Abstract
Synthetic genetic array (SGA) has been successfully used to identify genetic interactions in S. cerevisiae and S. pombe. In S. pombe, SGA methods use either cycloheximide (C) or heat shock (HS) to select double mutants before measuring colony size as a surrogate for fitness. Quantitative Fitness Analysis (QFA) is a different method for determining fitness of microbial strains. In QFA, liquid cultures are spotted onto solid agar and growth curves determined for each spot by photography and model fitting. Here, we compared the two S. pombe SGA methods and found that the HS method was more reproducible for us. We also developed a QFA procedure for S. pombe. We used QFA to identify genetic interactions affecting two temperature sensitive, telomere associated query mutations (taz1Δ and pot1-1). We identify exo1∆ and other gene deletions as suppressors or enhancers of S. pombe telomere defects. Our study identifies known and novel gene deletions affecting the fitness of strains with telomere defects. The interactions we identify may be relevant in human cells.
Introduction
Genetic interactions (GIs) arise when the function of one gene is affected by the function of another [1]. In budding yeast, synthetic genetic array (SGA) methodology has been used to characterise GIs on a genome-wide scale [2–4]. SGA uses large-scale robotic procedures for mating and sporulation carried out on solid agar media to generate double mutant colonies and to measure their size [5]. By comparing the size of double mutants it is possible to classify GIs as negative (where double mutant colonies are smaller than expected), positive (larger than expected) or neutral [6–10]. Genome-wide SGA screens performed using the budding yeast S. cerevisiae have categorised gene subsets based on functionality [5, 11–13]. Techniques similar to budding yeast SGA have been developed for E coli [14, 15] and S. pombe [16, 17].
Quantitative fitness analysis (QFA) is another high throughput method for measuring fitness phenotypes in budding yeast. In QFA, strains are cultured in liquid media, spotted onto solid agar plates and growth is monitored by time course photography. A logistic growth curve model is fitted to the data to infer fitness phenotypes such as maximum growth rate or maximum doubling potential [18–20]. QFA has been successfully used to establish GIs affecting telomere related query mutations [19].
Hundreds of mutations interacting both positively and negatively with mutations affecting telomere-binding proteins such as Cdc13 in budding yeast have been identified using QFA [19]. We were therefore interested to try to apply QFA to S. pombe telomere-defective strains to permit us to compare and contrast the genetic interactions we see in the two yeast species. Fission yeast is evolutionarily distant from budding yeast and the comparisons have the potential to identify interactions conserved in metazoans [21–23]. Telomere structure is similar in budding and fission yeast and key proteins associated with telomeres in these yeasts are functionally conserved in mammals [21, 24, 25]. For example, the single strand DNA-capping protein Pot1 in human and fission yeast contains OB-folds as do budding yeast Cdc13 and human CTC1. Furthermore fission yeast pot1-1 mutants confer similar phenotypes to budding yeast cdc13-1 mutants [26]. Telomere defects in human cells are relevant to ageing and carcinogenesis, for example telomere degradation and fusion events occur during carcinogenesis and ageing [27–32].
There are two published methods to generate S. pombe double mutant strains during SGA, using either heat shock (HS) at 42°C or cycloheximide (C) as a critical selective step [16, 17]. Cyclohexamide-based SGAs require the genetically engineered ‘pombe epistatic marker 2’ (PEM2) parental strain and, in this background, growth in presence of cyclohexamide serves as both anti-diploid and mating-type selection [16, 33–36]. The HS based method does not require a specific genetic background [17] but uses high temperature to kill vegetative cells and therefore select for spores. In this paper we set out to test the different methods for carrying out genome-wide SGA screens in S. pombe and to develop a QFA procedure for S. pombe. We used QFA to identify known and novel suppressors and enhancers of S. pombe telomere defects.
Results
Comparing SGA Methods
To evaluate the two different S. pombe SGA methods and to choose the best ‘neutral’ mutation, a gene deletion library (2936 S. pombe yfgΔ, your favourite gene deletions, version 3) [37] was crossed with query mutations using two SGA methodologies [3]. In principle, each of the two S. pombe SGA methods to generate double mutants should result in a similar pattern of GIs. To test if this is the case we first carried out SGAs with his3Δ, ura5Δ and his7Δ as comparatively ‘neutral’ query mutations using heat shock (HS) [17] or cyclohexamide (C) based methods [16, 38].
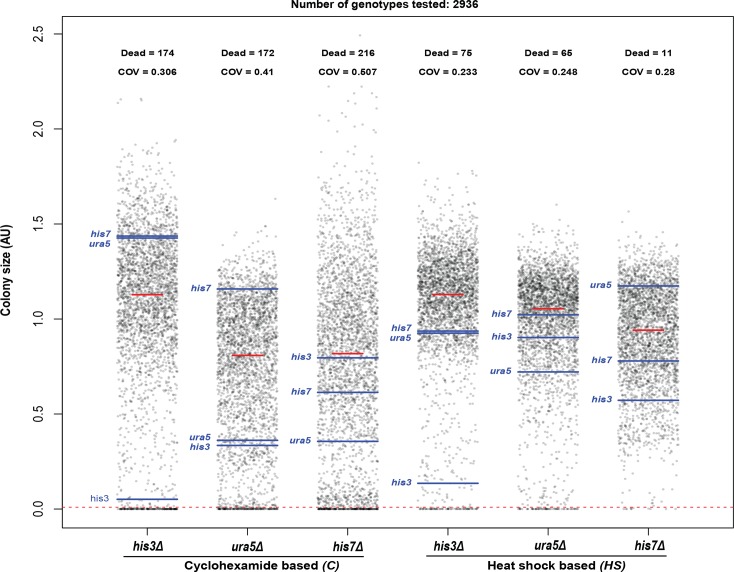
Fig 1 is a summary of the fitnesses we observed in six independent SGA experiments. Unexpectedly, we found that the fitness rankings for his3Δ, ura5Δ and his7Δ SGAs were different to each other in the rich media we use for SGA. Using either HS or C methods we observed that, on average, the his3∆ strains were fitter than ura5∆ or his7∆ strains. We also observed that overall colony size distribution was different between HS and C methods (Fig 1). Overall, colony size distribution in his3Δ, ura5Δ and his7Δ SGAs appeared tighter using the HS method compared to the C method (Fig 1). This tighter spread was reflected by lower coefficient of variation (COV) values for the his3Δ, ura5Δ and his7Δ SGAs with the HS method versus the C method. We also observed that the proportion of very low fitness, or "dead" double mutant strains was lower for the HS method in all cases (Fig 1).
