerase IV 102–104,107,109 topoisomerase II ATPase inhibitor kibdelomycin 107,112 and benzothiazole ethyl urea inhibitors of DNA gyrase GyrB and topoisomerase IV ParE ATPase 105 and 106.111
Dual action benzothiazole ethyl urea based compounds have been explored by Biota, inhibiting the DNA gyrase GyrB and topoisomerase IV ParE ATPase.111 Two compounds, 105 and 106, inhibited C. difficile 630 (ATCC 9689) at 0.03 and 0.015 μg/mL, respectively (Figure 11).111 Kibdelomycin, 107, is a natural product with broad-spectrum antimicrobial activity by inhibiting topoisomerase II ATPase. A 2014 study by Miesel et al. from Merck Research Laboratories examined 107 for in vitro activity against a panel of 168 C. difficile clinical isolates (MIC90 = 0.25 μg/mL) and other commensal anaerobic organisms.112 The compound was not orally absorbed and exhibited in vivo efficacy in a hamster model of infection using the toxigenic B1 C. difficile strain SM8-6865 (Figure 11).112
3.9. Peptidic Antimicrobials
Pexiganan 108 (Cytolex, MSI-78, Locilex) (structure in Table 2) is a 22-amino-acid antimicrobial peptide with broad-spectrum antimicrobial activity that has recently entered phase III clinical trials under a special protocol assessment as a topically applied treatment of mild infections of diabetic foot ulcers.113 During development this compound was evaluated for activity against C. difficile and was active against four strains of C. difficile from 0.5 to 4 μg/mL114 and again at 4 μg/mL against one other strain of C. difficile.115 A range of 16 cecropin–melittin hybrid peptides (CAMEL analogs) 109–124 were also tested against anaerobic bacteria including 10 C. difficile strains and were active between 1 and 4 μg/mL (structures in Table 2).116 However, peptidic antibiotics such as pexiganan 108 and the cecropin–melittin hybrid peptides 109–124 are unlikely to survive passage through the gastrointestinal tract.
Table 2. Pexiganan and Cecropin-Melittin Hybrid Peptidesa with Antimicrobial Activity against C. difficile.
| peptide | amino acid sequenceb |
|---|---|
| pexiganan, 108 | GIGKFLKKAKKFGKAFVKILKK-NH2 |
| CAMEL0, 109 | KWKLFKKIGAVLKVL |
| CAMEL9, 110 | KWRLFKNIGAVLKVL |
| CAMEL24, 111 | KWKLFKHIGAVLKVL |
| CAMEL42, 112 | HWKLFKKIGAVLKVL |
| CAMEL46, 113 | KWKLFKGIRAVLKVL |
| CAMEL48, 114 | KWKLGKKILAVLKVL |
| CAMEL48D, 115 | KWKLGKKILAVLKVL |
| CAMEL101, 116 | KWKLGKKILRVLKVL |
| CAMEL102, 117 | GWKLGKKILRVLKVL |
| CAMEL108, 118 | KWKLGKKILNVLKVL |
| CAMEL109, 119 | GWRLGKKILRVLKVL |
| CAMEL110, 120 | GWKLGKKILNVLKVL |
| CAMEL123, 121 | LWKLFKKIRRVLRVL |
| CAMEL129, 122 | LWKLFKKINRVLKVL |
| CAMEL135, 123 | GWRLIKKILRVFKGL |
| CAMEL136, 124 | VWRLIKKILRVFKGL |
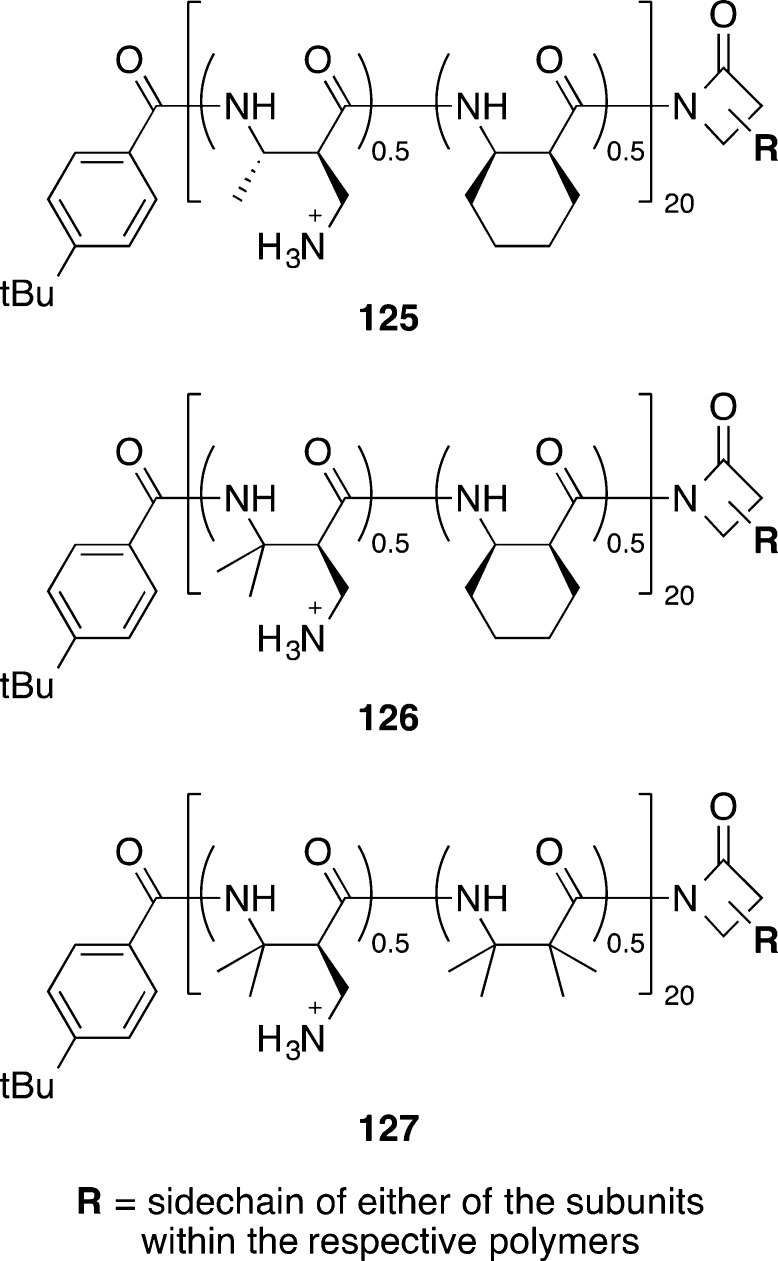
Recently, synthetic nylon-3 polymers (poly-β-peptides) 125–127 (Figure 12) designed to mimic host-defense peptides were shown to inhibit C. difficile growth (MICs of 12.5–25 μg/mL) and importantly also blocked spore outgrowth in two strains of C. difficile (MIC of spore outgrowth of 3.13–12.5 μg/mL).117 Compared to host-defense peptides, synthetic polymer mimics also offer potential advantages for the relative ease of synthesis and stability to proteolytic degradation.
Figure 12.
Nylon-3 polymers 125–127.117
3.10. Natural Products
Natural products have a history of use as antibiotics and treatment for C. difficile is no exception, with metronidazole, vancomycin, and fidaxomicin all having origins as natural products. A number of more recent, novel natural products investigated for antimicrobial activity against C. difficile are described in this subsection.
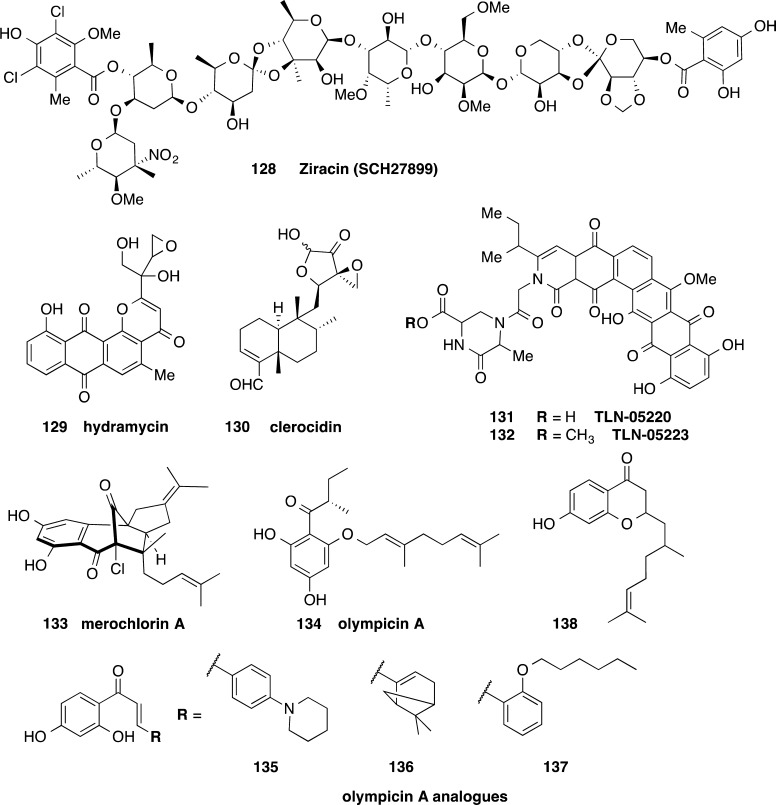
Ziracin 128 (SCH27899), an oligosaccharide antibiotic derivative of everninomicin, had potent activity against 27 strains of C. difficile with MICs ranging from 0.05 to 0.2 μg/mL (Figure 13).118 The MIC50 was 0.1 μg/mL, and MIC90 was 0.2 μg/mL.118 Compound 128 was subsequently tested for activity against a further 25 C. difficile strains, with the MIC ranging from 0.06 to 0.125 μg/mL.119 Although 128 was in development as an iv antibiotic for Gram-positive infections, Schering-Plough suspended development because of failure of efficacy versus safety end points. The two ortho ester linkages in 128 are unlikely to be stable under the acidic conditions of the stomach. Therefore, 128 in itself is not a good lead for oral treatment of CDI without additional measures such as enteric coated capsules.
Figure 13.
Structures of ziracin 128,118 hydramycin 129,120 clerocidin 130,121 echinosporamicin-type antibiotics 131 and 132,123 merochlorin A 133,124 and olympicin A 134,126 and analogs 135–138126 with activity against C. difficile.
Hydramycin 129, a pyranoanthraquinone isolated from Streptomyces violaceus, was shown to have antibiotic and antitumor properties by Bristol Myers Squibb (Figure 13).120 The activity against one strain of C. difficile was 0.4 μg/mL. The cytotoxicity of this antibiotic limits the likelihood of further development, although fidaxomicin also exhibits cytotoxicity.
The natural product clerocidin 130, a terpenoid antibiotic, displayed potent activity against one C. difficile strain with MIC = 0.2 μg/mL.121 Compound 130 was initially thought to be a DNA gyrase inhibitor,121 but it has since been shown to act as a DNA alkylating agent that targets single stranded DNA.122 This mechanism of action limits further use of 130 as an antimicrobial agent.
