Abstract
Lymphocyte antigen receptor engagement profoundly changes the cellular content of phosphoinositide lipids and soluble inositol phosphates. Among these, the phosphoinositides phosphatidylinositol 4,5-bisphosphate (PIP2) and phosphatidylinositol 3,4,5-trisphosphate (PIP3) play key signaling roles by acting as pleckstrin homology (PH) domain ligands that recruit signaling proteins to the plasma membrane. Moreover, PIP2 acts as a precursor for the second messenger molecules diacylglycerol and soluble inositol 1,4,5-trisphosphate (IP3), essential mediators of PKC, Ras/Erk, and Ca2+ signaling in lymphocytes. IP3 phosphorylation by IP3 3-kinases generates inositol 1,3,4,5-tetrakisphosphate (IP4), an essential soluble regulator of PH domain binding to PIP3 in developing T cells. Besides PIP2, PIP3, IP3, and IP4, lymphocytes produce multiple other phosphoinositides and soluble inositol phosphates that could have important physiological functions. To aid their analysis, detailed protocols that allow one to simultaneously measure the levels of multiple different phosphoinositide or inositol phosphate isomers in lymphocytes are provided here. They are based on thin layer, conventional and high-performance liquid chromatographic separation methods followed by radiolabeling or non-radioactive metal-dye detection. Finally, less broadly applicable nonchromatographic methods for detection of specific phosphoinositide or inositol phosphate isomers are discussed. Support protocols describe how to obtain pure unstimulated CD4+CD8+ thymocyte populations for analyses of inositol phosphate turnover during positive and negative selection, key steps in T cell development.
Keywords: lymphocyte, inositol, phosphoinositide, phospholipid, second messenger, T cell, thymocyte, signal transduction, IP3, IP4, IP5, IP6, PIP2, PIP3, HPLC, MDD-HPLC
INTRODUCTION
Lymphocyte antigen receptor engagement profoundly changes the cellular content of multiple different inositol phospholipids (phosphoinositides—PIs, including phosphatidylinositol phosphates, PIPs) and soluble inositol phosphates (IPs, inositol-monophosphates and polyphosphates) (Imboden and Stobo, 1985; Stewart et al., 1986, 1987; Imboden and Pattison, 1987; Zilberman et al., 1987; Guse and Emmrich, 1991, 1992; Guse et al., 1992, 1993; Pouillon et al., 2003; Wen et al., 2004; Huang et al., 2007). In particular, it induces activation of phospholipases Cγ (PLCγ1/2), which then hydrolyze membrane phosphatidylinositol 4,5-bisphosphate (PIP2) into the second messenger molecules diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3; Fig. 11.1.1; Sommers et al., 2004; Berg et al., 2005; Schwartzberg et al., 2005). The lipid DAG remains in cellular membranes, where it recruits and activates key signaling mediators such as the Ras activator RasGRP1 and protein kinases C (PKCs). In contrast, soluble IP3 binding to IP3-receptors mediates Ca2+ release from intracellular stores. This in turn triggers store-operated Ca2+ influx through plasma membrane channels, an essential component of antigen receptor signaling (Feske, 2007). IP3 can also be phosphorylated into inositol 1,3,4,5-tetrakisphosphate (IP4) through the action of IP3 3-kinases (IP3K, Itpk; Irvine et al., 1986; Irvine et al., 2006). Recently, it has been found that T cell receptor (TCR)– or B cell receptor (BCR)–induced IP4 production by IP3KB/ItpkB is essential for T and B cell development in mice (Pouillon et al., 2003;Wen et al., 2004; Huang et al., 2007; Marechal et al., 2007; Miller et al., 2007). Others found evidence for ItpkB/IP4 roles in neutrophilic granulocyte and, possibly, mast cell development or function (Cunha-Melo et al., 1987, 1988; Stokes et al., 2006; Jia et al., 2008). IP4 at least in part acts by regulating the recruitment of pleckstrin homology (PH) domain containing proteins to another important phosphoinositide, phosphatidylinositol 3,4,5-trisphosphate (PIP3). Following antigen receptor engagement, PIP3 is generated via PIP2 phosphorylation by phosphoinositide 3-kinase (PI3K). PIP3-metabolizing phosphoinositide phosphatases such as SHIP1/2 and PTEN control PIP3 levels by counteracting PI3K. In addition, IP4 can either promote or inhibit PIP3 binding to PH domains (Huang et al., 2007). This finding for the first time attributed in vivo relevance to soluble IPs as counterparts of their lipid relatives. In addition to PIP2, PIP3, IP3, and IP4, other PIs and soluble IPs are produced in lymphocytes (Figs. 11.1.1, 11.1.2, 11.1.4, and 11.1.6; Stewart et al., 1986, 1987; Guse and Emmrich, 1991, 1992; Guse et al., 1992, 1993; Pouillon et al., 2003) and could thus have important physiological functions. Recent PI and IP implication in regulating protein membrane recruitment, cytoskeletal dynamics, ion channel function, apoptosis, nuclear functions including mRNA export from the nucleus, transcriptional regulation, mRNA editing and chromatin remodeling, and in nonenzymatic protein phosphorylation (Irvine, 2001, 2003, 2006, 2007; Irvine and Schell, 2001; Saiardi et al., 2004; Irvine et al., 2006; Seeds and York, 2007; Shears, 2007; Alcazar-Roman and Wente, 2008; Majerus et al., 2008; Resnick and Saiardi, 2008) suggests that the elucidation of their functions in lymphocytes will be extremely interesting. To aid in these studies, the following protocols may be utilized.
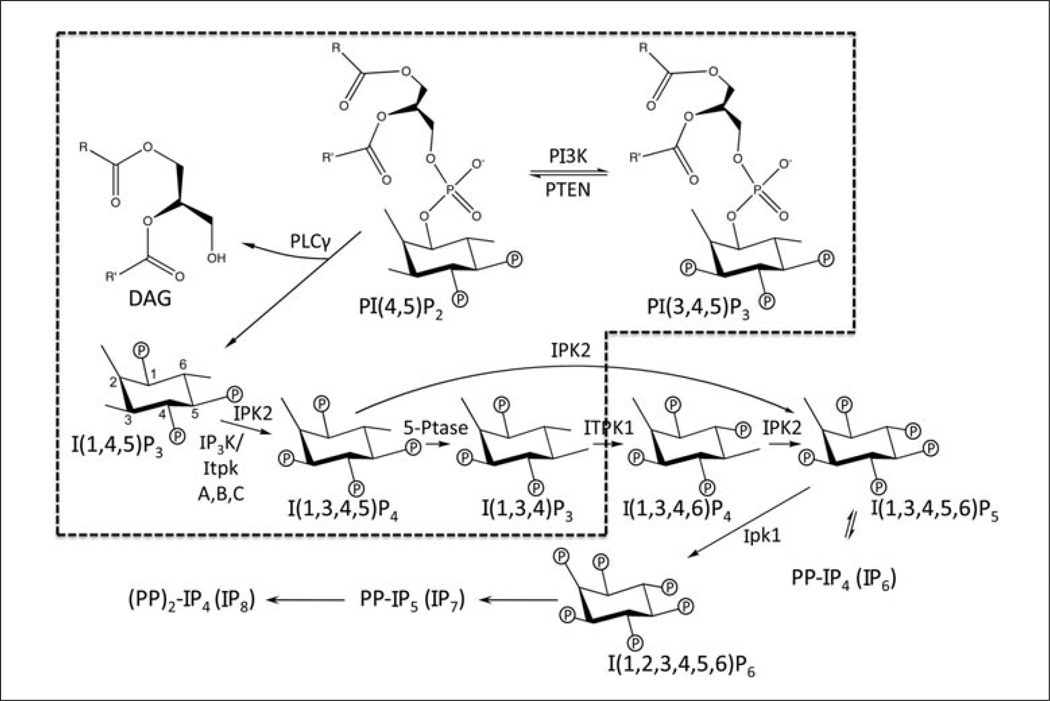
Figure 11.1.1.
Mammalian inositol phosphate metabolism. Simplified scheme of the known inositol phosphate metabolic pathway in mammalian cells. Circled P, phosphate moiety; R, R’, fatty acid side chains. The hatched box encloses pathway components for which genetic data suggest relevance in lymphocytes. For more details and discussions of the enzymes involved and of potential cellular inositol phosphate functions, see previously published works (Irvine, 2001, 2005, 2007; Irvine and Schell, 2001; Irvine et al., 2006; Rusten and Stenmark, 2006; Otto et al., 2007; Seeds et al., 2007; Miller et al., 2008; Alcazar-Roman and Wente, 2008; Huang et al., 2008; Lin et al., 2009). The membrane phospholipid phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2/PI(4,5)P2, PIP2) acts as a precursor for the phosphoinositide PI(3,4,5)P3 (PIP3), and for the second messenger molecules diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (Ins(1,4,5)P3/I(1,4,5)P3/IP3). In mammalian cells, I(1,4,5)P3 acts as a key precursor for multiple higher order, soluble inositol phosphates. An important step in the synthesis of several inositol phosphates is I(1,4,5)P3 phosphorylation into I(1,3,4,5)P4 (IP4) by either one of three IP3 3-kinases (IP3KA, B or C, also termed ItpkA, B, or C; Pouillon et al., 2003; Wen et al., 2004; Huang et al., 2007) or by IPK2/IPMK (Irvine, 2005; Irvine et al., 2006; Otto et al., 2007). Multiple higher order inositol phosphates have been reported in lymphocytes, including several of those shown here. The levels of some inositol phosphates are modulated after antigen receptor engagement (Imboden and Stobo, 1985; Stewart et al., 1986, 1987; Imboden and Pattison, 1987; Zilberman et al., 1987; Guse and Emmrich, 1991, 1992; Guse et al., 1992, 1993; Pouillon et al., 2003). Complementing known PIP3, IP3, and DAG functions in lymphocyte development and function (Starr et al., 2003; Fruman, 2004; Cante-Barrett et al., 2006; Jodi et al., 2008; Juntilla and Koretzky, 2008), it has been recently found that IP4 is essential for these processes through novel roles in antigen receptor signaling and myelopoiesis (Pouillon et al., 2003; Wen et al., 2004; Huang et al., 2007, 2008; Jia et al., 2007, 2008; Marechal et al., 2007; Miller et al., 2007). The protocols described here are thus optimized for analyses of IP3 and IP4 isomers.
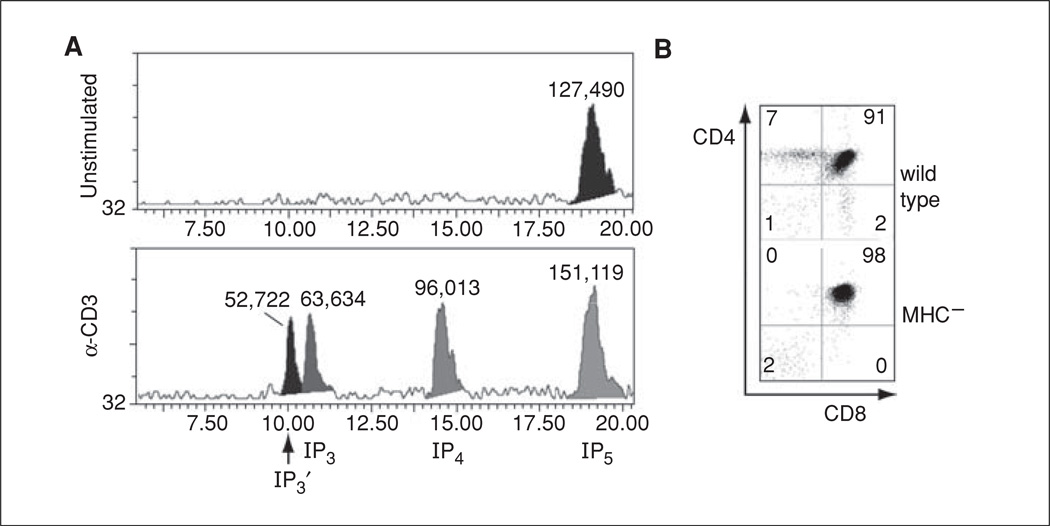
Figure 11.1.2.
Analysis of TCR induced inositol phosphate production in MCHI−MHCII− thymocytes. (A) HPLC elution profiles of extracts from unstimulated or αCD3-stimulated MHC− murine thymocytes. 2 × 108 cells were labeled overnight with 40 µCi myo-[3H] inositol, the precursor for all IPs. At 5 min post-stimulation with medium or 5 µg αCD3 (2C11), cells were lysed in 100 µl of 3% PCA and loaded onto a Whatman cartridge Col SAX PRTSPHR 15-cm HPLC column. [3H] IP content in the eluates was monitored with an IN/US systems Bram-4 in-line β-detector. IP3 or IP4 retention times were determined by spiking [3H] IP3 or [3H] IP4 into unlabeled cell extracts (not shown). IP3′ represents Ins(1,3,4)P3, an IP3 isomer originating from IP4 metabolism (Pouillon et al., 2003). IP5 represents a pool of IP5 isomers (Pouillon et al., 2003). (B) MHC− thymocytes contain ≥98% DP cells, shown by FACS analysis of CD4 and CD8 expression on total thymocytes from 6-week-old C57BL/6 wild type (wt) or MHCI−MHCII− (MHC−) mice. The two-dimensional plots indicate CD4 (y-axis) or CD8 (x-axis) fluorescence intensity for individual cells (dots). The numbers indicate % cells in the respective quadrant.
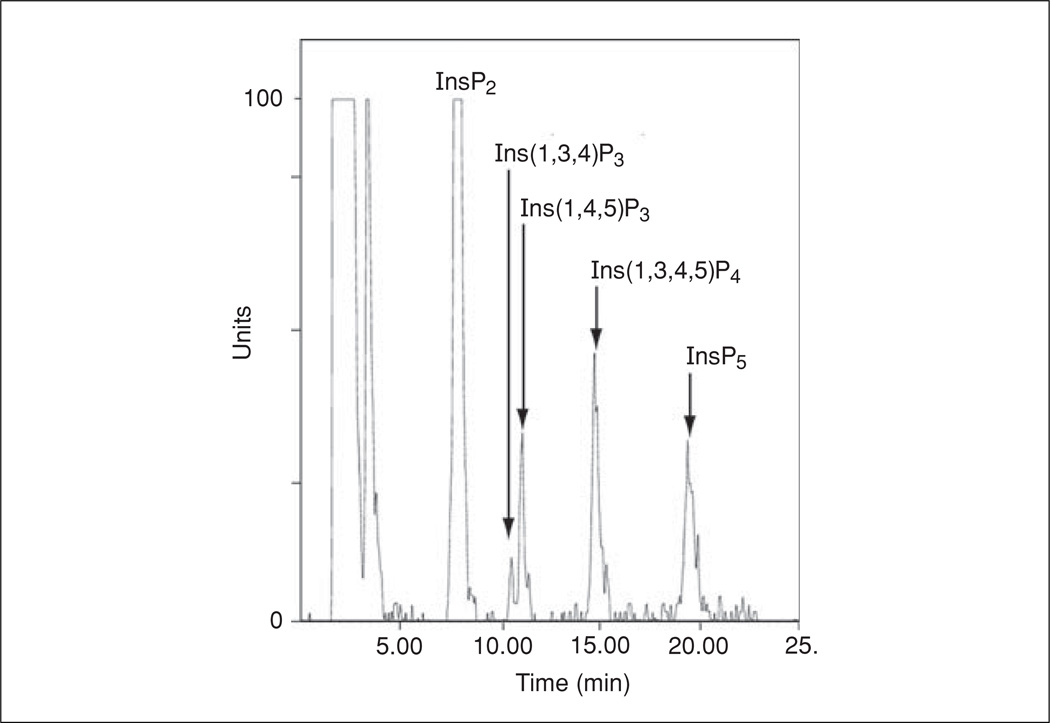
Figure 11.1.4.
A sample trace obtained from Jurkat cells labeled with myo-[3H] inositol and stimulated for 5 min with OKT3 and αCD28 (1 µg/ml). The inositol phosphate isomers detected are indicated. The peaks corresponding to Ins(1,4,5)P3 and Ins(1,3,4,5)P4 were verified with [3H]-labeled purified standards (Perkin-Elmer).
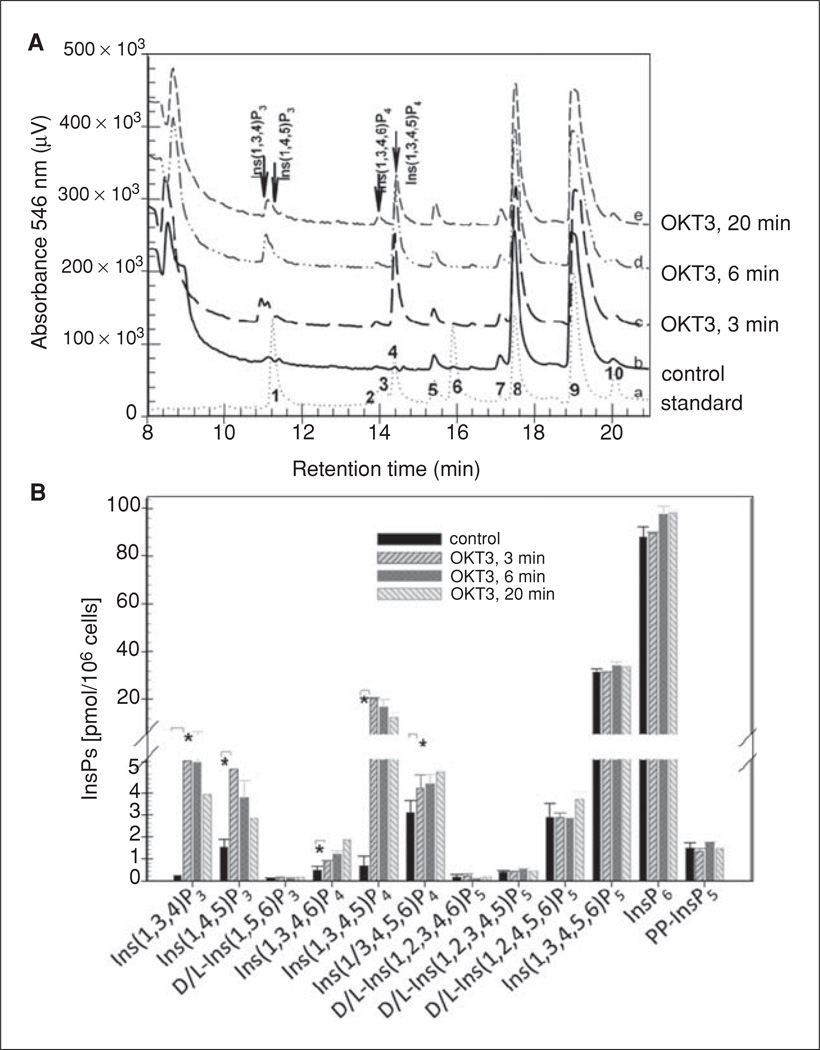
Figure 11.1.6.
MDD-HPLC analysis of soluble inositol phosphate isomers in Jurkat T cells. The MDD-HPLC method has been applied in a number of studies to analyze IPs in cells and tissues, including human Jurkat T cells (Guse et al., 1995a). In the example shown here, soluble IPs were extracted from ~5 × 107 unstimulated or 10 µg/ml OKT3-stimulated Jurkat T cells. (A) a, separation of an IP standard mixture containing 554 pmol I(1,4,5)P3 (peak 1), 43 pmol I(1,2,3,5)P4 (peak 2), 113 pmol I(1,3,4,6)P4 (peak 3), 217 pmol I(1,3,4,5)P4 (peak 4), 116 pmol I(1,4,5,6)P4 (peak 5), 300 pmol I(1,2,3,4,6)P5 (peak 6), 20 pmol I(1,2,4,5,6)P5 (peak 7), 415 pmol I(1,3,4,5,6)P5 (peak 8), 646 pmol IP6 (peak 9), and 215 pmol PP-IP5 (peak 10). b–e, samples from unstimulated (b), 3 (c), 6 (d), or 20 min (e) OKT3-stimulated Jurkat cells. (B) Quantified amounts of the indicated IPs in the Jurkat cell samples from A. *, p < 0.01, obtained via Student’s t-test.
Basic Protocol 1 describes how to radiolabel primary mouse thymocytes with [3H] myo-inositol, stimulate them, and prepare extracts for resolution and highly sensitive detection of radiolabeled inositol phosphates. Alternate Protocol 1 describes a simpler procedure for transformed lymphocyte cell lines.
Support Protocols 1 and 2 describe methods to enrich CD4+CD8+ double positive (DP) thymocytes, the most abundant thymocyte population whose TCR stimulation mediates positive/negative selection through PI/IP-dependent signaling processes (Pouillon et al., 2003; Wen et al., 2004; Huang et al., 2007).
Basic Protocol 2 describes [3H] inositol phosphate resolution by HPLC with an in-line β-detector, and Alternate Protocol 2 a variant without use of this costly device.
Basic Protocol 3 describes a non-radioactive metal dye detection (MDD) method for HPLC analysis of soluble IPs, while Alternate Protocol 3 is a modified version of this protocol for measuring inositol phospholipids. Compared to metabolic labeling, MDD may better represent total PI/IP mass and avoids complications associated with radioactivity.
Using appropriate separation columns and elution protocols, HPLC allows analyses of many soluble IP and lipid PI isomers, but requires costly equipment. To provide cost-effective alternatives that can readily be established in any laboratory and may suffice for low-resolution experiments, Dowex anion-exchange chromatography (see Basic Protocol 4) and thin-layer chromatography (TLC; see Basic Protocol 5) protocols are provided. Less broadly applicable non-chromatographic methods for PI and IP detection in cells or extracts are discussed in the Commentary.
CAUTION: Prior to using radioactive reagents, obtain all required institutional authorization, training, and protective and monitoring equipment. Establish appropriate waste handling and decontamination procedures.
NOTE: All reagents and equipment coming into contact with cells must be sterile, and proper sterile technique must be used.
NOTE: All animal housing, husbandry, handling, and experimental analysis must be approved by, and conducted in accordance with the rules and regulations set forth by the appropriate institutional, national, and state review boards.
STRATEGIC PLANNING
A major decision to be made before the onset of any studies is whether to use metabolic radiolabeling or metal dye detection. Their respective advantages and disadvantages are discussed in the Commentary. Radiolabeling requires institutional approval, training, and monitoring equipment, appropriate waste handling, and decontamination procedures as well as dedicated equipment that will be contaminated, including the HPLC system.
