Abstract
Microarray-based profiling represents an effective method to analyze cellular or tissue-specific gene expression on the genome-level. However, in comparative analyses between control and mutant samples, microarrays often identify a large number of differentially expressed genes, in turn making it challenging to isolate the select “high-priority candidates” that are most relevant to an observed mutant phenotype. Here, we describe an integrative approach for mouse mutant lens microarray gene expression analysis using publically accessible systems-level information such as wild-type mouse lens expression data in iSyTE (integrated Systems Tool for Eye gene discovery), protein–protein interaction data in public databases, gene ontology enrichment data, and transcription factor binding profile data. This strategy, when applied to small Maf Mafg −/−:Mafk +/− mouse lens microarray datasets (deposited in NCBI Gene Expression Omnibus database with accession number GSE65500) in Agrawal et al. 2015 [1], led to the effective prioritization of candidate genes linked to lens defects in these mutants. Indeed, from the original list of genes that are differentially expressed at ± 1.5-fold and p < 0.05 in Mafg −/−:Mafk +/− mutant lenses, this analysis led to the identification of thirty-six high-priority candidates, in turn reducing the number of genes for further study by approximately 1/3 of the total. Moreover, eight of these genes are linked to mammalian cataract in the published literature, validating the efficacy of this approach. Additionally, these high-priority candidates contribute valuable information for the assembly of a gene regulatory network in the lens. In sum, the pipeline outlined in this report represents an effective approach for initial as well as downstream microarray expression data analysis to identify genes important for lens biology and cataracts. We anticipate that this integrative strategy can be extended to prioritize phenotypically relevant candidate genes from microarray data in other cells and tissues.
Keywords: Cataract, Lens, Microarrays, iSyTE, Gene expression
| Specifications | |
|---|---|
| Organism/cell line/tissue | Mus musculus; 60 day post-natal lens tissue |
| Sex | N/A |
| Sequencer or array type | MouseWG-6 v2.0 Expression BeadChip arrays (Illumina) |
| Data format | Raw |
| Experimental factors | Mafg −/−:Mafk +/− (test) vs. Mafg +/−:Mafk +/− (control) (Both on mixed background, with contributions from the 129Sv/J, C57BL/6J, and ICR strains) |
| Experimental features | Identification of differentially expressed genes in the lens of mice lacking two copies of Mafg and one copy of Mafk that exhibit lens defects with age compared to lens of mice lacking one copy of Mafg and one copy of Mafk that are normal |
| Consent | N/A |
| Sample source location | Newark, Delaware, USA |
1. Direct link to deposited data
Deposited data can be found at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65500.
2. Experimental design, materials and methods
2.1. Experimental design
The eye gene discovery tool iSyTE [2] predicted that the small Maf transcription factor genes Mafg and Mafk are highly enriched and expressed, respectively, in the mouse lens, and may potentially function in its development or homeostasis. Indeed, we find that Mafg −/−:Mafk +/− compound mouse mutants exhibit lens defects including cataracts [1]. To investigate the molecular changes associated with this phenotype, we performed microarray-based gene expression profiling on lens tissue obtained from Mafg −/−:Mafk +/− (“test” animals that exhibit lens defects) and Mafg +/−:Mafk +/− (“control” animals that do not exhibit lens defects) mice. These microarray datasets were analyzed to identify differentially expressed genes (DEGs) that were subjected to further integrated analysis using a workflow pipeline that incorporates wild-type lens expression data from the iSyTE database (http://bioinformatics.udel.edu/Research/iSyTE), chromatin immunoprecipitation sequencing (ChIP-Seq) data on Mafg and its co-regulatory protein Nrf2, protein–protein interactions from open resource databases, as well as gene ontology enrichment and cis-regulatory motif analyses (Fig. 1). Together, this approach led to the prioritization of genes most relevant to the phenotype, and to the assembly of a small Maf gene regulatory network in the lens. Below, we describe these analytical procedures in detail.
Fig. 1.
Workflow of microarray design, data pre-processing and analysis of differentially expressed genes.
