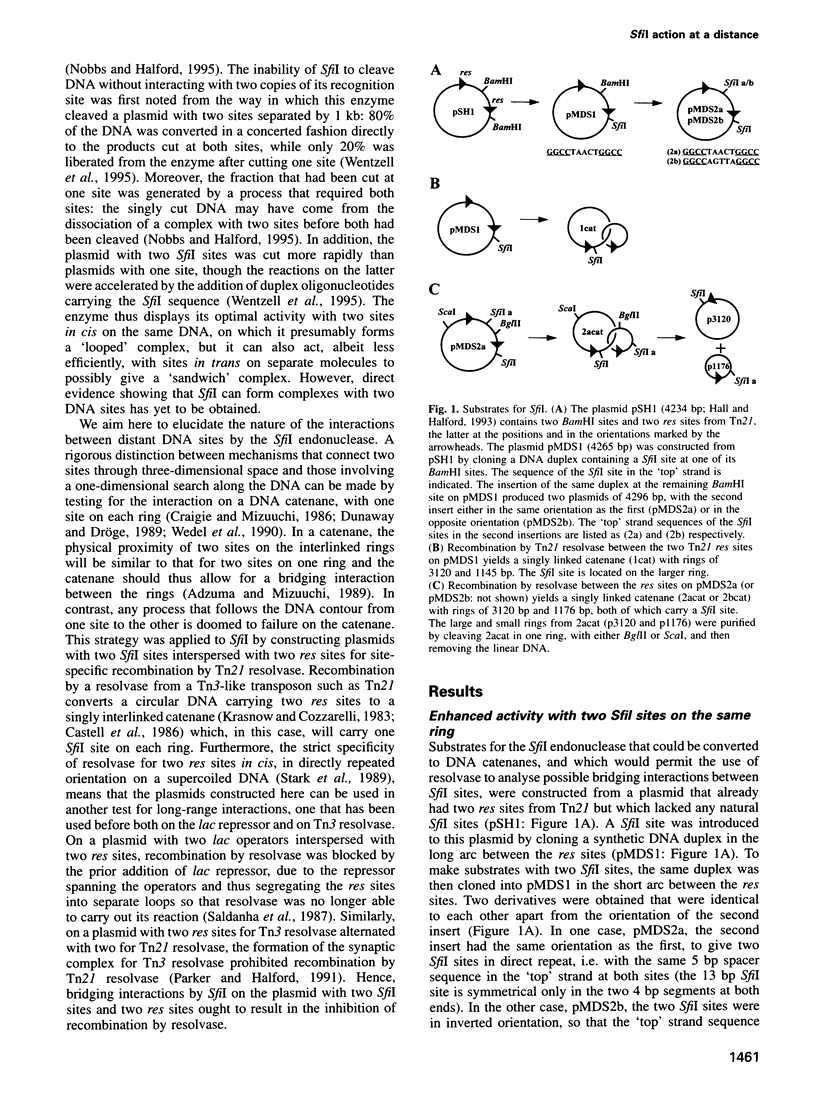
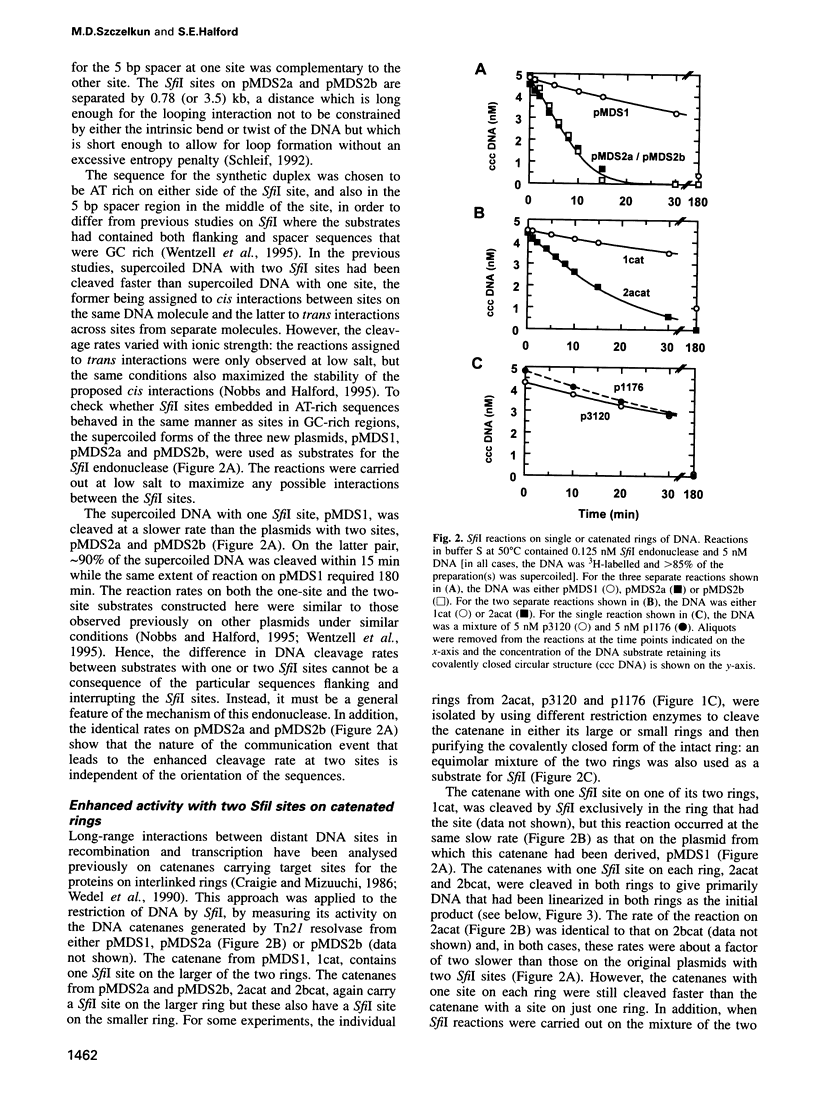
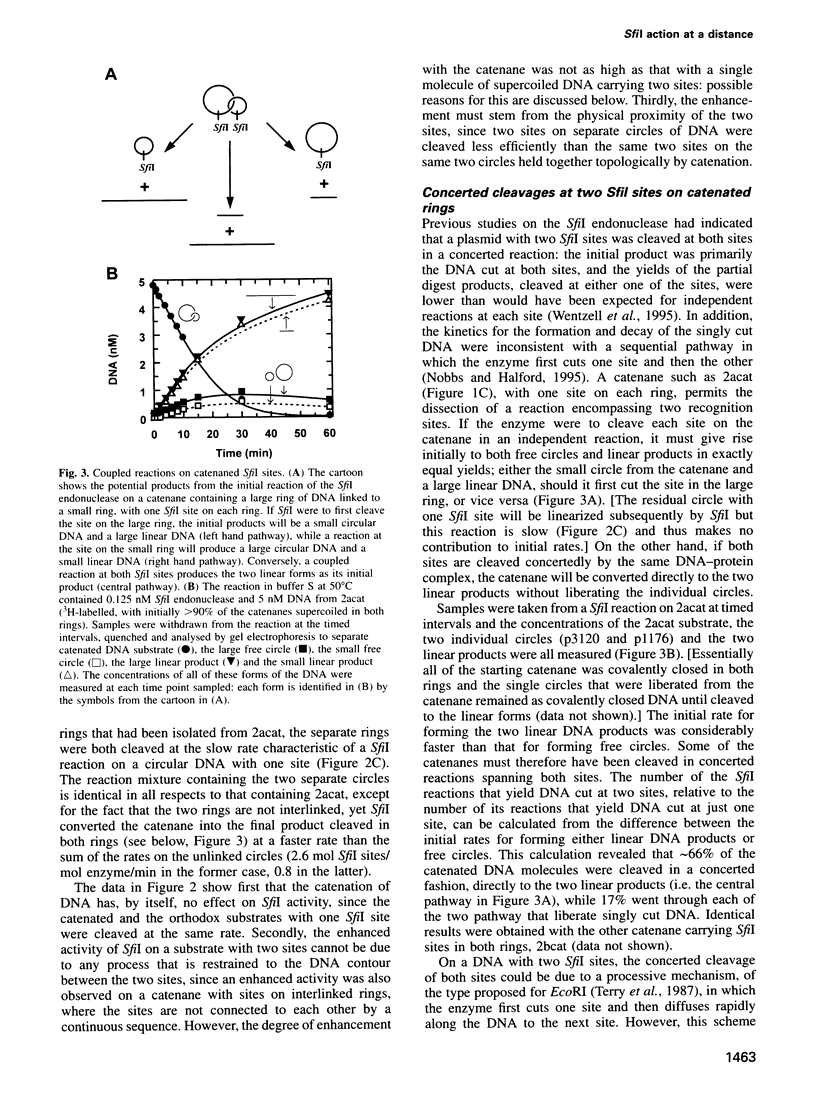
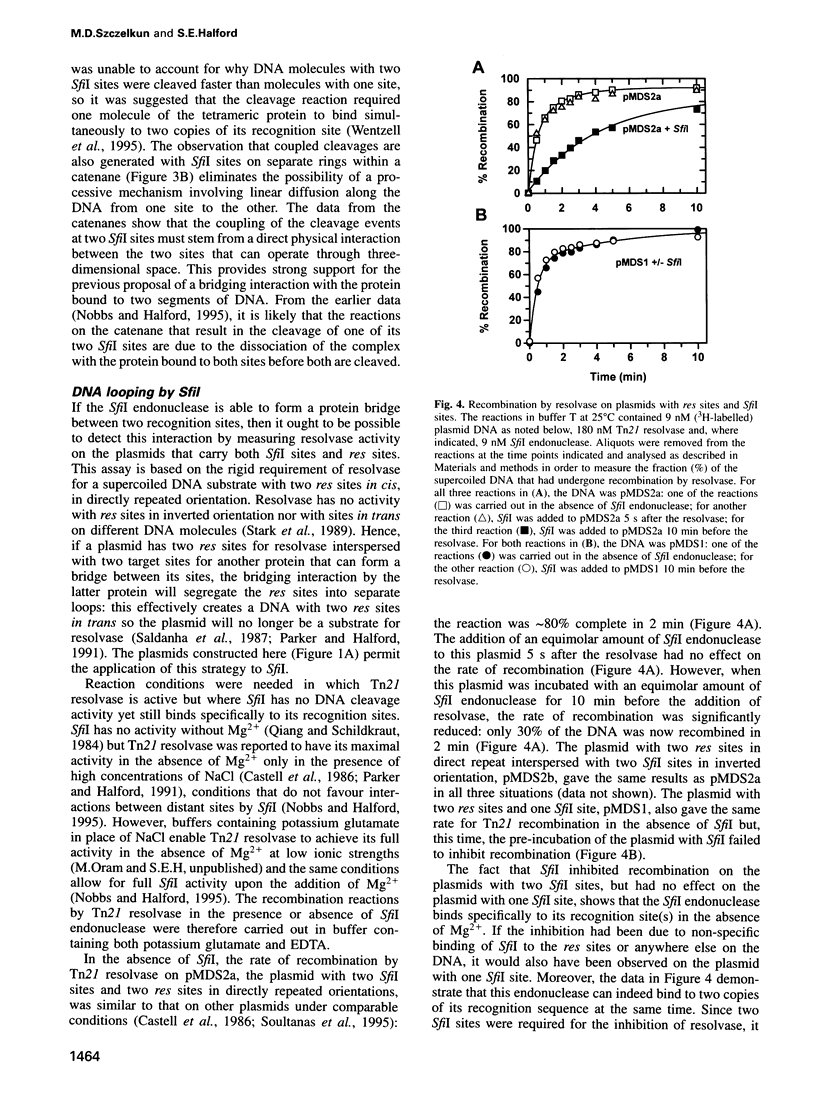
Abstract
The SfiI endonuclease differs from other type II restriction enzymes by cleaving DNA concertedly at two copies of its recognition site, its optimal activity being with two sites on the same DNA molecule. The nature of this communication event between distant DNA sites was analysed on plasmids with recognition sites for SfiI interspersed with recombination sites for resolvase. These were converted by resolvase to catenanes carrying one SfiI site on each ring. The catenanes were cleaved by SfiI almost as readily as a single ring with two sites, in contrast to the slow reactions on DNA rings with one SfiI site. Interactions between SfiI sites on the same DNA therefore cannot follow the DNA contour and, instead, must stem from their physical proximity. In buffer lacking Mg2+, where SfiI is inactive while resolvase is active, the addition of SfiI to a plasmid with target sites for both proteins blocked recombination by resolvase, due to the restriction enzyme bridging its sites and thus isolating the sites for resolvase into separate loops. The extent of DNA looping by SfiI matched its extent of DNA cleavage in the presence of Mg2+.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackroyd A. J., Avila P., Parker C. N., Halford S. E. Site-specific recombination by mutants of Tn21 resolvase with DNA recognition functions from Tn3 resolvase. J Mol Biol. 1990 Dec 5;216(3):633–643. doi: 10.1016/0022-2836(90)90388-3. [DOI] [PubMed] [Google Scholar]