Fig 1. Comparison of HS and C based SGA methods.
his3Δ, ura5Δ and his7Δ mutations were crossed with a deletion library either using C or HS method, and colony size measured. Strip charts comparing colony size distributions of double mutants on SGA final selection plates are shown. The horizontal blue lines represent library deletion strains and the red line is the median value of each experiment. Colony sizes were scaled relative to the median of the overall distribution across all six screens. The horizontal dotted red line is the threshold below which colonies were classified as "dead", and is based on a colony size less than 1% of the median across all 6 SGAs. The coefficient of variation (COV) for colony sizes above the cut-off is shown.
Finally, we looked for evidence of genetic linkage to his3Δ, ura5Δ or his7Δ to determine SGA success. If a haploid his3::KANMX strain is crossed with a his3::NATMX strain, haploid double deletion progeny should not arise after mating and sporulation. However we should be able to combine the his3∆ allele with all other unlinked gene deletions. The position of the his3Δ strains, as indicated by the blue line, is close to zero in both his3Δ SGA methods suggesting that these SGAs were most successful (Fig 1). Importantly, the his3Δ parental strain generated his3Δ ura5Δ double mutants and his3Δ his7Δ double mutants. The ura5∆ and his7∆ strains showed less strong evidence for genetic linkage (and by implication SGA success). We conclude that his3∆ is a better choice than his7∆ or ura5∆ to use as a neutral mutation for control SGAs. Overall, based on these six SGAs we also conclude that the HS method generates more viable double mutants than the C method, and that the double mutants within the HS method are more similar in fitness to each other (Fig 1).
We next performed a small scale SGA on a telomere-defective taz1Δ query strain and a neutral his3Δ control strain. For this experiment a small library of deletion mutations (n = 308, Worksheet B in S1 Tables), many of which were shown previously to interact with taz1Δ mutation, was used. At permissive temperatures (30°C) we generated 8 independent replicates of each genotype arranged on four separate plates. Fig 2A shows images of four SGA final selection media plates. Double mutants were arrayed in a 768 colony format with 308 deletion mutations arranged in pairs and surrounded by a neutral mutation (mug134Δ) in pairs. A true synthetic lethal interaction is inferred when each of a pair of replicate double deletion mutants does not form colonies. SGA artefacts can be inferred if colony pairs carrying the same mutations do not behave similarly on the same plate (viable or non-viable) or across plates (S1 Fig).
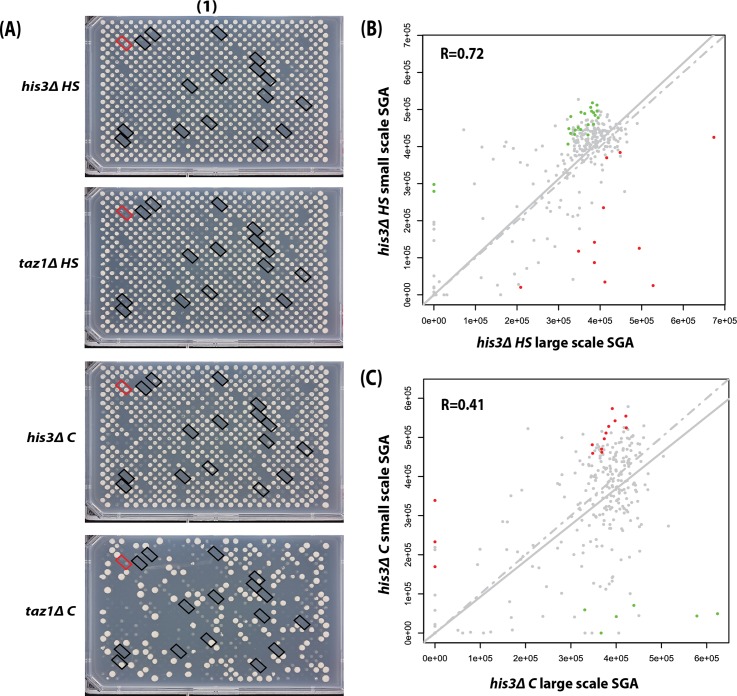
Fig 2. Small scale SGA demonstrates the taz1Δ query mutation survives the HS method better than the C method.
his3Δ and taz1Δ strains were crossed with a subset (n = 308, Worksheet B in S1 Tables; many telomere-related) of gene deletions from the deletion library using the two methods. (A) Images of final selection plates from the HS and C methods are shown. The double mutants were grown at 30°C after 3 days. One plate (of a total of 4) having two independent replicates was processed for each method. The 16 diagonal black rectangular boxes indicate a proportion of double deletion pairs showing growth defects in the four SGA plates. The red rectangular box highlights the sde2Δ location. (B) Correlation scatter plots comparing the his3Δ small scale SGA and the his3Δ large scale SGA using the HS method for 308 genes (C) Same as (B) but comparison made using the C method.