Two polycyclic antibiotics consisting of a backbone of six fused rings, 131 (TLN-05220) and 132 (TLN-05223), were isolated from Micromonospora echinospora ssp. challisensis (Figure 13).123 The echinosporamicin-type antibiotics were potent against various tumor cell lines (IC50 of <0.1 to 9.7 μM depending on the cell line), although 131 failed to show efficacy in an in vivo mouse human prostate tumor model.123 Both 131 and 132 were active against Gram-positive bacteria C. difficile, S. aureus, S. pneumoniae, and Enterococcus faecalis. Compound 131 with a free carboxylic acid was active at 0.06 μg/mL against C. difficile, while the methyl ester analogue 132 was less potent with MIC = 2 μg/mL.123 This compound class would only have use as an anti-C. difficile agent if the cytotoxic effects were avoided by poor oral bioavailability.
Merochlorin A 133, a meroterpenoid, has been investigated for antistaphylococcal and anticlostridial activity (Figure 13).124 Compound 133 was active at 0.3 and 0.15 μg/mL against two C. difficile strains, more potent than the activity against S. pneumoniae, multidrug resistant MRSA, and VRE (MICs of 1–4).124 No activity was observed (MIC of >64 μg/mL) against Gram-negative strains including Pseudomonas aeruginosa, E. coli, Enterobacter cloacae, Acinetobacter baumannii, and Klebsiella pneumoniae. However, 133 was cytotoxic against HeLa cells at close to the staphylococcal MIC after extended incubation (IC50 = 64 μg/mL at 24 h, IC50 = 2 μg/mL at 72 h) and also was inactivated by the presence of 20% human serum (S. aureus MIC > 64 μg/mL).124 A gram scale synthesis was recently completed, opening up the path for further studies.125
A collection of natural product flavonoid and related phytochemicals was assessed for their inhibitory action against C. difficile in 2014.126 Olympicin A, 134, was active at 1 μg/mL against a BI/NAP1/027 C. difficile strain, equivalent to vancomycin activity against this strain (Figure 13).126 Several analogs of 134, 135–138, containing the natural product chalcone motif, 135–137, or an alternative flavonone core, 138, were 1–2 dilutions less active against C. difficile (Figure 13).126 Mode of action studies in S. aureus showed that 135–137 disrupted membrane potential and also inhibited the macromolecular synthesis of proteins, RNA, and DNA.127 Further analogs have now been reported and evaluated for activity against Mycobacterium tuberculosis, E. faecalis, S. aureus, and E. coli but not C. difficile.127
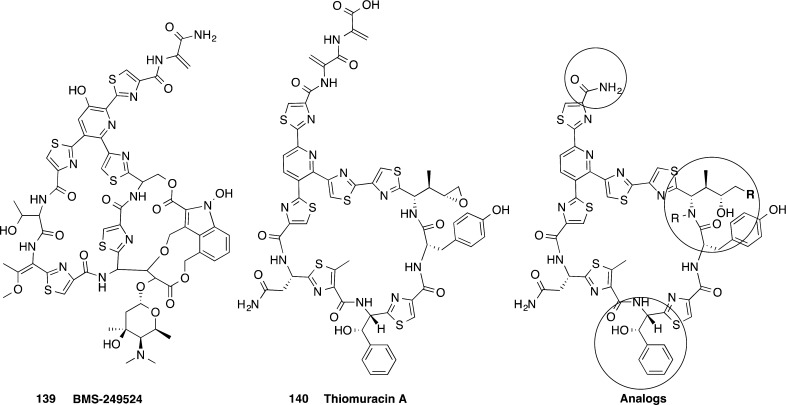
The cyclic thiazolyl derivative 139 (BMS-249524), part of the nocathiacin family, was potent against five strains of C. difficile with the MIC activity ranging from 0.06 to 1 μg/mL (Figure 14).128 Nine analogs of the related thiomuracin A 140, a thiazolyl actinomycetales metabolite, were also active against C. difficile with MICs from 0.003 to ≤0.008 μg/mL (Figure 14).137 Notably, these compounds are structurally related to 8, which is in phase II clinical trials as a treatment for C. difficile infection.65,129
Figure 14.
Cyclic thiazolyl peptidic antimicrobial compounds 139 and 140 similar to 8, which is in phase II clinical trials.128,129
3.11. C. difficile Specific Drug Targets and Inhibitors
Discovery of inhibitors with antimicrobial activity against new drug targets specific to C. difficile has so far been limited, in part because of historical difficulties in performing genetic manipulation. The development of the ClosTron gene-knockout system in 2007, which is used to inactive specific genes, has aided essential drug target validation.130 Potential drug targets for C. difficile have been compiled into a database by computational analysis of its genome, searching for choke point enzymes potentially necessary for cell survival, with exclusion of proteins found in human gut microbiota.131 More recently a curated C. difficile metabolic network has been established and used to predict essential targets and potential inhibitors.132 Validation of these predicted targets and inhibitors is still required. Despite C. difficile focused target based drug discovery being in the relative early stages, a number of studies have made progress in this area.
Dang et al. used activity based chemical probes to identify new drug targets in C. difficile.133 They identified Cwp84 as a protease that mediates cleavage of SlpA, required for formation of the surface layer that coats the C. difficile cell and important for host–pathogen interactions.133 No inhibitors have been reported to date.
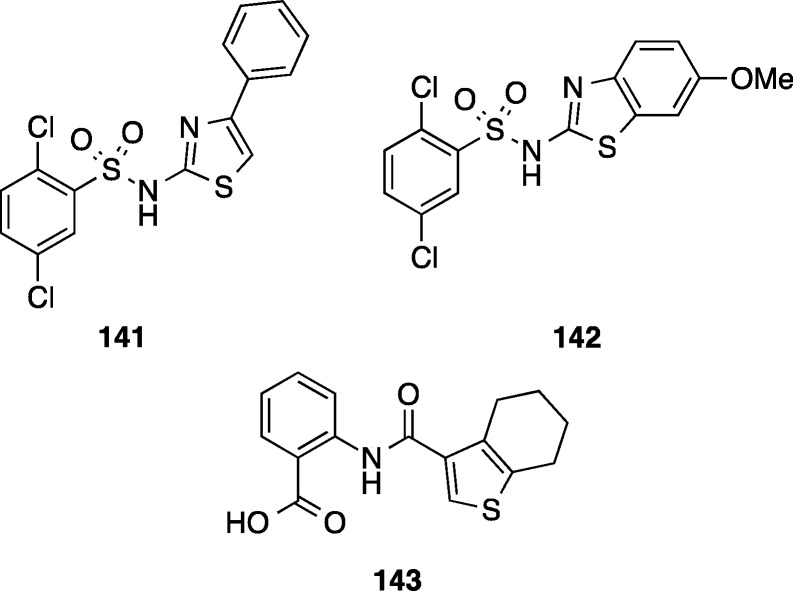
Ratia et al. attempted to develop selective inhibitors of C. difficile by targeting the enzyme dehydroquinate dehydratase (DHQD), which is involved in the shikimate pathway for biosynthesis of chorismate, a precursor required for biosynthesis of three aromatic amino acids.134 The shikimate pathway is not present in humans, but as DHQD is present in bacteria in two different subtypes, selective inhibitors could potentially be obtained by a target based drug discovery approach. The type I DHDQ is present in C. difficile, while the type II DHQD is present in commensal bacterial species such as Bacteroides thetaiotaomicron and Bifidobacterium longum.134 The type I C. difficile DHQD (cdDHQD) was selectively inhibited compared to the type II B. thetaiotaomicron DHDQ (btDHDQ) by three compounds 141–143 (cdDHQD IC50 = 31–35 μM, cf. btDHDQ IC50 > 200 μM) identified from high throughput screening efforts (Figure 15).134 However, the antimicrobial efficacy of the enzyme inhibitors has not yet been reported.
Figure 15.
Inhibitors of type I dehydroquinate dehydratase 141–143 (IC50 = 31–35 μM).134 Antimicrobial activity was not reported.
Inhibitors have been developed that inactivate the toxins TcdA and TcdB, but these do not possess antimicrobial activity.135,136 Inhibitors of the enzymatic component of binary toxin, CDT, that is expressed by some strains of C. difficile have also been investigated.137 This approach potentially mitigates the disease symptoms, since CDI is a toxin-based disease but is not likely to kill the bacteria themselves. However, by not applying a harsh selection pressure, development of resistance might be prevented.
C. difficile spores are a dormant reservoir within the patient or environment that can cause relapsing infection cycles. The C. difficile spores sense host signals such as bile salts and amino acid nutrients and germinate into vegetative bacteria that produce toxins and cause disease. Therefore, one potential strategy to prevent relapse is to activate the spores and then eradicate them with an antimicrobial agent. Identification of agents that can promote germination of the entire population of spores, including those in a hyperdormant state, is challenging. This strategy has been explored in vitro whereby osmotic salts were used to activate spores and the antibiotic nisin then inhibited bacterial outgrowth.138 The identification of agonists of master regulator proteins for spore germination would facilitate this approach. Paradoxically, the opposite approach may also be useful, since blocking germination may prevent colonization and subsequent infection. CspC, a protease, may represent such a target, since it was identified as a bile acid-recognizing germinant receptor and was required for virulence in a hamster model of C. difficile infection.139
3.12. Summary
Antimicrobial agents that previously were considered to lack broad spectrum activity, had poor oral availability, or had systemic toxicity liabilities may be good starting points for further development of new treatments for CDI. In contrast, research into C. difficile focused target based drug discovery and therapeutic approaches directed at neutralizing the disease-causing toxins or preventing spore persistence offer new directions to explore. However, there are challenges and limitations associated with the spore-forming and anaerobic nature of C. difficile that hamper drug discovery efforts focused on this pathogen. These in vitro and in vivo challenges and limitations, discussed below, must be overcome to progress C. difficile-directed new drug discovery.
4. Challenges and Limitations Facing New Drug Discovery
4.1. In Vitro Assays and Challenges
Discovery of new drugs against C. difficile is difficult because of a number of challenges. First, there are technical barriers to general research, as C. difficile must be grown under strictly anaerobic conditions, usually in an anaerobic chamber. Specialist equipment and reagents are therefore required to meet these basic growth conditions, with the outright cost of these items, together with maintenance and consumable costs, being prohibitive to a nonspecialist laboratory. Anaerobic microbiology skills are also required, although stepwise protocols for laboratory maintenance of C. difficile and prevention of culture contamination with spores when handling multiple strains are available.140,141 Industry–academic collaborations are providing a path for advancing compound development as difficulties, and costs associated with establishing the specialist infrastructure and in-house expertise can be overcome.
The awkwardness of manipulating cultures inside an anaerobic chamber limits throughput, as does the size of the incubator compared to standard large shaker incubators available for bacteria grown aerobically. The agar dilution technique remains the gold standard for anaerobic susceptibility testing of anaerobes recommended by NCCLS.142 However, agar dilution has several limitations for C. difficile-specific drug discovery. First, it limits the number of compounds that can be tested at any one time because of the large amount of incubator space required when each compound is tested at numerous concentrations. Agar dilution also requires a larger amount of test compound for preparing the agar, compared to high throughput 96-well or 384-well broth microdilution assays. This can be prohibitive when the compound is only available in milligram quantities or when compound libraries or many analogs need to be tested. Therefore, the agar dilution method favors investigation of a limited number of lead compounds, which can be tested against multiple strains spotted on the same agar plate concurrently, rather than allowing large compound libraries or a large number of synthesized analogs to be screened. A better alternative for this purpose is microdilution susceptibility testing which only uses a fraction of the amount of compound required for the agar dilution method. Note that the disk diffusion method tends not to be used as often as agar dilution or broth microdilution.
