Abstract
Synthesis of novel HIV-1 protease inhibitors incorporating dioxatriquinane-derived P2-ligands is described. The tricyclic ligand alcohol contains five contiguous chiral centers. The ligand alcohols were prepared in optically active form by an enzymatic asymmetrization of mesodiacetate, cascade radical cyclization, and Lewis acid catalyzed reduction as the key steps. Inhibitors with dioxatriquinane-derived P2-ligands exhibited low nanomolar HIV-1 protease activity.
Keywords: HIV-1 protease, Inhibitor, Cascade, Radical, Dioxatriquinane
The introduction of HIV-1 protease inhibitors into antiretroviral therapy (ART) has greatly improved the outlook for patients with HIV infection and AIDS.1,2 However, the emergence of multidrug resistant HIV-1 variants has raised major concerns about the prospects of long-term treatment options.3,4 Consequently, there is a critical need for more effective protease inhibitors for the treatment of the growing number of treatment-experienced HIV/AIDS patients carrying multidrug-resistant HIV-1 strains.[4,5] In an effort to combat drug resistance, our structure-based design strategy targeting the protein backbone has led to the design and discovery of a variety of novel HIV-1 protease inhibitors. This includes FDA approved inhibitor darunavir (1, Figure 1), which is an exceptionally potent inhibitor that exhibits broad-spectrum activity against multidrug-resistant HIV-1 variants.5-8
Figure 1.
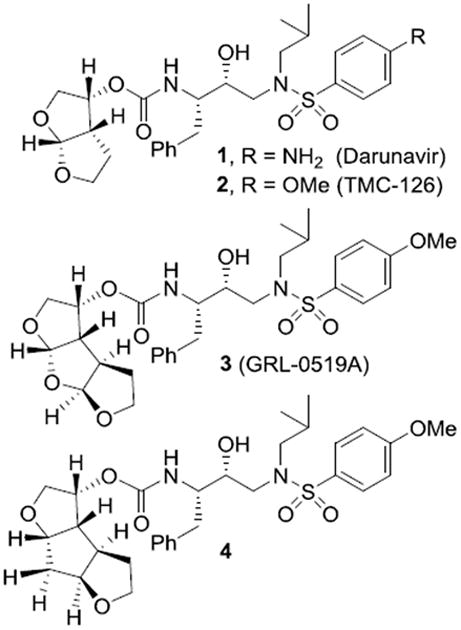
Structure of darunavir (1) and PIs 2-4.
Our inhibitor design strategy involves maximizing inhibitor-HIV-1 protease interactions in the active-site, particularly promoting extensive hydrogen bond interactions with the protein backbone atoms.9,10 The detailed X-ray structural analyses of darunavir-bound protease complexes revealed a series of conserved interactions between the inhibitor and key backbone atoms of HIV-1 protease. This backbone-binding molecular design strategy corroborated observed superior drug-resistance properties of darunavir and its P2' 4-methoxysulfonamide derivative (2, TMC-126) compared to other FDA approved inhibitors.8, 11
Subsequently, based upon examination of the protein-ligand X-ray structure of darunavir-bound HIV-1 protease, we have designed fused tris-tetrahydrofuan (tris-THF) as the P2-ligand and incorporated it in inhibitor 3 in order to promote additional interactions in the HIV-1 protease active site.12 Indeed, inhibitor 3 with a syn-anti-syn-fused tris-THF ligand exhibited nearly 10-fold improvement of potency against highly resistant clinical HIV-1 strains compared to darunavir. The corresponding inhibitor with a syn-syn-syn-fused tris-THF ligand also provided very potent inhibitor, however, it was somewhat less effective than inhibitor 3.12,13 Interestingly, our recent work indicated that the top oxygen of tris-THF may not be critical to the overall potency of the inhibitor.14 In an effort to ascertain the contribution of the middle oxygen of the tris-THF ligand toward the ligand binding site interactions, we sought to synthesize the corresponding syn-anti-syn- and syn-syn-syn-fused 1,6- dioxatriquinane derivatives and compare enzyme inhibitory activity of the analogs with inhibitor 3. Herein, we report our enantioselective syntheses of (3R,3aS,3bR,6aS,7aS)-octahydro-2H-cyclopenta[1,2-b:4,3-b′]-bis-furan-3-ol and (3R,3aS,3bS,6aR,7aS)-octahydro-2H-cyclopenta[1,2-b:4,3-b′]-bis-furan-3-ol using cascade radical cyclization and enzyme-catalyzed desymmetrization as the key steps. We have then incorporated these ligands into HIV-1 protease inhibitors and evaluated their inhibitory potency.