In the his3Δ HS SGA experiment, absence of particular his3Δ progeny pairs was largely concordant within each plate (Fig 2A, top row- the black diagonal boxes highlight all consistently ‘synthetic lethal’ interactions) or within the eight repeats across all four his3Δ HS SGA plates (S2 Fig, top row). The taz1Δ HS experiment generated a broadly similar synthetic lethal growth pattern (Fig 2A, second row) and was largely consistent when compared with the his3Δ HS SGA (correlation, R = 0.59; Panel B in S3 Fig). It has been reported that sde2∆ is synthetically lethal with taz1∆ [39] and reassuringly we confirmed this result (highlighted in red, Fig 2A, top 1 panels). We concluded that the HS based SGA method is reproducible and could identify meaningful genetic interactions.
We next conducted a taz1Δ SGA using the C method. We found that the his3Δ C experiment resulted in a higher proportion of inviability and inconsistency across the eight replicates in comparison with the HS method (Fig 2A, third panel and S2 Fig). Despite observing the synthetic lethality with sde2∆, the taz1Δ C method generated poorly growing or dead strains extremely frequently (Fig 2A, bottom panel).
Finally, the his3Δ small scale (Fig 2A) and large scale SGAs (Fig 1) were compared to test reproducibility across experiments performed at different times (Fig 2B and 2C). Replicates of screens carried out using the HS methods were consistent, with a strong correlation between colony sizes (R = 0.72) whereas a more moderate correlation (R = 0.41) was observed with the C method (Fig 2B and 2C). Overall, we found the HS method more reproducible and used it for standardising QFA in S. pombe.
QFA Identifies Known and Novel GIs among Telomere Defective Mutants
The SGA technique uses colony size to determine strain fitness [2, 11, 40]. Quantitative Fitness Analysis (QFA) is different and measures fitnesses by analysing growth curves [19, 20]. QFA growth curves are very similar to those observed in liquid culture, with clear exponential and saturation phases [18]. Using QFA many more cultures can be examined in parallel than is practical using liquid cultures. Furthermore, spotted QFA provides more accurate fitness measurements than can be measured using pinned cultures [19]. Therefore we wanted to assess QFA as a means of identifying and quantifying GIs for taz1Δ and pot1-1 telomere capping mutations in S. pombe.
QFA was first performed on double mutants obtained after crossing his3Δ and taz1Δ mutations to a small deletion library by SGA (n = 308, Worksheet B in S1 Tables). The taz1Δ strain is cold sensitive at 20°C [41, 42]. Therefore we first cultured double mutants in liquid under permissive conditions (30°C) before measuring fitness by QFA under restrictive conditions (20°C).
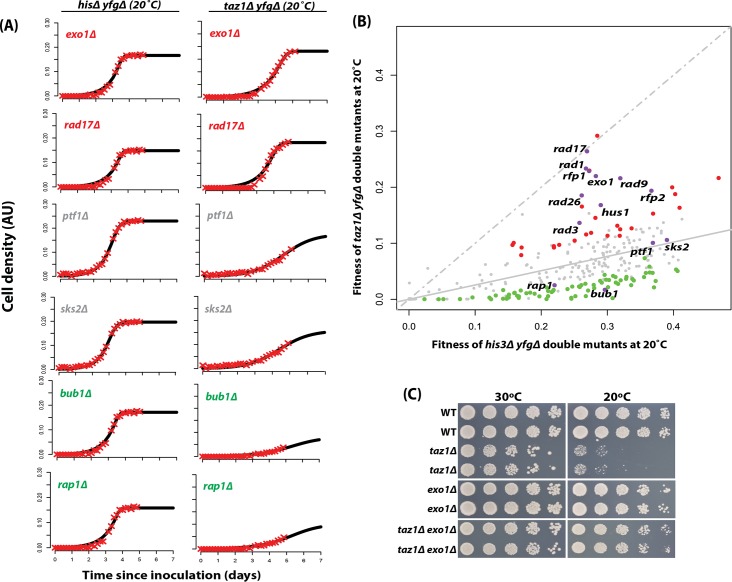
In Fig 3A, we show growth curves for some representative his3Δ yfgΔ and taz1Δ yfgΔ double mutant strains, some of which were previously known to interact with taz1 +. The taz1Δ exo1Δ and taz1Δ rad17Δ strains grew as well as the equivalent his3Δ strains at 20°C whereas taz1Δ bub1Δ and taz1Δ rap1Δ strains grew relatively poorly. The taz1Δ ptf1Δ and taz1Δ sks2Δ had intermediate fitnesses. Fig 3B shows fitnesses of all his3Δ and taz1Δ strains as a scatter plot. A comparison of the his3Δ yfgΔ and taz1Δ yfgΔ strain fitnesses at the permissive temperature (30°C) showed that taz1Δ mutants were as fit as the his3Δ mutants (the solid grey line superimposing over the line of equal fitness (S4 Fig)). At 20°C, as expected, taz1Δ mutants exhibited growth defects relative to his3Δ mutants [43]. Interestingly, a number of taz1Δ yfgΔ strains grew significantly better than expected, given the fitness of the equivalent his3Δ yfgΔ mutation at 20°C (red points, Fig 3B). These yfgΔ gene deletions can be classified as taz1Δ suppressors. There were also a proportion of taz1Δ yfgΔ strains that grew worse than expected and these can be classified as taz1Δ enhancers (green points, Fig 3B). The taz1Δ yfgΔ strains close to the regression line (neither suppressors nor enhancers) showed no evidence of genetic interaction. The fitness plot in Fig 3B highlights the positions of known suppressors of taz1Δ cold sensitivity, such as members of the 9-1-1 complex (rad9Δ, rad1Δ and hus1Δ), the clamp loader (rad17Δ); and gene deletions of the ATR kinase RAD3 and its interacting partner RAD26 [44]. Other known interactions with taz1 + (enhancers bub1Δ and rap1Δ) [41, 45] were also identified. Interestingly, deletion of the exonuclease gene EXO1 was identified as one of the significant suppressors of taz1Δ cold sensitivity (Fig 3B). Importantly, we confirmed this novel observation by spot tests of strains generated by tetrad dissection (Fig 3C). Therefore we conclude that QFA is useful for identifying suppressors and enhancers of the taz1∆ cold sensitive phenotype.