C. difficile spores are critical to C. difficile disease etiology, and the infection cycle and must be considered in the evaluation of potential of new drug candidates. It is reasonable to assume that to target vegetative and spore forms, different physicochemical properties for cellular entry and even different compound targets are likely to be required. During CDI, in an in vivo context, C. difficile exists as a mixed population of vegetative bacteria and spores. Therefore, testing that is confined to the vegetative bacterial form or isolated spores does not represent the situation in vivo. Adding to this complexity is the recent demonstration that C. difficile forms biofilms,143 which adds a further hurdle to effective antibiotic treatment and therapeutic development. Spore formation, germination, and outgrowth are highly regulated processes that shift according to growth phase, nutrient availability, and other host-specific selection pressures such as antibiotic treatment and the presence of competing bacteria. To address the effectiveness of compounds at each stage of the C. difficile life cycle, separate assays studying sporulation, germination, and vegetative cell outgrowth are required.144−146 However, performing each of these assays in a high throughput manner to keep up with medicinal chemistry efforts is challenging. Adding to this difficulty is the unreliable sporulation of C. difficile in liquid media, with significant difficulties encountered in attempts to reproducibly synchronize sporulation during in vitro growth.18
Given the physiological limitations of standard in vitro testing of organisms in a single microbe culture system that does not contain competing microbiota, microbial fermentation products, or bile salts, all of which influence C. difficile growth, sporulation, and germination, alternative models have been developed that more closely resemble the host environment. These in vitro gut models involve a single continuous culture reactor, a more complex three-tier system, or a scaled down minibioreactor. The bioreactor is inoculated with a human fecal suspension, treated with antibiotic(s) to model the initial infection susceptibility disease stage, and then challenged with C. difficile spores or vegetative cells to initial infection. Culture samples can be taken over the course of the growth period, and C. difficile total cell counts and spore counts are enumerated using selective agar media. A metagenomic sequencing approach can be used to study changes in the diversity of the microbial populations, which overcomes the problem posed by the presence of uncultivable microbes in this type of analysis.
The three-tier system models the increasing alkalinity found through the gastrointestinal tract in three stages. The effectiveness of oritavancin,1476,148 and cadazolid149 has been evaluated in this system, showing the effect of antibiotic treatment on vegetative and spore C. difficile bacterial numbers as well as the relative numbers of the indigenous bacteria over time.147,148,150 More recently, the three-tier system model has been modified to study C. difficile biofilm formation in the context of the gut microbiota environment and antibiotic treatment.151,152 One disadvantage of the three-tier model is the time required to complete experiments, which are of 6–8 weeks duration, thereby limiting compound throughput. Therefore, despite the potentially useful culture and microbiome information that this model can provide, this system is limited to the evaluation purposes conducted in the late stages of compound development and it is difficult to envision how it can be applied to high throughput screens of large numbers of analogs. The single reactor is a simplified version of the three-tier reactor, which may provide more utility for this purpose. Scaled down minibioreactors, located inside an anaerobic chamber, offer the advantage of being able to test multiple antibiotics at different concentrations at the same time. Once again, monitoring the microbiome community at the population level can be achieved using genome sequencing technologies.
4.2. Advantages and Limitations of Animal Infection Models
Animal infection models are critical for the assessment of drug candidate efficacy, since the contribution of many host factors to disease complexity, such as the immune response, simply cannot be reproduced in vitro. They also allow gross toxicities that may not be apparent through in vitro toxicity assays to be determined and are also essential for downstream regulatory body approval processes. The gold standard C. difficile infection model has been the hamster model (comprehensive review by Best et al).153 However, one disadvantage of the hamster model is that disease takes place at a different site of infection (cecum compared to colon for humans). Furthermore, disease symptoms do not closely mimic those of humans, since the infection does not typically cause diarrhea and almost always leads to death in hamsters as a result of their exquisite sensitivity to the C. difficile toxins. The typical human CDI disease spectrum is not represented in this model, and antimicrobial efficacy can only be assessed in the context of prevention of death. Nevertheless, a wide range of antimicrobials and drug candidates have been evaluated and shown to be effective against C. difficile in the hamster model including rifaximin,154 fidaxomicin,1555,156 cadazolid,157 ramoplanin,158 nitazoxanide,159 rifalazil,160 and oritavancin.161
The more recently developed mouse CDI models appear to correlate more closely with human CDI (recently reviewed by Hutton et al.)162 Importantly, mouse antibiotic treatment CDI induction regimes can be tailored to yield different disease outcomes. Treatment with single antibiotics, such as clindamycin, results in mice that reproducibly develop a mild, nonfatal disease upon infection. By contrast, mice treated with a mixture of broad-spectrum antibiotics develop a severe, often fatal disease following infection.162 The ability to modulate disease severity, presumably through differential effects on host microbiota depending on the antibiotic treatment regime, allows flexibility in the assessment of new therapeutics on disease stages representing the spectrum seen in human CDI. The smaller size of mice also means that less compound is required for testing, which is an important consideration in the early stages of compound development.
Additional advantages of using mouse rather than hamster infection models include the greater ease of scoring disease symptoms such as weight loss and diarrhea as well as the ability to use the diverse range of mouse-specific reagents available for immunohistological tissue or other assessments. Furthermore, disease transmission163 and relapse models164 have been developed that allow these aspects of disease to be studied. Although the latter model does not allow relapse versus reinfection to be distinguished, both of which are likely to be important in recurrent human infection, it does provide a tool that can be used to determine how effective a treatment is in eradicating recurrent infection within the host following a primary infection. The variation between human and animal gut microbiomes must also be considered when animal models are used to test new antimicrobial compounds, since CDI susceptibility hinges on the host microbiome. However, next generation sequencing provides a powerful tool to evaluate these differences and the effect of treatments on microbial populations, providing unique opportunities that will lead to a better understanding of the complex host–microbial interactions that occur during CDI. Such efforts are also focused on understanding what microbiome community structures constitute “healthy”, “susceptible”, and “disease” states, which will also provide insights into our understanding of CDI.
Finally, it is important to consider which C. difficile strains are used in animal infection and CDI treatment studies. Recent epidemiological and genome sequencing studies have shown that C. difficile is a genetically heterogeneous species and that new variants are detected regularly, some of which cause epidemics.7,165 For example, these studies demonstrated that the epidemic BI/NAP01/027 group comprises two independently arisen lineages of fluoroquinolone resistant strains that spread globally7 and that diverse strains of unknown origin cause CDI,165 illustrating how new technologies can provide important insights into disease transmission and evolution on a population scale.7 In the context of drug discovery, it is important to include a variety of strains in compound testing to assess their effectiveness across the range of C. difficile strain variants, especially at the lead optimization stage. These therapeutic agents must be effective against diverse strains, or selective forces will favor the proliferation and dissemination of strains against which the compound is least effective.
5. Conclusion
In the past 10 years C. difficile has become a significant threat to human health. Even with the recent launch of the C. difficile-targeted antibiotic fidaxomicin, there is still no antibiotic available that can completely prevent recurrent disease. There remains an unmet need for better treatments that prevent relapse and reinfection, as well as transmission. To address this need, the scientific and medical communities are developing new treatment agents from both small molecule and biotherapeutic approaches. Selective antibiotics that do not destroy the diversity of microbiota associated with gut homeostasis and that also effectively inhibit spore formation are required. Nonorally absorbed agents, stable to passage through the stomach and small intestine, are desirable to maintain high concentrations of antibiotic at the site of infection, thus minimizing systemic toxicity and reducing the possibility of treatment failure due to MIC creep associated with antimicrobial resistance. Reinvestigation of abandoned antimicrobial lead candidates and analogs for their ability to fit the target product profile above, as occurred to bring the long ago discovered fidaxomicin out of the shadows and to market in 2011, offers one avenue toward the development of new drugs to fight CDI. Meanwhile, as C. difficile gains ground as a superbug to be reckoned with, further investment into understanding the biology of this organism and the subsequent identification of new drug targets will provide opportunities for medicinal chemists to develop first-in-class antimicrobials using target based drug discovery approaches. Other nonantibiotic treatment approaches such as vaccines and monoclonal antibiotics in clinical trials offer hope for another strategy to combat CDI and if successful could change the landscape of C. difficile research and development. The restoration of damaged gut microbiota using bacteriotherapy, whether fecal transplant or microbial ecosystem replacement, and as a treatment, adjunct therapy, or prophylactic offers great promise as a nonantibiotic therapy. The growing understanding of the complex gut microbe interactions and identification of which bacterial populations need to be maintained for good health will influence the development of new drugs. While concerns remain over the long-term health effects of bacteriotherapy, C. difficile drug discovery remains challenging because of the complex disease etiology and the technical challenges inherent with recurrence and relapse in predictive in vivo animal models. Looking forward, a combination of selective antibiotic and microbial restorative treatment may prove a winning formula.
Acknowledgments
This paper was prepared with the support of NHMRC Grant APP1063214. A.M.J. is supported by an Australian Postgraduate Award (APA) Ph.D. scholarship and Queensland Government Smart Futures Scholarship. M.A.T.B. and T.K. were supported by Wellcome Trust Seeding Drug Discovery Award 094977/Z/10/Z. D.L. is supported by an ARC Future Fellowship (Grant FT120100779), and M.A.C. is supported by an NHMRC Principal Research Fellowship (Grant APP1059354).
Glossary
Abbreviations Used
- CDI
Clostridium difficile infection
- CA-CDI
community acquired C. difficile infection
- FMT
fecal microbiota transplantation
- TcdA
toxin A
- TcdB
toxin B
- CDT
binary toxin
Biographies
Angie M. Jarrad received her B.Sc. (Hons) from the University of Adelaide in 2011 and was awarded a University Medal. She is an Australian Postgraduate Award and Queensland Smart Futures Scholarship recipient at the University of Queensland. Her research is on the development of new antibiotics that are effective against C. difficile, focused on novel structural classes derived from existing vancomycin and metronidazole-based therapies. Ms. Jarrad has strong research interests in anti-infective agents and has expertise in medicinal chemistry, organic synthesis, and microbiology.
Tomislav Karoli has over 16 years experience in medicinal and process chemistry working in antibacterial, antiviral, and oncology therapeutic areas in a variety of industrial and academic organisations. He spent 5 years at the University of Queensland working on a number of anti-infective drug discovery programs, including modified glycopeptide derivatives for Gram-positive infections and new antitubercular drugs. Dr. Karoli is currently the Head of Process Chemistry at Progen Pharmaceuticals and is an inventor of PG545, currently in clinical trials in advanced cancer patients.
Mark A. T. Blaskovich has extensive medicinal chemistry expertise and over 15 years of industrial drug development experience, including research on peptidomimetic inhibitors of penicillin binding proteins (Molecumetics) and development of an melanocortin-5 receptor antagonist, currently in phase IIb trials for the treatment of acne (Mimetica). Since moving to the University of Queensland, Dr. Blaskovich has led programs focused on the development of vancomycin analogs for treating Gram-positive infections (including C. difficile), polymyxin analogs for Gram-negative infections, and derivatives of existing antibiotics as probes for mode of action studies.