Using primary mouse cells requires that enough animals of the desired genotype and appropriate controls be generated and genotyped before experiments can be conducted. For thymocyte studies, the mice should be 6 to 8 weeks old at the time of analysis. Thus, careful planning and preparation are required.
BASIC PROTOCOL 1
MYO-[3H] INOSITOL LABELING AND STIMULATION OF PRIMARY MURINE THYMOCYTES
This protocol is optimized to myo-[3H] inositol label and extract primary murine thymocytes for detection of IP3, IP4, and IP5 before and after TCR stimulation via HPLC with an in-line β-detector (see Basic Protocol 2). This protocol can be modified to measure IPs in other lymphoid cells, or using other modes of stimulation with or without addition of compounds that affect IP metabolism. An adaptation for immortalized lymphocyte cell lines is described in Alternate Protocol 1. Alternate protocols for [32P] labeling or for extraction and deacylation of radiolabeled PIs followed by HPLC separation and detection of the resulting glycero-inositol phosphates have been published elsewhere (Irvine, 1986; Imai and Gershengorn, 1987; Lapetina and Siess, 1987; Inokuchi and Imboden, 1990; Singh and Jiang, 1995; Kuksis, 2003; Berrie et al., 2007; Guillou et al., 2007b; Sergeant and McPhail, 2007). Support Protocols 1 and 2 describe genetic or flow cytometric methods for enriching unstimulated CD4+CD8+ DP thymocytes, respectively.
IMPORTANT NOTE: The radiolabeling must be conducted following all applicable governmental and institutional radiation safety procedures and regulations. Dedicated experimental equipment and personal protective devices are required, including appropriate monitoring, waste disposal, and decontamination procedures. For tritiated samples, liquid scintillation counters will be required to monitor wipe test results.
Materials
PBS and Ca2+-free PBS (APPENDIX 2A)
Fibronectin (lyophilized human plasma fibronectin; Invitrogen, cat. no. 33016-015 or equivalent; or fibronectin-like engineered protein polymer-plus; Sigma, cat. no. F8141)
1% bovine serum albumin (BSA; Fischer, cat. no. AC61191-0010 or equivalent) in PBS
Mice (6-week-old C57BL/6 mice) or purified DP cells
CO2 source
M199/FCS/HEPES (see recipe)
PharmLyse (BD Biosciences, cat. no. 555899), 1× dilution in double distilled water and filter sterilized, optional
Inositol-free DMEM/2.5% FCS (see recipe)
myo-[3H] inositol (~2.89 TBq/mmol; GE Healthcare, cat. no. TRK883)
Recombinant IL-7 (R&D Systems, cat. no. 407-ML)
0.5 mM EDTA in Ca2+-free PBS
HBSS/HEPES (see recipe), ice cold
Biotinylated anti-mouse CD3 antibodies, clones 145-2C11 or 500A2 (Invitrogen; CALTAG; BD Biosciences)
Unconjugated streptavidin (SA; Jackson Immunoresearch, cat. no. 016-000-113)
Concanavalin A type IV (ConA; Sigma-Aldrich, cat. no. C5275)
3% perchloric acid (PCA) in water
6- and 12-well tissue culture plastic plates, untreated
37°C incubator with and without 5% CO2
Mouse vivarium with appropriate euthanasia equipment in procedure room
60-mm petri dishes
40-µm nylon cell strainer
1- to 5-ml syringes
50-ml conical tubes
Hemacytometer or electronic cell counter
2-ml microcentrifuge tubes
37°C water bath or heating block
Additional reagents and equipment for thymi removal (UNIT 1.9)
Prepare fibronectin-coated plates
-
1Incubate an untreated 6-well tissue culture plastic plate 2 hr at 37°C with 0.5 ml PBS containing 25 µg/ml fibronectin. Then, remove excess liquid and incubate 30 min at 37°C with 1 ml of 1% BSA in PBS to block uncoated areas. Prior to cell addition, remove the PBS solution.Fibronectin allows otherwise non-adherent lymphocytes to adhere to the plate bottoms, forming a homogeneous monolayer. This warrants equal exposure of all cells to medium, oxygen, and labeling reagents. This procedure is helpful to obtain optimal cell labeling and maintain maximal thymocyte viability during the long incubation period required for equilibrium-labeling. The fibronectin may be replaced by genetically engineered fibronectin-like engineered protein polymer-plus (available from Sigma) to further improve cell adhesion (Esty, 1991). The plates can conveniently be coated while thymocytes are being prepared.
Prepare thymocytes
-
2Determine the total number of thymocytes (or purified DP cells) needed per genotype (or treatment group, if mice are treated with immune-modulating agents—typically, 5 × 107 to 1 × 108 thymocytes need to be labeled per sample). Then, determine how many mice are needed per genotype to provide this thymocyte number.For reference, thymi from 6-week-old C57BL/6 mice, the commonly used age and wildtype strain for biochemical analyses of TCR signaling in DP thymocytes undergoing positive/negative selection, typically contain ~2 × 108 thymocytes. From this amount, 83% to 90% are CD4+CD8+ DP cells (Figs. 11.1.2 and 11.1.3). Thymi from mutant mice may contain significantly less cells.
-
3
Per genotype/treatment group, euthanize the number of age- and, preferably, gender-matched mice required to provide the calculated number of thymocytes (or purified DP cells) via an appropriate, government and institutionally approved euthanasia method such as CO2 inhalation.
-
4Immediately, remove all thymi surgically as described in UNIT 1.9 and place into 10 ml of M199/FCS/HEPES (thymi of identical genotypes/treatment groups can be pooled).Process the thymi as quickly as possible to preserve thymocyte viability. M199 medium assures optimal thymocyte viability. Other cell types may require other media for optimal results. Neomycin and its analogs bind inositol lipids and should not be present.
-
5Pour 5 ml of the medium into a 60-mm petri dish and place a 40-µm nylon cell strainer into the plate such that the medium covers the mesh completely. Add a thymus and use the rubber end of a 1- to 5-ml syringe plunger to carefully disperse the thymocytes through the mesh into the medium. Transfer this cell suspension into a 50-ml conical tube and use 2 to 3 ml additional medium to wash remaining cells from the strainer into this cell stock. Identical genotypes/treatment groups can again be pooled.Thymocytes can also be dispersed by carefully “grinding” a thymus between the frosted sides of two microscope slides, followed by washing with medium. However, the cell strainer method removes more debris and yields more homogeneous cell suspensions.If significant red blood cell (RBC) contamination is present or lymphocyte suspensions from RBC-rich tissues (spleen, bone marrow) are prepared, lyse the RBCs at this point by centrifuging 5 min at 500 × g in a tabletop centrifuge, removing the supernatant and very carefully resuspending the cells in 1× PharmLyse (1 ml per thymus or bone marrow preparation, 2 ml per spleen). Incubate 2 to 3 min at room temperature and stop RBC lysis by adding 20 ml M199/FCS/HEPES and gently mixing via tube inversion. Proceed immediately to the next step, as prolonged incubation in RBC lysis buffer will lyse thymocytes and lymphocytes as well.
Figure 11.1.3.
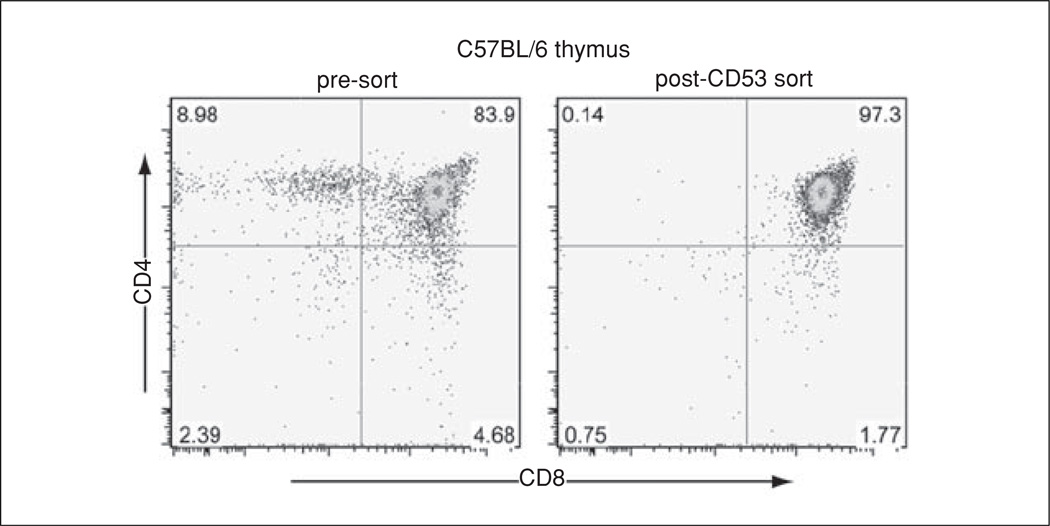
Analysis of thymocyte populations pre- and post-anti-CD53 AB sort. Two-dimensional plots showing CD4 (y-axis) and CD8 (x-axis) fluorescence intensities for individual thymocytes (dots) from 6-week-old C57BL/6 mice before (pre-sort) or after (post-CD53 sort) depletion of CD53+ cells. The numbers indicate percent cells in the respective quadrant.
Label with myo-[3H] inositol
-
6Centrifuge cells 5 min at 500 × g, room temperature, remove supernatant, and resuspend the cells in exactly 20 ml M199/FCS/HEPES. Count appropriately diluted cell aliquots using a hemacytometer (APPENDIX 3A & 3B) or electronic cell counter following the manufacturer’s instructions. Then, centrifuge cells as above and resuspend at a concentration of 2 × 107 cells/ml in inositol-free DMEM/2.5% FCS.Pooled thymocytes from ten 6-week-old C57BL/6 mice will need to be diluted ~1:20 for counting. The cells can be diluted in medium or in cell counter electrolyte. Trypan blue staining (APPENDIX 3B) can provide an estimate of cell viability if a hemacytometer is used. Depending on the cell type, cell densities can be optimized between 2 × 107 and 2 × 108 cells/ml.
-
7Add myo-[3H] inositol to a final activity of 20 µCi/ml and recombinant IL-7 to a final concentration of 5 ng/ml.The IL-7 improves cell viability during the following long incubation period. This and all subsequent procedures need to be conducted following all applicable governmental and institutional radiation safety guidelines and procedures. All wash solutions and other waste are to be considered radioactive and disposed of accordingly. myo-[3H] inositol activities can be optimized between 2 and 40 µCi/ml.
-
8Add 2.5 ml of the 2 × 107 cells/ml thymocyte suspension (5 × 107 thymocytes total) per well to the pre-prepared fibronectin plates (from step 1). Incubate 20 hr in a 37°C, 5% CO2 humidified incubator. After the first 3 hr, add 2.5 ml of inositol-free DMEM/2.5% FCS.The added medium supplies additional nutrients and buffer after an initial incubation in a higher myo-[3H] inositol concentration. FCS contains variable amounts of inositol and may need to be titrated for optimal labeling. If labeling is insufficient, reduce FCS content or increase labeling times. If viability is compromised, try adding more FCS, or adding non-radioactive inositol up to the concentration in normal DMEM, either immediately or up to several hours after an initial pulse in radioactive inositol only. See Critical Parameters and Troubleshooting.
-
9After a total of 20 hr incubation (17 hr after dilution), detach the cells by replacing the labeling medium with 1 ml of 0.5 mM EDTA in Ca2+-free PBS and incubating for 15 min at 37°C. Gently agitate the plates at the end of the incubation period to fully suspend the cells. Transfer the cells into a 2-ml microcentrifuge tube and centrifuge 5 min at 500 × g, room temperature, in a benchtop microcentrifuge. Aspirate the supernatant and resuspend the cells in 1 ml ice-cold HBSS/HEPES (naturally inositol free) or in inositol-free DMEM/2.5% FCS. Wash two additional times to remove all radioactivity from the medium.In microcentrifuge tubes, the cells can quickly become hypoxic due to impaired air exchange, which reduces viability. To prevent this, avoid longer storage in microcentrifuge tubes and work quickly but carefully as the cells are radioactive.
-
10After the final wash, gently suspend the cells in 1 ml ice-cold inositol-free DMEM/2.5% FCS in microcentrifuge tubes to a concentration of 5 × 107 cells/ml. Allow cells to equilibrate for 20 min at 37°C. Then place on ice for 5 min to slow cell metabolism.This step serves to reduce background activation, which may be caused by cell handling, shear stress, etc. If no further incubation with compounds (see below) is planned, LiCl can be added to the medium at a final concentration of 5 to 20 mM. By inhibiting cellular phosphatases that metabolize PIs and IPs, LiCl can enhance PI/IP detection. If PI/IP steady-state levels, which are the sum of their production by kinases and their dephosphorylation by phosphatases in the cell, are to be analyzed, LiCl should not be added.
Incubate with compounds and stimulate
The cells can be stimulated using different reagents and regimen. Frequently, this involves pre-binding of receptor-specific antibodies followed by their crosslinking through secondary reagents as the actual activating step. Alternatively, some receptors can be directly activated without secondary crosslinking. Moreover, modulating agents such as small molecule inhibitors of cellular signaling proteins or of PI/IP metabolizing enzymes may be applied, either during a pre-incubation period to allow them to enter the cells and possibly be converted into metabolically active species, or together with the stimulating agents in cases where cell penetration and modulation are known to occur rapidly. The following steps describe a generic regimen of reagent addition, using specific exemplary reagents to provide estimates for working concentrations and incubation times. When using other reagents, optimal concentrations, delivery carriers, and incubation times must be determined empirically.
-
11To pre-incubate the cells with compounds, centrifuge the cells as in step 9 and resuspend them in fresh ice-cold DMEM/2.5% FCS containing the respective compound at the desired concentration. Incubate the cells in 12-well plates for the desired time, typically 30 to 120 min, in a 37°C, 5% CO2 incubator to allow the compounds to enter the cells and, if needed, be converted into active derivatives. Then, place the cells on ice.For short incubation times <10 min, this can be done in microcentrifuge tubes. Otherwise, use 12-well plates to allow sufficient air exchange. If FCS binding of the compounds precludes achieving sufficiently high concentrations, try to reduce the FCS content in the medium. Shorter pre-incubation times may help to preserve cell viability. The optimal cell number required per sample depends on the cell type, the labeling efficiency/specific activity, and the response intensity achieved in each experiment. It needs to be determined empirically. As a guideline, try 5 × 106 to 5 × 107 cells in a 1-ml volume.
-
12To pre-bind receptor-specific antibodies (Abs) without activating the cells, add the Abs at pre-determined optimal concentrations and incubate 15 min on ice.The Abs should be added in a small volume followed by gentle agitation with a pipet tip or by gentle up and down pipetting to distribute the Abs evenly. If concentrated Ab or reagent stocks are unavailable, replace the entire medium with ice-cold medium containing the Abs (and above compounds if applicable) at optimized concentrations. For example, use 1.2 µg of biotinylated anti-CD3 Ab (clone 145-2C11)/5 × 107 cells.
-
13
If the cells are on plates, transfer them into microcentrifuge tubes. Wash cells two times with 1ml ice-cold HBSS/HEPES solution, centrifuging each time 5 min at 500 × g, room temperature. Aspirate all liquid.
-
14
For stimulation, gently resuspend the cells in 400 µl PBS (final concentration 1.25 × 108 cells/ml if 5 × 107 cells were stimulated), pre-warmed to 37°C and containing the desired reagents, for example, cross-linking secondary Abs, streptavidin (5 ng/ml final) for crosslinking biotinylated primary Abs, or directly stimulating agents such as concanavalin A (25 µg/ml final). Immediately close and place the microcentrifuge tubes into a 37°C water bath or heating block.
-
15At the desired time points, remove the tubes and add 900 µl of ice-cold PBS to stop the reaction. Immediately centrifuge samples 1 min at 500 × g, 4°C, in a benchtop centrifuge. Remove the supernatant and lyse the cells in 100 µl of 3% PCA in water through vigorous vortexing or pipetting up and down. Incubate 20 min on ice.Longer incubation times in acid, especially at higher temperatures, promote PIP head-group hydrolysis, IP isomerization, or IP hydrolysis and should thus be avoided (Singh and Jiang, 1995). Lymphocytes contain relatively high PIP and PIP2 amounts. Their acid hydrolysis can lead to superphysiological IP2 and IP3 levels.
-
16Centrifuge samples 5 min at 15,000 × g, 4°C, in a benchtop microcentrifuge, then transfer 100 µl of the supernatant into new microcentrifuge tubes. Use immediately for HPLC analysis of soluble inositol phosphates or neutralize and store up to 2 weeks frozen at −70°C.The PCA treatment serves to precipitate proteins, terminate metabolic processes, and dissolve IPs (Singh and Jiang, 1995). Alternatively, the cells can be lysed in chloroform/methanol/concentrated HCl or in 10% TCA (Berridge et al., 1983; Irvine et al., 1986; Berrie et al., 2007). Maximal IP yields were obtained when cells were lysed in 10% TCA, followed by a 20-min incubation on ice (to minimize IP hydrolysis), centrifugation for 10 min at 15,000 × g, 4 ° C, and TCA removal through three rounds of supernatant extraction with 2 vol diethyl ether, each round. PCA can be removed by equimolar KHCO3 addition (~0.5 M) and centrifugation (as above) to remove the insoluble K-perchlorate. However, the precipitate may contain trapped IP-metal complexes that form at a pH >7.0, reducing the IP yield in the supernatant (Lin et al., 2009).Addition of each 20 µg of unlabeled IP3 and IP4 may enhance extraction of these IPs, in particular of IP4 (Wreggett et al., 1987). If labeling efficiency is low, replicate samples can be pooled prior to extraction to increase extracted radioactivity. Typically, time points of 0, 0.5, 1, 3, 5, and 15 min allow one to monitor most TCR-modulated PIs/IPs (Imboden and Pattison, 1987; Budd et al., 1990; Inokuchi and Imboden, 1990; Guse and Emmrich, 1991, 1992; Guse et al., 1993; Pouillon et al., 2003).Because of increased IP hydrolysis and isomerization in an acidic environment (Singh and Jiang, 1995), cell pellet–free PCA or HCl extracts are only stable for ≤ 2 hr at room temperature. Moreover, creatine phosphate, which is present in the extracts, can lead to non-enzymatic IP-pyrophosphorylation in particular at pH >4.0 in the presence of bivalent metal ions (Lin et al., 2009). It can be destroyed via a brief, 15- to 20-min incubation of the acidic extracts at 30° to 35°C with typically <10% concurrent IP hydrolysis. For long-term storage, sample neutralization (via TCA ether extraction or KHCO3 precipitation of PCA) and storage at − 70°C are recommended.
ALTERNATE PROTOCOL 1
MYO-[3H] INOSITOL LABELING AND STIMULATION OF IMMORTALIZED T CELLS
This protocol describes simplified modifications of Basic Protocol 1 for myo-[3H] inositol labeling of immortalized cell lines, using human Jurkat αβT cells as an example. Immortalized cells grow more robustly and are metabolically more active, allowing for a simplified labeling procedure. If problems are encountered, follow Basic Protocol 1, starting from step 6.
Additional Materials (also see Basic Protocol 1)
Human Jurkat αβT cells (ATCC # TIB-152) or other cells of interest RPMI/10% FCS/PSG (see recipe)
- Grow cells in exponential culture (<107 cells/ml) in appropriate medium such as RPMI/10% FCS/PSG for Jurkat cells. Suspend via gentle shaking (Jurkat cells) or 2- to 10-min incubation at room temperature or 37°C with 5 ml (75-cm2 flask) of 2 mM EDTA followed by gentle shaking (adherent cells). Count cells as in Basic Protocol 1, centrifuge in a tabletop centrifuge 5 min at 553 × g, 4°C, and resuspend at 2–3 × 107 cells/ml in inositol-free RPMI or DMEM without FCS.About 107 cells per sample will be needed. For assay optimization, compare results for 5 × 106 to 5 × 107 cells per sample. Omit FCS during the initial labeling as FCS/FBS contains inositol. Jurkat cells can grow without FCS for 6 to 8 hr.
- Add myo-[3H] inositol to a final activity of 20 µCi/ml and transfer into an appropriately sized tissue culture flask.Myo-[3H] inositol levels can be optimized between 2 and 40 µCi/ml.
- Incubate 5 to 6 hr in a 37°C, 5% CO2 humidified incubator. Then, add 40 vol of complete medium (with inositol)/2.5% to 10% FCS, diluting the cells to 5 × 105/ml. Incubate overnight for a total of at least 20 hr in a 37°C, 5%CO2 humidified incubator.For assay optimization, evaluate incubation times up to 72 hr, monitoring labeling efficiency and viability. An initial incubation with medium in which all inositol is radio labeled, followed by longer incubation with additional non-radioactive inositol and FCS to augment cell viability, provided the best results. If viability is still low, try adding low amounts of FCS or of non-radioactive inositol during the first 6 hr of incubation. If labeling is suboptimal, try longer incubation with myo-[3H] inositol in the absence of cold inositol/FCS. See Critical Parameters and Troubleshooting.
- Transfer the cells into 50-ml conical tubes, centrifuge as in step 1, wash two times in RPMI/10% FCS, resuspend at 107 cells/ml in RPMI/10% FCS, equilibrate for 20 min at 37°C, and rest on ice.The wash and equilibration step serves to reduce non-specific activation due to cell handling, shear stress, etc. This step is recommended for optimal results but can be omitted without major loss of assay performance to save time. Refer to the Commentary, for discussion of the drawbacks in cell labeling with myo-[3H] inositol. Lymphocytes can also be stimulated in PBS, HBSS/HEPES, or other solutions with physiological salt concentrations, or in antibiotic-free tissue culture media (APPENDIX 2A). Neomycin and analogs bind inositol lipids and should be omitted.
- For stimulation, follow Basic Protocol 1, steps 11 to 16, but using appropriate stimulation reagents for cell type and species.Jurkat cells are larger than thymocytes. Thus, stimulation in a total volume of 1 ml containing 2 × 106 to 2 × 107 cells may give better results. The optimal cell number required per sample depends on the cell type, the labeling efficiency/specific activity and the response intensity achieved in each experiment, and must be determined empirically. For a discussion of potential LiCl addition to enhance sensitivity, see Basic Protocol 1, step 10 annotation.Suitable stimulation reagents for Jurkat cells are antibodies against human CD3ε (OKT3, 1 to 5 µg/ml final) and CD28 (1 µg/ml final), or ConA (2.5 µg/ml final).