2.2. Microarrays
Lenses were collected on post-natal day (P) 60 from Mafg −/−:Mafk +/− (test) and Mafg +/−:Mafk +/− (control) mice for microarray-based gene expression profiling. Selection of the stage for genome-wide expression profiling of mutant tissue represents an important step, because this data influences the proper interpretation of the phenotype as well as the directions of future experiments. For example, microarray profiling performed at a stage when the phenotype is already evident, and therefore well manifested, may provide differential expression data that is potentially “contaminated” with secondary gene expression changes. This presents a challenge to resolve primary expression changes from such secondary expression changes, in turn making it difficult to correlate genetic perturbations to the observed phenotype. To address this issue, the age of the animals in this analysis was selected as P60 (2 months) because at this stage, Mafg −/−:Mafk +/− compound mutants do not exhibit any overt abnormalities in the lens (Fig. 2A–B’), which are observed in these mutants at later stages (Fig. 2C–D’). Thus, analysis of lenses at P60 increases the likelihood of detecting changes in mutant gene expression that occur prior to the onset of the overt phenotype and therefore represent primary alterations. Total RNA was isolated from both test and control lenses in biological duplicates using the RNeasy Mini Kit (Qiagen). Microarrays were performed on MouseWG-6 v2.0 Expression BeadChip arrays (Illumina) using manufacturer recommended hybridization conditions. Microarray chips were scanned using the Illumina BeadArray reader.
Fig. 2.
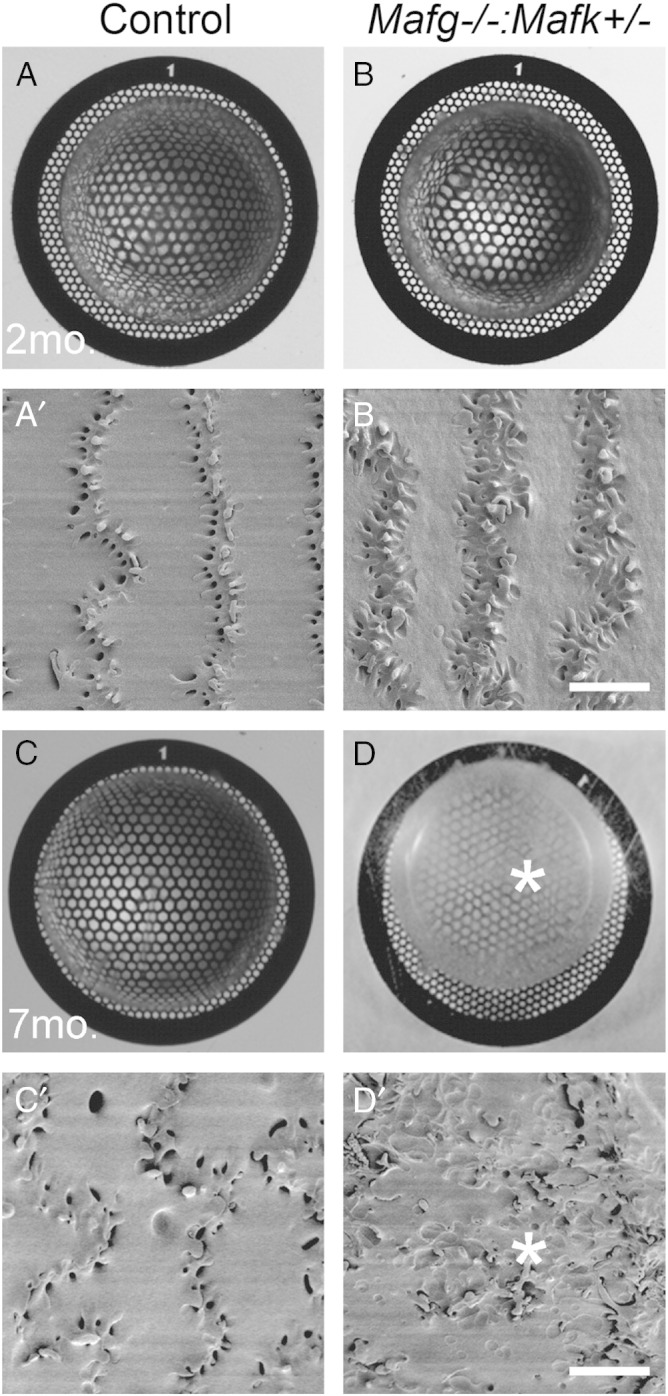
Selection of mutant stage for lens microarray analysis. Light microscopy based grid imaging of lenses from (A) control and (B) Mafg −/−:Mafk +/− compound mutant mice demonstrates no opacities at post-natal stage (P)60 or 2 months (2 mo.). High-resolution scanning electron microscopy (SEM) analysis of lenses from (A’) control and (B’) Mafg −/−:Mafk +/− compound mutant mice confirm the absence of overt abnormalities in mutant fiber cells at stage 2 mo. However, grid imaging analysis at age 7 months (7 mo.) demonstrates that while (C) control mice have transparent lenses, (D) Mafg −/−:Mafk +/− compound mutant mice exhibit lens opacities (asterisk). SEM analysis at age 7 months demonstrates that while (C’) Control lenses have normal fiber cells, (D’) Mafg −/−:Mafk +/− compound mutant mice exhibit severe fiber cell defects (asterisk). Based on this analysis, the age P60 (2 mo.), when Mafg −/−:Mafk +/− compound mutant mice do not exhibit overt lens defects, was selected as the stage to perform microarrays. This is based on the consideration that microarrays at P60 will increase the likelihood of detecting gene expression changes that reflect primary alterations prior to the manifestation of overt defects that occur with age. Scale bar in B’ and D’ is 10 μm.