Dena Lyras is an Australian Research Council Future Fellow investigating the molecular pathogenesis of C. difficile infections. Her work couples the genetics of C. difficile (antibiotic resistance, colonization, and virulence factors) with the mouse model of C. difficile infection. Her laboratory is the only Australian facility currently using an animal model to investigate C. difficile infections and has optimized this model to enable the study of colonization, acute disease, transmission, and disease recurrence.
Matthew A. Cooper is a translational research scientist in anti-infective drug discovery with a solid track record of innovation and intellectual property generation and product launches with 21 patents issued or pending. He is the founder of Cambridge Medical Innovations and a cofounder of Akubio Ltd. Following 9 years working in industry in England, Prof. Cooper returned to Australia to pursue translational research in antibiotic discovery and drug design with research programs focused on novel glycopeptides for Gram-positive therapies (including C. difficile infections), lipopeptides for emergent NDM-1 and ESBL Gram-negative bacteria, and a new class of antibiotics to kill MDR- and XDR-tuberculosis.
Supporting Information Available
Methods for clustering ChEMBL data and Table 1 listing summary of single drug candidates tested against C. difficile. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
§ T.K.: Progen Pharmaceuticals Limited, Darra, Queensland, Australia.
The authors declare no competing financial interest.
Supplementary Material
References
- Antibiotic Resistance Threats in the United States, 2013. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention: Atlanta, GA, 2013; pp 1–114. [Google Scholar]
- Carter G. P.; Rood J. I.; Lyras D. The Role of Toxin A and Toxin B in the Virulence of Clostridium difficile. Trends Microbiol. 2012, 20, 21–29. [DOI] [PubMed] [Google Scholar]
- Gerding D. N.; Johnson S.; Rupnik M.; Aktories K. Clostridium difficile Binary Toxin CDT: Mechanism, Epidemiology, and Potential Clinical Importance. Gut Microbes 2014, 5, 6–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds S. L.; Zapka C.; Kasper D.; Gerber R.; McCormack R.; Macinga D.; Johnson S.; Sambol S.; Fricker C.; Arbogast J.; Gerding D. N. Effectiveness of Hand Hygiene for Removal of Clostridium difficile Spores From Hands. Infect. Control Hosp. Epidemiol. 2013, 34, 302–305. [DOI] [PubMed] [Google Scholar]
- Sunkesula V. C. K.; Kundrapu S.; Jury L. A.; Deshpande A.; Sethi A. K.; Donskey C. J. Potential for Transmission of Spores by Patients Awaiting Laboratory Testing To Confirm Suspected Clostridium difficile Infection. Infect. Control Hosp. Epidemiol. 2013, 34, 306–308. [DOI] [PubMed] [Google Scholar]
- Warny M.; Pepin J.; Fang A.; Killgore G.; Thompson A.; Brazier J.; Frost E.; McDonald L. C. Toxin Production by an Emerging Strain of Clostridium difficile Associated with Outbreaks of Severe Disease in North America and Europe. Lancet 2005, 366, 1079–1084. [DOI] [PubMed] [Google Scholar]
- He M.; Miyajima F.; Roberts P.; Ellison L.; Pickard D. J.; Martin M. J.; Connor T. R.; Harris S. R.; Fairley D.; Bamford K. B.; D’Arc S.; Brazier J.; Brown D.; Coia J. E.; Douce G.; Gerding D.; Kim H. J.; Koh T. H.; Kato H.; Senoh M.; Louie T.; Michell S.; Butt E.; Peacock S. J.; Brown N. M.; Riley T.; Songer G.; Wilcox M.; Pirmohamed M.; Kuijper E.; Hawkey P.; Wren B. W.; Dougan G.; Parkhill J.; Lawley T. D. Emergence and Global Spread of Epidemic Healthcare-Associated Clostridium difficile. Nat. Genet. 2012, 45, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald L. C.; Killgore G. E.; Thompson A.; Owens R. C. Jr.; Kazakova S. V.; Sambol S. P.; Johnson S.; Gerding D. N. An Epidemic, Toxin Gene–Variant Strain of Clostridium difficile. N. Engl. J. Med. 2005, 353, 2433–2441. [DOI] [PubMed] [Google Scholar]
- Loo V. G.; Poirier L.; Miller M. A.; Oughton M.; Libman M. D.; Michaud S.; Bourgault A.-M.; Nguyen T.; Frenette C.; Kelly M. A Predominantly Clonal Multi-Institutional Outbreak of Clostridium difficile-Associated Diarrhea with High Morbidity and Mortality. N. Engl. J. Med. 2005, 353, 2442–2449. [DOI] [PubMed] [Google Scholar]
- Khanna S.; Pardi D. S.; Aronson S. L.; Kammer P. P.; Baddour L. M. Outcomes in Community-Acquired Clostridium difficile Infection. Aliment. Pharmacol. Ther. 2012, 35, 613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine D. P. Vancomycin: a History. Clin. Infect. Dis. 2006, 42, S5–S12. [DOI] [PubMed] [Google Scholar]
- Baines E. J. Metronidazole: Its Past, Present and Future. J. Antimicrob. Chemother. 1978, 4, 97–111. [DOI] [PubMed] [Google Scholar]
- Vardakas K. Z.; Polyzos K. A.; Patouni K.; Rafailidis P. I.; Samonis G.; Falagas M. E. Treatment Failure and Recurrence of Clostridium difficile Infection Following Treatment with Vancomycin or Metronidazole: A Systematic Review of the Evidence. Int. J. Antimicrob. Agents 2012, 40, 1–8. [DOI] [PubMed] [Google Scholar]
- Venugopal A. A.; Johnson S. Current State of Clostridium difficile Treatment Options. Clinical Infectious Diseases 2012, 55Suppl. 2S71–S76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornely O. A.; Miller M. A.; Louie T. J.; Crook D. W.; Gorbach S. L. Treatment of First Recurrence of Clostridium difficile Infection: Fidaxomicin Versus Vancomycin. Clin. Infect. Dis. 2012, 55Suppl. 2S154–S161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie T. J.; Cannon K.; Byrne B.; Emery J.; Ward L.; Eyben M.; Krulicki W. Fidaxomicin Preserves the Intestinal Microbiome during and after Treatment of Clostridium difficile Infection (CDI) and Reduces Both Toxin Reexpression and Recurrence of CDI. Clin. Infect. Dis. 2012, 55Suppl. 2S132–S142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold S. M.; Molitoris D.; Vaisanen M. L.; Song Y.; Liu C.; Bolanos M. In Vitro Activities of OPT-80 and Comparator Drugs against Intestinal Bacteria. Antimicrob. Agents Chemother. 2004, 48, 4898–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babakhani F.; Bouillaut L.; Gomez A.; Sears P.; Nguyen L.; Sonenshein A. L. Fidaxomicin Inhibits Spore Production in Clostridium difficile. Clin. Infect. Dis. 2012, 55Suppl. 2S162–S169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M. Mode of Action of Metronidazole on Anaerobic Bacteria and Protozoa. Surgery 1983, 93, 165–171. [PubMed] [Google Scholar]
- Edwards D. I. Nitroimidazole Drugs—Action and Resistance Mechanisms. II. Mechanisms of Resistance. J. Antimicrob. Chemother. 1993, 31, 201–210. [DOI] [PubMed] [Google Scholar]
- Bolton R. P.; Culshaw M. A. Faecal Metronidazole Concentrations during Oral and Intravenous Therapy for Antibiotic Associated Colitis Due to Clostridium difficile. Gut 1986, 27, 1169–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin J.; Alary M. E.; Valiquette L.; Raiche E.; Ruel J.; Fulop K.; Godin D.; Bourassa C. Increasing Risk of Relapse after Treatment of Clostridium difficile Colitis in Quebec, Canada. Clin. Infect. Dis. 2005, 40, 1591–1597. [DOI] [PubMed] [Google Scholar]
- Loll P. J.; Axelsen P. H. The Structural Biology of Molecular Recognition by Vancomycin. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 265–289. [DOI] [PubMed] [Google Scholar]
- Beauregard D. A.; Williams D. H.; Gwynn M. N.; Knowles D. J. Dimerization and Membrane Anchors in Extracellular Targeting of Vancomycin Group Antibiotics. Antimicrob. Agents Chemother. 1995, 39, 781–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold S. M.; Molitoris D.; Vaisanen M. L. Study of the in Vitro Activities of Rifaximin and Comparator Agents against 536 Anaerobic Intestinal Bacteria from the Perspective of Potential Utility in Pathology Involving Bowel Flora. Antimicrob. Agents Chemother. 2008, 53, 281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z. D.; Ke S.; Palazzini E.; Riopel L.; Dupont H. In Vitro Activity and Fecal Concentration of Rifaximin after Oral Administration. Antimicrob. Agents Chemother. 2000, 44, 2205–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New Drug To Fight Hospital Superbug Infection. http://www.specialisedtherapeutics.com.au/index.php?q=new-drug-to-fight-hospital-superbug-infection.html (accessed Mar 20, 2013).