ENRICHING UNSTIMULATED CD4+CD8+ DP THYMOCYTES
Total thymocytes include multiple subsets of developing T cells (Starr et al., 2003). Differential cell surface expression of the co-receptor molecules CD4 and CD8 can be used to distinguish four populations. CD4−CD8− double negative (DN) cells comprise <3% of total thymocytes. They include several early developmental stages. Only the most mature DN subsets express a pre-TCR and CD3 and can respond to CD3 stimulation. DN cells mature to CD4+CD8+ double-positive (DP) cells, which comprise ≥83% of all thymocytes. DP cells express a functional TCR/CD3 complex and represent the majority of cells that respond to TCR or CD3 stimulation. TCR stimulation on DP cells mediates positive and negative selection, key processes that govern DP cell maturation into functionally competent, self-tolerant mature CD4+ or CD8+ single-positive (SP) T cells, which comprise ~8% and ~4% of all thymocytes, respectively. The sensitivity of DP cells to TCR stimulation is higher than that of mature SP T cells. Because of this heterogeneity, analyzing bulk thymocyte PI/IP responses can reflect differential contributions by several subpopulations that are responsive to the stimulus applied. The very low total DN and SP cell numbers usually preclude biochemical assays in these populations, unless very large numbers of thymi are pooled. However, these minor populations can affect results of measuring DP cell responses, in particular when different mouse mutants or mice treated with different compounds are compared. For example, a comparison of TCR-induced PI/IP modulation in wild-type mice with ItpkB/IP3KB−/− mice that lack SP thymocytes (Pouillon et al., 2003; Wen et al., 2004; Huang et al., 2007) could be affected by SP cell contributions in the wild-type controls. Moreover, a significant proportion of DP cells engage in TCR interactions with endogenous ligands presented by major histocompatibility complex (MHC) class I and class II proteins on thymic stromal cells. Their responses to ectopic stimulation in the PI/IP assay may differ from those of DP cells that have not yet received TCR stimuli (Starr et al., 2003). For studies of TCR signaling in DP cells, it is thus important to use purified DP cells that have not undergone TCR stimulation. This can be achieved genetically (see Support Protocol 1) or through immuno-affinity sorting with magnetic beads (see Support Protocol 2).
SUPPORT PROTOCOL 1
MHCI−MHCII− (MHC−) Mice
B6.129-H2-Ab1tm1Gru B2mtm1Jae mice (Taconic) express no detectable MHCII and very low MHCI protein levels. As a result, no ligands can be presented to DP cells, resulting in impaired selection and absence of SP cells (Grusby et al., 1993). The thymi of these MHCI−MHCII− mice contain ~98% DP cells whose TCR has never been engaged by ligand (Fig. 11.1.2). Since most DN cells do not respond to TCR stimulation, MHCI−MHCII− mice provide a convenient model to investigate TCR signaling in highly enriched “unsignaled” DP cells. Their breeding to a mouse mutant or transgenic mouse of interest allows analyses of the contributions of specific genes such as ItpkB/IP3KB to TCR signaling and PI/IP production in DP cells and in the context of thymocyte positive/negative selection without the need for expensive subset purification (Fig. 11.1.2; Huang et al., 2007). Potential drawbacks are breeding costs and possible DP cell hypersensitization due to lack of TCR stimulation and positive selection in MHCI−MHCII− mice (Starr et al., 2003; Huang et al., 2007). Breeding three mutations (mutation under study, MHCI−, and MHCII−) to homozygosity using these mice will take two generations, typically requiring 7 to 9 months, and genotyping of many F2 animals as only 1/64 will be homozygous for all three mutations. Establishing the triple mutant through intercross to generate enough animals for analysis will take at least one additional generation, resulting in a total time of ~1 year. FACS sorting of CD53− DP cells from the mutant of interest (see Support Protocol 2) provides a much faster alternative but requires expensive reagents and FACS sorting time.
Breeding and maintenance of B6.129-H2-Ab1tm1Gru B2mtm1Jae mice (MHCI−MHCII− mice) should follow standard procedures (Chapter 1) and should be conducted in a specific pathogen–free (SPF) vivarium, as these mice lack peripheral T cells and are therefore immunodeficient.
Materials
B6.129-H2-Ab1tm1Gru B2mtm1Jae mice [Taconic, cat. no. 004080-MM-F (females) or -M (males)]
PharmLyse (BD Biosciences, cat. no. 555899): dilute to 1× in double distilled water and then filter sterilize
PBS containing 2% FCS
Fc block: anti-mouse CD16/32 antibody
FACS antibodies: H-2Kb-FITC (BD Pharmingen, cat. no. 553569); I-Ab-PE (BD Pharmingen, cat. no. 553552); CD4 (L3T4) APC (BD Pharmingen, cat. no. 553051); CD8a (Ly-2) PE-Cy7 (BD Pharmingen, cat. no. 552877)
Tail digestion buffer (see recipe)
25:24:1 (v/v) phenol/chloroform/isoamyl alcohol (Invitrogen, cat. no. 15593-031)
Genotyping primers: B2m primers [oIMR0160 (mutant):
TCTggACgAAgAgCATCAggg; oIMR0184 (common):
TATCAgTCTCAgTgggggTg; oIMR0185 (wild type):
CTgAgCTCTgTTTTCgTCTg]
I-Ab primers [oIMR5241 (mutant): gTgTTgggTCgTTTgTTCg; oIMR5239 (common): AgggAggTgTgggTCTCC; oIMR5240 (wild type):
gTACCAgTTCATgggCgAgT]
96-well U-bottom plate
Additional reagents and equipment for care and handling of laboratory animals (Chapter 1); flow cytometry (Chapter 5)
Identify MHCI−MHCII− mice via peripheral blood cell FACS analysis
-
1
Because MHCI−MHCII− mice lack peripheral T cells, they can be identified via FACS analysis of peripheral blood T cell numbers, unless the specific mouse mutant, transgenic mouse, or compound treatment under study results in peripheral T cell deficiency. For this procedure, obtain 50 µl peripheral blood via retro-orbital or tail vein bleeding as described in UNIT 1.7.
-
2Transfer peripheral blood samples to a 96-well U-bottom plate. Centrifuge plate 5 min at 500 × g, room temperature, and remove supernatant. Lyse red blood cells by resuspending cell pellet in 150 µl of 1× PharmLyse. Incubate 5 min at room temperature. Centrifuge plate 5 min at 500 × g, room temperature, and remove supernatant. Resuspend cell pellet in 30 µl PBS containing 2% FCS and 1 µl Fc block. Incubate 5 min at room temperature. Stain cells by adding 30 µl PBS containing 2% FCS and antibodies specific for H-2Kb-FITC, I-Ab-PE, CD4-APC, and CD8-PECy7. Add 100 µl PBS containing 2% FCS. Centrifuge 5 min at 500× g, room temperature. Remove supernatant and repeat wash with 200 µl PBS containing 2% FCS. Resuspend in 200 µl PBS containing 2% FCS and analyze by flow cytometry as described in Chapter 5.Abs need to be titrated for optimized detection by flow cytometry.For analysis of MHC I and II on genetic backgrounds other than C57BL/6, use general or haplotype-specific anti-MHCI/II antibodies.
Identify MHCI−MHCII− mice by tail DNA genotyping
-
3
To extract genomic DNA, digest 0.5 to 1 cm tail tips by incubating in 300 µl tail digestion buffer containing proteinase K for 1.5 hr at 55°C with agitation. Following digestion, add an equal volume of 25:24:1 (v/v) phenol/chloroform/isoamyl alcohol. Vortex to mix and centrifuge for 2 min at 15,000 × g, room temperature, in a microcentrifuge to separate aqueous and organic phases. Use 1 to 2 µl of the aqueous phase as the PCR template.
-
4
Conduct PCR reactions as stated at http://jaxmice.jax.org/pub-cgi/protocols/protocols.sh?objtype=protocol&protocol id=1138 and http://jaxmice.jax.org/pubcgi/protocols/protocols.sh?objtype=protocol&protocol id=1580.
SUPPORT PROTOCOL 2
CD53−CD4+CD8+ DP Thymocyte Enrichment by Magnetic Bead Immunoaffinity Cell Sorting (MACS)
Depletion of CD53+ cells from total thymocyte preparations yields >97% pure DP cells whose lack of CD69 expression indicates that these cells have not received a TCR stimulus (Puls et al., 2002; Huang et al., 2007).This allows analyses of TCR-induced PI/IP turnover in very pure “unsignaled” DP cells in any mouse line without the need for time-and resource-consuming breeding, but at the cost of expensive reagents (Fig. 11.1.3).
Materials
6-week-old C57BL/6 mice
MACS staining buffer (see recipe)
Anti-CD53 Ab (OX-79) (BD Biosciences, cat. no. 559364)
Biotinylated anti-rat IgG (Jackson Immunoresearch, cat. no. 112-065-167)
Anti-biotin microbeads (Miltenyi Biotec, cat. no. 130-042-401)
MidiMACS separation unit (Miltenyi Biotec, cat. no. 130-042-302)
Additional reagents and equipment for harvesting thymocytes (see Basic Protocol 1); flow cytometry (see Chapter 5)
Harvest thymocytes from 6-week-old C57BL/6 mice as in Basic Protocol 1. Resuspend in MACS staining buffer at a concentration of 2 × 108 cells/ml.
Incubate thymocytes with anti-CD53 Ab (75 µl/ml) 15 min on ice. Add 20 ml MACS staining buffer and centrifuge 5 min at 500 × g, 4°C. Aspirate off all of the MACS staining buffer.
Resuspend in MACS staining buffer at a concentration of 2 × 108 cells/ml. Incubate thymocytes with biotinylated anti-rat IgG (75 µl/ml) for 15 min on ice. Add 20 ml MACS staining buffer and centrifuge 5 min at 500 × g, 4°C. Aspirate off all of the MACS staining buffer.
Resuspend in MACS staining buffer at a concentration of 2 × 108 cells/ml. Add 150 µl anti-biotin microbeads and incubate 15 min on ice with periodic agitation. Add 20 ml MACS staining buffer and centrifuge 5 min at 500 × g, 4°C. Aspirate off all of the MACS staining buffer.
Resuspend in 2 ml MACS staining buffer and load up to 5 × 108 thymocytes on a Miltenyi MACS LS column prepared according to manufacturer’s instructions. Elute column with 3 ml MACS staining buffer.
- Check thymocyte purity by flow cytometry (see Chapter 5). If purity is sub-optimal, empirically determine the optimal amounts of anti-CD53 Ab and biotinylated anti-rat IgG.Complete aspiration of MACS washing buffer between antibody incubations is critical for optimal purity.
BASIC PROTOCOL 2
[3H] INOSITOL PHOSPHATE RESOLUTION BY HPLC WITH IN-LINE β-DETECTOR
This protocol is optimized to resolve Ins(1,4,5)P3, Ins(1,3,4)P3, and Ins(1,3,4,5)P4 from biochemical or cellular extracts using anion exchange HPLC with an in-line β-detector (Fig. 11.1.4). This method requires ~25 min to resolve a single sample and an additional 25 min to re-equilibrate the column for the next sample.
Materials
HPLC-grade (NH4)H2PO4
Phosphoric acid
NaN3
[3H]-Labeled inositol phosphate standard solutions, e.g.,:
d-myo-inositol-1,4,5-P3, [inositol-1-3H(N)] (Perkin Elmer, cat. no. NET-911001UC)
d-myo-inositol-1,3,4,5-P4, [inositol-1-3H(N)] (Perkin Elmer, cat. no. NET-941002UC)
Scintillation fluid compatible with high-salt solutions (Uniscint National Diagnostics, cat. no. LS-276, or equivalent)
HPLC system compatible with aqueous solutions
Inline β-detector (β-RAM-RHPLC detector from IN/US with a 500-µl flow cell or equivalent)
Partisphere strong anion exchange (SAX) column (12.5 cm × 4.6–mm; Whatman, cat. no. 4621-0505 or equivalent) or a 25-cm column (Whatman, cat. no. 4621-1507)
Prepare 1 liter of each 10 mM and 2 M (NH4)H2PO4 (buffers A and B, respectively). Adjust pH to ~3.35 with phosphoric acid. Filter solutions through a 0.45-µm filter and supplement with 0.005% NaN3.
Adjust the flowrate on the HPLC to 1 ml/min and equilibrate column for 30 min with buffer A.
- Inject sample containing [3H]-labeled IPs, activate gradient andβ-detector, and record elution profile (the ratio of scintillant to sample should be 3:1).
- Gradient:
- 0 to 12.5 min, 0% to 100% buffer B
- 12.5 to 25 min, 100% buffer B
- 25 to 30 min, 0% to 100% buffer A (β-detector can be turned off at this point)
- 30 to 50 min, 100% buffer A.
Protocols for preparation of [3H]-labeled inositol phosphate standards are provided in Berrie et al. (2007) and Otto et al. (2007). - Calculate the area under the curve (AUC) of each peak following HPLC software instructions.In cases of T cell stimulation, total IP5 can be used as a loading control as its amount is unaffected by short-term TCR stimulation.
-
The following can be done to enhance performance:
Increase throughput-
aAdd an extra HPLC column to run in parallel while the first column is being re-equilibrated to double the throughput.
-
bShorten the gradient time to increase throughput—this will result in some loss of resolution.
-
cAdd an auto sampler to automate running and greatly increase throughput.Neutralized extracts are relatively stable for >24 hr at room temperature, allowing long queues (see Basic Protocol 1, step 16 annotation).
Increase resolution-
dUse a 25-cm column.
-
eDecrease HPLC flow rate.Increasing sensitivity can most easily be achieved by using a larger capacity flow cell on the β-RAM; however, this will decrease resolution. Detailed explanations of the setup of ion-exchange liquid chromatography and HPLC—including guides to column matrices and buffers—and critical discussions of PI/IP extraction protocols can be found elsewhere (Dean and Beaven, 1989; Guse et al., 1995b; Jenkinson, 1995; Singh and Jiang, 1995; Williams and Frasca, 1998; Kuksis, 2003; Azevedo and Saiardi, 2006; Berrie et al., 2007). [3H]-Labeled IP standards can be obtained commercially or produced enzymatically (Berrie et al., 2007; Otto et al., 2007).
-
a
ALTERNATE PROTOCOL 2
[3H]-INOSITOL PHOSPHATE RESOLUTION BY HPLC WITHOUT IN-LINE β-DETECTOR
The steps in this protocol are identical to those detailed in Basic Protocol 2 except that the in-line β-detector is replaced with a fraction collector. The volume of the fractions collected is directly related to resolution and sensitivity. Scintillation fluid compatible with high-salt solutions (e.g., Uniscint, National Diagnostics, cat. no. LS-276) is added to the fractions to yield a 3:1 ratio of scintillant/sample. Samples are then read on an appropriate liquid scintillation counter.
BASIC PROTOCOL 3
SOLUBLE INOSITOL PHOSPHATE RESOLUTION BY HPLC WITH METAL DYE DETECTION (MDD)
This protocol describes a non-radioactive high-performance anion-exchange chromatographic method for the separation and quantification of picomole amounts of inositol bis- to poly-phosphates with gradient elution and visible absorbance detection after post-column derivatization (Mayr, 1988). Isomer-selective detection and quantification is possible with minimally ~20 to 50 pmol of individual IP isomers. Detection sensitivity increases with the number of phosphates per inositol ring. This technique uses metal-dye detection (MDD; Table 11.1.1), a complexometric dye- and transition-metal-based detection system for polyanions. It is based on the ability of yttrium (Y), a trivalent transition metal ion, to bind to both cation-specific dyes such as PAR and to polyanions such as IPs with high affinity. Mixing of PAR and yttrium ions with HPLC-resolved IPs in the reaction/detection chamber changes the absorbance of the yttrium-PAR complex at 546 nm, allowing quantitative IP measurements after calibration with purified external standards. MDD permits the direct, highly isomer-selective IP determination in the picomolar range from milligram amounts of cells or tissue specimens that are not readily amenable to analysis by radioisotopic techniques (Lorke et al., 2004).
Table 11.1.1.
Current Methods for Measuring Phosphoinositide (PI) Lipids or Soluble Inositol Phosphates (IP) in Cells or Extracts
| Assay | Advantages | Disadvantages | Used for HTS | References |
|---|---|---|---|---|
| Metabolic radiolabeling followed by anion exchange chromatographic, TLC or HPLC analysis of intact IPs or deacylated PIs | Quantitative. HPLC separation can allow analysis of all PI and IP isomers and is sensitive (picomolar range). High-throughput anion-exchange chromatography on 96-well plates available. | Requires radioactivity and time-consuming metabolic labeling. May not appropriately distinguish between PI/IP mass and phosphorylation. TLC has limited resolution. Anion exchange chromatography and TLC do not sufficiently resolve PI/IP isomers. HPLC is logistically challenging and time consuming, requires expensive equipment. | Yes | Irvine (1986, 1990); Irvine et al. (1986); Imboden and Pattison (1987); Takazawa et al. (1990); Wreggett et al. (1990); Jenkinson (1995); Singh and Jiang (1995); Chengalvala et al. (1999); Kuksis (2003); Pouillon et al. (2003); Benjamin et al. (2004); Wen et al. (2004); Rusten and Stenmark (2006); Skippen et al. (2006); Stevenson-Paulik et al. (2006); Berrie et al. (2007); Guillou et al. (2007b); Otto et al. (2007); Sergeant and McPhail (2007) |
| MDD-HPLC | Quantitative. Allows analysis of many PI and IP isomers. Very sensitive (<50 pmol). | HPLC is logistically challenging and time consuming, and requires expensive equipment | Guse et al. (1995a,b); Singh and Jiang (1995); Adelt et al. (1999); Casals et al. (2002); Kuksis (2003); Lorke et al. (2004); Lin et al. (2009) | |
| HPLC with suppressed conductivity detection | Quantitative. Non-radioactive. Can separate most isomers. | Less sensitive than radioactivity based detection and MS. HPLC is logistically challenging and time consuming, requires expensive equipment. | Nasuhoglu et al. (2002); Rusten and Stenmark (2006) | |
| Liquid chromatography - mass spectrometry (LC-MS) or HPLC-MS | Non-radioactive. No post-column derivatization required. Sensitive (100 fmol to 100 pmol scale). Can analyze multiple PIs in a mixture, including some regioisomers. Can yield information about the PI fatty acid components. | Requires sophisticated, expensive equipment and specific expertise. Requires specialized MS equipment such as a fast ion trap mass spectrometer for isomer identification. Unclear if suitable for higher order PIs/IPs. | Singh and Jiang (1995); Pettitt et al. (2006); Vats et al. (2008) | |
| Electrospray-ionization tandem mass spectrometry (ESI-MS-MS) | Sensitive and quantitative. Non-radioactive. | Does not distinguish between isomers. May require prior PI/IP isomer separation by one of the other techniques. Requires expensive equipment. Sophisticated. | Wenk et al. (2003); Rusten and Stenmark (2006); Berrie et al. (2007) | |
| High-performance thin-layer chromatography (HPTLC) with molybdate staining | Simple, rapid, sensitive (100 to 200 pmol), non-radioactive, can monitor several PIs and IPs | Low resolution, requires HPLC confirmation | Hatzack and Rasmussen (1999); Kuksis (2003) | |
| Radioreceptor/competitive radioligand displacement assays, radio-immunoassays (RIA); scintillation proximity (SPA) assays | Homogeneous, sensitive (high femtomolar to low picomolar range), simple, fast. Commercial kits available. More accurate IP mass measurements than via metabolic labeling. Can quantify specific IPs/PIs in tissue or cell extracts with little purification. Allow kinetic analyses of PI/IP levels in cell extracts or in vitro, measuring PI/IP-kinase or -phosphatase activities | Radioactive. Limited to single analytes such as PIP3, IP3, or IP4. In extracts, may read out combined activities of PIPs and corresponding IPs. | Yes | Amersham, Perkin Elmer; Challiss et al. (1988); Donie and Reiser (1989); Park et al. (1997); van der Kaay et al. (1997, 1998); Chang et al. (2002); Brandish et al. (2003); Kuksis (2003); Liu et al. (2003); Wen et al. (2004); Zheng et al. (2004); Sergeant and McPhail (2007); Mueller et al. (2008) |
| Luminescent oxygen channeling (LOCI), amplified luminescent proximity homogeneous assay (AlphaScreen), fluorescence polarization (FP) or fluorescence resonance energy transfer (FRET) assays | Homogeneous, non-radioactive, sensitive. Quantitative. Commercial kits available. Can quantify specific IPs/PIs in tissue or cell extracts with little purification. Allow kinetic analyses of PI/IP levels in cell extracts or in vitro, measuring PI/IP-kinase or -phosphatase activities. | Limited to single analytes such as PIP3, IP3, or IP4. In extracts, may read out combined activities of PIPs and corresponding IPs. LOCI/AlphaScreen sensitive to reactive oxygen scavengers. | Yes | Discoverex, Amersham, Echelon, Perkin Elmer; Ullman et al. (1996); Drees et al. (2003); Gray et al. (2003); Sato et al. (2003); Cicchetti et al. (2004); Prestwich (2004, 2005); Boldyreff et al. (2008) |
| Enzyme-linked immunosorbent assays (ELISA) | Quantitative. Commercial kits available. | Limited to single analytes such as PIP3, IP3, or IP4. In extracts, may read out combined activities of PIPs and corresponding IPs. | (Yes) | Rusten and Stenmark (2006) |
| PI extraction followed by spotting on nitrocellulose filters and antibody or PI-binding protein lipid overlay detection | Non-radioactive. Simple. | Low sensitivity, limited to few analytes. Nonquantitative. | Guillou et al. (2007b); Johnson et al. (2008); Rusten and Stenmark (2006) | |
| Ectopic expression of fluorophor-tagged PI or IP binding protein domain fusion proteins in cells followed by immunofluorescent, FRET or electron microscopic analysis. Examples: PLCδ1 PH domain-EGFP fusion protein for PI(4,5)P2 detection, GRP1, Btk or ARNO PH domain-EGFP fusion proteins for PI(3,4,5)P3, PKCε C1 domain-EGFP fusion protein for diacylglycerol. | Allows in situ imaging of PI/IP levels in live or fixed cells. In life cells, kinetic analyses are possible. Probes for multiple different PIs and IPs available. Domain point mutants can provide specificity controls. | Semiquantitative. Requires transient cell transfection or stably transfected cell line. Probe overexpression may interfere with normal cell functions. FRET requires rigorous controls. Probes may bind to several different PIs and IPs. Limited to specific PI isomers. The probe for a given PI may also bind to and be modulated by its soluble IP analog, I(1,3,4,5)P4 in the case of PI(3,4,5)P3, for example (Huang et al., 2007). Thus, the assay reads out the combined action of both rather than absolute levels of one specific analyte. Physiologically, however, this is likely more relevant than individual analyte levels. Thus, this “integrated detection” can be advantageous in biological systems. | Yes, requires high-throughput/content imaging technology. | Oatey et al. (1999); Zeidman et al. (1999); Holz et al. (2000); Balla and Varnai (2002); Parmryd et al. (2003); Sato et al. (2003); Cicchetti et al. (2004); Rusten and Stenmark (2006); Guillou et al. (2007a); Sakaguchi et al. (2009) |
| Direct fixed cell labeling with recombinant PI/IP binding protein domains or antibodies | Does not require transfection. Does not interfere with cell function. Non-radioactive. | Limited to fixed cells, where PI and in particular IP preservation is difficult. May not detect protein-bound IPs/PIs. | Rusten and Stenmark (2006) | |
| Enzymatic detection | Sensitive, non-radioactive, quantitative | Requires prior IP separation by HPLC or other techniques. Indirect multi-step process, requiring IP de-phosphorylation to myo-inositol which is then oxidized, followed by enzymatic measurement of oxidation products, or by measuring of the released inorganic phosphate. Salt interferes with IP dephosphorylation. Dephosphorylation rates differ for different IPs. IP5/IP6 are very poorly dephosphorylated. Thus, enzymological detection is not ideal for analyses of multiple different IPs/PIs and may be limited by elution reagent or buffer conditions. | Singh and Jiang (1995); Kuksis (2003) |
Compared to isotopic labeling, MDD HPLC has the following advantages: it is nonradioactive, facilitating handling, does not require radiolabeling of the cells with its potential complications regarding cell viability and labeling efficiency, can be used to analyze tissue specimens or primary cells that are difficult or impossible to radiolabel, and warrants high isomer selectivity and sensitivity. With acidic elution, MDD anion-exchange separation could resolve 26 inositol tris- to hexakis-phosphates (excluding enantiomers, 12 IP3, 9 IP4, and 4 IP5 isomers as well as IP6) into 14 peaks within 25 min. Furthermore, an in-house reference standard solution was produced by hydrolysis of phytic acid under autoclaving conditions to allow analyses for which standards are unavailable or very expensive.