- Adzuma K., Mizuuchi K. Interaction of proteins located at a distance along DNA: mechanism of target immunity in the Mu DNA strand-transfer reaction. Cell. 1989 Apr 7;57(1):41–47. doi: 10.1016/0092-8674(89)90170-0. [DOI] [PubMed] [Google Scholar]
- Au K. G., Welsh K., Modrich P. Initiation of methyl-directed mismatch repair. J Biol Chem. 1992 Jun 15;267(17):12142–12148. [PubMed] [Google Scholar]
- Benjamin H. W., Cozzarelli N. R. Geometric arrangements of Tn3 resolvase sites. J Biol Chem. 1990 Apr 15;265(11):6441–6447. [PubMed] [Google Scholar]
- Castell S. E., Jordan S. L., Halford S. E. Site-specific recombination and topoisomerization by Tn21 resolvase: role of metal ions. Nucleic Acids Res. 1986 Sep 25;14(18):7213–7226. doi: 10.1093/nar/14.18.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Role of DNA topology in Mu transposition: mechanism of sensing the relative orientation of two DNA segments. Cell. 1986 Jun 20;45(6):793–800. doi: 10.1016/0092-8674(86)90554-4. [DOI] [PubMed] [Google Scholar]
- Dunaway M., Dröge P. Transactivation of the Xenopus rRNA gene promoter by its enhancer. Nature. 1989 Oct 19;341(6243):657–659. doi: 10.1038/341657a0. [DOI] [PubMed] [Google Scholar]
- Friedman A. M., Fischmann T. O., Steitz T. A. Crystal structure of lac repressor core tetramer and its implications for DNA looping. Science. 1995 Jun 23;268(5218):1721–1727. doi: 10.1126/science.7792597. [DOI] [PubMed] [Google Scholar]
- Gabbara S., Bhagwat A. S. Interaction of EcoRII endonuclease with DNA substrates containing single recognition sites. J Biol Chem. 1992 Sep 15;267(26):18623–18630. [PubMed] [Google Scholar]
- Halford S. E., Taylor J. D., Vermote C. L., Vipond I. B. Assays for restriction endonucleases using plasmid substrates. Methods Mol Biol. 1994;30:385–396. doi: 10.1385/0-89603-256-6:385. [DOI] [PubMed] [Google Scholar]