Fig 3. Small scale QFA identifies previously known telomere associated genes as suppressors of taz1Δ cold sensitivity.
(A) QFA growth curve comparisons of representative his3Δ yfgΔ and taz1Δ yfgΔ strains at 20°C for 5 days. (B) Four replicates of double mutants from the his3Δ and taz1Δ small scale SGA using the HS method were first cultured in liquid at 30°C and then spotted. Growth of taz1Δ yfgΔ and the his3Δ yfgΔ double mutants was measured at 20°C. Numerical area under the curve (nAUC) values were used as measures of double mutant fitness. Gene deletions that significantly enhance (green) or suppress (red) the taz1Δ defect, in comparison with the his3Δ mutation are indicated. The line of equal growth (dashed grey) and a population model of expected fitness (solid grey) are indicated. (C) The exo1Δ (independent of the Bioneer library) was combined with a taz1Δ mutation and assessed for growth by manual spot test. The strains were cultured to saturation in 2ml YE5S media at 30°C, a five-fold serial dilution generated and spotted on YE5S plates. Strains were incubated in the same incubator at the same time and at indicated temperatures for 3–6 days before being photographed.
To gain further insights into the S. pombe telomere cap, we performed a large scale QFA on the temperature sensitive mutant pot1-1 (protection of telomere) [26]. Double mutants were cultured at 30°C for two days before measuring their fitness at 37°C. This temperature (37°C) was chosen because the fitness of pot1-1 mutants is about half of wild type (S5 Fig). We also assessed the value of a pinned QFA experiment, to determine if we could observe temperature dependent fitness defects of the pot1-1 strains after pinning, but we observed no strong differences between pot1-1 and wild type strains between 38°C and 41°C (S6 Fig). This suggests that spotted culture QFA is necessary to observe the temperature sensitive fitness defect in pot1-1 strains. We did observe that all POT1 and pot1-1 strains decreased in fitness as temperatures increased (S6 Fig).
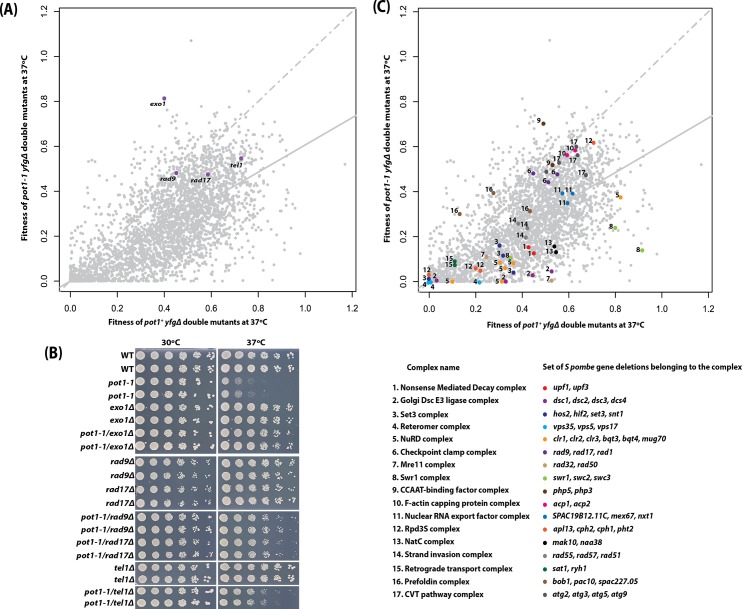
As expected at 37°C, the pot1-1 mutation caused a growth defect relative to pot1 + strains (Fig 4A). We identified exo1Δ as one of the strongest suppressors of pot1-1 temperature sensitivity (Fig 4A). An analogous result is observed in budding yeast where exo1Δ is a strong suppressor of cdc13-1 temperature sensitivity [19, 46]. In order to confirm the results from the pot1-1 QFA screen, spot tests were performed using manually derived double mutants, where the parental single deletions were constructed independently from those within the deletion library. We confirmed that exo1Δ suppresses pot1-1 temperature sensitivity as do the deletion of genes within the 9-1-1 complex (Fig 4B). Other telomere associated gene deletions (rad17Δ, tel1Δ) were also identified as suppressors of the pot1-1 query mutation. However, the QFA data was comparatively noisy in this pot1-1 experiment compared with the taz1∆ experiment. For example, there was a wide range of fitness values observed even in the control pot1 + strains (the x axis). One hypothesis to explain the noisy data is that there is poor growth for many S pombe mutant strains at 37°C. To test if this might be the case, we chose to highlight the positions of different gene deletions that each affected the same functionally related complexes. Importantly, we found that many gene deletions affecting the same functional complexes [10] clustered together in the data plot (Fig 4C). For example, gene deletions affecting retrograde transport (Complex 15, Fig 4C) grew poorly at 37°C irrespective of pot1-1/ pot1 + status. Overall, the clustering of gene deletions affecting similar processes in Fig 4C suggest that the data are representative of the true effects of each gene deletion on the growth of both pot1 + and pot1-1 strains at 37°C.
Fig 4. QFA identifies exo1Δ and other gene deletions as suppressors or enhancers of pot1-1 temperature sensitivity.
(A) The S. pombe genome deletion library collection was crossed to the pot1-1 mutation, or a POT1 control mutation. Four replicate crosses were carried out for each and the double mutants were first cultured in liquid medium at 30°C and then spotted. The fitness (numerical area under the curve values) of all double mutant cultures measured at 37°C. The line of equal growth (dashed grey) and a population model of expected fitness (solid grey) are indicated. (B) Validation of the pot1-1 QFA by independent spot tests as described in Fig 3C. (C) Genes grouped based on the hierarchical clustering of GI profiles [10], each number represents a specific complex and the corresponding colour code is a set of genes within that complex.