Materials
Thymocytes (see Basic Protocol 1) or T cell lines (see Alternate Protocol 1)
PBS (APPENDIX 2A)
Lysis buffer (see recipe), ice cold
Water-saturated diethyl ether: prepared by vigorously mixing 1 vol deionized water with 2 vol diethyl ether for at least 2 min
1 M triethanolamine (TEA, p.a., >99% purity; see recipe)
Charcoal suspension (see recipe)
0.1 M NaCl (APPENDIX 2A)
Methanol (LiChrosolv)
10% (w/v) trichloroacetic acid (TCA) solution, 4°C
0.2 M EDTA (APPENDIX 2A)
0.1 M NaF (see recipe)
MDD-HPLC eluent A (see recipe)
MDD-HPLC eluent B (see recipe)
Post-column reagent C (see recipe)
HPLC-injection solution (see recipe)
Degassed, filtered HPLC-grade water
30% analytical-grade HCl (suprapure)
Phytic acid
10 mM 4-(2-pyridylazo)-resorcinol monosodium salt monohydrate (PAR; see recipe)
18 mM yttrium trichloride (YCl3; see recipe)
0.5 M sodium acetate (see recipe)
Cell scraper
12- and 14-ml polypropylene tubes with caps (Greiner Bio-One, cat. no. 187262)
35°C water bath
2-ml microcentrifuge tubes
SpeedVac
Ultra-Turrax homogenizer
0.22-µm pore size membrane filters (Millipore, type GV) in a Pyrex glass filtration device
Vacuum pump
2-ml Pyrex glass vials
HPLC auto-sampler with a 1-ml injection loop and a 2.5-ml loading syringe (inert valve made from titanium or PEEK; HPLC auto-sampler 560, Kontron; loading syringe available from Hamilton)
MiniQ PC 3.2/3 column (3-µm bead diameter, GE Healthcare/Pharmacia Biotech, Uppsala)
Hand-made knitted coil from a 40-cm 1/16-in. × 0.5-mm i.d. PTFE capillary with 7 knots (CS-Chromatographie Service)
Two inert HPLC pumps for gradient elution (with titanium or PEEK pump-head; Pump 422, Kontron)
Pump for post-column dye reagent addition (Shimadzu, cat. no. LC-10AD)
UV/Vis recorder for absorbance recording (with titanium or PEEK flow cell; Shimadzu, cat. no. SPD-10Avvp)
Chromatography data system for control and data processing (e.g., Galaxie chromatography data system, Varian)
Graphing/interpolation software (e.g., GraphPad Prism)
Prepare IP samples
Prepare IP samples from cell lysates
-
1a
Prepare, incubate with compounds (if desired), and stimulate thymocytes as described in Basic Protocol 1, or T cell lines as described in Alternate Protocol 1, but omit the myo-[3H] inositol labeling steps.
-
2aWash cells three times with ice-cold PBS centrifuging each time for 5 min at 500 × g, 4°C.For fast stimulation times, this step may be omitted if media supernatants are quantitatively removed from the cells.
-
3a
Add 1 ml ice-cold lysis buffer, scrape off the cells with a cell scraper, pipet the lysates into 14-ml polypropylene tubes, and freeze/thaw two times in liquid nitrogen. Keep on ice after thawing.
-
4aWithin <20 min, centrifuge 10 min at 4000 × g, 4°C. Collect supernatants into new 14-ml polypropylene tubes for soluble inositol phosphate analysis as described below. If desired, save the pellets for inositol phospholipid analysis (see Alternate Protocol 3).After suspension in 10 vol methanol to reduce acidity, the pellets can be flash-frozen in liquid nitrogen or on dry ice and saved for up to 2 weeks at − 70°C.
-
5a
Incubate the supernatants 20 min at 35°C to destroy creatine phosphate.
-
6a
Cool the supernatants on ice and extract three times, each time with 3 ml of ice-cold water-saturated diethyl ether, to remove the TCA. Always leave a narrow layer of ether above the lower aqueous phase which contains the IPs.
-
7aAdjust pH of the aqueous phase to ~6 with ~10 µl of 1 M triethanolamine base and at least partially lyophilize in a Speed Vac to reduce sample volume to <500 µl.This step is essential to remove ether traces, which would disturb HPLC detection by forming gas bubbles.
-
8a
Transfer sample into a 2-ml microcentrifuge tube and add water to a final volume of 500 µl. To remove nucleotides, add 25 µl freshly resuspended 20% (w/v) charcoal suspension per 25 mg wet weight of tissue or cells and thoroughly vortex five times over a 15-min period. Centrifuge 3 min at 15,000 × g, room temperature, and treat the supernatants an additional time with the same amount of charcoal. Re-extract potentially carried over IPs from the two charcoal pellets with 1 ml of 0.1 M NaCl. Combine the re-extracts with the samples for micro-MDD-HPLC. Proceed to step 9.
Prepare IP samples from tissues
-
1b
Weigh the tissue and record the wet weight (at least 50 mg per sample is needed).
-
2b
Add 1 ml of 10% (w/v) TCA, 12 µl of 0.2 M EDTA (2.4 µmol), and 10 µl of 0.1 M NaF (1 µmol) per sample.
-
3b
Homogenize on ice in 12-ml polypropylene-tubes using an Ultra-Turrax homogenizer with a 7-mm diameter rotating at 9500 rpm, conduct 4 strokes of 10 sec each.
-
4bIncubate homogenates 20 min on ice. Centrifuge 5 min at 3500 × g, 4°C and collect supernatant. If desired, save the pellets for inositol phospholipid analysis (see Alternate Protocol 3).After suspension in 10 vol methanol to reduce acidity, the pellets can be flash-frozen in liquid nitrogen or on dry ice and saved for at least 2 weeks at − 70°C.
-
5b
Perform steps 5a through 8a. Proceed to step 9.
Separate inositol phosphates by Micro-MDD HPLC
-
9
Prepare MDD-HPLC eluent A, MDD-HPLC eluent B, and post-column reagent C. Prior to use, filter and degas these reagents by vacuum filtration through inert 0.22-µm pore size membrane filters in a Pyrex glass filtration device.
-
10Prior to sample analysis, determine the elution profiles of relevant standards. For loading, first adjust all samples to precisely a 1.3-ml volume by diluting with HPLC-injection solution in 2-ml Pyrex glass vials, filter through 0.2-µm membranes, and then inject using an auto-sampler with a 1-ml injection volume.An IP isomer control standard mix can be prepared by acidic partial hydrolysis of inositol hexakisphosphate (phytic acid, IP6) and purified either on a Q-Sepharose or a MonoQ column (Phillippy et al., 1987). Briefly, dissolve 20 g IP6 (dodecasodium salt hydrate) in 200 ml water and adjust the pH to 4.0 with HCl. Autoclave for 60 min at 121°C. Cool down and dilute to 500 ml with water. After neutralization with NaOH, this solution can be stored for at least 10 years at − 20°C.
-
11With a flow rate of 0.5 ml/min throughout the run, wash the column with degassed, filtered HPLC-grade water for 10 min prior to starting automated gradient analysis. Change pump A to eluant A, pump B to eluant B, and pump C to dye-reagent C, and equilibrate the column with eluant A for another 10 min. Pump C is providing the dye-reagent at 0.25 ml/min, mixed with the eluant gradient solution through a mixing T and a knitted coil of 125 µl internal volume. Use the following gradient from 0.2 mM to 0.5MHCl over 25 min to elute inositol phosphates: 0 to 1.9 min, 3% MDD-HPLC eluent B in eluent A; 1.9 to 2.9 min, 3% to 5% B; 2.9 to 3.6 min, 5% to 7% B; 3.6 to 4.1 min, 7% to 9% B; 4.1 to 7.4 min, 9% to 10% B; 7.4 to 7.7 min, 10% to 11% B; 7.7 to 7.9 min, 11% to 13% B; 7.9 to 8.2 min, 13% to 15% B; 8.2 to 8.6 min, 15% to 17% B; 8.6 to 9.2 min, 17% to 18% B; 9.2 to 10.2 min, 18% to 19% B; 10.2 to 11.4 min, 19% to 24% B; 11.4 to 11.9 min, 24% to 28% B; 11.9 to 12.7 min, 28% to 35% B; 12.7 to 14.5 min, 35% to 45% B; 14.5 to 15.9 min, 45% to 60% B; 15.9 to 17 min, 60% to 97% B; 17 to 21.5 min, 97% B; 21.5 to 21.6 min, 97% to 3% B; and 21.6 to 25 min, 3% B. YCl3 does not form complexes with phospho-compounds unless the pH is brought to >4.5. To achieve this, mix the column eluate in-line with 1/2 volume of post-column reagent C and record the degree of post-column complex formation by measuring absorbance at 546 nm.The following is a description of the basic setup of an automatized HPLC system with post-column metal dye detection. The MDD-HPLC system consists of two pumps for gradient elution, an additional pump for post-column dye reagent, an HPLC auto-sampler 560 (Kontron) with a 1-ml volume injection loop and a 2.5-ml loading syringe, a UV/V is recorder for absorbance recording, and a Galaxie chromatography data system for control and data processing. Data are stored in a polarity-switched format (multiplied by − 1) and exported into Excel for processing.For delivery of both solvent and post-column reagent, true double-piston pumps with (identical) stroke volumes <80µl are essential. All eluent-wetted parts in this or analogous systems have to be inert (i.e., from titanium, sapphire, PEEK, and Teflon). In addition, all fittings, filter units, column inlets and column frits are made from inert polymer such as PEEK or Teflon. HPLC is always conducted at a stable temperature below 25°C to avoid precipitation of the PAR solution.A representative HPLC analysis of hydrolyzed products of phytic acid (IP6) is shown in Figure 11.1.5, where 15 inositol phosphate isomers were separated and detected.
-
12After use, wash the column for 30 min with water prior to regeneration and storage.Column performance deteriorates after ~50 runs. However, the column can be cleaned and performance fully restored by sequential washing in reverse flow direction at a rate of 0.1 ml/min with 1 ml each of 1 M NaCl, 1 M NaOH, 1 M HCl, and again 1 M NaCl, and by rinsing with 0.5 ml water between each step. Normally, five loops are used to restore column performance. For details, see manufacturer’s instructions.
Figure 11.1.5.
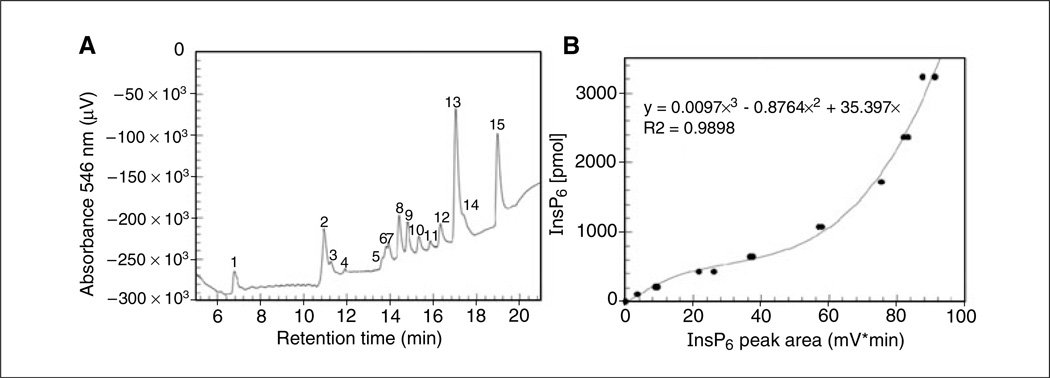
MDD-HPLC analysis of phytic acid hydrolysis products. (A) Elution profile. Peak identities were determined by comparison with the retention times for external standards (not shown). Peak 1 (retention time of 6.77 min) contains IP2 isomers, peak 2 (10.9 min) contains I(1,3,4)P3 and I(1,4,5)P3, peak 3 (11.23 min) contains D/L-I(1,5,6)P3, peak 4 (11.89 min) contains I(4,5,6)P3, peak 5 (13.63 min) contains I(1,2,3,5)P4 and I(1,2,4,6)P4, peak 6 (13.79 min) contains I(1,2,3,4)P4 and I(1,3,4,6)P4, peak 7 (13.91 min) contains I(1,2,4,5)P4 and I(1,3,4,5)P4, peak 8 (14.41 min) contains I(1,2,5,6)P4, peak 9 (14.78 min) contains I(2,4,5,6)P4, peak 10 (15.31 min) contains I(1/3,4,5,6)P4, peak 11 (15.83 min) contains D/L I(1,2,3,4,6)P5, peak 12 (16.31 min) contains D/L I(1,2,3,4,3)P5, peak 13 (17.04 min) contains D/L I(1,2,4,5,6)P5, peak 14 (17.32 min) contains I(1,3,4,5,6)P5, and peak 15 (18.97 min) contains I(1,2,3,4,5,6)P6 (unhydrolyzed phytic acid). (B) Calibration curve obtained with known amounts of an IP6 external standard.
Quantify inositol phosphates
-
13Quantify inositol phosphate isomers by comparing with external standards. Using a dilution series of a known amount of each IP standard, record a calibration curve that plots standard amount over area under the respective absorbance peak. Unknown sample amounts for an IP isomer of interest can then be calculated from the observed absorption peak areas using the corresponding standard curve and graphing/ interpolation software such as Graph-Pad Prism.As an example, Figure 11.1.5B shows a calibration curve of IP6. Representative Jurkat cell data are shown in Figure 11.1.6.
ALTERNATE PROTOCOL 3
INOSITOL PHOSPHOLIPID SEPARATION AND QUANTIFICATION BY HPLC WITH METAL DYE DETECTION (MDD)
Organic lipid extraction followed by phosphoinositide deacylation and fatty acid removal allows isomer-specific MDD-HPLC analysis of the resulting glycerophospho-inositol phosphates with the same advantages and sensitivity as described for IP MDD-HPLC analysis in Basic Protocol 3 and Table 11.1.1 (Mayr, 1988; Weernink et al., 2000; Horn et al., 2004). In particular, MDD-HPLC analysis of deacylated phosphoinositides allows detection and quantitative analysis of PIP, PIP2, and PIP3 isomers without radioactive pre-labeling. It makes tissue samples of 100 mg wet weight or more amenable to isomer selective PIPx determinations. All glycerophosphoinositol phosphates with a phosphate content of two to four are well separated by the MDD-HPLC system. The only phosphoinositide that cannot be sufficiently determined by MDD-HPLC is PI. PI is de-acylated into glycerophosphoinositol, whose phosphodiester group is detected by MDD with only very low sensitivity, <1% of that of free phosphomonoesters. Its selective detection is also subject to interference by glycerophosphate formed through deacylation of phosphatidic acid (PA). MDD detection sensitivity for glycerophosphate is much higher than that for glycerophosphoinositol.
Additional Materials (also see Basic Protocol 3)
PIP2, PIP3 standards
Methanol (LiChrosolv)
Chloroform (CHCl3; LiChrosolv)
0.1 M HCl
3:48:47 (v/v/v) chloroform/methanol/0.6 M HCl
n-Butanol
33% methylamine in ethanol
n-Propanol
20:4:1 (v/v/v) butanol/petroleum-ether/ethyl formate
53°C water bath or heating block
Prepare phospholipids from cell pellets
-
1Grow, treat, lyse, and centrifuge cells as described in Basic Protocol 3, steps 1a to 4a. Collect the precipitates that contain cellular membranes, lipids, and other insoluble components.If frozen pellets are used, briefly thaw in a 37°C water bath or heating block.
-
2To extract lipids, add 1.5 ml methanol if the cell number is <5 × 107, or 2.5 ml if the cell number is higher.Use 14-ml polypropylene centrifuge tubes with tight sitting caps.
-
3
Vortex thoroughly, then add 2 ml CHCl3 and 1 ml of 0.1 M HCl, mix by vigorously shaking by hand for 30 sec while strongly pressing the cap onto the tube. Centrifuge 5 min at 4000 × g, 4°C. Discard the upper, acidic water/methanol layer by careful aspiration.
-
4
Add 3 ml chloroform mixture (3:48:47 CHCl3/methanol/0.6 M HCl). Vortex, centrifuge as in step 3, and discard the upper layer and the interphase.
-
5Repeat step 4 but with only 1.5 ml chloroform mixture.Steps 2 to 5 serve to purify cellular lipids in a Folch extract (Folch, 1949).
-
6Evaporate the solvents 1 to 2 hr in a Speedvac.The lipids will precipitate as a thin film on the vial wall.
CAUTION: Chloroform vapor is harmful. To avoid human exposure, equip the Speedvac with an appropriate charcoal filter or conduct steps 2 to 6 in an externally vented fume hood.
-
7
To deacylate the lipids, add 200 µl n-butanol, vortex, then add 600 µl of 33% methylamine reagent solution and incubate 45 min at 53°C.
-
8
Cool to room temperature, add 300 µl n-propanol, vortex, and evaporate solvents as in step 6.
-
9
Dissolve dried pellet in 500 µl water, extract with 600 µl of 20:4:1 (v/v/v) butanol/petroleum ether/ethyl formate, vortex, and centrifuge 5 min at 4000 × g, 4°C. Discard the upper, organic layer.
-
10To the lower layer, add 600 µl water and 1.2 ml of 20:4:1 butanol/petroleum ether/ethyl formate, vortex, centrifuge as in step 9, discard the upper layer, and vacuum-dry the lower layer for ~4 hr as in step 6.The extractions in steps 9 and 10 serve to remove fatty acids, which dissolve in the upper, organic phase. The lower, aqueous phase contains the deacylated phospholipids as glycerophospho-inositol phosphates.
-
11
Dissolve the pellet in 500 µl water.
Micro-MDD HPLC analysis
-
12
Determine the elution profiles of relevant standards prior to analyzing the samples (see Basic Protocol 3, step 10).
-
13
Use the following gradient from 0.2 mM to 0.25 M HCl over 21 min to separate and elute the glycerophospho-inositol phosphates, following the general HPLC procedure described in Basic Protocol 3, step 11: 0 to 0.1 min, 0% to 3% MDD-HPLC eluent B in eluent A; 0.1 to 2 min, 3% to 4% B; 2 to 3 min, 4% to 5% B; 3 to 3.8 min, 5% B; 3.8 to 5 min, 5% to 7% B; 5 to 5.8 min, 7% to 8% B; 5.8 to 6.4 min, 8% to 11% B; 6.4 to 6.7 min, 11% to 15% B; 6.7 to 7 min, 15% to 18% B; 7 to 10.6 min, 18% to 22% B; 10.6 to 11.3 min, 22% to 37% B; 11.3 to 11.6 min, 37% to 44% B; 11.6 to 15.4 min, 44% to 47% B; 15.4 to 15.6 min, 47% to 49% B; 15.6 to 15.8 min, 49% to 51% B; 15.8 to 16 min, 51% to 100% B; 16 to 18.5 min, 100% B; 18.5 to 18.7 min, 100% to 0% B; and 18.7 to 21 min, 0% B.
BASIC PROTOCOL 4
[3H] INOSITOL PHOSPHATE RESOLUTION BY DOWEX ANION-EXCHANGE CHROMATOGRAPHY
For investigators without HPLC access, this protocol describes use of a Dowex anion exchange protocol. This protocol is best suited for small-scale experiments if limited sensitivity and IP isomer resolution are of no concern. In brief, extracts from myo-[3H] inositol–labeled cells (see Basic Protocol 1) are loaded onto Dowex columns. After extensive washing to remove free [3H] inositol, [3H] IP1, [3H] IP2, and [3H] IP3 are sequentially eluted by stepwise additions of increasing concentrations of ammonium formate in 0.1 M formic acid. [3H] inositol phosphates are quantified by liquid scintillation spectroscopy (LSC). Resolution of IP3 and IP4 isomers usually requires HPLC, described in Basic Protocol 1 and Alternate Protocol 1. Higher ammonium formate concentrations and gradient elution can allow IP3, IP4, IP5, and IP6 isomer detection. For detailed discussions of PI/IP extraction methods and of anion-exchange chromatography, including different column matrices, elution buffer systems, and suitable standards, see Basic Protocols 1 and 2 and the references listed therein.