- Hall S. C., Halford S. E. Specificity of DNA recognition in the nucleoprotein complex for site-specific recombination by Tn21 resolvase. Nucleic Acids Res. 1993 Dec 11;21(24):5712–5719. doi: 10.1093/nar/21.24.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo K., Topal M. D. DNA topoisomerase and recombinase activities in Nae I restriction endonuclease. Science. 1995 Mar 24;267(5205):1817–1820. doi: 10.1126/science.7892605. [DOI] [PubMed] [Google Scholar]
- Krasnow M. A., Cozzarelli N. R. Site-specific relaxation and recombination by the Tn3 resolvase: recognition of the DNA path between oriented res sites. Cell. 1983 Apr;32(4):1313–1324. doi: 10.1016/0092-8674(83)90312-4. [DOI] [PubMed] [Google Scholar]
- Krämer H., Niemöller M., Amouyal M., Revet B., von Wilcken-Bergmann B., Müller-Hill B. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987 May;6(5):1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger D. H., Barcak G. J., Reuter M., Smith H. O. EcoRII can be activated to cleave refractory DNA recognition sites. Nucleic Acids Res. 1988 May 11;16(9):3997–4008. doi: 10.1093/nar/16.9.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A. Mechanistic and structural complexity in the site-specific recombination pathways of Int and FLP. Curr Opin Genet Dev. 1993 Oct;3(5):699–707. doi: 10.1016/s0959-437x(05)80086-3. [DOI] [PubMed] [Google Scholar]
- Levene S. D., Donahue C., Boles T. C., Cozzarelli N. R. Analysis of the structure of dimeric DNA catenanes by electron microscopy. Biophys J. 1995 Sep;69(3):1036–1045. doi: 10.1016/S0006-3495(95)79978-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell R. B., Schleif R. F. DNA looping and unlooping by AraC protein. Science. 1990 Oct 26;250(4980):528–532. doi: 10.1126/science.2237403. [DOI] [PubMed] [Google Scholar]