For the target inhibitor 4, the synthesis of the corresponding syn-anti-syn-fused 1,6-dioxatriquinane structural template, (3R,3aS,3bR,6aS,7aS)-octahydro-2H-cyclopenta-[1,2-b:4,3-b′]-bis-furan-3-ol, is shown in Scheme 1. This tricyclic ligand alcohol contains five contiguous chiral centers. We planned to utilize a cascade radical cylization for the synthesis of this tricyclic framework with defined configuration. The efficiency of such related cascade radical cylization was previously demonstrated by Curran and Rakiewicz during the synthesis of hirsutine, a triquinane system.15,16 The synthesis of a related 1,6-dioxatriquinane derivative was also reported by Hanessian and Leger using a radical cyclization.17 For our present work, we planned to employ a haloacetal radical cyclization strategy pioneered by Stork and co-workers.18,19 Our synthesis began with the preparation of monoacetate 5 in multigram quantity using enzymatic asymmetrization of meso-diacetate with acetyl cholinesterase as described previously.20 Formation of the Mosher ester of 5 revealed that the enantiomeric purity of 5 was 95% ee.21,22 The hydroxyl group was protected as the TBS-ether with TBSCl in the presence of imidazole in THF in near quantitative yield. Hydrolysis of the resulting acetate afforded the alcohol 6 in quantitative yield. To set the bottom tetrahydrofuran stereochemistry, alcohol 6 was subjected to Mitsunobu inversion23 with diisopropyl azodicarboxylate (DIAD), triphenylphosphine, and p-nitrobenzoic acid to provide the corresponding nitrobenzoate derivative. Ester hydrolysis with K2CO3 in methanol afforded alcohol 7 in 91% yield in two steps.
Scheme 1. Synthesis of syn-anti-syn-fused dioxatriquinane ring system.
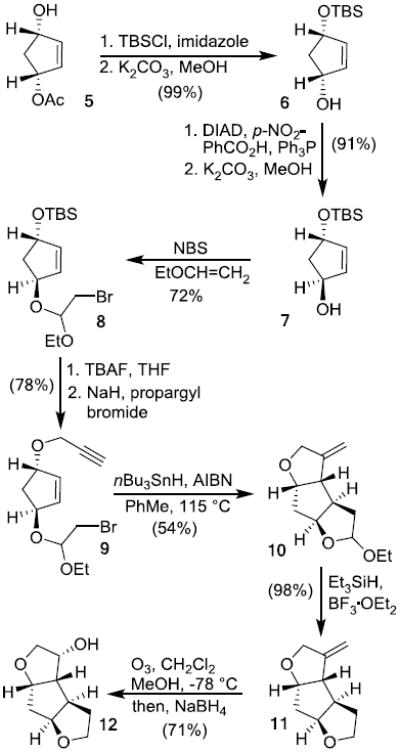
Treatment of alcohol 7 with NBS and ethyl vinyl ether in CH2Cl2 afforded bromo acetal 8 as a mixture (1:1) of diastereomers in 72% yield. Removal of the TBS group with nBu4N+F- (TBAF) in THF provided the corresponding alcohol. Treatment of the resulting mixture of alcohol with NaH and propargyl bromide in the presence of nBu4N+I- (TBAI) furnished the radical cyclization precursor 9 in 78% yield in two steps. The cascade cyclization using tri-n-butyltin hydride in refluxing toluene in the presence of AIBN resulted in a mixture (1:1) of tricyclic alkene derivative 10 in 54% yield. Reduction of acetal 10 with Et3SiH in the presence of BF3.OEt2 afforded tricyclic alkene 11 as a single isomer in 98% yield.24 Ozonolytic cleavage of 11 at -78 °C followed by reduction of the resulting ketone with NaBH4 at -15 °C furnished endo alcohol 12 as single isomer in 71% yield.
The synthesis of 1,6-dioxatriquinane alcohol with syn- syn-syn-fused structural motif is shown in Scheme 2. The overall strategy is similar to the synthesis of alcohol 12. As shown, optically active TBS-alcohol 6 was converted to bromo acetal 13 utilizing ethyl vinyl ether and NBS in CH2Cl2. Removal of the TBS-group followed by treatment of the resulting alcohol with NaH and propargyl bromide in the presence of TBAI gave the cyclization precursor 14 in 85% yield in two steps. The cascade cyclization using tri-n-butyltin hydride as described above generated the tricyclic alkene 15 in 79% yield. Acetal reduction with Et3SiH in the presence of BF3.OEt2 provided the alkene 16 in 76% yield.24 Ozonolytic cleavage followed by NaBH4 reduction afforded optically active syn-syn-syn-fused (3R,3aS,3bS, 6aR,7aS)-octahydro-2H-cyclopenta[1,2-b:4,3-b′]-bis-furan-3-ol (17) in 85% yield.