2.3. Microarray data quality assessment
To assess the quality of microarray expression data, we used inbuilt ‘R’ functions and performed Principle Component Analysis (PCA), derived boxplots, and analyzed histograms of the array intensities of raw unprocessed and normalized processed (see below) data (Fig. 3). We observed high level of background noise in raw mutant and control datasets in all quality plots. PCA plot from processed data showed clear separation of control and mutant datasets (Fig. 3A–B). Similar results were obtained with histograms (Fig. 3C–D) and boxplots (Fig. 3E–F) analysis of raw and processed data.
Fig. 3.
Microarray expression data quality assessment plots. Comparisons of raw unprocessed and normalized processed expression intensities between arrays of mutant and control datasets. PCA plots from (A) raw and (B) processed datasets. Histograms for (C) raw and (D) processed datasets. Boxplot for (E) raw and (F) processed datasets. Key to samples in each type of analysis is given below.
2.4. Microarray data preprocessing and analysis
Microarray data processing and analysis was performed under the ‘R’ statistical environment (http://www.r-project.org/) using lumi package for Illumina microarray data, available through Bioconductor (www.bioconductor.org). The workflow for microarray data analysis is described in detail (Fig. 1). As first step, raw output files from Illumina Bead Studio toolkit were read using lumi [3] to generate a lumiBatch object through lumiR.batch function. These imported files were then preprocessed using lumi built-in methods, bgAdjust which corrects raw Illumina probe intensities followed by rankinvariant for normalizing these corrected intensities. The choice of rankinvariant was made because only a small number of genes were expected to be differentially expressed, and Illumina recommends this normalizing method for such experiments. After pre-processing, presence–absence calls were generated using built-in function of lumi, and probe sets with detection p-value of ≤ 0.05 in at least two samples were considered as significantly present and used for further downstream analysis. Finally, probe set-level experiment was converted to gene-level by selecting a single probe set with the highest median expression, from all the probe sets representing a gene.
2.5. Identification of differentially expressed genes
Limma package available through Bioconductor (www.bioconductor.org) was used to identify genes that are differentially expressed (DEGs) between a mutant and control pair. To identify DEGs, a design matrix including samples from mutant and control, representing two-group microarray experiment was generated through R model.matrix function. This design matrix was then used to fit linear model to the expression data for each gene through built-in lmfit function. Next, a mutant vs. control contrast matrix was constructed using makeContrasts function, followed by computation of estimated coefficients and standard errors through contrast.fit function. Built-in ebayes method was used to compute moderated t-statistics, moderated F-statistic, and log-odds of differential expression. Finally, write.fit function was used to export results after adjusting t-static p-values by Benjamini and Hochberg method [4]. The final list of DEGs in Mafg −/−:Mafk +/− compound mouse mutants (± 1.5-fold and p < 0.05, total n = 97) is presented in Supplementary Table 1. These DEGs were further investigated to narrow down the list to a gene-set most relevant to our study, using publically available resources and tools as described below.
2.6. Gene ontology enrichment analysis
To extract biological relevance from the differentially expressed genes in Mafg −/−:Mafk +/− mutant lenses, we used a bioinformatics tool — DAVID (Database for Annotation, Visualization and Integrated Discovery; http://david.abcc.ncifcrf.gov), which provides functional interpretation of genes based on gene ontology enrichment analysis, among other criteria [5]. Up- (n = 42) and down-regulated (n = 55) genes were separately analyzed by functional annotation tool using default parameters. Statistically enriched terms (p-value ≤ 0.02) with at least two gene members were considered for both up- and down-regulated genes. Several terms relevant to the lens phenotype like “acetylation”, “extracellular region”, and “peptidase inhibitor activity” were found to be significantly associated with down-regulated genes in Mafg −/−:Mafk +/− mutant lenses (Supplementary Table 2). The up-regulated gene set was found to be enriched for genes belonging to “lipid biosynthetic process”, “sterol biosynthetic process”, “chaperone”, “response to oxidative stress”, “unfolded protein binding” and “negative regulation of transcription factor activity” (Supplementary Table 2). Together this analysis provided a preliminary insight into the function of genes that are mis-regulated in the Mafg −/−:Mafk +/− mutant lenses.