- Meisel A., Mackeldanz P., Bickle T. A., Krüger D. H., Schroeder C. Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J. 1995 Jun 15;14(12):2958–2966. doi: 10.1002/j.1460-2075.1995.tb07296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T., Kusano K., Kobayashi I. Selfish behavior of restriction-modification systems. Science. 1995 Feb 10;267(5199):897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- Nobbs T. J., Halford S. E. DNA cleavage at two recognition sites by the SfiI restriction endonuclease: salt dependence of cis and trans interactions between distant DNA sites. J Mol Biol. 1995 Sep 29;252(4):399–411. doi: 10.1006/jmbi.1995.0506. [DOI] [PubMed] [Google Scholar]
- Saldanha R., Flanagan P., Fennewald M. Recombination by resolvase is inhibited by lac repressor simultaneously binding operators between res sites. J Mol Biol. 1987 Aug 5;196(3):505–516. doi: 10.1016/0022-2836(87)90028-3. [DOI] [PubMed] [Google Scholar]
- Schleif R. DNA looping. Annu Rev Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- Soultanas P., Oram M., Halford S. E. Site-specific recombination at res sites containing DNA-binding sequences for both Tn21 and Tn3 resolvases. J Mol Biol. 1995 Jan 20;245(3):208–218. doi: 10.1006/jmbi.1994.0017. [DOI] [PubMed] [Google Scholar]
- Stark W. M., Boocock M. R., Sherratt D. J. Site-specific recombination by Tn3 resolvase. Trends Genet. 1989 Sep;5(9):304–309. doi: 10.1016/0168-9525(89)90113-3. [DOI] [PubMed] [Google Scholar]
- Terry B. J., Jack W. E., Modrich P. Mechanism of specific site location and DNA cleavage by EcoR I endonuclease. Gene Amplif Anal. 1987;5:103–118. [PubMed] [Google Scholar]