- Crook D. W.; Walker A. S.; Kean Y.; Weiss K.; Cornely O. A.; Miller M. A.; Esposito R.; Louie T. J.; Stoesser N. E.; Young B. C.; Angus B. J.; Gorbach S. L.; Peto T. E. A.; Fidaxomicin Versus Vancomycin for Clostridium difficile Infection: Meta-Analysis of Pivotal Randomized Controlled Trials. Clin. Infect. Dis. 2012, 55, S93–S103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb W.; Zhu J. From Natural Product to Marketed Drug: the Tiacumicin Odyssey. Nat. Prod. Rep. 2013, 30, 161–174. [DOI] [PubMed] [Google Scholar]
- Lynch T.; Chong P.; Zhang J.; Hizon R.; Du T.; Graham M. R.; Beniac D. R.; Booth T. F.; Kibsey P.; Miller M.; Gravel D.; Mulvey M. R. Canadian Nosocomial Infectious Surveillance Program (CNISP). Characterization of a Stable, Metronidazole-Resistant Clostridium difficile Clinical Isolate. PLoS One 2013, 8, e53757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snydman D. R.; Jacobus N. V.; McDermott L. A. Activity of a Novel Cyclic Lipopeptide, CB-183,315, against Resistant Clostridium difficile and Other Gram-Positive Aerobic and Anaerobic Intestinal Pathogens. Antimicrob. Agents Chemother. 2012, 56, 3448–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman J.; Bauer M. P.; Baines S. D.; Corver J.; Fawley W. N.; Goorhuis B.; Kuijper E. J.; Wilcox M. H. The Changing Epidemiology of Clostridium difficile Infections. Clin. Microbiol. Rev. 2010, 23, 529–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.; Weintraub A.; Fang H.; Nord C. E. Antimicrobial Resistance in Clostridium difficile. Int. J. Antimicrob. Agents 2009, 34, 516–522. [DOI] [PubMed] [Google Scholar]
- Peláez T.; Cercenado E.; Alcalá L.; Marín M.; Martín-López A.; Martínez-Alarcón J.; Catalán P.; Sánchez-Somolinos M.; Bouza E. Metronidazole Resistance in Clostridium difficile Is Heterogeneous. J. Clin. Microbiol. 2008, 46, 3028–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein E. J. C.; Citron D. M.; Sears P.; Babakhani F.; Sambol S. P.; Gerding D. N. Comparative Susceptibilities to Fidaxomicin (OPT-80) of Isolates Collected at Baseline, Recurrence, and Failure from Patients in Two Phase III Trials of Fidaxomicin against Clostridium difficile Infection. Antimicrob. Agents Chemother. 2011, 55, 5194–5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant E. M. Fidaxomicin: New Therapy for Clostridium difficile-Associated Diarrhea. Formulary 2011, 46, 297–308. [Google Scholar]
- O’Connor J. R.; Galang M. A.; Sambol S. P.; Hecht D. W.; Vedantam G.; Gerding D. N.; Johnson S. Rifampin and Rifaximin Resistance in Clinical Isolates of Clostridium difficile. Antimicrob. Agents Chemother. 2008, 52, 2813–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S.; Homann S. R.; Bettin K. M.; Quick J. N.; Clabots C. R.; Peterson L. R.; Gerding D. N. Treatment of Asymptomatic Clostridium difficile Carriers (Fecal Excretors) with Vancomycin or Metronidazole. A Randomized, Placebo-Controlled Trial. Ann. Intern. Med. 1992, 117, 297–302. [DOI] [PubMed] [Google Scholar]
- Sears P.; Crook D. W.; Louie T. J.; Miller M. A.; Weiss K. Fidaxomicin Attains High Fecal Concentrations with Minimal Plasma Concentrations Following Oral Administration in Patients with Clostridium difficile Infection. Clin. Infect. Dis. 2012, 55Suppl. 2S116–S120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly C. P. Fecal Microbiota Transplantation—An Old Therapy Comes of Age. N. Engl. J. Med. 2013, 368, 474–475. [DOI] [PubMed] [Google Scholar]
- Van Nood E.; Vrieze A.; Nieuwdorp M.; Fuentes S.; Zoetendal E. G.; de Vos W. M.; Visser C. E.; Kuijper E. J.; Bartelsman J. F. W. M.; Tijssen J. G. P.; Speelman P.; Dijkgraaf M. G. W.; Keller J. J. Duodenal Infusion of Donor Feces for Recurrent Clostridium difficile. N. Engl. J. Med. 2013, 368, 407–415. [DOI] [PubMed] [Google Scholar]
- Gough E.; Shaikh H.; Manges A. R. Systematic Review of Intestinal Microbiota Transplantation (Fecal Bacteriotherapy) for Recurrent Clostridium difficile Infection. Clin. Infect. Dis. 2011, 53, 994–1002. [DOI] [PubMed] [Google Scholar]
- Zipursky J. S.; Sidorsky T. I.; Freedman C. A.; Sidorsky M. N.; Kirkland K. B. Patient Attitudes toward the Use of Fecal Microbiota Transplantation in the Treatment of Recurrent Clostridium difficile Infection. Clin. Infect. Dis. 2012, 55, 1652–1658. [DOI] [PubMed] [Google Scholar]
- Kahn S. A.; Gorawara Bhat R.; Rubin D. T. Fecal Bacteriotherapy for Ulcerative Colitis: Patients Are Ready, Are We?. Inflammatory Bowel Dis. 2011, 18, 676–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G.Dongjin Dynasty. Zhou Hou Bei Ji Fang; Tianjin Science & Technology Press: Tianjin, China, 2000. [Google Scholar]
- Zhang F.; Luo W.; Shi Y.; Fan Z.; Ji G. Should We Standardize the 1,700-Year-Old Fecal Microbiota Transplantation?. Am. J. Gastroenterol. 2012, 107, 1755–1755. [DOI] [PubMed] [Google Scholar]
- Leake I. Clostridium difficile Infection: Real or Fake-Faecal Matter Does the Trick for Recurrent CDI. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 129. [DOI] [PubMed] [Google Scholar]
- Hamilton M. J.; Weingarden A. R.; Sadowsky M. J.; Khoruts A. Standardized Frozen Preparation for Transplantation of Fecal Microbiota for Recurrent Clostridium difficile Infection. Am. J. Gastroenterol. 2012, 107, 761–767. [DOI] [PubMed] [Google Scholar]
- Youngster I.; Russell G. H.; Pindar C.; Ziv-Baran T.; Sauk J.; Hohmann E. L. Oral, Capsulized, Frozen Fecal Microbiota Transplantation for Relapsing Clostridium difficile Infection. JAMA, J. Am. Med. Assoc. 2014, 312, 1772–1778. [DOI] [PubMed] [Google Scholar]
- Lawley T. D.; Clare S.; Walker A. W.; Stares M. D.; Connor T. R.; Raisen C.; Goulding D.; Rad R.; Schreiber F.; Brandt C. Targeted Restoration of the Intestinal Microbiota with a Simple, Defined Bacteriotherapy Resolves Relapsing Clostridium difficile Disease in Mice. PLoS Pathog. 2012, 8, e1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken J. S.; Borody T.; Brandt L. J.; Brill J. V.; Demarco D. C.; Franzos M. A.; Kelly C.; Khoruts A.; Louie T.; Martinelli L. P.; Moore T. A.; Russell G.; Surawicz C. Treating Clostridium difficile Infection with Fecal Microbiota Transplantation. Clin. Gastroenterol. Hepatol. 2011, 9, 1044–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapel N.; Thomas M.; Corcos O.; Mayeur C.; Barbot-Trystram L.; Bouhnik Y.; Joly F. Practical Implementation of Faecal Transplantation. Clin. Microbiol. Infect. 2014, 20, 1098–1105. [DOI] [PubMed] [Google Scholar]
- The Power of Poo: DIY Instructions. http://thepowerofpoo.blogspot.com.au/p/diy-instructions.html?m=1 (accessed Aug 18, 2013).
- American Gastroenterological Association. AGA Confirms IND Is Required for Fecal Microbiota Transplantation http://www.gastro.org/advocacy-regulation/regulatory-issues/fecal-microbiota-transplant/aga-confirms-ind-is-required-for-fecal-microbiota-transplantation (accessed Oct 29, 2014).
- Guidance for Industry: Enforcement Policy Regarding Investigational New Drug Requirements for Use of Fecal Microbiota for Transplantation To Treat Clostridium difficile Infection Not Responsive to Standard Therapies. http://www.fda.gov/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Guidances/Vaccines/ucm361379.htm (accessed Aug 17, 2013).
- Ratner M. Fecal Transplantation Poses Dilemma for FDA. Nat. Biotechnol. 2014, 32, 401–402. [DOI] [PubMed] [Google Scholar]
- Smith M. B.; Kelly C.; Alm E. J. Policy: How To Regulate Faecal Transplants. Nature 2014, 506, 290–291. [DOI] [PubMed] [Google Scholar]
- Tsutsumi L. S.; Owusu Y. B.; Hurdle J. G.; Sun D. Progress in the Discovery of Treatments for C. difficile Infection: A Clinical and Medicinal Chemistry Review. Curr. Top. Med. Chem. (Sharjah, United Arab Emirates) 2014, 14, 152–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phase I Trial of a Single Dose of CRS3123. Identifier NCT01551004. http://clinicaltrials.gov/show/NCT01551004 (accessed Mar 21, 2013).
- Novacta Biosystems Limited completes phase I study of NVB302 against C. difficile infection in healthy volunteers. http://www.novactabio.com/news.php (accessed Mar 18, 2014).
- A Study of SMT19969 Compared with Vancomycin for the Treatment of Clostridium difficile-Associated Diarrhoea (CDAD). Identifier NCT02092935. http://clinicaltrials.gov/show/NCT02092935 (accessed Jan 18, 2015).
- LaMarche M. J.; Leeds J. A.; Amaral A.; Brewer J. T.; Bushell S. M.; Deng G.; Dewhurst J. M.; Ding J.; Dzink-Fox J.; Gamber G.; Jain A.; Lee K.; Lee L.; Lister T.; McKenney D.; Mullin S.; Osborne C.; Palestrant D.; Patane M. A.; Rann E. M.; Sachdeva M.; Shao J.; Tiamfook S.; Trzasko A.; Whitehead L.; Yifru A.; Yu D.; Yan W.; Zhu Q. Discovery of LFF571: An Investigational Agent for Clostridium difficile Infection. J. Med. Chem. 2012, 55, 2376–2387. [DOI] [PubMed] [Google Scholar]
- Safety and Efficacy of Multiple Daily Dosing of Oral LFF571 in Patients with Moderate Clostridium difficile Infections. Identifier NCT01232595. http://www.clinicaltrials.gov/ct2/show/NCT01232595?term=clostridium+difficile&rank=14 (accessed Mar 20, 2013).
- Phase 3 Study with Cadazolid in CDAD. Identifier NCT01987895. https://clinicaltrials.gov/ct2/show/NCT01987895?term=cadazolid&rank=2 (accessed May 21, 2014).
- Business Wire. Nanotherapeutics Acquires Two Late Stage Clinical Programs for Alzheimer’s Treatment and CDA Disease. http://www.businesswire.com/news/home/20091216005124/en/Nanotherapeutics-Acquires-Late-Stage-Clinical-Programs-Alzheimer%E2%80%99s#.UyetsVzoXjI (accessed Mar 18, 2014).
- Nanotherapeutics. NTI-851 (Ramoplanin). http://www.nanotherapeutics.com/ramoplanin/ (accessed Oct 29, 2014).
- McLoughlin F.CB-315 Fact Sheet, 2013; pp 1–3.
- Mascio C. T. M.; Mortin L. I.; Howland K. T.; Van Praagh A. D. G.; Zhang S.; Arya A.; Chuong C. L.; Kang C.; Li T.; Silverman J. A. In Vitro and In Vivo Characterization of CB-183,315, a Novel Lipopeptide Antibiotic for Treatment of Clostridium difficile. Antimicrob. Agents Chemother. 2012, 56, 5023–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharmacokinetics and Safety of Tigecycline in the Treatment of C. difficile Associated Diarrhea (CDAD). Identifier NCT01401023. http://clinicaltrials.gov/ct2/show/NCT01401023 (accessed Mar 21, 2013).
- Study of Nitazoxanide in the Treatment of Clostridium difficile Colitis. Identifier NCT00417872. http://www.clinicaltrials.gov/ct2/show/NCT00417872?term=clostridium+difficile&rank=22 (accessed Mar 20, 2013).
- Study of a Candidate Clostridium difficile Toxoid Vaccine in Subjects at Risk for C. difficile Infection. Identifier NCT01887912. http://clinicaltrials.gov/ct2/show/NCT01887912?term=c+difficile+and+vaccine&rank=2 (accessed Oct 23, 2014).
- An Open-Label Study Assessing Safety, Immunogenicity and Dose Response of IC84. Identifier NCT01296386. http://clinicaltrials.gov/ct2/show/NCT01296386?term=IC84&rank=1 (accessed Oct 23, 2014).
- Johnson S.; Louie T. J.; Gerding D. N.; Cornely O. A.; Chasan-Taber S.; Fitts D.; Gelone S. P.; Broom C.; Davidson D. M. Polymer Alternative for CDI Treatment (PACT) investigators. Vancomycin, Metronidazole, or Tolevamer for Clostridium difficile Infection: Results From Two Multinational, Randomized, Controlled Trials. Clin. Infect. Dis. 2014, 59, 345–354. [DOI] [PubMed] [Google Scholar]
- A Study of MK-3415, MK-6072, and MK-3415A in Participants Receiving Antibiotic Therapy for Clostridium difficile Infection (MK-3415A-001 AM2). Identifier NCT01241552. http://www.clinicaltrials.gov/ct2/show/NCT01241552?term=clostridium+difficile&rank=6 (accessed Mar 20, 2013).