Discussion
GIs have been measured in different genetic model systems to help understand how cells and organisms function [10, 47–54]. GIs in S. cerevisiae have been extensively studied [3, 11, 13, 19, 55, 56]. The development of SGA in S. pombe allows productive comparisons between the genetic networks in budding and fission yeasts [9]. There are two published methods in S. pombe. The more widely used C based method did not work well for us, for reasons that are not clear, while the HS method was more reproducible.
QFA measures fitness by analysing growth curves potentially allowing the measurement of more subtle genetic interactions than can be detected by SGA [19, 20]. QFA has been developed in budding yeast to identify subtle suppressor or enhancer GIs in the context of telomere defects. We have established an S. pombe QFA protocol based on budding yeast protocols. Interestingly, the two yeasts respond differently to the SGA/QFA protocols. For example S. pombe cell fitnesses are reduced more strongly by storage on agar plates at 4°C, time spent in stationary phase (in 96 well liquid cultures) and antibiotic treatments. Therefore we think further optimisation of the QFA protocols are likely necessary. However, even at this stage, QFA can be used to compare genome- wide genetic interactions in fission and budding yeast and this has the potential to identify conserved genetic interactions that are important across all eukaryotic organisms.
Using QFA in S. pombe we confirmed many known GIs for the taz1Δ query mutation such as with gene deletions affecting the 9-1-1 complex [44]. Other novel interactions such as with exo1Δ were also identified (Fig 3). Recent studies have demonstrated the role of sumoylation in telomere length regulation [57, 58]. Interestingly, we also identified SUMO-targeted ubiquitin-protein ligases rfp1 + and rfp2 + as suppressors of taz1Δ cold sensitivity. The human orthologue RNF4 has been shown to play critical roles in DNA damage response and genome stability [59–62]. Elucidating the roles of exo1 +, rfp1 + and rfp2 + at fission yeast telomeres will require further experiments.
Interestingly our S. pombe QFA identified exo1∆ as a suppressor of both taz1Δ and pot1-1 induced telomere defects. In budding yeast exo1∆ suppresses telomere defects because Exo1 is involved in generating ssDNA at uncapped telomeres [19, 46, 63]. The suppression of taz1Δ growth defects by exo1∆ is seemingly at odds with previous findings which indicate that Dna2, not Exo1 is not involved in resecting telomere ends in taz1Δ cells [64]. However, in budding yeast there is complex interplay between Exo1 and Dna2 and other factors to control resection at uncapped telomeres [63]. Further experiments will be necessary to clarify the roles of Exo1, Dna2 and other factors at uncapped telomeres in S. pombe.
Our S. pombe experiments also showed that deletions affecting the 9-1-1 complex suppressed pot1-1 temperature sensitivity and these observations correlate with analysis of budding yeast cdc13-1 mutants [19]. Interestingly, gene deletions affecting the NMD pathway suppress cdc13-1 [19] but enhance pot1-1 temperature sensitivity (Fig 4).
POT1 has been recently identified as a major susceptibility gene for familial melanoma, and is somatically inactivated in chronic lymphocytic leukemia [65–67]. The S. pombe pot1-1 QFA identifies numerous gene deletions which may help in generating exciting hypotheses about roles of individual genes in cells with defective telomeres and potentially therapeutic targets.
Materials and Methods
Yeast Strains
All the strains used in this study are listed in Worksheet A in S1 Tables. The strains were generated and grown using standard protocols [68–70]. For the pot1-1 thermosensitive strain, a marker switch of the KanMX6 module to the HphMX6 module was achieved by transforming the G418R strains with an HphMX6 cassette amplified from pFA6a-HphMX6 plasmid [71]. Positive clones were selected based on growth on YE5S with Hygromycin and inability to grow on YE5S with G418; and tested for thermosensitivity [26].
SGA Methods
The HS and C SGA methods were performed as previously described [16, 17]. The Bioneer deletion library collection was used for SGAs (http://us.bioneer.com/products/spombe/spombeoverview.aspx). The deletion library (768 format, 384x2) was grown on YE5S (Yeast extract, 5 supplements) rectangular agar plates with G418. For S. pombe SGA, a 768 pinning format was used. The cells arrayed on rectangular plates are read from left to right to identify genotypes (column 1, row1 is the top most left corner), two nicks within the plate are towards the top and bottom left corners (S1 Fig). The YE5S agar plates used in SGA had the following drug concentrations; G418 (Geneticin-100 μg/ml), clonNAT (Nourseothricin-100 μg/ml), Hyg (Hygromycin-300 μg/ml, CycH (Cycloheximide-100 μg/ml). EMM1/2G (Edinburgh Minimal Media, 0.5 Glutamate; ForMedium, PMD1210) media was used for sporulation.
S. pombe QFA
The S. pombe QFA was developed from S. cerevisiae QFA as previously described [19, 20]. Drug concentrations in media varied depending on whether cells were grown on solid or in liquid media. We observed that 96 well liquid cultures with usual drug concentrations (100 μg/ml G418; 300 μg/ml Hygromycin; 100 μg/ml Nat) grew poorly. Therefore double mutants were cultured in 96-well plates with each well containing 200μl YE5S liquid media with 6.25 μg/ml each (for G418, clonNAT), and 18.75 μg/ml (for Hyg) as final drug concentration were used (G418+Nat for taz1Δ QFA or G418+Hyg for pot1-1 QFA). Liquid cultures were incubated at 30°C for two days without shaking and 384-format robotic spot tests were performed using a Biomek FX robot (Beckman Coulter (UK) Limited, High Wycombe, UK) equipped with a pintool magnetic mount and a 96-pin (2 mm diameter) pintool (V&P Scientific, Inc., San Diego, CA, USA). Photography, image analysis and modelling of fitness were performed as described previously [19]. Strip charts were generated using the stripchart function from the statistical programming software R [72].