Scheme 2. Synthesis of syn-syn-syn-fused ring system.
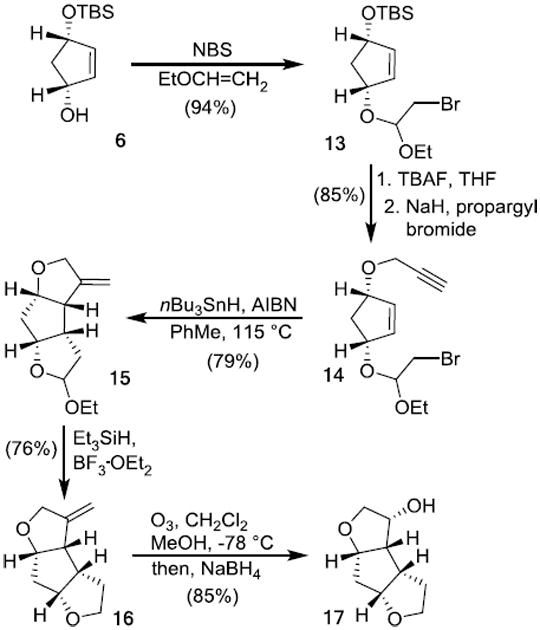
The stereochemical assignment of alcohols 12 and 17 was supported by 1H-NMR NOESY experiments (Figure 2). The observed NOE between Ha-Hb, Hb-Hc, and Hd-He for compound 12 is in line with the assigned syn-anti-syn-fused tricyclic ring system. Similarly, the observed NOE between Ha-Hb, Hb-Hc, Hd-He, and Hc-He supported the syn-syn-syn- fused tricyclic ring system for compound 17.
Figure 2. 1H NOESY analysis of alcohols 12 and 17.
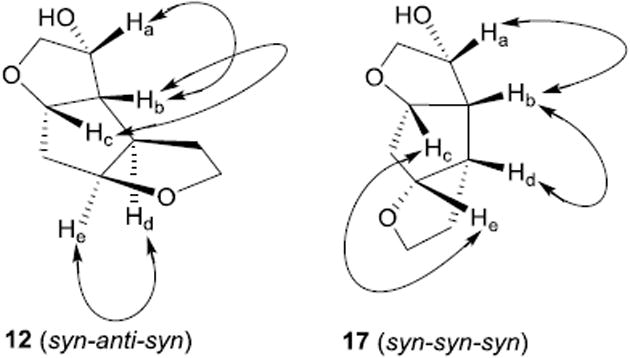
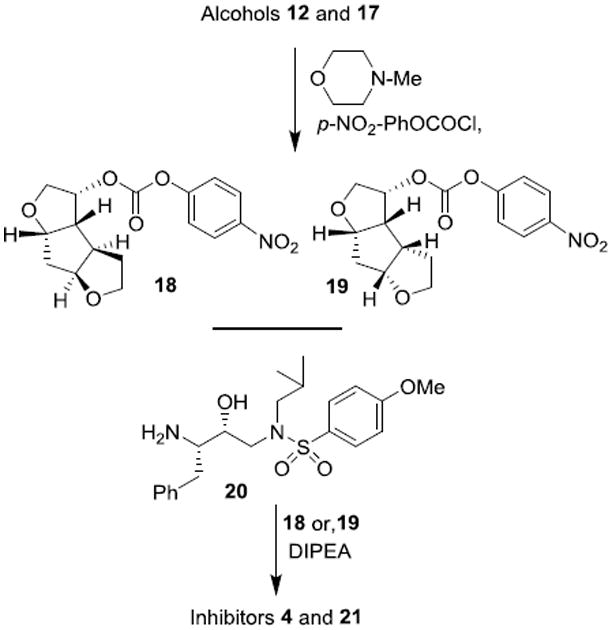
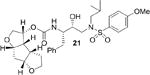
The syntheses of inhibitors 4 and 21 are outlined in Scheme 3. Optically active ligand alcohols 12 and 17 were reacted with p-nitrophenyl chloroformate in the presence of N-methyl morpholine in CH2Cl2 at 23°C for 12 h to provide carbonates 18 and 19 in 60% and 62% yield respectively.14 Reaction of these activated carbonates with amine 20 in the presence of diisopropylethylamine (DIPEA) in CH2Cl2 at 23°C for 12 h furnished inhibitors 4 and 21 in 65% yield.25 These compounds showed satisfactory analytical data. Inhibitor purity was determined by reverse HPLC analysis and purity was >98% as determined by HPLC assay.26
Scheme 3. Synthesis of inhibitors 4 and 21.