- Safety and Efficacy Study of VP20621 for Prevention of Recurrent Clostridium difficile Infection. Identifier NCT01259726. http://clinicaltrials.gov/ct2/show/NCT01259726?term=VP20621&rank=1 (accessed Jan 21, 2015).
- Brouwer M. S. M.; Roberts A. P.; Hussain H.; Williams R. J.; Allan E.; Mullany P. Horizontal Gene Transfer Converts Non-Toxigenic Clostridium difficile Strains into Toxin Producers. Nat. Commun. 2013, 4, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villano S. A.; Seiberling M.; Tatarowicz W.; Monnot-Chase E.; Gerding D. N. Evaluation of an Oral Suspension of VP20621, Spores of Nontoxigenic Clostridium difficile Strain M3, in Healthy Subjects. Antimicrob. Agents Chemother. 2012, 56, 5224–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie C. G.; Bucci V.; Stein R. R.; McKenney P. T.; Ling L.; Gobourne A.; No D.; Liu H.; Kinnebrew M.; Viale A.; Littmann E.; van den Brink M. R. M.; Jenq R. R.; Taur Y.; Sander C.; Cross J.; Toussaint N. C.; Xavier J. B.; Pamer E. G. Precision Microbiome Reconstitution Restores Bile Acid Mediated Resistance to Clostridium difficile. Nature 2015, 517, 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. W.; Mubasher M.; Fang C. Y.; Reifer C.; Miller L. E. Dose-Response Efficacy of a Proprietary Probiotic Formula of Lactobacillus acidophilus Cl1285 and Lactobacillus casei Lbc80r for Antibiotic-Associated Diarrhea and Clostridium difficile-Associated Diarrhea Prophylaxis in Adult Patients. Am. J. Gastroenterol. 2010, 105, 1636–1641. [DOI] [PubMed] [Google Scholar]
- Microbiota Restoration Therapy for Recurrent Clostridium difficile-associated Diarrhea. Identifier NCT01925417. https://clinicaltrials.gov/ct2/show/NCT01925417?term=FMT&phase=1&rank=17 (accessed Oct 28, 2014).
- Monarch Labs. Monarch Labs and BTER Foundation collaborate to develop fecal transplantion products http://www.monarchlabs.com/Monarch-Labs-and-BTER-Foundation-collaborate-to-develop-fecal-transplantion-products (accessed Aug 18, 2013).
- Gaulton A.; Bellis L. J.; Bento A. P.; Chambers J.; Davies M.; Hersey A.; Light Y.; McGlinchey S.; Michalovich D.; Al-Lazikani B.; Overington J. P. ChEMBL: A Large-Scale Bioactivity Database for Drug Discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard T. E.; Wang X.; Olekhnovich I.; Koerner T.; Seymour C.; Hoffman P. S.; Macdonald T. L. Biological Activity of Modified and Exchanged 2-Amino-5-Nitrothiazole Amide Analogues of Nitazoxanide. Bioorg. Med. Chem. Lett. 2010, 20, 3537–3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard T. E.; Wang X.; Olekhnovich I.; Koerner T.; Seymour C.; Salamoun J.; Warthan M.; Hoffman P. S.; Macdonald T. L. Synthesis and Antimicrobial Evaluation of Nitazoxanide-Based Analogues: Identification of Selective and Broad Spectrum Activity. ChemMedChem 2011, 6, 362–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura Y.; Nishino S.; Ohno M.; Sakane K.; Matsumoto Y.; Morinaga C.; Ishikawa H.; Takasugi H. Anti-Helicobacter pylori Agents. 3. 2-[(Arylalkyl)Guanidino]-4-furylthiazoles. J. Med. Chem. 1999, 42, 2920–2926. [DOI] [PubMed] [Google Scholar]
- Katsura Y.; Nishino S.; Tomishi T.; Sakane K.; Matsumoto Y.; Ishikawa H.; Takasugi H. Anti-Helicobacter pylori Agents. 2. Structure Activity Relationships in a New Series of 2-Alkylguanidino-4-furylthiazoles. Bioorg. Med. Chem. Lett. 1998, 8, 1307–1312. [DOI] [PubMed] [Google Scholar]
- Emerging Drugs for Active Tuberculosis. http://www.thieme-connect.de/DOI/DOI?10.1055/s-0028-1085706 (accessed Apr 9, 2014).
- Pankuch G. A.; Appelbaum P. C. Activities of Tizoxanide and Nitazoxanide Compared to Those of Five Other Thiazolides and Three Other Agents against Anaerobic Species. Antimicrob. Agents Chemother. 2006, 50, 1112–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello C.; Karpanen T.; Lambert P. A.; Mistry P.; Parker K. J.; Rathbone D. L.; Ren J.; Wheeldon L.; Worthington T. Thiosemicarbazones Active Against Clostridium difficile. Bioorg. Med. Chem. Lett. 2008, 18, 1708–1711. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y.; Kalisiak J.; Korthals K.; Lauwaet T.; Cheung D. Y.; Lozano R.; Cobo E. R.; Upcroft P.; Upcroft J. A.; Berg D. E.; Gillin F. D.; Fokin V. V.; Sharpless K. B.; Eckmann L. Expanded Therapeutic Potential in Activity Space of Next-Generation 5-Nitroimidazole Antimicrobials with Broad Structural Diversity. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 17564–17569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J. Y.; Mukhopadhyay A. K.; Akada J. K.; Dailidiene D.; Hoffman P. S.; Berg D. E. Roles of FrxA and RdxA Nitroreductases of Helicobacter pylori in Susceptibility and Resistance to Metronidazole. J. Bacteriol. 2001, 183, 5155–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segreti J.; Goodman L. J.; Trenholme G. M. In Vitro Activity of New Quinoxaline Compounds against Campylobacter Species and Clostridium difficile. Diagn. Microbiol. Infect. Dis. 1993, 17, 177–179. [DOI] [PubMed] [Google Scholar]
- Zhang S.-J.; Yang Q.; Xu L.; Chang J.; Sun X. Synthesis and Antibacterial Activity against Clostridium difficile of Novel Demethylvancomycin Derivatives. Bioorg. Med. Chem. Lett. 2012, 22, 4942–4945. [DOI] [PubMed] [Google Scholar]
- Kirst H. A.; Toth J. E.; Debono M.; Willard K. E.; Truedell B. A.; Ott J. L.; Counter F. T.; Felty-Duckworth A. M.; Pekarek R. S. Synthesis and Evaluation of Tylosin-Related Macrolides Modified at the Aldehyde Function: A New Series of Orally Effective Antibiotics. J. Med. Chem. 1988, 31, 1631–1641. [DOI] [PubMed] [Google Scholar]
- Terasawa T.; Watanabe M.; Okubo T.; Mitsuhashi S. In Vitro Activity of YM133, a New Semisynthesized Macrolide. Antimicrob. Agents Chemother. 1991, 35, 1370–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez J. P.; Gogliotti R. D.; Domagala J. M.; Gracheck S. J.; Huband M. D.; Sesnie J. A.; Cohen M. A.; Shapiro M. A. The Synthesis, Structure-Activity, and Structure-Side Effect Relationships of a Series of 8-Alkoxy- and 5-Amino-8-alkoxyquinolone Antibacterial Agents. J. Med. Chem. 1995, 38, 4478–4487. [DOI] [PubMed] [Google Scholar]
- Rosen T.; Fernandes P. B.; Marovich M. A.; Shen L.; Mao J.; Pernet A. G. Aromatic Dienoyl Tetramic Acids. Novel Antibacterial Agents with Activity against Anaerobes and Staphylococci. J. Med. Chem. 1989, 32, 1062–1069. [DOI] [PubMed] [Google Scholar]
- Ueda C.; Tateda K.; Horikawa M.; Kimura S.; Ishii Y.; Nomura K.; Yamada K.; Suematsu T.; Inoue Y.; Ishiguro M.; Miyairi S.; Yamaguchi K. Anti-Clostridium difficile Potential of Tetramic Acid Derivatives from Pseudomonas Aeruginosa Quorum-Sensing Autoinducers. Antimicrob. Agents Chemother. 2010, 54, 683–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwowski J. P.; Jackson M.; Theriault R. J.; Barlow G. J.; Coen L.; Hensey D. M.; Humphrey P. E. Tirandalydigin, a Novel Tetramic Acid of the Tirandamycin-Streptolydigin Type. I. Taxonomy of the Producing Organism, Fermentation and Biological Activity. J. Antibiot. 1992, 45, 1125–1132. [DOI] [PubMed] [Google Scholar]
- Shoji J.; Sakazaki R.; Hattori T.; Matsumoto K.; Uotani N.; Yoshida T. Isolation and Characterization of Agglomerins A, B, C and D. J. Antibiot. 1989, 42, 1729–1733. [DOI] [PubMed] [Google Scholar]
- Terui Y.; Sakazaki R.; Shoji J. Structures of Agglomerins. J. Antibiot. 1990, 43, 1245–1253. [DOI] [PubMed] [Google Scholar]
- Butler M. M.; Williams J. D.; Peet N. P.; Moir D. T.; Panchal R. G.; Bavari S.; Shinabarger D. L.; Bowlin T. L. Comparative in Vitro Activity Profiles of Novel Bis-Indole Antibacterials against Gram-Positive and Gram-Negative Clinical Isolates. Antimicrob. Agents Chemother. 2010, 54, 3974–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal R. G.; Ulrich R. L.; Lane D.; Butler M. M.; Houseweart C.; Opperman T.; Williams J. D.; Peet N. P.; Moir D. T.; Nguyen T.; Gussio R.; Bowlin T.; Bavari S. Novel Broad-Spectrum Bis-(imidazolinylindole) Derivatives with Potent Antibacterial Activities against Antibiotic-Resistant Strains. Antimicrob. Agents Chemother. 2009, 53, 4283–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrus J. I.; Kelso M. J.; Bremner J. B.; Ball A. R.; Casadei G.; Lewis K. Structure-Activity Relationships of 2-Aryl-1H-Indole Inhibitors of the NorA Efflux Pump in Staphylococcus aureus. Bioorg. Med. Chem. Lett. 2008, 18, 4294–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternansky R. J.; Jordan C. L.; Eudaly J. A.; Kasher J. S. Synthesis and Anaerobic Activity of Novel 1-Carba-1-dethiacephalosporins. J. Med. Chem. 1993, 36, 2332–2334. [DOI] [PubMed] [Google Scholar]
- Corraz A. J.; Dax S. L.; Dunlap N. K.; Georgopapadakou N. H.; Keith D. D.; Pruess D. L.; Rossman P. L.; Then R.; Unowsky J.; Wei C. C. Dual-Action Penems and Carbapenems. J. Med. Chem. 1992, 35, 1828–1839. [DOI] [PubMed] [Google Scholar]
- Mani N.; Gross C. H.; Parsons J. D.; Hanzelka B.; Müh U.; Mullin S.; Liao Y.; Grillot A.-L.; Stamos D.; Charifson P. S.; Grossman T. H. In Vitro Characterization of the Antibacterial Spectrum of Novel Bacterial Type II Topoisomerase Inhibitors of the Aminobenzimidazole Class. Antimicrob. Agents Chemother. 2006, 50, 1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charifson P. S.; Grillot A.-L.; Grossman T. H.; Parsons J. D.; Badia M.; Bellon S.; Deininger D. D.; Drumm J. E.; Gross C. H.; LeTiran A.; Liao Y.; Mani N.; Nicolau D. P.; Perola E.; Ronkin S.; Shannon D.; Swenson L. L.; Tang Q.; Tessier P. R.; Tian S.-K.; Trudeau M.; Wang T.; Wei Y.; Zhang H.; Stamos D. Novel Dual-Targeting Benzimidazole Urea Inhibitors of DNA Gyrase and Topoisomerase IV Possessing Potent Antibacterial Activity: Intelligent Design and Evolution through the Judicious Use of Structure-Guided Design and Structure-Activity Relationships. J. Med. Chem. 2008, 51, 5243–5263. [DOI] [PubMed] [Google Scholar]
- Grillot A.-L.; Tiran A. L.; Shannon D.; Krueger E.; Liao Y.; O’Dowd H.; Tang Q.; Ronkin S.; Wang T.; Waal N.; Li P.; Lauffer D.; Sizensky E.; Tanoury J.; Perola E.; Grossman T. H.; Doyle T.; Hanzelka B.; Jones S.; Dixit V.; Ewing N.; Liao S.; Boucher B.; Jacobs M.; Bennani Y.; Charifson P. S. Second-Generation Antibacterial Benzimidazole Ureas: Discovery of a Preclinical Candidate with Reduced Metabolic Liability. J. Med. Chem. 2014, 57, 8792–8816. [DOI] [PubMed] [Google Scholar]
- Perola E.; Stamos D.; Grillot A.-L.; Ronkin S.; Wang T.; LeTiran A.; Tang Q.; Deininger D. D.; Liao Y.; Tian S.-K.; Drumm J. E.; Nicolau D. P.; Tessier P. R.; Mani N.; Grossman T. H.; Charifson P. S. Successful Application of Serum Shift Prediction Models to the Design of Dual Targeting Inhibitors of Bacterial Gyrase B and Topoisomerase IV with Improved in Vivo Efficacy. Bioorg. Med. Chem. Lett. 2014, 24, 2177–2181. [DOI] [PubMed] [Google Scholar]
- Stokes N. R.; Thomaides-Brears H. B.; Barker S.; Bennett J. M.; Berry J.; Collins I.; Czaplewski L. G.; Gamble V.; Lancett P.; Logan A.; Lunniss C. J.; Peasley H.; Pommier S.; Price D.; Smee C.; Haydon D. J. Biological Evaluation of Benzothiazole Ethyl Urea Inhibitors of Bacterial Type II Topoisomerases. Antimicrob. Agents Chemother. 2013, 57, 5977–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesel L.; Hecht D. W.; Osmolski J. R.; Gerding D.; Flattery A.; Li F.; Lan J.; Lipari P.; Polishook J. D.; Liang L.; Liu J.; Olsen D. B.; Singh S. B. Kibdelomycin Is a Potent and Selective Agent against Toxigenic Clostridium difficile. Antimicrob. Agents Chemother. 2014, 58, 2387–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipexium Pharmaceuticals, Inc. Clinical Trials: Mild Infections of Diabetic Foot Ulcers http://www.dipexiumpharmaceuticals.com/locilex/clinical-trials (accessed Jan 21, 2015).