Manual Growth Assays
The strains were cultured to saturation in 2ml YE5S media with rolling at 30°C. A five or six-fold serial dilution using distilled water was then generated and spotted on YE5S plates. Strains were incubated at the indicated temperatures for 3–6 days before being photographed.
Supporting Information
Double mutants are arrayed in a 768 colony format with 308 deletion mutations arranged in pairs and surrounded by a neutral mutation (mug134Δ) in pairs (in blue). The green rectangular boxes are putative synthetic lethal interactions when pairs of double deletion mutations do not form colonies. Single yellow colonies are either pinning artefacts or an issue arising in an SGA.
(TIF)
Images of final selection plates from the HS and C methods are shown. The double mutants were photographed after growing them at 30°C for 3 days. Four plates (eight independent replicates, each row) were examined. The 16 diagonal black rectangular boxes indicate a proportion of double deletion pairs showing growth defects in all SGA plates. The red rectangular box highlights the sde2Δ location.
(TIF)
(B) Same as (A) but for the HS method.
(TIF)
(TIF)
(TIF)
Boxplots summarising quantitative fitness distributions for pot1-1 query strains and the wild-type surrogate strains (pot1 +) at 38°C, 39°C, 40°C and 41°C (N = 44).
(TIF)
The raw data below can be used to replicate all of the plots and statistical analysis presented in the manuscript.
(PDF)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
(ZIP)
(ZIP)
(ZIP)
(ZIP)
(ZIP)
(ZIP)
(ZIP)
(ZIP)
Excel spreadsheet containing the following tables as worksheets. Worksheet A: Strains used in the study. Worksheet B: List of gene deletions in the small scale library. Worksheet C: Raw data from the taz1Δ small scale QFA screen. Worksheet D: Raw data from the pot1-1 QFA screen.
(XLSX)
Acknowledgments
We very gratefully acknowledge Julia Cooper and Sophie Zaaijer (National Cancer Institute, NIH, Bethesda, MD 20892) for generously providing strains, advice and comments on the manuscript. Special thanks to Alan Leake for making reagents/media for robotic work.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Funding provided by C23629/A13314 (http://www.cancerresearchuk.org/).
References
- 1. Boone C. Yeast Systems Biology: Our Best Shot at Modeling a Cell. Genetics. 2014;198(2):435–7. 10.1534/genetics.114.169128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tong AHY, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294(5550):2364–8. [DOI] [PubMed] [Google Scholar]
- 3. Tong AHY, Lesage G, Bader GD, Ding H, Xu H, Xin X, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303(5659):808–13. [DOI] [PubMed] [Google Scholar]
- 4. Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, et al. The genetic landscape of a cell. Science. 2010;327(5964):425–31. 10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dixon SJ, Costanzo M, Baryshnikova A, Andrews B, Boone C. Systematic mapping of genetic interaction networks. Annu Rev Genet. 2009;43:601–25. 10.1146/annurev.genet.39.073003.114751 [DOI] [PubMed] [Google Scholar]
- 6. St Onge RP, Mani R, Oh J, Proctor M, Fung E, Davis RW, et al. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet. 2007;39(2):199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mani R, Onge RPS, Hartman JL, Giaever G, Roth FP. Defining genetic interaction. Proc Natl Acad Sci U S A. 2008;105(9):3461–6. 10.1073/pnas.0712255105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods. 2008;5(8):711–8. 10.1038/nmeth.1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baryshnikova A, Costanzo M, Kim Y, Ding H, Koh J, Toufighi K, et al. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Methods. 2010;7(12):1017–24. 10.1038/nmeth.1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ryan CJ, Roguev A, Patrick K, Xu J, Jahari H, Tong Z, et al. Hierarchical modularity and the evolution of genetic interactomes across species. Mol Cell. 2012;46(5):691–704. 10.1016/j.molcel.2012.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446(7137):806–10. 10.1038/nature05649 [DOI] [PubMed] [Google Scholar]
- 12. Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123(3):507–19. [DOI] [PubMed] [Google Scholar]
- 13. Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, DeVay RM, Lackner LL, et al. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol. 2011;195(2):323–40. 10.1083/jcb.201107053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Typas A, Nichols RJ, Siegele DA, Shales M, Collins SR, Lim B, et al. High-throughput, quantitative analyses of genetic interactions in E. coli. Nat Methods. 2008;5(9):781–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Butland G, Babu M, Díaz-Mejía JJ, Bohdana F, Phanse S, Gold B, et al. eSGA: E. coli synthetic genetic array analysis. Nat Methods. 2008;5(9):789–95. 10.1038/nmeth.1239 [DOI] [PubMed] [Google Scholar]
- 16. Roguev A, Wiren M, Weissman JS, Krogan NJ. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat Methods. 2007;4(10):861–6. 10.1038/nmeth1098 [DOI] [PubMed] [Google Scholar]