Materials
Cells
Medium supplemented with 2 to 20 µCi/ml myo-[3H] inositol
Balanced salt solution (see recipe)
Stimulus
10% (w/v) trichloroacetic acid (TCA), ice cold
Diethyl ether, water-saturated
1:100 (v/v) dilution of concentrated ammonia
0.5 g/ml Dowex 1-X8 resin (100 to 200 mesh; formate form; Bio-Rad, Sigma-Aldrich, GFS Chemicals, Serva Electrophoresis; formate form may require custom production), slurry in water
60 mM sodium formate/5 mM disodium tetraborate
0.2 M ammonium formate/0.1 M formic acid
0.4 M ammonium formate/0.1 M formic acid
0.8 M ammonium formate/0.1 M formic acid
Scintillation fluor cocktail (compatible with aqueous samples)
13 × 100–mm glass tubes
Pasteur pipets
0.6-cm diameter disposable columns
Liquid scintillation counter
Label 2 × 106 to 2 × 107 cells/sample in medium supplemented with 2 to 20 µCi/ml myo-[3H] inositol for 72 hr as described in Basic Protocol 1 for primary cells, and in Alternate Protocol 1 for transformed tissue culture cells.
- Wash the cells three times by centrifuging in a tabletop centrifuge 5 min at 500 × g, room temperature, discarding the supernatant and resuspending in balanced salt solution.Refer to Basic Protocol 1 and Alternate Protocol 1 and to the Commentary for discussions of the pitfalls in labeling cells with [3H] inositol, and for alternative labeling media.
- Stimulate the cells in a total volume of 200 µl containing ~2 × 106 to 5 × 107 cells as described in Basic Protocol 1 and Alternate Protocol 1, respectively.The number of cells required per sample depends upon the specific activity achieved during labeling, as well as on the magnitude of the PI/IP response. See Basic Protocol 1 and Alternate Protocol 1 for discussions on how to optimize cell number and stimulation, and of potential LiCl addition to enhance PI/IP detection. If interactions between two cell types are being studied, pelleting the cells at the initiation of the experiment will promote cell-cell contact and may facilitate detection of a response.
- At the appropriate time points, lyse cells by adding 1ml of ice-cold 10% TCA. Vortex and incubate 20 min on ice.Longer incubation times in acid, especially at higher temperatures, promote PIP head-group hydrolysis, IP isomerization, or IP hydrolysis and should thus be avoided (Singh and Jiang, 1995). Lymphocytes contain relatively high PIP and PIP2 amounts. Their acid hydrolysis can in particular lead to superphysiological IP2 and IP3 levels.Addition of each 20 µg of unlabeled IP3 and IP4 may enhance extraction of these IPs, in particular of IP4 (Wreggett et al., 1987). If labeling efficiency is low, replicate samples can be pooled prior to TCA extraction to increase extracted radioactivity. Typically, time points of 0, 0.5, 1, 3, 5, and 15 min allow one to monitor most TCR modulated PIs/IPs (Imboden and Pattison, 1987; Budd et al., 1990; Inokuchi and Imboden, 1990; Guse and Emmrich, 1991, 1992; Guse et al., 1993; Pouillon et al., 2003).
Microcentrifuge 10 min at 12,000 × g, 4°C.
Transfer supernatant to a 13 × 100–mm glass tube. In a fume hood, add 1 ml water-saturated diethyl ether to extract acid and save the aqueous lower layer using a Pasteur pipet. Repeat extraction four to five times.
- Neutralize samples with a 1:100 dilution of concentrated ammonia, then add 4 ml water.The amount of dilute ammonia added will vary, depending on the specific sample. Measure neutralization using pH paper. At this point, samples are stable for at least several days at 4°C. For long-term storage, freezing at − 70°C is recommended.
- Prepare column by pipetting 1.2 ml of 0.5 g/ml Dowex slurry into 0.6-cm diameter column. Apply sample.Before analyzing experimental samples, it is essential to confirm that [3H] IP standards elute in the order [3H] IP1, [3H] IP2, and [3H] IP3. Some adjustments in elution volumes may be necessary. In initial studies of experimental samples, sequential 2-ml fractions should be collected to confirm the location of peaks and the adequacies of the washes (see steps 10 to 12). Thereafter, the radioactivity in 2-ml aliquots of batch-eluted peaks can be determined, and the radioactivity in the peak can then be calculated (this permits substantial savings in terms of scintillation vials and cocktail).If the formate form of the resin is not available, it can be generated from other forms of the resin as described in Berrie et al. (2007).
- Wash column with 30 ml of 60 mM sodium formate/5 mM disodium tetraborate. Collect the final 1 ml of eluate for scintillation counting.This wash elutes free [3H] inositol and [3H] glycerophosphatidylinositol. Because the amount of [3H] inositol radioactivity in the samples will far exceed that of the [3H] IPs, it is important to ascertain that the eluted radioactivity has returned to background levels prior to elution of the [3H] IPs. Typically, a 30-ml wash is sufficient, but different cell systems or numbers may require larger or smaller wash volumes.
Elute [3H] IP1 with 6 ml of 0.2 M ammonium formate/0.1 M formic acid. Wash the column with 2 ml of the same buffer.
Elute [3H] IP2 with 16 ml of 0.4 M ammonium formate/0.1 M formic acid. Wash the column with 6 ml of the same buffer.
Elute [3H] IP3 in 10 ml of 0.8 M ammonium formate/0.1 M formic acid.
Add 16 ml scintillation fluor cocktail to 2 ml of eluate and measure counts per minute for 5 min in a liquid scintillation counter.
BASIC PROTOCOL 5
[3H] INOSITOL PHOSPHOLIPID RESOLUTION BY THIN LAYER CHROMATOGRAPHY
Thin layer chromatography (TLC) provides an inexpensive and simple alternative to HPLC for analysis of inositol phospholipid and IP levels in cell extracts or in in vitro reactions (Table 11.1.1). Compared to HPLC, its sensitivity, isomer resolution, and throughput are lower. Different variants of this technique, including its use for IP separation, are discussed elsewhere (Singh and Jiang, 1995; Hatzack and Rasmussen, 1999; Kuksis, 2003; Berrie et al., 2007; Otto et al., 2007; Sergeant and McPhail, 2007). Lipids are extracted from cells labeled with either [32P] orthophosphate or myo-[3H] inositol, dried under nitrogen, redissolved in chloroform/methanol, and then applied to a silica-gel plate that has been pre-run in potassium oxalate and heat-activated. Cell labeling with myo-[3H] inositol is described in Basic Protocol 1, Alternate Protocol 1, and Basic Protocol 4. Reliable one-dimensional separation of phosphatidylinositol (PI), phosphatidylinositol phosphate (PIP), phosphatidylinositol bisphosphate (PIP2), and phosphatidic acid (PA) from one another and from other phospholipids (e.g., phosphatidylcholine, phosphatidylethanolamine, and phosphatidylserine) is achieved by the TLC method presented here. Labeled lipids are visualized by autoradiography, scraped from the glass plates, and quantified by liquid scintillation spectroscopy. A high-performance TLC variant that allowed detection of 100 to 200 pmol of IP1–IP6 via molybdate staining is described in Hatzack and Rasmussen (1999) and Kuksis (2003). Low resolution of this technique requires HPLC confirmation, in particular, for reliable isomer separation.
Materials
1% potassium oxalate/2 mM EDTA
Solvent: 60:20:23:18:12 (v/v/v/v/v) chloroform/methanol/acetone/acetic acid/H2O
Cells (see Critical Parameters, Basic Protocol 1, and Alternate Protocol 1)
Phosphate-free medium (e.g., GIBCO/BRL) containing 10% heat-inactivated fetal calf serum (FCS), dialyzed against HBSS or TBS (APPENDIX 2A) to remove phosphate
Carrier-free [32P] orthophosphate (296 mBq/ml, 8 mCi/ml)
Stimulus (see Commentary)
50:100:1 (v/v/v) chloroform/methanol/concentrated HCl
100 mM EDTA, pH 7.4 (APPENDIX 2A)
100 mM KCl
Chloroform
Nitrogen source
2:1 (v/v) chloroform/methanol
PI, PIP, PIP2, and PA standards (Sigma)
Iodine
Silica-gel plates (Baker Si250, J.T. Baker)
TLC tanks
100° and 110°C ovens
Filter paper
2-ml microcentrifuge tubes
37°C water bath or heating block
Kodak X-Omat film
EN3HANCE spray surface autoradiography enhancer (Perkin Elmer)
Prepare TLC plates
-
1
Pre-run silica-gel plate overnight in 1% potassium oxalate/2 mM EDTA and allow to air dry. Just prior to use, heat-activate plates by heating for 30 min at 110°C.
-
2Line TLC tank with filter paper and allow it to equilibrate with solvent for ≥6 hr.When the plate is added to the chamber, solvent should cover the bottom 0.5 cm of the plate.An alternative solvent system is 45:35:10 chloroform/methanol/4 N NH4OH (Lapetina and Siess, 1987). This results in excellent separation of PI, PIP, and PIP2 but should only be used with [3H] inositol-labeled samples (PI is not well-separated from PA; the latter can be labeled with 32P but not with [3H] inositol).
Non-equilibrium label cells with [32P] orthophosphate
Steps 3 and 4 are carried out only for non-equilibrium labeling with [32P] orthophosphate. myo-[3H] inositol–labeled cells are obtained from Basic Protocol 1, step 14, or from Basic Protocol 4, step 3. If cell viability is a problem, try incubation on fibronectin-coated plates or addition of FCS/cytokines/growth factors as described in Basic Protocol 1 and Critical Parameters.
-
3
Wash ~2×106 to 5×107 cells in phosphate-free medium/10% FCS, and resuspend in this medium. Divide into 0.2-ml aliquots in 2-ml microcentrifuge tubes and incubate at 37°C.
-
4Add [32P] orthophosphate to 0.1 to 1.0 mCi/ml final. Incubate 3 min at 37°C then add stimulus or diluent control.CAUTION: All experiments employing [32P] should be performed behind appropriate Plexiglas shielding, and investigators should wear a dosimeter ring.In [32P]-labeling experiments, it is important to avoid inadvertent cold chases by adding stimuli in phosphate-containing solutions (e.g., monoclonal antibodies in hybridoma culture medium). It is also important to be precise in timing and to use replicate cell samples (e.g., triplicates). Even in unstimulated cells, there is rapid incorporation of [32P] orthophosphate into 4- and 5-position phosphates of PIP and PIP2 for 20 min, but <40 min. Equilibrium between the ATP pool and the 4- and 5-phosphates does not occur within 30 min.
Extract and resolve inositol phospholipids from labeled cells
-
5At the desired time points post-stimulation, lyse the cells (obtained from step 4 or from Basic Protocol 1, step 14, or from Basic Protocol 4, step 3) by adding 0.8 ml of 50:100:1 chloroform/methanol/HCl.For [3H] inositol-labeled cells, it is important to achieve steady-state labeling of the inositol lipids as described in Basic Protocol 1 and Commentary. For studies of [3H] inositol lipids, it is recommended that cells be stimulated in the labeling medium (i.e., in the presence of [3H] inositol) to avoid any changes in specific activity during the assay.CAUTION: [32P]-labeled samples should be kept in a Plexiglas block or similar shield to reduce radiation exposure.
-
6
Add 0.03 ml of 100 mM EDTA, pH 7.4, 0.1 ml of 100 mM KCl, 0.15 ml chloroform, 0.15 ml water, and vortex for several seconds. Microcentrifuge 2 min at 15,000 × g, 4°C, and remove the lower phase (containing the lipids) and set aside.
-
7Add 0.4 ml chloroform to the remaining upper phase, vortex, and microcentrifuge 2 min at 15,000 × g, 4°C. Remove the lower phase and combine it with the lower phase from step 6. Dry under nitrogen.For drying, very carefully blow a slow stream of nitrogen gas across the surface of the chloroform extract until all liquid has evaporated.CAUTION: This step must be performed in an externally vented fume hood with all appropriate radiation protection, including Plexiglas shielding.The dried samples can be stored capped overnight at 4°C.
-
8Redissolve the lipids in 50 µl of 2:1 chloroform/methanol and promptly spot 20 µl onto the silica-gel plate (from step 1). Run PI, PIP, PIP2, and PA standards (2 to 4 µg each) on each plate. Allow to air dry at room temperature.The lipids should be applied 1 cm from the bottom of the plate (marked in pencil).
-
9
Place plate in the pre-equilibrated TLC chamber (from step 2) and allow the plate to develop to a height of 15 cm (~1 hr).
-
10
Dry plate for 3 min at 100°C.
-
11In a fume hood, pre-equilibrate a TLC tank with iodine vapor for several minutes. Visualize standards by placing the plate for 3 to 5 min in the preequilibrated TLC tank; mark these by pencil.The standards will be stained yellow-orange but will fade. There should also be sufficient PI (but not PIP and PIP2) in the samples to be visualized.
-
12Autoradiograph the plate using Kodak X-Omat or similar film. If [3H] inositol labeling has been used, spray the plates with EN3HANCE and air-dry overnight in a fume hood prior to autoradiography.A several-hour exposure time is usually sufficient for 32P-labeled samples. Visualization of [3H] PIP2 may require exposure up to 2 weeks at − 80°C.
-
13
Scrape the lipids of interest from the plate and quantify the incorporated radioactivity by liquid scintillation spectroscopy.
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see APPENDIX 2A; for suppliers, see APPENDIX 5.
Balanced salt solution
135 mM NaCl (APPENDIX 2A)
5 mM KCl
1 mM Na2HPO4
5.6 mM glucose
1 mM CaCl2
0.5 mM MgCl2 (APPENDIX 2A)
10 mM HEPES, pH 7.4
0.1% bovine serum albumin (BSA)
Filter-sterilize through a 0.22-µm filter, store up to 1 week at room temperature (short term) or up to 2 months at 4°C (long term).
Charcoal suspension
Prepare a 20% (w/v) suspension of acid-washed Norit A in 0.1 M NaCl (APPENDIX 2A), 1 mM EDTA (APPENDIX 2A), and 50 mM sodium acetate, pH 4.0 (APPENDIX 2A). Store up to 3 months at 4°C.
HBSS/HEPES
To 500 ml of 1× Hank’s buffered salt solution (HBSS; APPENDIX 2A), add 10 ml of 1 M HEPES solution, pH 7.4. Filter sterilize through a 0.22-µm filter and store up to 2 weeks at 4°C.
HPLC-injection solution
Prepare a 2 mM sodium acetate/2 mM NaF solution as follows:
4 ml 0.5 M sodium acetate (APPENDIX 2A)
4 ml 0.5 M NaF (see recipe)
Bring up to 1 liter with water
Filter sterilize through a 0.22-µm filter
Store up to 1 month at 4°C
Inositol-free DMEM/2.5% FCS
To 500 ml inositol-free DMEM (Chemicon, cat. no. SLM-100-B or other vendors), add:
5 ml penicillin/streptomycin/glutamine solution (GIBCO/Invitrogen, cat. no. 10378-016)
100 µl 100×non-essential amino acids (NEAA; GIBCO/Invitrogen, cat. no. 11140- 050 or equivalent)
13 ml FCS (2.5% of the total final volume; HyCLone U.S. standard FBS, cat. no. SH30088.03HI)
Adjust pH to 7.4 with 0.1 N HCl or NaOH
Filter sterilize through a 0.22-µm filter
Store up to 2 weeks at 4°C
Lysis buffer
10% (w/v) trichloroacetic acid (TCA) solution
10 mM EDTA (APPENDIX 2A)
10 mM NaF (see recipe)
Store up to 1 month at 4°C
M199/FCS/HEPES
To 500 ml sterile medium 199 (M199; see recipe), add:
10.4 ml FCS (2.0% of the total final volume; HyCLone U.S. standard FBS, cat. no. SH30088.03HI)
10 ml 1 M HEPES solution, pH 7.4
Filter sterilize through a 0.22-µm filter
Store up to 2 weeks at 4°C
To 500 ml sterile medium 199 (M199; see recipe), add:
10.4 ml FCS (2.0% of the total final volume; HyCLone U.S. standard FBS, cat. no. SH30088.03HI)
10 ml 1 M HEPES solution, pH 7.4
Filter sterilize through a 0.22-µm filter
Store up to 2 weeks at 4°C
M199 medium, 1×
Medium 199 liquid contains Earle’s salts, L-glutamine, and 2200 mg/liter sodium bicarbonate (GIBCO/Invitrogen, cat. no. 11150-059 or other vendors). Store up to 2 weeks at 4°C.
MACS staining buffer
Prepare DPBS containing 0.5% BSA and 1 mM EDTA (APPENDIX 2A). Filter sterilize through a 0.22-µm filter and degas. Store up to 2 months at 4°C.
MDD-HPLC eluent A (0.2 mM HCl/15 µM YCl3)
21 µl 30% HCl (APPENDIX 2A)
833 µl 18 mM YCl3 (see recipe)
Bring up to 1 liter with water
Prepare fresh immediately before running HPLC
MDD-HPLC eluent B (0.5 M HCl, 15 µM YCl3)
52.9 ml 30% HCl (APPENDIX 2A)
833 µl 18 mM YCl3 (see recipe)
Bring up to 1 liter with water
Prepare fresh immediately before running HPLC
NaF, 0.5 M
Dissolve 2.10 g NaF in 100 ml deionized water and store up to 3 months at 4°C.
4-(2-Pyridylazo)-resorcinol monosodium salt monohydrate (PAR) stock solution, 10 mM
Dissolve 2.55 g 4-(2-pyridylazo)-resorcinol monosodium salt monohydrate (PAR; Sigma-Aldrich; Acros Organics) in 1 liter methanol and store in a plastic bottle up to 1 year at −20°C.
Post-column reagent C (pH 9.0)
Dissolve 238.7 g triethanolamine (TEA; 1.6 M final) in 900 ml of deionized water, add 30 ml of 10 mM PAR (300 µM final; see recipe), adjust pH to 9 with 30% HCl (~11 ml), then bring up to 1 liter with water. Store in a brown bottle up to 1 week at 4°C.
RPMI/10% FCS/PSG
To 500 ml inositol-free RPMI, add 5 ml penicillin/streptomycin/glutamine solution (GIBCO/Invitrogen, cat. no. 10378-016) and 50 ml FCS (HyCLone U.S. standard FBS, cat. no. SH30088.03HI). Store up to 2 weeks at 4°C.
Sodium acetate, 0.5 M
Dissolve 4.10 g sodium acetate in 100 ml deionized water and store up to 1 month at 4°C.
Tail digestion buffer
50 mM Tris·Cl, pH 8 (APPENDIX 2A)
100 mM NaCl (APPENDIX 2A)
5 mM EDTA (APPENDIX 2A)
0.5% SDS supplemented with fresh proteinase K (100 µg/ml final)
Prepare fresh
Triethanolamine, 1 M
Dissolve 7.45 g triethanolamine (TEA, p.a., >99% purity) in 50 ml of deionized water. Store up to 1 week at 4°C.
Yttrium trichloride, 18 mM
Dissolve 1.09 g yttrium trichloride (YCl3; Aldrich) in 200 ml of deionized water. Store in a dark brown bottle up to 6 months at 4°C.
COMMENTARY
Background Information
Phosphorylation of the membrane phosphoinositide (PI) lipid PIP2 into the PH domain ligand PIP3, and PIP2 hydrolysis into the second messengers Ins(1,4,5)P3 (IP3) and diacylglycerol (DAG) are both pivotal signaling events downstream of multiple receptors in many cell types. In lymphocytes, they are triggered by antigen receptor engagement, which results in activation of PI3K and phospholipase Cγ1/2 (PLCγ1/2; Fig. 11.1.1; Sommers et al., 2004; Berg et al., 2005; Schwartzberg et al., 2005). The lipid DAG remains in cellular membranes, where it recruits and activates key signaling mediators such as the Ras activator RasGRP1 and protein kinases C (PKCs). In contrast, the inositol phosphate (IP) IP3 is soluble and can diffuse through the cytoplasm. By binding to IP3-receptors, IP3 mediates Ca2+ release from intracellular stores. This in turn triggers store-operated Ca2+ influx through plasma membrane channels, an essential component of antigen receptor signaling (Feske, 2007). IP3 can also be phosphorylated into inositol 1,3,4,5-tetrakisphosphate (IP4) through the action of IP3 3-kinases (IP3K, Itpk). Recently, it has been found that T cell receptor (TCR)- or B cell receptor (BCR)-induced IP4 production by IP3KB/ItpkB is essential for T and B cell development in mice through physiologically important roles for IP4 in regulating PH domain interactions with PIP3, in regulating store-operated Ca2+ influx, and possibly through other, little understood IP4 functions (Irvine, 2001, 2007; Irvine and Schell, 2001; Pouillon et al., 2003; Wen et al., 2004; Huang et al., 2007; Marechal et al., 2007; Miller et al., 2007). Others found evidence for IP4 roles in neutrophilic granulocyte and, possibly, mast cell development or function (Cunha-Melo et al., 1987, 1988; Stokes et al., 2006; Jia et al., 2008). The specific roles of other inositol phosphates in lymphocytes (Figs. 11.1.1, 11.1.2, 11.1.4, and 11.1.6; Stewart et al., 1986, 1987; Guse and Emmrich, 1991, 1992; Guse et al., 1992, 1993; Pouillon et al., 2003) are unknown, but represent an exciting area for future research. Recent PI and IP implication in regulating protein membrane recruitment, cytoskeletal dynamics, ion channel function, and apoptosis, in nuclear functions including mRNA export from the nucleus, transcriptional regulation, mRNA editing and chromatin remodeling, and in nonenzymatic protein phosphorylation (Irvine, 2001, 2003, 2006, 2007; Irvine and Schell, 2001; Saiardi et al., 2004; Irvine et al., 2006; Seeds and York, 2007; Shears, 2007; Alcazar-Roman and Wente, 2008; Majerus et al., 2008; Resnick and Saiardi, 2008) suggests that the elucidation of their functions in lymphcoytes will be very exciting. To promote these studies, several methods for measuring PIs and soluble IPs in lymphocytes are described in this unit.
The focus is on methods that allow the simultaneous quantitative analysis of multiple PI or IP isomers: metabolic myo-[3H] inositol or [32P] phosphate labeling (see Basic Protocol 1 and Alternate Protocol 1), followed by extraction of PIs or IPs, and by PI deacylation and anion exchange chromatographic or HPLC separation and detection of PI-derivatives or IPs (see Basic Protocol 2, Alternate Protocol 2, and Basic Protocol 4), or by TLC separation and detection (see Basic Protocol 5) (Skippen et al., 2006; Berrie et al., 2007; Otto et al., 2007). However, these radioactivity based methods are relatively demanding in terms of logistics, equipment cost, and time. Moreover, they may not reliably distinguish changes in IP/PI mass/concentration from changes in radiolabel incorporation due to variability in metabolic labeling efficiency, in particular, under non-equilibrium labeling conditions (Singh and Jiang, 1995; Kuksis, 2003; Rusten and Stenmark, 2006). Metal dye detection (MDD) HPLC (see Basic Protocol 3 and Alternate Protocol 3; Guse et al., 1995a; Lin et al., 2009) can combine the advantages of isomer resolution and better representation of IP/PI mass but is still logistically demanding, in particular for small laboratories without HPLC equipment.