Inhibitors incorporating dioxatriquinane-derived P2-ligands exhibited very potent HIV-1 protease inhibitory activity. Inhibitor structures and activity are shown in Table 1. We utilized the enzyme inhibitory assay protocol developed by Toth and Marshall27 and the Ki values denote the mean values of at least six determinations. As can be seen, inhibitor 4, with a stereochemically defined syn-anti-syn- fused (3R,3aS,3bR,6aS,7aS)-octahydro-2H-cyclopenta-[1,2- b:4,3-b′]-bis-furan-3-ol-derived urethane as the P2-ligand and p-methoxysulfonamide as the P2′-ligand, showed enzymatic Ki value of 1.39 nM. Inhibitor 21 with syn-syn-syn-fused (3R,3aS,3bS,6aR,7aS)-octahydro-2H-cyclopenta[1,2-b:4,3-b′]-bis-furan-3-ol-derived urethane also displayed a comparable enzymatic Ki value of 1.43 nM. Both compounds are very potent, however the inhibitory activity is significantly lower than bis-THF-derived inhibitor 2 (TMC-126) or syn-anti-syn-fused tris-THF-derived inhibitor 3. The difference in activity is likely due to the absence of oxygen in the dioxatriquinane-derived P2-ligands.28 While the P2-ligands in inhibitors 3 and 4 have similar steric features, the hydrogen bonding abilty of the second oxygen in the tris-THF is critical to potency.12,13
Table 1. HIV-1 Protease Inhibitory Activity of compounds.
| Entry | Inhibitor | Ki (nM) |
|---|---|---|
| 1 |
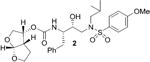
|
0.014 |
| 2 |
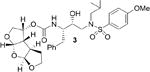
|
0.006 |
| 3 |
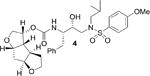
|
1.39 |
| 4 |
|
1.43 |
| (Darunavir displayed Ki = 0.016 nM in this assay) |
In conclusion, we have reported novel HIV-1 protease inhibitors incorporating dioxatriquinane-derived P2-ligands. Synthesis of the tricyclic ligand alcohols is carried out stereoselectively in an optically active form. The synthesis features an enzymatic asymmetrization of mesodiacetate, a highly effienct cascade radical cyclization, and Lewis acid catalyzed reduction as the key steps. Inhibitors containing dioxatriquinane-derived ligands showed very good HIV-1 protease inhibitory activity, however the observed activity is significantly lower than syn-anti-syn-fused tris-THF- derived inhibitor 3. The data demonstrates the importance of the ligand oxygen in ligand-binding site interactions. Further chemical modifications are currently underway in our laboratory.
Supplementary Material
Acknowledgments
Financial support by the National Institutes of Health (GM53386) is gratefully acknowledged.
Footnotes
This paper is dedicated to the memory of Professor Harry Wasserman of Yale University, a beloved teacher, scholar, artist, and editor who made a great difference in so many of our lives.