- Topal M. D., Thresher R. J., Conrad M., Griffith J. NaeI endonuclease binding to pBR322 DNA induces looping. Biochemistry. 1991 Feb 19;30(7):2006–2010. doi: 10.1021/bi00221a038. [DOI] [PubMed] [Google Scholar]
- Vipond I. B., Halford S. E. Specific DNA recognition by EcoRV restriction endonuclease induced by calcium ions. Biochemistry. 1995 Jan 31;34(4):1113–1119. doi: 10.1021/bi00004a002. [DOI] [PubMed] [Google Scholar]
- Vipond I. B., Halford S. E. Structure-function correlation for the EcoRV restriction enzyme: from non-specific binding to specific DNA cleavage. Mol Microbiol. 1993 Jul;9(2):225–231. doi: 10.1111/j.1365-2958.1993.tb01685.x. [DOI] [PubMed] [Google Scholar]
- Vologodskii A. V., Cozzarelli N. R. Conformational and thermodynamic properties of supercoiled DNA. Annu Rev Biophys Biomol Struct. 1994;23:609–643. doi: 10.1146/annurev.bb.23.060194.003141. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Giaever G. N. Action at a distance along a DNA. Science. 1988 Apr 15;240(4850):300–304. doi: 10.1126/science.3281259. [DOI] [PubMed] [Google Scholar]
- Wedel A., Weiss D. S., Popham D., Dröge P., Kustu S. A bacterial enhancer functions to tether a transcriptional activator near a promoter. Science. 1990 Apr 27;248(4954):486–490. doi: 10.1126/science.1970441. [DOI] [PubMed] [Google Scholar]
- Wentzell L. M., Nobbs T. J., Halford S. E. The SfiI restriction endonuclease makes a four-strand DNA break at two copies of its recognition sequence. J Mol Biol. 1995 May 5;248(3):581–595. doi: 10.1006/jmbi.1995.0244. [DOI] [PubMed] [Google Scholar]
- Yuan R., Hamilton D. L., Burckhardt J. DNA translocation by the restriction enzyme from E. coli K. Cell. 1980 May;20(1):237–244. doi: 10.1016/0092-8674(80)90251-2. [DOI] [PubMed] [Google Scholar]