Many alternative assays are available that allow cheaper, faster, simpler, or more efficient high-throughput PI or IP analyses. However, most of them are limited to analyses of one isomer at a time, rely on indirect detection techniques, or lack specificity for one PI or IP isomer. Since several of these techniques have not been used in published studies with lymphocytes, their detailed discussion is beyond the scope of this unit. Instead, the most common techniques are listed in Table 11.1.1 and the reader is referred to excellent reviews for details (Palmer and Wakelam, 1989; Wreggett et al., 1990; Jenkinson, 1995; Singh and Jiang, 1995; van der Kaay et al., 1997, 1998; Balla and Varnai, 2002; Casals et al., 2002; Kuksis, 2003; Prestwich, 2004, 2005; Rusten and Stenmark, 2006; Skippen et al., 2006; Stevenson-Paulik et al., 2006; Berrie et al., 2007; Guillou et al., 2007a,b; Otto et al., 2007; Sergeant and McPhail, 2007). Assays for the identification of PI/IP-binding proteins and quantification of their PI/IP interactions are described in Prestwich (2004, 2005) and Rusten and Stenmark (2006). Assays for PI/IP-metabolizing enzyme activities include many of those listed in Table 11.1.1, and are discussed in more detail in Prestwich (2004, 2005), Rusten and Stenmark (2006), and Sergeant and McPhail (2007).
The choice of method and optimization of separation and detection conditions depends on the specific goal of each experiment. The following points may help in choosing the most appropriate protocol.
Molybdate staining can replace the need for a radiolabel for TLC studies (Andrews and Conn, 1987; Singh and Jiang, 1995; Hatzack and Rasmussen, 1999; Kuksis, 2003; Berrie et al., 2007; Otto et al., 2007; Sergeant and McPhail, 2007). However, low stereo- and regioisomer resolution limits the use of TLC or Dowex anion exchange chromatography to studies where only bulk IP3, IP4, IP5, etc., levels are monitored. TCR stimulation leads to rapid PIP2 conversion into PIP3, I(1,4,5)P3 and its products I(1,3,4,5)P4 and I(1,3,4)P3. Total IP5 and IP6 levels are relatively unaffected (Figs. 11.1.1 and 11.1.6; Stewart et al., 1986, 1987; Guse and Emmrich, 1991, 1992; Guse et al., 1992, 1993; Pouillon et al., 2003). Thus, Dowex anion exchange chromatography may suffice for measuring bulk IP3 and IP4 accumulation as a relatively specific indicator of PIP2 hydrolysis and, by inference, PLCγ activation. However, immunoblot or flow-cytometric analysis of PLCγ phosphorylation provides an inexpensive, faster, and simpler assay for PLCγ activation in lymphocytes (Huang et al., 2007; Barouch-Bentov et al., 2009).
Fluorescent PIP- or IP-binding protein membrane recruitment, monitored microscopically or via FRET, or competitive PIP/IP ligand protein binding assays allow one to indirectly monitor PIP/IP turnover in whole cells or in relatively unpurified cell or tissue lysates with high stereoselectivity and sensitivity (Table 11.1.1). In particular, competitive protein-binding assays for I(1,4,5)P3 are commercially available, and similar I(1,3,4,5)P4 assays have been published (Table 11.1.1). An advantageous feature of the homogeneous assays is that they can allow kinetic analyses of IP3 or IP4 turnover in cell extracts or in vitro, and monitoring of IP-kinase or -phosphatase activities (Chang et al., 2002;Wen et al., 2004). However, protein binding assays are limited to analysis of one PIP/IP isomer at a time. Moreover, a protein ligand probe for a specific PIP will likely bind its IP analog via the same binding site, complicating the results. For example, the Itk PH domain binds either PI(3,4,5)P3 or I(1,3,4,5)P4. Depending on its concentration, I(1,3,4,5)P4 may either augment or inhibit PI(3,4,5)P3 binding. Similar results were observed for several other PI(3,4,5)P3 binding PH domains (Huang et al., 2007). While disabling precise analyses of specific PI or IP isomer mass through protein binding or imaging assays, this does allow for a more accurate determination of physiologically regulated protein-PI interactions (Huang et al., 2007).
Specific analyses of IP isomer levels are best conducted via HPLC. The HPLC protocols described here clearly separate [3H] labeled IP3 isomers, IP4 and IP5 (Figs. 11.1.2 and 11.1.4; Irvine, 1986, 1990; Irvine et al., 1986; Imboden and Pattison, 1987; Pouillon et al., 2003). Micro metal-dye detection (MDD) HPLC (see Basic Protocol 3 and Alternate Protocol 3) allows separation and highly sensitive detection of multiple different IP3, IP4, and IP5 isomers, and of IP6 and PP-IP5 (Fig. 11.1.6; Guse et al., 1995a; Adelt et al., 1999; Lin et al., 2009). Several excellent recent reviews provide in-depth discussions of different PI/IP extraction procedures, anion exchange chromatography and HPLC column matrices, buffers, elution strategies, and radiolabeled IP standard preparation as starting points for further optimization or troubleshooting (Dean and Beaven, 1989; Guse et al., 1995b; Jenkinson, 1995; Singh and Jiang, 1995; Williams and Frasca, 1998; Kuksis, 2003; Azevedo and Saiardi, 2006; Berrie et al., 2007; Otto et al., 2007). Berrie et al. (2007) also describe methods for post-HPLC desalting, extraction, and further analysis of the purified IPs. The disadvantages of HPLC analysis include relatively high equipment costs and long processing times per sample. Shorter protocols can increase throughput but will result in a loss of IP isomer resolution. At an additional cost, addition of a second HPLC column to run while the first column is being re-equilibrated, use of an auto-sampler, which allows automated running, or the use of multiple HPLC systems in parallel can greatly increase HPLC throughput. Resolution can be increased by using longer columns, decreasing flow rates, or further optimizing elution buffer composition, pH, and gradient. Generally, shallower gradients or stepwise gradient increments provide better IP isomer resolution but increase run-times. Low pH may improve resolution but can hydrolyze or isomerize IPs. Sensitivity can be increased by using larger capacity flow cells, but at a cost of reduced resolution.
Compared to HPLC separation of radiolabeled IPs, MDD-HPLC analysis (see Basic Protocol 3 and Alternate Protocol 3; Table 11.1.1) could, in principle, allow better isomer resolution and provide a more direct measure of PI/IP mass, as it does not require time consuming and potentially incomplete radiolabeling of the cellular phosphoinositide pool (Mayr, 1988; Guse et al., 1995b; Kuksis, 2003). This technique uses a complexometric dye- and transition-metal-based post-column detection system for polyanions. MDD permits the direct, highly isomer-selective determination of IPs in the picomolar range from milligram amounts of cells or tissue specimens, which are not readily amenable to analysis by radioisotopic techniques (Lorke et al., 2004). The absence of a need for overnight metabolic radiolabeling facilitates handling, avoids potential complications regarding cell viability and labeling efficiency, and warrants high isomer selectivity and sensitivity. With acidic elution, MDD anion-exchange separation could resolve 26 inositol tris- to hexakisphosphates (excluding enantiomers, 12 IP3, 9 IP4, and 4 IP5 isomers as well as IP6) into 14 peaks within 25 min (Fig. 11.1.5; Guse et al., 1995a,b; Lorke et al., 2004; Lin et al., 2009). However, MDD-HPLC requires expensive equipment and more extensive sample processing than HPLC analysis of isotopically labeled samples. Thus, time savings are limited. Incomplete lipid deacylation and fatty acid extraction can limit quantitative analyses of the cellular phosphoinositide levels, because these may not be quantitatively represented in the glycerophospho-inositol phosphates that are ultimately loaded onto the column. Resolution and throughput can be optimized as described above for HPLC IP analysis. Other non-radioactive IP detection techniques such as amperometric or conductivity detection, gas chromatography (GC), mass spectrometry (MS), enzymatic assays, inductively coupled plasma optical emission spectroscopy, fluorescence resonance energy transfer (FRET), or synthetic pore fluorimetry (Table 11.1.1) could possibly provide alternatives to MDD-detection but require further technological development, specific expertise, or additional sophisticated or expensive equipment (Singh and Jiang, 1995; Kuksis, 2003; Amaro et al., 2004; Rusten and Stenmark, 2006; Berrie et al., 2007; Butterfield et al., 2008).
Quantitative analysis of receptor-mediated changes in inositol phospholipids is relatively complicated. Ideally, one would like to know the mass of the lipid, its turnover, the routes of its metabolism, and how each of these parameters changes following receptor stimulation. No single assay provides all this information at the same time. Nonequilibrium labeling studies measuring [32P] orthophosphate incorporation are commonly used as an index of turnover. Indeed, [32P] labeling of phosphatidic acid and phosphatidyl-inositol are quite sensitive indicators of receptor-mediated PI turnover and are useful adjuncts to studies of [3H] IP generation. Enhanced turnover of PIP and PIP2, however, is far more difficult to measure (see Critical Parameters).
In metabolic labeling approaches, inositol lipid mass is measured indirectly by labeling (usually with [3H] inositol) to isotopic equilibrium. In terms of mass, PIP and PIP2 are minor phospholipids and can be difficult to detect by labeling with [3H] inositol. After calibration with appropriate standards, direct PI/IP detection techniques such as MDD-HPLC, competitive radioligand binding assays, fluorescent detectors, or any of the other direct assays listed in Table 11.1.1 can allow more precise mass determination. Moreover, substantial receptor-mediated increases in the rate of flux through an inositol lipid (particularly phosphatidyl-inositol) often occur with little or no change in mass. Thus, isolated studies of inositol lipids in cells that have been labeled to isotopic equilibrium are relatively difficult and may not be informative. The measurements of mass can be combined with nonequilibrium-labeling studies to provide a more accurate index of PI/IP turnover (Inokuchi and Imboden, 1990). Independent analyses of the activities of the PI/IP kinases and phosphatases involved using immunoblot or FACS assays with activation-state specific antibodies, the assays in Table 11.1.1 or other approaches provide valuable complements.
Critical Parameters
Commonly used HPLC methods separate IPs on anion exchange columns. Their separation depends on the type and size of the column, the composition of the solvent mixture, and the gradient of the solvents. In addition to Table 11.1.1 and the Commentary section, several excellent recent reviews provide in-depth discussions of PI/IP extraction methods, anion exchange chromatography and HPLC column matrices, buffers, and elution strategies for troubleshooting and as starting points for further optimization (Dean and Beaven, 1989; Guse et al., 1995b; Jenkinson, 1995; Singh and Jiang, 1995; Williams and Frasca, 1998; Kuksis, 2003; Azevedo and Saiardi, 2006; Berrie et al., 2007).
It is important to bear in mind that in studies of IP levels in radiolabeled cell extracts, the [3H]-label incorporation is taken as a measure of mass. However, labeling of inositol phospholipid precursors to isotopic equilibrium is a slow process, taking 48 to 72 hr. Different phospholipids or IPs may require different incubation periods to reach isotopic equilibrium, requiring extensive analyses to optimize the method for specific IPs (Singh and Jiang, 1995). This notwithstanding, equilibrium labeling should always be used in studies of cell lines, hybridomas, and clones where long labeling times can be used without compromising cell viability. However, this may not be feasible in studies of freshly isolated lymphocytes. Optimized serum and air exposure through incubation on fibronectin-coated plates and addition of FCS, cytokines, and other growth or survival factors can possibly allow longer labeling through enhanced survival, but lengthy labeling times and/or the reagents themselves may also affect cell behavior, in particular, responses to receptor stimulation, possibly compromising assay results. If equilibrium labeling is not achieved, changes in the [3H]-label incorporation may not simply reflect changes in mass. It is not clear to what extent this actually leads to misleading results. It is very unlikely that the use of nonequilibrium labeling can produce an increase in [3H] IP without a concomitant increase in IP mass. Rather, nonequilibrium labeling may underestimate the magnitude and duration of an IP response. For example, in nonlymphoid cells, the same stimulus triggered a transient increase in [3H] I(1,4,5)P3 in nonequilibrium-labeled cells but a sustained increase in I(1,4,5)P3 mass, as measured by a competitive protein-binding assay. The most likely explanation for these discordant results is that while the initial I(1,4,5)P3 increase includes generation from [3H]-labeled lipid precursors, these are turned over and replaced by unlabeled precursors. As a consequence, after several minutes of stimulation, I(1,4,5)P3 is generated primarily from unlabeled precursors and the fall in [3H] I(1,4,5)P3 measured via HPLC reflects a decrease in specific activity, not a change in mass.
Poor [3H] inositol incorporation into inositol phospholipid precursors is the most commonly encountered difficulty in studies of receptor-mediated PI/IP generation in lymphocytes. When labeling cells with [3H] inositol, it is important to realize that culture media vary substantially in the concentration of inositol. RPMI-1640, for example, contains 190 µM myo-inositol (35 mg/liter), Dulbecco’s MEM contains 40 µM (7.2 mg/liter), medium 199 (M199) contains only 0.3 µM (0.05 mg/liter, Sigma-Aldrich formulations). For other media, see APPENDIX 2A, vendor information, or Ham and McKeehan (1979). The inositol concentration in fetal calf serum (FCS) is ~550 µM. Although inositol is an obligate growth factor for many cell lines, lowering the inositol concentration can enhance the incorporation of [3H] inositol into the lipid pool without compromising growth. This is more economical than simply increasing the concentration of [3H] inositol. For example, Jurkat cells can tolerate 6 to 8 hr in FCS- and inositol-free RPMI and grow well in inositol-free RPMI supplemented with 2.5% to 10% FCS (final inositol concentration, 14 to 55 µM). The typically resulting four-fold increase in specific activity is crucial for measuring [3H] PIP2. With occasional exceptions, tumor lines, hybridomas, cell clones, and lectin-stimulated cells label adequately with [3H] inositol. Insufficient [3H] inositol incorporation into phospholipid pools however is a major obstacle in studies of freshly isolated lymphocytes. Labeling in the absence of cold inositol substantially increases [3H] inositol uptake into resting lymphocytes, permitting detectable IP responses after several hours of labeling. Adherence to fibronectin-coated plates for optimal medium and air exposure, and addition of FCS, cytokines (IL-7), or growth factors can augment cell viability and improve labeling. Murine thymocytes show good viability in M199, which contains only 0.3 µM myo-inositol, making M199 a medium of choice for thymocyte labeling. However, this approach presents several problems. First, equilibrium labeling is not necessarily achieved. Second, inositol deprivation may have adverse effects on inositol lipid metabolism and can prevent phytohemagglutinin (PHA)-induced T cell mitogenesis (Mustelin et al., 1986). Thus, it might be best not to rely solely on metabolic radiolabeling when measuring PI/IP turnover in primary lymphocytes. Direct detection methods such as MDD-HPLC (see Basic Protocol 3 and Alternate Protocol 3), competitive radioligand binding or other homogeneous assays in cell extracts, or fluorescent detectors avoid these complications and appear best suited for analyses of PI or IP mass in primary lymphocytes. Their advantages and disadvantages are discussed in the Commentary section and compiled in Table 11.1.1, which also lists key references and suppliers. In particular, binding assays are likely to yield results of combined effects of PIs and their respective modulating IPs, exemplified by the modulation of PI(3,4,5)P3 binding to certain PH domains by soluble I(1,3,4,5)P4 in vitro and in vivo (Huang et al., 2007).
[32P]-orthophosphate incorporation into inositol lipids can be used to detect enhanced inositol lipid turnover. To be valid, this approach requires that any changes in the mass of the inositol phospholipid have not appreciably influenced labeling; that the label in the lipid is not in equilibrium with the label in the ATP pool; and that the agonist has not altered the specific activity of the ATP pool. However, TCR stimulation and, in particular, co-stimulation profoundly impact energy metabolism, ATP production, and consumption (Krauss et al., 2001; Fox et al., 2005; Jones and Thompson, 2007; Schenk et al., 2008). Although TCR stimulation was reported not to alter the specific activity of the ATP pool in Jurkat cells (Inokuchi and Imboden, 1990), it induces ATP production and accumulation in primary naive T cells (Schenk et al., 2008). In primary lymphocytes, “tonic” TCR signaling prevents a loss of ATP and death by “neglect” (Rathmell et al., 2000). In platelets, receptor-induced changes in the ATP pool have complicated studies of PIP and PIP2 (Verhoeven et al., 1987). [32P]-labeling of PIP and PIP2 in short-term studies is almost exclusively in the 4-position phosphate of PIP and in the 4- and 5-position phosphates of PIP2. Even in unstimulated cells, these phosphates are highly labile and reach equilibrium far faster than that of the 1-position phosphate of PI. Studies of Jurkat cells and of peripheral T cells indicate that >93% of the label (either 32P or 3H) is incorporated into the 4- and 4,5-isomers of PIP and PIP2, respectively. All these observations render quantitative analyses of the turnover, i.e., the combined production and metabolism, of specific PI/IP isomers in lymphocytes non-trivial, in particular, if other phosphate positions are involved, or if the IPs under study are not derived from PIP or PIP2 (Fig. 11.1.1). This notwithstanding, the protocols and alternative approaches discussed in this unit have been used successfully to analyze stimulation-induced changes in the masses of multiple different PI/IP isomers in wild-type and mutant lymphocytes in many studies and proven highly valuable to investigate their physiological functions and those of the enzymes involved (Imboden and Stobo, 1985; Stewart et al., 1986, 1987; Cunha-Melo et al., 1987, 1988; Imboden et al., 1987; Imboden and Pattison, 1987; Zilberman et al., 1987; Inokuchi and Imboden, 1990; Guse and Emmrich, 1991, 1992; Guse et al., 1992, 1993, 1995b; Irvine, 2001, 2007; Irvine and Schell, 2001; Pouillon et al., 2003; Wen et al., 2004; Stokes et al., 2006; Huang et al., 2007; Marechal et al., 2007; Miller et al., 2007; Jia et al., 2008).
Troubleshooting
Potential problems, possible causes, and solutions are summarized in Table 11.1.2.
Table 11.1.2.
Troubleshootinga
| Problem | Possible cause | Solution |
|---|---|---|
| Poor [3H] or [32P] labeling | Low cell viability | Determine whether cell viability is compromised via Trypan blue stain and microscopy, or via AnnexinV/7-AAD stain and FACS analysis (Barouch-Bentov et al., 2009). Add cytokines (IL-7) or other growth factors, or add low unlabeled myo-inositol amounts or more FCS (which contains myo-inositol, ~55 µM final for 10% FCS), possibly after an initial short 2- to 4-hr labeling period without exogenous inositol. Avoid extended cell incubation in microvials, where cells can become hypoxic. Increase medium and air exposure of the cells by promoting monolayer formation on the plate bottom by coating it with fibronectin or genetically engineered fibronectin-like engineered protein polymer-plus to further improve cell adhesion (Esty, 1991). Decrease labeling time. If using thymocytes, use inositol-free M199 medium to improve overall thymocyte viability. Lymphocytes can also be stimulated in PBS, HBSS/HEPES, or other solutions with physiological salt concentrations, or in antibiotic-free tissue culture media (APPENDIX 2A). Optimize red blood cell lysis by varying incubation times or lysis reagent to minimize effects on the lymphocytes. Optimize cell densities between 2 × 107 and 2 × 108 cells/ml. |
| Poor [3H] or [32P] labeling | Low myo-[3H] inositol uptake | Increase radiolabel amount or labeling time. Optimize cell density during labeling. Make sure that neomycin or other inositol lipid binding agents are absent from the medium. Add up to 20 mM LiCl, which inhibits phosphoinositide and IP phosphatases. |
| Poor [3H] or [32P] labeling | Inefficient extraction from the labeled cells | Use alternative lysis/extraction reagents such as chloroform/methanol/concentrated HCl or 10% TCA (Berridge et al., 1983; Irvine et al., 1986; Berrie et al., 2007). Maximal IP yields were obtained when lysing the cells in 10% TCA, followed by a 20-min incubation on ice (to minimize IP hydrolysis), 10-min centrifugation at 15,000 × g, 4°C, and TCA removal through three rounds of supernatant extraction with 2 vol diethyl-ether each. PCA can be removed by equimolar KHCO3 addition (~0.5 M) and centrifugal removal (as above) of the insoluble K-perchlorate. However, the precipitate may contain trapped IP-metal complexes, which form at a pH > 7.0, reducing the IP yield in the supernatant (Lin et al., 2009). Addition of each 20 µg of unlabeled IP3 and IP4 may enhance extraction of these IPs, in particular of IP4 (Wreggett et al., 1987). Replicate samples can be pooled prior to extraction to increase extracted radioactivity. |
| Poor [3H] or [32P] labeling | Inadvertent cold chase | In 32P-labeling experiments, it is important to avoid inadvertent cold chases by adding stimuli in phosphate-containing solutions (e.g., monoclonal antibodies in hybridoma culture medium). For studies of [3H] inositol lipids, it is recommended that cells be stimulated in the labeling medium (i.e., in the presence of [3H] inositol) to avoid any changes in specific activity during the assay. |
| Cell activation modulating control compounds have no or little effect | Reduced effective concentration | Reduce FCS in the medium, which may bind compound and reduce its effective concentration. Titrate compound. Change carrier/solvent. |
| Cell activation modulating control compounds have no or little effect | Compound may not enter cell or undergo fast metabolization/inactivation within the cell | Add detergents such as Pluronic to the compound to improve uptake. Use less polar compound derivatives such as AM/PM esters that are hydrolyzed into the active species by cellular enzymes to improve uptake and, possibly, stability of the compound. Use caged compounds that are taken up and then released very quickly and homogeneously via UV-irradiation, resulting in a compound burst. Or use alternative compounds with improved uptake and stability. |
| Insufficient stimulation | Stimulation regimen not optimized | Optimize stimulation reagent, its concentration, and application time |
| Insufficient stimulation | Compromised cell viability/fitness | Improve cell viability/fitness as described above |
| Insufficient stimulation | High background activity | Rest cells in medium for 15 to 30 min or longer at 25° to 37°C before stimulation |
| Low HPLC sensitivity for detecting [3H] PIs or IPs | Poor cell viability or labeling | Troubleshoot cell viability and labeling as described above |
| Low HPLC sensitivity for detecting [3H] PIs or IPs | Low capacity flow cell | Use larger capacity flow cell; however, this may decrease resolution |
| Low HPLC sensitivity for detecting [3H] PIs or IPs | LSC analysis too insensitive | Use in-line β-detector (see Basic Protocol 2) |
| Low HPLC sensitivity for detecting [3H] PIs or IPs | Radio-detection too insensitive for given cell amount and labeling efficiency | Use an alternate detection method, such as MDD-HPLC (see Basic Protocol 3 and Alternate Protocol 3; Table 11.1.1). |
| Low HPLC resolution | Column overloaded or too short | Use larger or longer column (25–cm; Whatman no. 4621-1507). Vary column matrices or separation gradient buffer. HPLC anion exchange matrices, buffers and other aspects of the method are reviewed in detail in Wreggett et al. (1990); Guse et al. (1995b); Singh and Jiang (1995); Williams and Frasca (1998); Kuksis (2003); Azevedo and Saiardi (2006); and Berrie et al. (2007). |
| Low HPLC resolution | Gradient time too short | Increase gradient time |
| Low HPLC resolution | Gradient steps too steep | Decrease steepness |
| Low HPLC resolution | Flow rate too fast | Decrease flow rate |
| Low HPLC throughput | Number of HPLC columns limiting | Add a second column to run while the first column is re-equilibrated to double throughput |
| Low HPLC throughput | Gradient time longer than needed | Shorten gradient time, but this may reduce resolution |
| Low HPLC throughput | Manual sample exchange too slow | Use auto-sampler, which allows automated non-stop running. Neutralized samples are relatively stable for >24 hr at room temperature, allowing long queues. |
| HPLC resolution deteriorates | Column matrix deteriorates after ~50 runs | Regenerate/re-equilibrate column as described in the respective HPLC protocols and in the respective column manufacturer’s instructions |
| Insufficient TLC resolution | Suboptimal solvent | Try alternative solvents. For example, 45:35:10 chloroform/methanol/4 N NH4OH (Lapetina and Siess, 1987) allows excellent separation of PI, PIP, and PIP2 but should only be used with [3H] inositol-labeled samples (PI is not well-separated from PA; the latter can be labeled with [32P] but not with [3H] inositol). For additional discussions of TLC, see Singh and Jiang (1995); Hatzack and Rasmussen (1999); Kuksis (2003); Berrie et al. (2007); Otto et al. (2007); Sergeant and McPhail (2007). |
For additional HPLC troubleshooting, see Wreggett et al. (1990); Singh and Jiang (1995); Kuksis (2003); Azevedo and Saiardi (2006); and Berrie et al. (2007).