- Fuchs P. C.; Barry A. L.; Brown S. D. In Vitro Antimicrobial Activity of MSI-78, a Magainin Analog. Antimicrob. Agents Chemother. 1998, 42, 1213–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y.; MacDonald D. L.; Holroyd K. J.; Thornsberry C.; Wexler H.; Zasloff M. In Vitro Antibacterial Properties of Pexiganan, an Analog of Magainin. Antimicrob. Agents Chemother. 1999, 43, 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H.; Hedberg M.; Wade D.; Edlund C. Activities of Synthetic Hybrid Peptides against Anaerobic Bacteria: Aspects of Methodology and Stability. Antimicrob. Agents Chemother. 2000, 44, 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R.; Suárez J. M.; Weisblum B.; Gellman S. H.; McBride S. M. Synthetic Polymers Active against Clostridium difficile Vegetative Cell Growth and Spore Outgrowth. J. Am. Chem. Soc. 2014, 136, 14498–14504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashio S.; Iwasawa H.; Dun F. Y.; Kanemitsu K.; Shimada J. Everninomicin, a New Oligosaccharide Antibiotic: Its Antimicrobial Activity, Post-Antibiotic Effect and Synergistic Bactericidal Activity. Drugs Exp. Clin. Res. 1995, 21, 7–16. [PubMed] [Google Scholar]
- Tanaka K.; Kato N.; Watanabe K. In Vitro Activity of an Evernimicin Derivative, SCH27899, against Anaerobic Bacteria and Propionibacterium Acnes. J. Antimicrob. Chemother. 2000, 46, 465–469. [DOI] [PubMed] [Google Scholar]
- Hanada M.; Kaneta K.; Nishiyama Y.; Hoshino Y.; Konishi M.; Oki T. Hydramycin, a New Antitumor Antibiotic. Taxonomy, Isolation, Physico-Chemical Properties, Structure and Biological Activity. J. Antibiot. 1991, 44, 824–831. [DOI] [PubMed] [Google Scholar]
- McCullough J. E.; Muller M. T.; Howells A. J.; Maxwell A.; O’Sullivan J.; Summerill R. S.; Parker W. L.; Wells J. S.; Bonner D. P.; Fernandes P. B. Clerocidin, a Terpenoid Antibiotic, Inhibits Bacterial DNA Gyrase. J. Antibiot. 1993, 46, 526–530. [DOI] [PubMed] [Google Scholar]
- Nadai M.; Palù G.; Palumbo M.; Richter S. N. Differential Targeting of Unpaired Bases within Duplex DNA by the Natural Compound Clerocidin: A Valuable Tool To Dissect DNA Secondary Structure. PLoS One 2012, 7, e52994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banskota A. H.; Aouidate M.; rensen D. S. O.; Ibrahim A.; Piraee M.; Zazopoulos E.; Alarco A.-M.; Gourdeau H.; Mellon C.; Farnet C. M.; Falardeau P.; McAlpine J. B. TLN-05220, TLN-05223, New Echinosporamicin-Type Antibiotics, and Proposed Revision of the Structure of Bravomicins. J. Antibiot. 2009, 62, 565–570. [DOI] [PubMed] [Google Scholar]
- Sakoulas G.; Nam S.-J.; Loesgen S.; Fenical W.; Jensen P. R.; Nizet V.; Hensler M. Novel Bacterial Metabolite Merochlorin a Demonstrates in Vitro Activity against Multi-Drug Resistant Methicillin-Resistant Staphylococcus aureus. PLoS One 2012, 7, e29439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper H. P.; George J. H. Biomimetic Total Synthesis of (±)-Merochlorin A. Angew. Chem., Int. Ed. 2013, 52, 12170–12173. [DOI] [PubMed] [Google Scholar]
- Wu X.; Alam M. Z.; Feng L.; Tsutsumi L. S.; Sun D.; Hurdle J. G. Prospects for Flavonoid and Related Phytochemicals as Nature-Inspired Treatments for Clostridium difficile Infection. J. Appl. Microbiol. 2014, 116, 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L.; Maddox M. M.; Alam M. Z.; Tsutsumi L. S.; Narula G.; Bruhn D. F.; Wu X.; Sandhaus S.; Lee R. B.; Simmons C. J.; Tse-Dinh Y.-C.; Hurdle J. G.; Lee R. E.; Sun D. Synthesis, Structure-Activity Relationship Studies, and Antibacterial Evaluation of 4-Chromanones and Chalcones, as Well as Olympicin a and Derivatives. J. Med. Chem. 2014, 57, 8398–8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucci M. J.; Bronson J. J.; Barrett J. F.; DenBleyker K. L.; Discotto L. F.; Fung-Tomc J. C.; Ueda Y. Antimicrobial Evaluation of Nocathiacins, a Thiazole Peptide Class of Antibiotics. Antimicrob. Agents Chemother. 2004, 48, 3697–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMarche M. J.; Leeds J. A.; Dzink-Fox J.; Gangl E.; Krastel P.; Neckermann G.; Palestrant D.; Patane M. A.; Rann E. M.; Tiamfook S.; Yu D. Antibiotic Optimization and Chemical Structure Stabilization of Thiomuracin A. J. Med. Chem. 2012, 55, 6934–6941. [DOI] [PubMed] [Google Scholar]
- Heap J. T.; Pennington O. J.; Cartman S. T.; Carter G. P.; Minton N. P. The ClosTron: A Universal Gene Knock-Out System for the Genus Clostridium. J. Microbiol. Methods 2007, 70, 452–464. [DOI] [PubMed] [Google Scholar]
- Jadhav A.; Ezhilarasan V.; Prakash Sharma O.; Pan A. Clostridium-DTDB: A Comprehensive Database for Potential Drug Targets of Clostridium difficile. Comput. Biol. Med. 2013, 43, 362–367. [DOI] [PubMed] [Google Scholar]
- Larocque M.; Chénard T.; Najmanovich R. A Curated C. difficile Strain 630 Metabolic Network: Prediction of Essential Targets and Inhibitors. BMC Syst. Biol. 2014, 8, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang T. H. T.; de la Riva L.; Fagan R. P.; Storck E. M.; Heal W. P.; Janoir C.; Fairweather N. F.; Tate E. W. Chemical Probes of Surface Layer Biogenesis in Clostridium difficile. ACS Chem. Biol. 2010, 5, 279–285. [DOI] [PubMed] [Google Scholar]
- Ratia K.; Light S. H.; Antanasijevic A.; Anderson W. F.; Caffrey M.; Lavie A. Discovery of Selective Inhibitors of the Clostridium difficile Dehydroquinate Dehydratase. PLoS One 2014, 9, e89356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri A. W.; Lupardus P. J.; Deu E.; Albrow V. E.; Garcia K. C.; Bogyo M.; Shen A. Rational Design of Inhibitors and Activity-Based Probes Targeting Clostridium difficile Virulence Factor TcdB. Chem. Biol. 2010, 17, 1201–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdeen S. J.; Swett R. J.; Feig A. L. Peptide Inhibitors Targeting Clostridium difficile Toxins A and B. ACS Chem. Biol. 2010, 5, 1097–1103. [DOI] [PubMed] [Google Scholar]
- Maurer B.; Mathias U.; Papatheodorou P.; Shekfeh S.; Orth J.; Jank T.; Schwan C.; Sippl W.; Aktories K.; Jung M. From Cosubstrate Similarity to Inhibitor Diversity—Inhibitors of ADP-Ribosyltransferases from Kinase Inhibitor Screening. Mol. BioSyst. 2011, 7, 799–808. [DOI] [PubMed] [Google Scholar]
- Nerandzic M. M.; Donskey C. J. Activate to Eradicate: Inhibition of Clostridium difficile Spore Outgrowth by the Synergistic Effects of Osmotic Activation and Nisin. PLoS One 2013, 8, e54740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis M. B.; Allen C. A.; Shrestha R.; Sorg J. A. Bile Acid Recognition by the Clostridium difficile Germinant Receptor, CspC, Is Important for Establishing Infection. PLoS Pathog. 2013, 9, e1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg J. A.; Dineen S. S.. Laboratory Maintenance of Clostridium difficile. Curr. Protoc. Microbiol. 2009, Chapter 9, Unit 9A.1. [DOI] [PubMed] [Google Scholar]
- Edwards A. N.; Suárez J. M.; McBride S. M. Culturing and Maintaining Clostridium difficile in an Anaerobic Environment. J. Visualized Exp. 2013, e50787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria, 8th ed.; CLSI M11-A8 ; Clinical and Laboratory Standards Institute: Wayne, PA, 2012; pp 1–56. [Google Scholar]
- Dawson L. F.; Valiente E.; Faulds-Pain A.; Donahue E. H.; Wren B. W. Characterisation of Clostridium difficile Biofilm Formation, a Role for Spo0A. PLoS One 2012, 7, e50527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen C. A.; Babakhani F.; Sears P.; Nguyen L.; Sorg J. A. Both Fidaxomicin and Vancomycin Inhibit Outgrowth of Clostridium difficile Spores. Antimicrob. Agents Chemother. 2013, 57, 664–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton C. H.; Freeman J.; Baines S. D.; Crowther G. S.; Nicholson S.; Wilcox M. H. Evaluation of the Effect of Oritavancin on Clostridium difficile Spore Germination, Outgrowth and Recovery. J. Antimicrob. Chemother. 2013, 68, 2078–2082. [DOI] [PubMed] [Google Scholar]
- Francis M. B.; Allen C. A.; Sorg J. A. Muricholic Acids Inhibit Clostridium difficile Spore Germination and Growth. PLoS One 2013, 8, e73653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines S. D.; O’Connor R.; Saxton K.; Freeman J.; Wilcox M. H. Comparison of Oritavancin Versus Vancomycin as Treatments for Clindamycin-Induced Clostridium difficile PCR Ribotype 027 Infection in a Human Gut Model. J. Antimicrob. Chemother. 2008, 62, 1078–1085. [DOI] [PubMed] [Google Scholar]