Supplementary Data: Supplementary data associated with this article can be found in the online version.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sepkowitz KA. N Engl J Med. 2001;344:1764–1772. doi: 10.1056/NEJM200106073442306. [DOI] [PubMed] [Google Scholar]
- 2.Flexner C. N Engl J Med. 1998;338:1281–1292. doi: 10.1056/NEJM199804303381808. [DOI] [PubMed] [Google Scholar]
- 3.Waters L, Nelson M. Int J Clin Pract. 2007;61:983–990. doi: 10.1111/j.1742-1241.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- 4.Yerly S, Kaiser L, Race E, Bru JP, Clavel F, Perrin L. Lancet. 1999;354:729–733. doi: 10.1016/S0140-6736(98)12262-6. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh AK, Dawson ZL, Mitsuya H. Bioorg Med Chem. 2007;15:7576–7580. doi: 10.1016/j.bmc.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh AK, Kincaid JF, Cho W, Walters DE, Krishnan K, Hussain KA, Koo Y, Cho H, Rudall C, Holland L, Buthod J. Bioorg Med Chem Lett. 1998;8:687–690. doi: 10.1016/s0960-894x(98)00098-5. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh AK, Sridhar PR, Kumaragurubaran N, Koh Y, Weber IT, Mitsuya H. ChemMedChem. 2006;1:939–950. doi: 10.1002/cmdc.200600103. [DOI] [PubMed] [Google Scholar]
- 8.Yoshimura K, Kato R, Kavlick MF, Nguyen A, Maroun V, Maeda K, Hussain KA, Ghosh AK, Gulnik SV, Erickson JW, Mitsuya H. J Virol. 2002;76:1349–1358. doi: 10.1128/JVI.76.3.1349-1358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh AK, Chapsal BD, Weber IT, Mitsuya H. Acc Chem Res. 2008;41:78–86. doi: 10.1021/ar7001232. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh AK, Anderson DD, Weber IT, Mitsuya H. Angew Chem Int Ed. 2012;51:1778–1802. doi: 10.1002/anie.201102762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koh Y, Nakata H, Maeda K, Ogata H, Bilcer G, Devasamudram T, Kincaid JF, Boross P, Wang YF, Tie Y, Volarath P, Gaddis L, Harrison RW, Weber IT, Ghosh AK, Mitsuya H. Antimicrob Agents Chemother. 2003;47:3123–3129. doi: 10.1128/AAC.47.10.3123-3129.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh AK, Xu CX, Rao KV, Baldridge A, Agniswamy J, Wang YF, Weber IT, Aoki M, Miguel SGP, Amano M, Mitsuya H. ChemMedChem. 2010;5:1850–1854. doi: 10.1002/cmdc.201000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amano M, Tojo Y, Salcedo-Gomez PM, Campbell JR, Das D, Aoki M, Xu CX, Rao KV, Ghosh AK, Mitsuya H. Antimicrob Agents Chemother. 2013;57:2036–2046. doi: 10.1128/AAC.02189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh AK, Parham GL, Martyr CD, Nyalapatla PR, Osswald HL, Agniswamy J, Wang YF, Amano M, Weber IT, Mitsuya H. J Med Chem. 2013;56:6792–6802. doi: 10.1021/jm400768f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curran DP, Rakiewicz DM. J Am Chem Soc. 1985;107:1448–1449. [Google Scholar]
- 16.Curran DP, Rakiewicz DM. Tetrahedron. 1985;41:3943–3958. [Google Scholar]
- 17.Hanessian S, Leger R. J Am Chem Soc. 1992;114:3115–3117. [Google Scholar]
- 18.Stork G, Mook R. J Am Chem Soc. 1983;105:3720–3722. [Google Scholar]
- 19.Stork G, Mook R, Biller SA, Rychnovsky SD. J Am Chem Soc. 1983;105:3741–3742. [Google Scholar]
- 20.Deardorff DR, Windham CQ, Craney CL. Org Synth. 1998;9:487–492. [Google Scholar]
- 21.The Mosher ester was formed by reaction of Mosher acid and alcohol 5 with EDCI in the presence of DMAP. The 19F-NMR analysis of Mosher ester19 revealed an enantiomeric purity >95%.
- 22.Dale JA, Dull DL, Mosher HS. J Org Chem. 1969;34:2543–2549. [Google Scholar]
- 23.Martin SF, Dodge JA. Tetrahedron Lett. 1991;32:3017–3020. [Google Scholar]
- 24.Ghosh AK, Sridhar PR, Leshchenko S, Hussain AK, Li J, Kovalevsky AY, Walters DE, Wedekind JE, Grum-Tokars V, Das D, Koh Y, Maeda K, Gatanaga H, Weber IT, Mitsuya H. J Med Chem. 2006;49:5252–5261. doi: 10.1021/jm060561m. [DOI] [PubMed] [Google Scholar]
- 25.For more details, please see supporting information section.
- 26.HPLC conditions:column, YMC pack ODS-A;flow rate, 1 mL/min; 25°C, 256 nM detection, solvent, MeCN:H2O 90:10; retention time for compound 4, 13.40 min; retention time for compound 21, 14.85 min.
- 27.Toth MV, Marshall GR. Int J Pep Prot Res. 1990;36:544–550. doi: 10.1111/j.1399-3011.1990.tb00994.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Wang YF, Shen CH, Agniswamy J, Rao KV, Xu CX, Ghosh AK, Harrison RW, Weber IT. J Med Chem. 2013;56:1074–1083. doi: 10.1021/jm301519z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