Anticipated Results
Magnitude and duration of receptor-induced responses vary with agonist and cell type. Measured via [3H]-labeling and HPLC, TCR engagement on Jurkat cells generates an about five-fold increase in [3H] I(1,4,5)P3 and over a six-fold increase in I(1,3,4,5)P4 (Imboden and Pattison, 1987). The response is maximal at 45 to 60 sec and sustained for >30 min. Increases in I(1,3,4,5)P4, I(1,3,4)P3, IP2, and IP1 lag behind those of I(1,4,5)P3. I(1,3,4,5)P4 levels remain elevated for >20 min. MDD-detection revealed a three-to four-fold increase in I(1,4,5)P3 to ~5 pmols/106 cells 3 min post-OKT3-stimulation. I(1,3,4,5)P4 increased ~40-fold to ~20 pmols/106 cells and I(1,3,4)P3 ~20-fold to ~5 pmols/106 cells (Fig. 11.1.6). All methods showed relatively minor IP5 and IP6 mass changes. Published analyses showed qualitatively similar results whose variation likely reflects different stimulation conditions and times, and differences in the specific labeling protocols and/or HPLC setups and detection procedures (Imboden and Stobo, 1985; Stewart et al., 1986, 1987; Imboden et al., 1987; Imboden and Pattison, 1987; Inokuchi and Imboden, 1990; Guse and Emmrich, 1991, 1992; Guse et al., 1992, 1993, 1995b). Similar results were obtained via myo-[3H] inositol labeling and HPLC in primary mouse thymocytes (Fig. 11.1.2; Zilberman et al., 1987; Pouillon et al., 2003) and in a limited MDD-HPLC analysis in rat thymocytes (Guse et al., 1993). Confirmation of the Jurkat cell data in primary T cells is important because Jurkat cells contain elevated basal PI(3,4,5)P3 levels and show perturbed PI(3,4,5)P3 downstream signaling compared to primary human peripheral blood leukocytes due to deficiency in the PI(3,4,5)P3 metabolizing phosphatases PTEN and SHIP-1 (Fig. 11.1.1; Shan et al., 2000; Astoul et al., 2001; Freeburn et al., 2002).
Time Considerations
Preparation of primary mouse lymphocytes requires ~1 to 2 hr depending on the number of mice and tissues prepared, and on institutional vivarium entry/exit procedures. Cell subset enrichment via MACS requires ~2 hr plus an additional ~1 hr for purity analysis via FACS.
Myo-[3H] inositol equilibrium labeling of lymphocytes requires ~1 hr, followed by an ~20-hr incubation (if necessary, with additional medium addition after 3 to 6 hr) and cell washing (~1 hr). Optional compound incubation requires 1 to 3 hr, cell stimulation, washing, and lysis require ~1 hr. Neutralized, cell-free IP extracts can be frozen and stored for several weeks at −70°C. HPLC analysis requires ~2 hr per sample plus set-up time and is conveniently done unsupervised using an auto-loader and in-line β-detector if multiple samples are to be analyzed. Neutralized IP extracts are relatively stable for >24 hr at room temperature, allowing long queues.
Cell or tissue preparation for MDD requires ~1 to 3 hr for compound incubation, and 3 to 4 hr for stimulation and inositol phosphate extraction. Cell pellets or neutralized, cell-free IP extracts can be stored at −70°C. Phospholipid extraction, deacetylation and preparation for MDD-HPLC requires 7 to 10 hr. Micro-MDD HPLC requires ~2 hr per sample and is conveniently done unsupervised using an auto-loader.
[3H]-IP extraction and separation by Dowex anion exchange chromatography can be completed in 4 to 5 hr. HPLC analysis of each sample requires ~2 hr. Two days should be allocated to the analysis of inositol phospholipids by TLC.
Acknowledgements
The authors thank Jiaxue Huang, Steven Soonthornvacharin, Catherine Tan, and Todd Christopher White for their help in establishing some of the assays and procedures described here. Part of this work was funded by NIH/NIAID grant AI070845, NIH/NIGMS grant GM088647 or by the Genomics Institute of the Novartis Research Foundation (GNF).
Literature Cited
- Adelt S, Plettenburg O, Stricker R, Reiser G, Altenbach HJ, Vogel G. Enzyme-assisted total synthesis of the optical antipodes d-myo-inositol 3,4,5-trisphosphate and d-myo-inositol 1,5,6-trisphosphate: Aspects of their structure-activity relationship to biologically active inositol phosphates. J. Med. Chem. 1999;42:1262–1273. doi: 10.1021/jm981113k. [DOI] [PubMed] [Google Scholar]
- Alcazar-Roman AR, Wente SR. Inositol polyphosphates: A new frontier for regulating gene expression. Chromosoma. 2008;117:1–13. doi: 10.1007/s00412-007-0126-4. [DOI] [PubMed] [Google Scholar]
- Amaro R, Escalona A, Murillo M. HPLC with inductively coupled plasma optical emission spectrometric detection for the analysis of inositol phosphates. J. Chromatogr. Sci. 2004;42:491–494. doi: 10.1093/chromsci/42.9.491. [DOI] [PubMed] [Google Scholar]
- Andrews WV, Conn PM. Measurement of inositol phospholipid metabolites by one-dimensional thin-layer chromatography. Methods Enzymol. 1987;141:156–168. doi: 10.1016/0076-6879(87)41064-1. [DOI] [PubMed] [Google Scholar]
- Astoul E, Edmunds C, Cantrell DA, Ward SG. PI 3-K and T-cell activation: Limitations of T-leukemic cell lines as signaling models. Trends Immunol. 2001;22:490–496. doi: 10.1016/s1471-4906(01)01973-1. [DOI] [PubMed] [Google Scholar]
- Azevedo C, Saiardi A. Extraction and analysis of soluble inositol polyphosphates from yeast. Nat. Protoc. 2006;1:2416–2422. doi: 10.1038/nprot.2006.337. [DOI] [PubMed] [Google Scholar]
- Balla T, Varnai P. Visualizing cellular phosphoinositide pools with GFP-fused protein-modules. Sci. STKE. 2002;2002:PL3. doi: 10.1126/stke.2002.125.pl3. [DOI] [PubMed] [Google Scholar]
- Barouch-Bentov R, Che J, Lee CC, Yang Y, Herman A, Jia Y, Velentza A, Watson J, Sternberg L, Kim S, Ziaee N, Miller A, Jackson C, Fujimoto M, Young M, Batalov S, Liu Y, Warmuth M, Wiltshire T, Cooke MP, Sauer K. A conserved salt bridge in the G loop of multiple protein kinases is important for catalysis and for in vivo lyn function. Mol. Cell. 2009;33:43–52. doi: 10.1016/j.molcel.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin ER, Haftl SL, Xanthos DN, Crumley G, Hachicha M, Valenzano KJ. A miniaturized column chromatography method for measuring receptor-mediated inositol phosphate accumulation. J. Biomol. Screen. 2004;9:343–353. doi: 10.1177/1087057103262841. [DOI] [PubMed] [Google Scholar]
- Berg LJ, Finkelstein LD, Lucas JA, Schwartzberg PL. Tec family kinases in T lymphocyte development and function. Annu. Rev. Immunol. 2005;23:549–600. doi: 10.1146/annurev.immunol.22.012703.104743. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Dawson RM, Downes CP, Heslop JP, Irvine RF. Changes in the levels of inositol phosphates after agonist-dependent hydrolysis of membrane phosphoinositides. Biochem. J. 1983;212:473–482. doi: 10.1042/bj2120473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrie CP, Iurisci C, Piccolo E, Bagnati R, Corda D. Analysis of phosphoinositides and their aqueous metabolites. Methods Enzymol. 2007;434:187–232. doi: 10.1016/S0076-6879(07)34011-1. [DOI] [PubMed] [Google Scholar]
- Boldyreff B, Rasmussen TL, Jensen HH, Cloutier A, Beaudet L, Roby P, Issinger OG. Expression and purification of PI3 kinase {alpha} and development of an ATP depletion and an AlphaScreen PI3 kinase activity assay. J. Biomol. Screen. 2008;13:1035–1040. doi: 10.1177/1087057108326079. [DOI] [PubMed] [Google Scholar]
- Brandish PE, Hill LA, Zheng W, Scolnick EM. Scintillation proximity assay of inositol phosphates in cell extracts: High-throughput measurement of G-protein-coupled receptor activation. Anal. Biochem. 2003;313:311–318. doi: 10.1016/s0003-2697(02)00630-9. [DOI] [PubMed] [Google Scholar]
- Budd RC, Winslow G, Inokuchi S, Imboden JB. Intact antigen receptor-mediated generation of inositol phosphates and increased intracellular calcium in CD4 CD8 T lymphocytes from MRL lpr mice. J. Immunol. 1990;145:2862–2872. [PubMed] [Google Scholar]
- Butterfield SM, Tran DH, Zhang H, Prestwich GD, Matile S. Fluorometric detection of inositol phosphates and the activity of their enzymes with synthetic pores: Discrimination of IP7 and IP6 and phytate sensing in complex matrices. J. Am. Chem. Soc. 2008;130:3270–3271. doi: 10.1021/ja710186e. [DOI] [PubMed] [Google Scholar]
- Cante-Barrett K, Gallo EM, Winslow MM, Crabtree GR. Thymocyte negative selection is mediated by protein kinase C- and Ca2+-dependent transcriptional induction of bim. J. Immunol. 2006;176:2299–2306. doi: 10.4049/jimmunol.176.4.2299. [DOI] [PubMed] [Google Scholar]
- Casals I, Villar JL, Riera-Codina M. A straightforward method for analysis of highly phosphorylated inositols in blood cells by high-performance liquid chromatography. Anal. Biochem. 2002;300:69–76. doi: 10.1006/abio.2001.5449. [DOI] [PubMed] [Google Scholar]
- Challiss RA, Batty IH, Nahorski SR. Mass measurements of inositol (1,4,5)trisphosphate in rat cerebral cortex slices using a radioreceptor assay: Effects of neurotransmitters and depolarization. Biochem. Biophys. Res. Commun. 1988;157:684–691. doi: 10.1016/s0006-291x(88)80304-8. [DOI] [PubMed] [Google Scholar]
- Chang YT, Choi G, Bae YS, Burdett M, Moon HS, Lee JW, Gray NS, Schultz PG, Meijer L, Chung SK, Choi KY, Suh PG, Ryu SH. Purine-based inhibitors of inositol-1,4,5-trisphosphate-3-kinase. Chem biochem. 2002;3:897–901. doi: 10.1002/1439-7633(20020902)3:9<897::AID-CBIC897>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Chengalvala M, Kostek B, Frail DE. A multi-well filtration assay for quantitation of inositol phosphates in biological samples. J. Biochem. Biophys. Methods. 1999;38:163–170. doi: 10.1016/s0165-022x(98)00046-3. [DOI] [PubMed] [Google Scholar]
- Cicchetti G, Biernacki M, Farquharson J, Allen PG. A ratiometric expressible FRET sensor for phosphoinositides displays a signal change in highly dynamic membrane structures in fibroblasts. Biochemistry. 2004;43:1939–1949. doi: 10.1021/bi035480w. [DOI] [PubMed] [Google Scholar]
- Cunha-Melo JR, Dean NM, Moyer JD, Maeyama K, Beaven MA. The kinetics of phosphoinositide hydrolysis in rat basophilic leukemia (RBL-2H3) cells varies with the type of IgE receptor cross-linking agent used. J. Biol. Chem. 1987;262:11455–11463. [PubMed] [Google Scholar]
- Cunha-Melo JR, Dean NM, Ali H, Beaven MA. Formation of inositol 1,4,5-trisphosphate and inositol 1,3,4-trisphosphate from inositol 1,3,4,5-tetrakisphosphate and their pathways of degradation in RBL-2H3 cells. J. Biol. Chem. 1988;263:14245–14250. [PubMed] [Google Scholar]
- Dean NM, Beaven MA. Methods for the analysis of inositol phosphates. Anal. Biochem. 1989;183:199–209. doi: 10.1016/0003-2697(89)90468-5. [DOI] [PubMed] [Google Scholar]
- Donie F, Reiser G. A novel, specific binding protein assay for quantitation of intracellular inositol 1,3,4,5-tetrakisphosphate (InsP4) using a high-affinity InsP4 receptor from cerebellum. FEBS Lett. 1989;254:155–158. doi: 10.1016/0014-5793(89)81029-4. [DOI] [PubMed] [Google Scholar]
- Drees BE, Weipert A, Hudson H, Ferguson CG, Chakravarty L, Prestwich GD. Competitive fluorescence polarization assays for the detection of phosphoinositide kinase and phosphatase activity. Comb. Chem. High Throughput Screen. 2003;6:321–330. doi: 10.2174/138620703106298572. [DOI] [PubMed] [Google Scholar]
- Esty A. Receptor-specific serum-free cell attachment using a highly stable engineered protein polymer. Am. Biotechnol. Lab. 1991;9:44. [PubMed] [Google Scholar]
- Feske S. Calcium signaling in lymphocyte activation and disease. Nat. Rev. Immunol. 2007;7:690–702. doi: 10.1038/nri2152. [DOI] [PubMed] [Google Scholar]
- Folch J. Complete fractionation of brain cephalin; isolation from it of phosphatidyl serine, phosphatidyl ethanolamine, and diphosphoinositide. J. Biol. Chem. 1949;177:497–504. [PubMed] [Google Scholar]
- Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: Energy metabolism and the T-cell response. Nat. Rev. Immunol. 2005;5:844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- Freeburn RW, Wright KL, Burgess SJ, Astoul E, Cantrell DA, Ward SG. Evidence that SHIP-1 contributes to phosphatidylinositol 3,4,5-trisphosphate metabolism in T lymphocytes and can regulate novel phosphoinositide 3-kinase effectors. J. Immunol. 2002;169:5441–5450. doi: 10.4049/jimmunol.169.10.5441. [DOI] [PubMed] [Google Scholar]
- Fruman DA. Phosphoinositide 3-kinase and its targets in B-cell and T-cell signaling. Curr. Opin. Immunol. 2004;16:314–320. doi: 10.1016/j.coi.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Gray A, Olsson H, Batty IH, Priganica L, Peter Downes C. Nonradioactive methods for the assay of phosphoinositide 3-kinases and phosphoinositide phosphatases and selective detection of signaling lipids in cell and tissue extracts. Anal. Biochem. 2003;313:234–245. doi: 10.1016/s0003-2697(02)00607-3. [DOI] [PubMed] [Google Scholar]
- Grusby MJ, Auchincloss H, Jr, Lee R, Johnson RS, Spencer JP, Zijlstra M, Jaenisch R, Papaioannou VE, Glimcher LH. Mice lacking major histocompatibility complex class I and class II molecules. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3913–3917. doi: 10.1073/pnas.90.9.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou H, Lecureuil C, Anderson KE, Suire S, Ferguson GJ, Ellson CD, Gray A, Divecha N, Hawkins PT, Stephens LR. Use of the GRP1 PH domain as a tool to measure the relative levels of PtdIns(3,4,5)P3 through a protein-lipid overlay approach. J. Lipid Res. 2007a;48:726–732. doi: 10.1194/jlr.D600038-JLR200. [DOI] [PubMed] [Google Scholar]
- Guillou H, Stephens LR, Hawkins PT. Quantitative measurement of phosphatidylinositol 3,4,5-trisphosphate. Methods Enzymol. 2007b;434:117–130. doi: 10.1016/S0076-6879(07)34007-X. [DOI] [PubMed] [Google Scholar]
- Guse AH, Emmrich F. T cell receptor–mediated metabolism of inositol polyphosphates in Jurkat T-lymphocytes. Identification of a d-myo-inositol 1,2,3,4,6-pentakisphosphate-2-phosphomonoesterase activity, a d-myo-inositol 1,3,4,5,6-pentakisphosphate-1/3-phosphatase activity and a d/l-myo-inositol 1,2,4,5,6-pentakisphosphate-1/3-kinase activity. J. Biol. Chem. 1991;266:24498–24502. [PubMed] [Google Scholar]
- Guse AH, Emmrich F. Determination of inositol polyphosphates from human T-lymphocyte cell lines by anion-exchange high-performance liquid chromatography and post-column derivatization. J. Chromatogr. 1992;593:157–163. doi: 10.1016/0021-9673(92)80281-x. [DOI] [PubMed] [Google Scholar]
- Guse AH, Roth E, Broker BM, Emmrich F. Complex inositol polyphosphate response induced by co-cross-linking of CD4 and Fc gamma receptors in the human monocytoid cell line U937. J. Immunol. 1992;149:2452–2458. [PubMed] [Google Scholar]
- Guse AH, Greiner E, Emmrich F, Brand K. Mass changes of inositol 1,3,4,5,6-pentakisphosphate and inositol hexakisphosphate during cell cycle progression in rat thymocytes. J. Biol. Chem. 1993;268:7129–7133. [PubMed] [Google Scholar]
- Guse AH, da Silva CP, Emmrich F, Ashamu GA, Potter BV, Mayr GW. Characterization of cyclic adenosine diphosphateribose-induced Ca2+ release in T lymphocyte cell lines. J. Immunol. 1995a;155:3353–3359. [PubMed] [Google Scholar]
- Guse AH, Goldwich A, Weber K, Mayr GW. Non-radioactive, isomer-specific inositol phosphate mass determinations: High-performance liquid chromatography-micro-metal-dye detection strongly improves speed and sensitivity of analyses from cells and microenzyme assays. J. Chromatogr. B Biomed. Appl. 1995b;672:189–198. doi: 10.1016/0378-4347(95)00219-9. [DOI] [PubMed] [Google Scholar]
- Ham RG, McKeehan WL. Media and growth requirements. Methods Enzymol. 1979;58:44–93. doi: 10.1016/s0076-6879(79)58126-9. [DOI] [PubMed] [Google Scholar]
- Hatzack F, Rasmussen SK. High-performance thin-layer chromatography method for inositol phosphate analysis. J. Chromatogr. B Biomed. Sci. Appl. 1999;736:221–229. doi: 10.1016/s0378-4347(99)00465-x. [DOI] [PubMed] [Google Scholar]
- Holz RW, Hlubek MD, Sorensen SD, Fisher SK, Balla T, Ozaki S, Prestwich GD, Stuenkel EL, Bittner MA. A pleckstrin homology domain specific for phosphatidylinositol 4, 5-bisphosphate (PtdIns-4,5-P2) and fused to green fluorescent protein identifies plasma membrane PtdIns-4,5-P2 as being important in exocytosis. J. Biol. Chem. 2000;275:17878–17885. doi: 10.1074/jbc.M000925200. [DOI] [PubMed] [Google Scholar]
- Horn S, Endl E, Fehse B, Weck MM, Mayr GW, Jucker M. Restoration of SHIP activity in a human leukemia cell line downregulates constitutively activated phosphatidylinositol 3-kinase/Akt/GSK-3b signaling and leads to an increased transit time through the G1 phase of the cell cycle. Leukemia. 2004;18:1839–1849. doi: 10.1038/sj.leu.2403529. [DOI] [PubMed] [Google Scholar]
- Huang YH, Grasis JA, Miller AT, Xu R, Soonthornvacharin S, Andreotti AH, Tsoukas CD, Cooke MP, Sauer K. Positive regulation of Itk PH domain function by soluble IP4. Science. 2007;316:886–889. doi: 10.1126/science.1138684. [DOI] [PubMed] [Google Scholar]
- Huang YH, Hoebe K, Sauer K. New therapeutic targets in immune disorders: ItpkB, Orai1 and UNC93B. Expert Opin. Ther. Targets. 2008;12:391–413. doi: 10.1517/14728222.12.4.391. [DOI] [PubMed] [Google Scholar]
- Imai A, Gershengorn MC. Measurement of lipid turnover in response to thyrotropin-releasing hormone. Methods Enzymol. 1987;141:100–101. doi: 10.1016/0076-6879(87)41059-8. [DOI] [PubMed] [Google Scholar]
- Imboden JB, Pattison G. Regulation of inositol 1,4,5-trisphosphate kinase activity after stimulation of human T cell antigen receptor. J. Clin. Invest. 1987;79:1538–1541. doi: 10.1172/JCI112986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imboden JB, Stobo JD. Transmembrane signaling by the T cell antigen receptor. Perturbation of the T3-antigen receptor complex generates inositol phosphates and releases calcium ions from intracellular stores. J. Exp. Med. 1985;161:446–456. doi: 10.1084/jem.161.3.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imboden J, Weyand C, Goronzy J. Antigen recognition by a human T cell clone leads to increases in inositol trisphosphate. J. Immunol. 1987;138:1322–1324. [PubMed] [Google Scholar]
- Inokuchi S, Imboden JB. Antigen receptor-mediated regulation of sustained polyphosphoinositide turnover in a human T cell line. Evidence for a receptor-regulated pathway for production of phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 1990;265:5983–5989. [PubMed] [Google Scholar]
- Irvine RF. The structure, metabolism, and analysis of inositol lipids and inositol phosphates. In: Putney JW, editor. Receptor Biochemistry and Methodology. New York: Wiley-Liss; 1986. pp. 89–108. [Google Scholar]
- Irvine RF. Methods in Inositide Research. New York: Raven Press; 1990. [Google Scholar]
- Irvine RF. Inositol phosphates: Does IP(4) run a protection racket? Curr. Biol. 2001;11:R172–R174. doi: 10.1016/s0960-9822(01)00086-0. [DOI] [PubMed] [Google Scholar]