- 17. Dixon SJ, Fedyshyn Y, Koh JLY, Prasad TSK, Chahwan C, Chua G, et al. Significant conservation of synthetic lethal genetic interaction networks between distantly related eukaryotes. Proceedings of the National Academy of Sciences. 2008;105(43):16653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lawless C, Wilkinson DJ, Young A, Addinall SG, Lydall DA. Colonyzer: automated quantification of micro-organism growth characteristics on solid agar. BMC Bioinformatics. 2010;11(1):287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Addinall SG, Holstein EM, Lawless C, Yu M, Chapman K, Banks AP, et al. Quantitative fitness analysis shows that NMD proteins and many other protein complexes suppress or enhance distinct telomere cap defects. PLoS genetics. 2011;7(4):e1001362 10.1371/journal.pgen.1001362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Banks AP, Lawless C, Lydall DA. A quantitative fitness analysis workflow. Journal of visualized experiments: JoVE. 2012;(66). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lewis KA, Wuttke DS. Telomerase and telomere-associated proteins: structural insights into mechanism and evolution. Structure. 2012;20(1):28–39. 10.1016/j.str.2011.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sipiczki M. Where does fission yeast sit on the tree of life. Genome Biol. 2000;1(2):1011.1-.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Linger BR, Price CM. Conservation of telomere protein complexes: shuffling through evolution. Crit Rev Biochem Mol Biol. 2009;44(6):434–46. 10.3109/10409230903307329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nandakumar J, Cech TR. Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol. 2013;14(2):69–82. 10.1038/nrm3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jain D, Cooper JP. Telomeric strategies: means to an end. Annu Rev Genet. 2010;44:243–69. 10.1146/annurev-genet-102108-134841 [DOI] [PubMed] [Google Scholar]
- 26. Pitt CW, Cooper JP. Pot1 inactivation leads to rampant telomere resection and loss in one cell cycle. Nucleic Acids Res. 2010;38(20):6968–75. 10.1093/nar/gkq580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31(1):9–18. 10.1093/carcin/bgp268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Günes C, Rudolph KL. The role of telomeres in stem cells and cancer. Cell. 2013;152(3):390–3. 10.1016/j.cell.2013.01.010 [DOI] [PubMed] [Google Scholar]
- 29. Aubert G, Lansdorp PM. Telomeres and aging. Physiol Rev. 2008;88(2):557–79. 10.1152/physrev.00026.2007 [DOI] [PubMed] [Google Scholar]
- 30. Nault JC, Mallet M, Pilati C, Calderaro J, Bioulac-Sage P, Laurent C, et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun. 2013;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–9. 10.1126/science.1229259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–61. 10.1126/science.1230062 [DOI] [PubMed] [Google Scholar]
- 33. Stevens DR, Atteia A, Franzen LG, Purton S. Cycloheximide resistance conferred by novel mutations in ribosomal protein L41 of Chlamydomonas reinhardtii. Mol Genet Genomics. 2001;264(6):790–5. [DOI] [PubMed] [Google Scholar]
- 34. Kondo K, Saito T, Kajiwara S, Takagi M, Misawa N. A transformation system for the yeast Candida utilis: use of a modified endogenous ribosomal protein gene as a drug-resistant marker and ribosomal DNA as an integration target for vector DNA. Journal of bacteriology. 1995;177(24):7171–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kawai S, Murao S, Mochizuki M, Shibuya I, Yano K, Takagi M. Drastic alteration of cycloheximide sensitivity by substitution of one amino acid in the L41 ribosomal protein of yeasts. J Bacteriol. 1992;174(1):254–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Varma A, Kwon‐Chung KJ. Characterization of the L41 gene in Cryptococcus neoformans: its application as a selectable transformation marker for cycloheximide resistance. Yeast. 2000;16(15):1397–403. [DOI] [PubMed] [Google Scholar]
- 37. Kim D-U, Hayles J, Kim D, Wood V, Park H-O, Won M, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotech. 2010;28(6):617–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322(5900):405–10. 10.1126/science.1162609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sugioka-Sugiyama R, Sugiyama T. Sde2: A novel nuclear protein essential for telomeric silencing and genomic stability in Schizosaccharomyces pombe. Biochem Biophys Res Commun. 2011;406(3):444–8. 10.1016/j.bbrc.2011.02.068 [DOI] [PubMed] [Google Scholar]
- 40. Tong AHY, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae In Xiao W editor, 2nd ed. Yeast Protocols: Springer; 2006. pp. 171–91. [DOI] [PubMed] [Google Scholar]
- 41. Miller KM, Cooper JP. The telomere protein Taz1 is required to prevent and repair genomic DNA breaks. Mol Cell. 2003;11(2):303–13. [DOI] [PubMed] [Google Scholar]
- 42. Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440(7085):824–8. [DOI] [PubMed] [Google Scholar]
- 43. Miller KM, Cooper JP. The telomere protein Taz1 is required to prevent and repair genomic DNA breaks. Mol Cell. 2003;11(2):303–13. [DOI] [PubMed] [Google Scholar]
- 44. Nakamura TM, Moser BA, Russell P. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics. 2002;161(4):1437–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miller KM, Ferreira MG, Cooper JP. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J 2005;24(17):3128–35. 10.1038/sj.emboj.7600779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev. 2002;16(15):1919–33. 10.1101/gad.225102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tosti E, Katakowski JA, Schaetzlein S, Kim HS, Ryan CJ, Shales M, et al. Evolutionarily conserved genetic interactions with budding and fission yeast MutS identify orthologous relationships in mismatch repair-deficient cancer cells. Genome Med. 2014;6(9):68 10.1186/s13073-014-0068-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Babu M, Gagarinova A, Emili A. Array-based synthetic genetic screens to map bacterial pathways and functional networks in Escherichia coli In James A William editor. Strain Engineering Methods in Molecular Biology: Springer; 2011. p. 125–153. [DOI] [PubMed] [Google Scholar]
- 49. Lehner B. Modelling genotype–phenotype relationships and human disease with genetic interaction networks. J Exp Biol. 2007;210(9):1559–66. [DOI] [PubMed] [Google Scholar]