- Crowther G. S.; Baines S. D.; Todhunter S. L.; Freeman J.; Chilton C. H.; Wilcox M. H. Evaluation of NVB302 Versus Vancomycin Activity in an in Vitro Human Gut Model of Clostridium difficile Infection. J. Antimicrob. Chemother. 2013, 68, 168–176. [DOI] [PubMed] [Google Scholar]
- Chilton C. H.; Crowther G. S.; Baines S. D.; Todhunter S. L.; Freeman J.; Locher H. H.; Athanasiou A.; Wilcox M. H. In Vitro Activity of Cadazolid against Clinically Relevant Clostridium difficile Isolates and in an in Vitro Gut Model of C. difficile Infection. J. Antimicrob. Chemother. 2014, 69, 697–705. [DOI] [PubMed] [Google Scholar]
- Chilton C. H.; Freeman J.; Crowther G. S.; Todhunter S. L.; Wilcox M. H. Effectiveness of a Short (4 Day) Course of Oritavancin in the Treatment of Simulated Clostridium difficile Infection Using a Human Gut Model. J. Antimicrob. Chemother. 2012, 67, 2434–2437. [DOI] [PubMed] [Google Scholar]
- Crowther G. S.; Chilton C. H.; Todhunter S. L.; Nicholson S.; Freeman J.; Baines S. D.; Wilcox M. H. Development and Validation of a Chemostat Gut Model To Study Both Planktonic and Biofilm Modes of Growth of Clostridium difficile and Human Microbiota. PLoS One 2014, 9, e88396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther G. S.; Chilton C. H.; Todhunter S. L.; Nicholson S.; Freeman J.; Baines S. D.; Wilcox M. H. Comparison of Planktonic and Biofilm-Associated Communities of Clostridium difficile and Indigenous Gut Microbiota in a Triple-Stage Chemostat Gut Model. J. Antimicrob. Chemother. 2014, 69, 2137–2147. [DOI] [PubMed] [Google Scholar]
- Best E. L.; Freeman J.; Wilcox M. H. Models for the Study of Clostridium difficile Infection. Gut Microbes 2012, 3, 145–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkotou E.; Moss A. C.; Michos A.; Espinoza D.; Cloud J. W.; Mustafa N.; O’Brien M.; Pothoulakis C.; Kelly C. P. Comparative Efficacies of Rifaximin and Vancomycin for Treatment of Clostridium difficile-Associated Diarrhea and Prevention of Disease Recurrence in Hamsters. Antimicrob. Agents Chemother. 2008, 52, 1121–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R. N.; Hardy D. J.; Shipkowitz N. L.; Hanson C. W.; Ramer N. C.; Fernandes P. B.; Clement J. J. In Vitro and in Vivo Evaluation of Tiacumicins B and C against Clostridium difficile. Antimicrob. Agents Chemother. 1991, 35, 1108–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner U. A.; Bell S. J.; O’Leary A. L.; Hoang T.; Stone K. C.; Young C. L.; Critchley I. A.; Janjic N. Inhibitory Effect of REP3123 on Toxin and Spore Formation in Clostridium difficile, and in Vivo Efficacy in a Hamster Gastrointestinal Infection Model. J. Antimicrob. Chemother. 2009, 63, 964–971. [DOI] [PubMed] [Google Scholar]
- Locher H. H.; Seiler P.; Chen X.; Schroeder S.; Pfaff P.; Enderlin M.; Klenk A.; Fournier E.; Hubschwerlen C.; Ritz D.; Kelly C. P.; Keck W. In Vitro and in Vivo Antibacterial Evaluation of Cadazolid, a New Antibiotic for Treatment of Clostridium difficile Infections. Antimicrob. Agents Chemother. 2014, 58, 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman J.; Baines S. D.; Jabes D.; Wilcox M. H. Comparison of the Efficacy of Ramoplanin and Vancomycin in Both in Vitro and in Vivo Models of Clindamycin-Induced Clostridium difficile Infection. J. Antimicrob. Chemother. 2005, 56, 717–725. [DOI] [PubMed] [Google Scholar]
- McVay C. S.; Rolfe R. D. In Vitro and in Vivo Activities of Nitazoxanide against Clostridium difficile. Antimicrob. Agents Chemother. 2000, 44, 2254–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton P. M.; O’Brien M.; Kokkotou E.; Eisenstein A. M.; Rothstein D. M.; Paraschos S.; Kelly C. P.; Pothoulakis C. Rifalazil Treats and Prevents Relapse of Clostridium difficile-Associated Diarrhea in Hamsters. Antimicrob. Agents Chemother. 2004, 48, 3975–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman J.; Marquis M.; Crowther G. S.; Todhunter S. L.; Fawley W. N.; Chilton C. H.; Moeck G.; Lehoux D.; Wilcox M. H. Oritavancin Does Not Induce Clostridium difficile Germination and Toxin Production in Hamsters or a Human Gut Model. J. Antimicrob. Chemother. 2012, 67, 2919–2926. [DOI] [PubMed] [Google Scholar]
- Hutton M. L.; Mackin K. E.; Chakravorty A.; Lyras D. Small Animal Models for the Study of Clostridium difficile Disease Pathogenesis. FEMS Microbiol. Lett. 2014, 352, 140–149. [DOI] [PubMed] [Google Scholar]
- Deakin L. J.; Clare S.; Fagan R. P.; Dawson L. F.; Pickard D. J.; West M. R.; Wren B. W.; Fairweather N. F.; Dougan G.; Lawley T. D. Clostridium difficile spo0A Gene Is a Persistence and Transmission Factor. Infect. Immun. 2012, 80, 2704–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.; Wang H.; Zhang Y.; Chen K.; Davis B.; Feng H. Mouse Relapse Model of Clostridium difficile Infection. Infect. Immun. 2011, 79, 2856–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre D. W.; Cule M. L.; Wilson D. J.; Griffiths D.; Vaughan A.; O’Connor L.; Ip C. L. C.; Golubchik T.; Batty E. M.; Finney J. M.; Wyllie D. H.; Didelot X.; Piazza P.; Bowden R.; Dingle K. E.; Harding R. M.; Crook D. W.; Wilcox M. H.; Peto T. E. A.; Walker A. S. Diverse Sources of C. difficile Infection Identified on Whole-Genome Sequencing. N. Engl. J. Med. 2013, 369, 1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A Trial To Compare Xifaxan to Vancomycin for the Treatment of Clostridium difficile-Associated Diarrhea (CDAD). Identifier NCT00269399. https://clinicaltrials.gov/ct2/show/NCT00269399?term=NCT00269399&rank=1 (accessed Oct 21, 2014).
- Fecal Microbiota Transplant for Relapsing Clostridium difficile Infection in Adults and Children Using a Frozen Encapsulated Inoculum. Identifier NCT01914731. http://clinicaltrials.gov/ct2/show/NCT01914731?term=FMT&phase=0&rank=9.
- Youngster I.; Sauk J.; Pindar C.; Wilson R. G.; Kaplan J. L.; Smith M. B.; Alm E. J.; Gevers D.; Russell G. H.; Hohmann E. L. Fecal Microbiota Transplant for Relapsing Clostridium difficile Infection Using a Frozen Inoculum rrom Unrelated Donors: A Randomized, Open-Label, Controlled Pilot Study. Clin. Infect. Dis. 2014, 58, 1515–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defined Fecal Microbiota Transplantation for Clostridium difficile Diarrhea. Identifier NCT01868373. https://clinicaltrials.gov/ct2/show/NCT01868373?term=FMT&phase=0&rank=10 (accessed Oct 28, 2014).
- A Study To Investigate a Clostridium difficile Vaccine in Healthy Adults Aged 50 to 85 Years, Who Will Each Receive 3 Doses of Vaccine. Identifier NCT02117570. http://clinicaltrials.gov/ct2/show/NCT02117570?term=vaccine+difficile&rank=4 (accessed Oct 23, 2014).
- Lowy I.; Molrine D. C.; Leav B. A.; Blair B. M.; Baxter R.; Nichol G.; Thomas W. D. Jr.; Leney M.; Sloan S.; Hay C. A.; Ambrosino D. M. Treatment with Monoclonal Antibodies against Clostridium difficile Toxins. N. Engl. J. Med. 2010, 362, 197–205. [DOI] [PubMed] [Google Scholar]
- FMT Versus Antimicrobials for Initial Treatment of Recurrent CDI. Identifier NCT02255305. https://clinicaltrials.gov/ct2/show/NCT02255305?term=FMT&phase=1&rank=1 (accessed Oct 28, 2014).
- Stool Transplant in Pediatric Patients with Recurring C. difficile Infection. Identifier NCT01972334. http://clinicaltrials.gov/ct2/show/NCT01972334?term=FMT&phase=1&rank=5 (accessed Jan 20, 2014).
- Fecal Transplant for Relapsing C. difficile Infection. Identifier NCT01703494. http://clinicaltrials.gov/ct2/show/NCT01703494 (accessed Oct 28, 2014).
- Fecal Microbiota Transplantation by Colonoscopy for Recurrent C. difficile Infection. NCT02148601. https://clinicaltrials.gov/ct2/show/NCT02148601?term=FMT&phase=1&rank=15 (accessed Oct 28, 2014). [DOI] [PMC free article] [PubMed]
- FMT Delivered by Capsule Versus Colonoscopy for Recurrent C. diff. Identifier NCT02254811. http://clinicaltrials.gov/ct2/show/NCT02254811?term=FMT&phase=2&rank=1 (accessed Oct 23, 2014).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.