- Irvine RF. Nuclear lipid signaling. Nat. Rev. Mol. Cell Biol. 2003;4:349–360. doi: 10.1038/nrm1100. [DOI] [PubMed] [Google Scholar]
- Irvine RF. Inositide evolution—Towards turtle domination? J. Physiol. 2005;566:295–300. doi: 10.1113/jphysiol.2005.087387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine RF. Nuclear inositide signaling—Expansion, structures and clarification. Biochim. Biophys. Acta. 2006;1761:505–508. doi: 10.1016/j.bbalip.2006.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine RF. Cell signaling. The art of the soluble. Science. 2007;316:845–846. doi: 10.1126/science.1143339. [DOI] [PubMed] [Google Scholar]
- Irvine RF, Schell MJ. Back in the water: The return of the inositol phosphates. Nat. Rev. Mol. Cell Biol. 2001;2:327–338. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- Irvine RF, Letcher AJ, Heslop JP, Berridge MJ. The inositol tris/tetrakisphosphate pathway—Demonstration of Ins(1,4,5)P3 3-kinase activity in animal tissues. Nature. 1986;320:631–634. doi: 10.1038/320631a0. [DOI] [PubMed] [Google Scholar]
- Irvine RF, Lloyd-Burton SM, Yu JC, Letcher AJ, Schell MJ. The regulation and function of inositol 1,4,5-trisphosphate 3-kinases. Adv. Enzyme Regul. 2006;46:314–323. doi: 10.1016/j.advenzreg.2006.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson S. Separation of labeled inositol phosphate isomers by high-pressure liquid chromatography (HPLC) Methods Mol. Biol. 1995;41:151–165. doi: 10.1385/0-89603-298-1:151. [DOI] [PubMed] [Google Scholar]
- Jia Y, Subramanian KK, Erneux C, Pouillon V, Hattori H, Jo H, You J, Zhu D, Schurmans S, Luo HR. Inositol 1,3,4,5-tetrakisphosphate negatively regulates phosphatidylinositol-3,4,5-trisphosphate signaling in neutrophils. Immunity. 2007;27:453–467. doi: 10.1016/j.immuni.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Loison F, Hattori H, Li Y, Erneux C, Park SY, Gao C, Chai L, Silberstein LE, Schurmans S, Luo HR. Inositol trisphosphate 3-kinase B (InsP3KB) as a physiological modulator of myelopoiesis. Proc. Natl. Acad. Sci. U.S.A. 2008;105:4739–4744. doi: 10.1073/pnas.0800218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodi L, Buckler XL, Laurence A, Turka LA. Regulation of T cell responses by PTEN. Immunol. Rev. 2008;224:239–248. doi: 10.1111/j.1600-065X.2008.00650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Chichili GR, Rodgers W. Compartmentalization of phosphatidylinositol 4,5-bisphosphate signaling evidenced using targeted phosphatases. J. Biol. Chem. 2008;283:29920–29928. doi: 10.1074/jbc.M805921200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RG, Thompson CB. Revving the engine: Signal transduction fuels T cell activation. Immunity. 2007;27:173–178. doi: 10.1016/j.immuni.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Juntilla MM, Koretzky GA. Critical roles of the PI3K/Akt signaling pathway in T cell development. Immunol. Lett. 2008;116:104–110. doi: 10.1016/j.imlet.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S, Brand MD, Buttgereit F. Signaling takes a breath—New quantitative perspectives on bioenergetics and signal transduction. Immunity. 2001;15:497–502. doi: 10.1016/s1074-7613(01)00205-9. [DOI] [PubMed] [Google Scholar]
- Kuksis A. Inositol Phospholipid Metabolism and Phosphatidyl Inositol Kinases. Vol. 30. New York: Elsevier; 2003. [Google Scholar]
- Lapetina EG, Siess W. Measurement of inositol phospholipid turnover in platelets. Methods Enzymol. 1987;141:176–192. doi: 10.1016/0076-6879(87)41066-5. [DOI] [PubMed] [Google Scholar]
- Lin H, Fridy PC, Ribeiro AA, Choi JH, Barma DK, Vogel G, Falck JR, Shears SB, York JD, Mayr GW. Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J. Biol. Chem. 2009;284:1863–1872. doi: 10.1074/jbc.M805686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Hartman DS, Bostwick JR. An immobilized metal ion affinity adsorption and scintillation proximity assay for receptor-stimulated phosphoinositide hydrolysis. Anal. Biochem. 2003;318:91–99. doi: 10.1016/s0003-2697(03)00159-3. [DOI] [PubMed] [Google Scholar]
- Lorke DE, Gustke H, Mayr GW. An optimized fixation and extraction technique for high resolution of inositol phosphate signals in rodent brain. Neurochem. Res. 2004;29:1887–1896. doi: 10.1023/b:nere.0000042216.86633.71. [DOI] [PubMed] [Google Scholar]
- Majerus PW, Zou J, Marjanovic J, Kisseleva MV, Wilson MP. The role of inositol signaling in the control of apoptosis. Adv. Enzyme Regul. 2008;48:10–17. doi: 10.1016/j.advenzreg.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal Y, Pesesse X, Jia Y, Pouillon V, Perez-Morga D, Daniel J, Izui S, Cullen PJ, Leo O, Luo HR, Erneux C, Schurmans S. Inositol 1,3,4,5-tetrakisphosphate controls proapoptotic Bim gene expression and survival in B cells. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13978–13983. doi: 10.1073/pnas.0704312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr GW. A novel metal-dye detection system permits picomolar-range HPLC analysis of inositol polyphosphates from non-radioactively labeled cell or tissue specimens. Biochem. J. 1988;254:585–591. doi: 10.1042/bj2540585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AT, Sandberg M, Huang YH, Young M, Sutton S, Sauer K, Cooke MP. Production of Ins(1,3,4,5)P(4) mediated by the kinase Itpkb inhibits store-operated calcium channels and regulates B cell selection and activation. Nat. Immunol. 2007;8:514–521. doi: 10.1038/ni1458. [DOI] [PubMed] [Google Scholar]
- Miller AT, Chamberlain PP, Cooke MP. Beyond IP3: Roles for higher order inositol phosphates in immune cell signaling. Cell Cycle. 2008;7:463–467. doi: 10.4161/cc.7.4.5518. [DOI] [PubMed] [Google Scholar]
- Mueller P, Massner J, Jayachandran R, Combaluzier B, Albrecht I, Gatfield J, Blum C, Ceredig R, Rodewald HR, Rolink AG, Pieters J. Regulation of T cell survival through coronin-1-mediated generation of inositol-1,4,5-trisphosphate and calcium mobilization after T cell receptor triggering. Nat. Immunol. 2008;9:424–431. doi: 10.1038/ni1570. [DOI] [PubMed] [Google Scholar]
- Mustelin T, Poso H, Iivanainen A, Andersson LC. myo-Inositol reverses Li+-induced inhibition of phosphoinositide turnover and ornithine decarboxylase induction during early lymphocyte activation. Eur. J. Immunol. 1986;16:859–861. doi: 10.1002/eji.1830160724. [DOI] [PubMed] [Google Scholar]
- Nasuhoglu C, Feng S, Mao J, Yamamoto M, Yin HL, Earnest S, Barylko B, Albanesi JP, Hilgemann DW. Nonradioactive analysis of phosphatidylinositides and other anionic phospholipids by anion-exchange high-performance liquid chromatography with suppressed conductivity detection. Anal. Biochem. 2002;301:243–254. doi: 10.1006/abio.2001.5489. [DOI] [PubMed] [Google Scholar]
- Oatey PB, Venkateswarlu K, Williams AG, Fletcher LM, Foulstone EJ, Cullen PJ, Tavare JM. Confocal imaging of the subcellular distribution of phosphatidylinositol 3,4,5-trisphosphate in insulin- and PDGF-stimulated 3T3-L1 adipocytes. Biochem. J. 1999;344:511–518. [PMC free article] [PubMed] [Google Scholar]
- Otto JC, Mulugu S, Fridy PC, Chiou ST, Armbruster BN, Ribeiro AA, York JD. Biochemical analysis of inositol phosphate kinases. Methods Enzymol. 2007;434:171–185. doi: 10.1016/S0076-6879(07)34010-X. [DOI] [PubMed] [Google Scholar]
- Palmer S, Wakelam MJ. Mass measurement of inositol phosphates. Biochim. Biophys. Acta. 1989;1014:239–246. doi: 10.1016/0167-4889(89)90219-x. [DOI] [PubMed] [Google Scholar]
- Park TJ, Song SK, Kim KT. A2A adenosine receptors inhibit ATP-induced Ca2+ influx in PC12 cells by involving protein kinase A. J. Neurochem. 1997;68:2177–2185. doi: 10.1046/j.1471-4159.1997.68052177.x. [DOI] [PubMed] [Google Scholar]
- Parmryd I, Adler J, Patel R, Magee AI. Imaging metabolism of phosphatidylinositol 4,5-bisphosphate in T cell GM1-enriched domains containing Ras proteins. Exp. Cell Res. 2003;285:27–38. doi: 10.1016/s0014-4827(02)00048-4. [DOI] [PubMed] [Google Scholar]
- Pettitt TR, Dove SK, Lubben A, Calaminus SD, Wakelam MJ. Analysis of intact phosphoinositides in biological samples. J. Lipid Res. 2006;47:1588–1596. doi: 10.1194/jlr.D600004-JLR200. [DOI] [PubMed] [Google Scholar]
- Phillippy BQ, White KD, Johnston MR, Tao SH, Fox MR. Preparation of inositol phosphates from sodium phytate by enzymatic and nonenzymatic hydrolysis. Anal. Biochem. 1987;162:115–121. doi: 10.1016/0003-2697(87)90015-7. [DOI] [PubMed] [Google Scholar]
- Pouillon V, Hascakova-Bartova R, Pajak B, Adam E, Bex F, Dewaste V, Van Lint C, Leo O, Erneux C, Schurmans S. Inositol 1,3,4,5-tetrakisphosphate is essential for T lymphocyte development. Nat. Immunol. 2003;4:1136–1143. doi: 10.1038/ni980. [DOI] [PubMed] [Google Scholar]
- Prestwich GD. Phosphoinositide signaling: From affinity probes to pharmaceutical targets. Chem. Biol. 2004;11:619–637. doi: 10.1016/j.chembiol.2004.03.025. [DOI] [PubMed] [Google Scholar]
- Prestwich GD. Visualization and perturbation of phosphoinositide and phospholipid signaling. Prostaglandins Other Lipid Mediat. 2005;77:168–178. doi: 10.1016/j.prostaglandins.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Puls KL, Hogquist KA, Reilly N, Wright MD. CD53, a thymocyte selection marker whose induction requires a lower affinity TCR-MHC interaction than CD69, but is up-regulated with slower kinetics. Int. Immunol. 2002;14:249–258. doi: 10.1093/intimm/14.3.249. [DOI] [PubMed] [Google Scholar]
- Rathmell JC, Vander Heiden MG, Harris MH, Frauwirth KA, Thompson CB. In the absence of extrinsic signals, nutrient utilization by lymphocytes is insufficient to maintain either cell size or viability. Mol. Cell. 2000;6:683–692. doi: 10.1016/s1097-2765(00)00066-6. [DOI] [PubMed] [Google Scholar]
- Resnick AC, Saiardi A. Inositol polyphosphate multikinase: Metabolic architect of nuclear inositides. Front. Biosci. 2008;13:856–866. doi: 10.2741/2726. [DOI] [PubMed] [Google Scholar]
- Rusten TE, Stenmark H. Analyzing phosphoinositides and their interacting proteins. Nat. Methods. 2006;3:251–258. doi: 10.1038/nmeth867. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Bhandari R, Resnick AC, Snowman AM, Snyder SH. Phosphorylation of proteins by inositol pyrophosphates. Science. 2004;306:2101–2105. doi: 10.1126/science.1103344. [DOI] [PubMed] [Google Scholar]
- Sakaguchi R, Endoh T, Yamamoto S, Tainaka K, Sugimoto K, Fujieda N, Kiyonaka S, Mori Y, Morii T. A single circularly permuted GFP sensor for inositol-1,3,4,5-tetrakisphosphate based on a split PH domain. Bioorg. Med. Chem. 2009;17:7381–7386. doi: 10.1016/j.bmc.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Sato M, Ueda Y, Takagi T, Umezawa Y. Production of PtdInsP3 at endomembranes is triggered by receptor endocytosis. Nat. Cell Biol. 2003;5:1016–1022. doi: 10.1038/ncb1054. [DOI] [PubMed] [Google Scholar]
- Schenk U, Westendorf AM, Radaelli E, Casati A, Ferro M, Fumagalli M, Verderio C, Buer J, Scanziani E, Grassi F. Purinergic control of T cell activation by ATP released through pannexin-1 hemichannels. Sci. Signal. 2008;1:ra6. doi: 10.1126/scisignal.1160583. [DOI] [PubMed] [Google Scholar]
- Schwartzberg PL, Finkelstein LD, Readinger JA. TEC-family kinases: Regulators of T-helper-cell differentiation. Nat. Rev. Immunol. 2005;5:284–295. doi: 10.1038/nri1591. [DOI] [PubMed] [Google Scholar]
- Seeds AM, York JD. Inositol polyphosphate kinases: Regulators of nuclear function. Biochem. Soc. Symp. 2007;2007:183–197. doi: 10.1042/BSS0740183. [DOI] [PubMed] [Google Scholar]
- Seeds AM, Frederick JP, Tsui MM, York JD. Roles for inositol polyphosphate kinases in the regulation of nuclear processes and developmental biology. Adv. Enzyme Regul. 2007;47:10–25. doi: 10.1016/j.advenzreg.2006.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant S, McPhail LC. Measurement of phospholipid metabolism in intact neutrophils. Methods Mol. Biol. 2007;412:69–83. doi: 10.1007/978-1-59745-467-4_6. [DOI] [PubMed] [Google Scholar]
- Shan X, Czar MJ, Bunnell SC, Liu P, Liu Y, Schwartzberg PL, Wange RL. Deficiency of PTEN in Jurkat T cells causes constitutive localization of Itk to the plasma membrane and hyperresponsiveness to CD3 stimulation. Mol. Cell Biol. 2000;20:6945–6957. doi: 10.1128/mcb.20.18.6945-6957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB. Understanding the biological significance of diphosphoinositol polyphosphates (‘inositol pyrophosphates’) Biochem. Soc. Symp. 2007;2007:211–221. doi: 10.1042/BSS0740211. [DOI] [PubMed] [Google Scholar]
- Singh AK, Jiang Y. Quantitative chromatographic analysis of inositol phospholipids and related compounds. J. Chromatogr. B Biomed. Appl. 1995;671:255–280. doi: 10.1016/0378-4347(94)00558-m. [DOI] [PubMed] [Google Scholar]
- Skippen A, Swigart P, Cockcroft S. Measurement of phospholipase C by monitoring inositol phosphates using [3H]-inositol-labeling protocols in permeabilized cells. Methods Mol. Biol. 2006;312:183–193. [PubMed] [Google Scholar]
- Sommers CL, Samelson LE, Love PE. LAT: A T lymphocyte adapter protein that couples the antigen receptor to downstream signaling pathways. Bioessays. 2004;26:61–67. doi: 10.1002/bies.10384. [DOI] [PubMed] [Google Scholar]
- Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu. Rev. Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- Stevenson-Paulik J, Chiou ST, Frederick JP, dela Cruz J, Seeds AM, Otto JC, York JD. Inositol phosphate metabolomics: Merging genetic perturbation with modernized radiolabeling methods. Methods. 2006;39:112–121. doi: 10.1016/j.ymeth.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Stewart SJ, Prpic V, Powers FS, Bocckino SB, Isaacks RE, Exton JH. Perturbation of the human T cell antigen receptor-T3 complex leads to the production of inositol tetrakisphosphate: Evidence for conversion from inositol trisphosphate. Proc. Natl. Acad. Sci. U.S.A. 1986;83:6098–6102. doi: 10.1073/pnas.83.16.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SJ, Kelley LL, Powers FS. Production of inositol pentakisphosphate in a human T lymphocyte cell line. Biochem. Biophys. Res. Commun. 1987;145:895–902. doi: 10.1016/0006-291x(87)91049-7. [DOI] [PubMed] [Google Scholar]
- Stokes AJ, Shimoda LM, Lee JW, Rillero C, Chang YT, Turner H. Fcepsilon RI control of Ras via inositol (1,4,5) trisphosphate 3-kinase and inositol tetrakisphosphate. Cell Signal. 2006;18:640–651. doi: 10.1016/j.cellsig.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Takazawa K, Lemos M, Delvaux A, Lejeune C, Dumont JE, Erneux C. Rat brain inositol 1,4,5-trisphosphate 3-kinase. Ca2(+)-sensitivity, purification and antibody production. Biochem. J. 1990;268:213–217. doi: 10.1042/bj2680213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman EF, Kirakossian H, Switchenko AC, Ishkanian J, Ericson M, Wartchow CA, Pirio M, Pease J, Irvin BR, Singh S, Singh R, Patel R, Dafforn A, Davalian D, Skold C, Kurn N, Wagner DB. Luminescent oxygen channeling assay (LOCI): Sensitive, broadly applicable homogeneous immunoassay method. Clin.Chem. 1996;42:1518–1526. [PubMed] [Google Scholar]
- van der Kaay J, Batty IH, Cross DA, Watt PW, Downes CP. Anovel, rapid, and highly sensitive mass assay for phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3) and its application tomeasure insulin-stimulated PtdIns(3,4,5)P3 production in rat skeletal muscle in vivo. J. Biol. Chem. 1997;272:5477–5481. doi: 10.1074/jbc.272.9.5477. [DOI] [PubMed] [Google Scholar]
- van der Kaay J, Cullen PJ, Downes CP. Phosphatidylinositol(3,4,5)trisphosphate (Ptdins(3,4,5)P3) mass measurement using a radioligand displacement assay. Methods Mol. Biol. 1998;105:109–125. doi: 10.1385/0-89603-491-7:109. [DOI] [PubMed] [Google Scholar]
- Vats P, Bhushan B, Chakarborti AK, Banerjee UC. Separation and identification of enzymatically prepared dephosphorylated products of myo-inositolhexakisphosphate using LC-MS. J. Sep. Sci. 2008;31:3829–3833. doi: 10.1002/jssc.200800372. [DOI] [PubMed] [Google Scholar]
- Verhoeven AJ, Tysnes OB, Horvli O, Cook CA, Holmsen H. Stimulation of phosphate uptake in human platelets by thrombin and collagen. Changes in specific 32P labeling of metabolic ATP and polyphosphoinositides. J. Biol. Chem. 1987;262:7047–7052. [PubMed] [Google Scholar]
- Weernink PAO, Schulte P, Guo Y, Wetzel J, Amano M, Kaibuchi K, Haverland S, Vo M, Schmidt M, Mayr GW, Jakobs KH. Stimulation of phosphatidylinositol-4-phosphate 5-kinase by rho-kinase. J. Biol. Chem. 2000;275:10168–10174. doi: 10.1074/jbc.275.14.10168. [DOI] [PubMed] [Google Scholar]
- Wen BG, Pletcher MT, Warashina M, Choe SH, Ziaee N, Wiltshire T, Sauer K, Cooke MP. Inositol (1,4,5) trisphosphate 3 kinase B controls positive selection of T cells and modulates Erk activity. Proc. Natl. Acad. Sci. U.S.A. 2004;101:5604–5609. doi: 10.1073/pnas.0306907101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk MR, Lucast L, Di Paolo G, Romanelli AJ, Suchy SF, Nussbaum RL, Cline GW, Shulman GI, McMurray W, De Camilli P. Phosphoinositide profiling in complex lipid mixtures using electrospray ionization mass spectrometry. Nat. Biotechnol. 2003;21:813–817. doi: 10.1038/nbt837. [DOI] [PubMed] [Google Scholar]
- Williams A, Frasca V. Ion-exchange chromatography. Curr. Protoc. Mol. Biol. 1998;44:10.10.1–10.10.30. doi: 10.1002/0471142727.mb1010s44. [DOI] [PubMed] [Google Scholar]
- Wreggett KA, Howe LR, Moore JP, Irvine RF. Extraction and recovery of inositol phosphates from tissues. Biochem. J. 1987;245:933–934. doi: 10.1042/bj2450933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreggett KA, Lander DJ, Irvine RF. Two-stage analysis of radiolabeled inositol phosphate isomers. Methods Enzymol. 1990;191:707–718. doi: 10.1016/0076-6879(90)91043-6. [DOI] [PubMed] [Google Scholar]
- Zeidman R, Lofgren B, Pahlman S, Larsson C. PKCepsilon, via its regulatory domain and independently of its catalytic domain, induces neurite-like processes in neuroblastoma cells. J. Cell Biol. 1999;145:713–726. doi: 10.1083/jcb.145.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Brandish PE, Kolodin DG, Scolnick EM, Strulovici B. High-throughput cell-based screening using scintillation proximity assay for the discovery of inositol phosphatase inhibitors. J. Biomol. Screen. 2004;9:132–140. doi: 10.1177/1087057103261039. [DOI] [PubMed] [Google Scholar]
- Zilberman Y, Howe LR, Moore JP, Hesketh TR, Metcalfe JC. Calcium regulates inositol 1,3,4,5-tetrakisphosphate production in lysed thymocytes and in intact cells stimulated with concanavalin A. Embo J. 1987;6:957–962. doi: 10.1002/j.1460-2075.1987.tb04845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]