- 50. Park S, Lehner B. Epigenetic epistatic interactions constrain the evolution of gene expression. Mol Sys Biol. 2013;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roguev A, Talbot D, Negri GL, Shales M, Cagney G, Bandyopadhyay S, et al. Quantitative genetic-interaction mapping in mammalian cells. Nat Methods. 2013;10(5):432–7. 10.1038/nmeth.2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boucher B, Jenna S. Genetic interaction networks: better understand to better predict. Front Genet. 2013;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Beltrao P, Cagney G, Krogan NJ. Quantitative genetic interactions reveal biological modularity. Cell. 2010;141(5):739–45. 10.1016/j.cell.2010.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gibson TJ. Cell regulation: determined to signal discrete cooperation. Trends Biochem Sci. 2009;34(10):471–82. 10.1016/j.tibs.2009.06.007 [DOI] [PubMed] [Google Scholar]
- 55. Fiedler D, Braberg H, Mehta M, Chechik G, Cagney G, Mukherjee P, et al. Functional Organization of the S. cerevisiae Phosphorylation Network. Cell. 2009;136(5):952–63. 10.1016/j.cell.2008.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323(5922):1693–7. 10.1126/science.1167983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Garg M, Gurung RL, Mansoubi S, Ahmed JO, Davé A, Watts FZ, et al. Tpz1TPP1 SUMOylation reveals evolutionary conservation of SUMO‐dependent Stn1 telomere association. EMBO Rep. 2014;15(8):871–7. 10.15252/embr.201438919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Miyagawa K, Low RS, Santosa V, Tsuji H, Moser BA, Fujisawa S, et al. SUMOylation regulates telomere length by targeting the shelterin subunit Tpz1Tpp1 to modulate shelterin–Stn1 interaction in fission yeast. Proc Natl Acad Sci U S A. 2014;111(16):5950–5. 10.1073/pnas.1401359111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nagai S, Davoodi N, Gasser SM. Nuclear organization in genome stability: SUMO connections. Cell Res. 2011;21(3):474–85. 10.1038/cr.2011.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gibbs-Seymour I, Oka Y, Rajendra E, Weinert BT, Passmore LA, Patel KJ, et al. Ubiquitin-SUMO Circuitry Controls Activated Fanconi Anemia ID Complex Dosage in Response to DNA Damage. Mol Cell. 2014. [DOI] [PMC free article] [PubMed]
- 61. Galanty Y, Belotserkovskaya R, Coates J, Jackson SP. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 2012;26(11):1179–95. 10.1101/gad.188284.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cremona CA, Sarangi P, Zhao X. SUMOylation and the DNA damage response. Biomolecules. 2012;2(3):376–88. 10.3390/biom2030376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ngo GHP, Balakrishnan L, Dubarry M, Campbell JL, Lydall D. The 9-1-1 checkpoint clamp stimulates DNA resection by Dna2-Sgs1 and Exo1. Nucleic Acids Res. 2014:gku746. [DOI] [PMC free article] [PubMed]
- 64. Tomita K, Kibe T, Kang H-Y, Seo Y-S, Uritani M, Ushimaru T, et al. Fission yeast Dna2 is required for generation of the telomeric single-strand overhang. J Mol Cell Biol. 2004;24(21):9557–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi J, Yang XR, Ballew B, Rotunno M, Calista D, Fargnoli MC, et al. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat Genet. 2014. [DOI] [PMC free article] [PubMed]
- 66. Ramsay AJ, Quesada V, Foronda M, Conde L, Martínez-Trillos A, Villamor N, et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet. 2013;45(5):526–30. 10.1038/ng.2584 [DOI] [PubMed] [Google Scholar]
- 67. Robles-Espinoza CD, Harland M, Ramsay AJ, Aoude LG, Quesada V, Ding Z, et al. POT1 loss-of-function variants predispose to familial melanoma. Nat Genet. 2014;46(5):478–81. 10.1038/ng.2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Forsburg SL. Growth and manipulation of S. pombe. Curr Protoc Mol Biol. 2003:13.6. 1-.6. 7. [DOI] [PubMed]
- 69. Sabatinos SA, Forsburg SL. Molecular Genetics of Schizosaccharomyces pombe . Methods Enzymol. 2010;470:759–95. 10.1016/S0076-6879(10)70032-X [DOI] [PubMed] [Google Scholar]
- 70. Sunnerhagen P. PCR used to determine mating type in S. pombe. Biotechniques. 1993;14(1):18 [PubMed] [Google Scholar]
- 71. Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast. 2005;22(13):1013–9. [DOI] [PubMed] [Google Scholar]
- 72. RCoreTeam. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria: 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Double mutants are arrayed in a 768 colony format with 308 deletion mutations arranged in pairs and surrounded by a neutral mutation (mug134Δ) in pairs (in blue). The green rectangular boxes are putative synthetic lethal interactions when pairs of double deletion mutations do not form colonies. Single yellow colonies are either pinning artefacts or an issue arising in an SGA.
(TIF)
Images of final selection plates from the HS and C methods are shown. The double mutants were photographed after growing them at 30°C for 3 days. Four plates (eight independent replicates, each row) were examined. The 16 diagonal black rectangular boxes indicate a proportion of double deletion pairs showing growth defects in all SGA plates. The red rectangular box highlights the sde2Δ location.
(TIF)
(B) Same as (A) but for the HS method.
(TIF)
(TIF)
(TIF)
Boxplots summarising quantitative fitness distributions for pot1-1 query strains and the wild-type surrogate strains (pot1 +) at 38°C, 39°C, 40°C and 41°C (N = 44).
(TIF)
The raw data below can be used to replicate all of the plots and statistical analysis presented in the manuscript.
(PDF)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
(TXT)
(ZIP)
(ZIP)
(ZIP)
(ZIP)
(ZIP)
(ZIP)
(ZIP)
(ZIP)
Excel spreadsheet containing the following tables as worksheets. Worksheet A: Strains used in the study. Worksheet B: List of gene deletions in the small scale library. Worksheet C: Raw data from the taz1Δ small scale QFA screen. Worksheet D: Raw data from the pot1-1 QFA screen.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.