2.7. iSyTE-based analysis for prioritization of candidates with potential function in the lens
Next, to prioritize the DEGs based on their relevance to lens biology, we investigated whether they are significantly expressed or enriched in the lens using the eye gene discovery tool iSyTE. iSyTE is a resource for lens gene expression and provides an estimate of “lens-enriched” expression based on t-statistic computed from in silico comparative analysis of mouse lens microarray datasets against whole body embryonic tissue (without lens) microarray datasets [2]. iSyTE has led to the identification of several new genes linked to cataract [1], [2], [6], [7], [8], and to the characterization of regulators of lens development [9], [10] as well as to the development of resources for lens studies [11], [12], [13]. The iSyTE analysis for DEGs revealed that 84% (n = 55) of down-regulated genes are lens enriched, while 64% (n = 42) of up-regulated genes are not lens-enriched in Mafg −/−:Mafk +/− mutant lenses. A chi-square test was performed to examine whether the difference in iSyTE-identified lens-enriched and non-enriched genes is significant between the up-regulated and down-regulated genes in Mafg −/−:Mafk +/− mutants. A chi-square value of 425.92 at two-tailed p-value < 0.0001 indicated that the iSyTE assigned lens-enriched genes are significantly greater in number in the down-regulated dataset compared to the up-regulated dataset of DEGs in the mutant lenses. This analysis led to the identification of 46/55 down-regulated genes that are lens-enriched according to iSyTE, and therefore represent candidates for future investigations.
2.8. Predictions of functional connectivity of small Mafs in the lens based on chromatin immunoprecipitation (ChIP) data analysis
Small Maf proteins are basic leucine zipper (bZIP) transcription factors that heterodimerize with cap and collar (CNC) family proteins and regulate gene expression by directly binding to cis-regulatory motifs [14]. We hypothesized that it may be possible to make specific predictions of direct targets of small Mafs in the lens, based on the following logic and use of resources. First, small Maf regulatory protein partners that are relevant to the lens can be identified using iSyTE. Second, ChIP-Seq data on small Mafs or their co-regulatory proteins that have been performed on non-lens cells can be analyzed in the context of the lens using iSyTE. Third, the Mafg −/−:Mafk +/− microarray datasets that potentially contain both direct and indirect small Maf target genes, can be further analyzed in the context of the above analyses. Thus, a comparative analysis between known direct targets in previous ChIP-Seq assays on Mafg or its lens-expressed partner protein Nrf2 and the Mafg −/−:Mafk +/− lens DEGs identify candidates common to these datasets and therefore are putative direct small Maf targets in the lens. We performed this analysis by examining ChIP data for Mafg and its partner Nrf2 obtained from previous studies [15], [16], [17] and identified 36 high-priority candidates from the original 97 differentially expressed genes in Mafg −/−:Mafk +/− mutant lenses (Supplementary Table 1). Importantly, literature-based analysis showed that 8 of these 36 high-priority candidates are linked with cataract, in turn validating this approach.
2.9. Cis-regulatory motif analysis
To gain further insight into putative direct targets of small Mafs in the lens, we next examined Mafg −/−:Mafk +/− DEGs for the presence of potential bZIP transcription factor binding motifs such as ARE core, NF-E2, and MARE in the 2.5 kb upstream region from the transcription start site. This analysis was also performed on the lens-relevant ChIP targets identified above. Transcription factor specific position-weight matrices (PWMs) were fetched from MotifDb R package (version 1.6.0), which maintains an updated and comprehensive collection of publicly available motif sites. These PWMs were then used as an input for motif search through sequence matching by matchPWM algorithm [18], with a sequence match threshold of ≥ 80%. This led to the identification of putative bZIP transcription factor targets in the DEGs (Supplementary Table 1).
2.10. Integration of protein–protein and other interaction-based data
Next, to extract known potential protein-level associations between Mafg −/−:Mafk +/− differentially expressed genes, we analyzed molecular interactions from experimental and curated data stored in the STRING database (http://string-db.org/) [19]. For this analysis, we developed an in-house Python script to obtain high confidence interactions (score > 0.7) between the input set consisting of the 97 DEGs (1.5-fold change, p-value < 0.05) from raw (flat) files. Protein interactions belonging to different modes such as activation, binding, and post-transcriptional modification, among others, led to the identification of new edges and ultimately contributed to assembly of small Maf regulatory network in the lens. The derived network (see below) was visualized in Cytoscape (http://www.cytoscape.org/) [20].
3. Discussion
3.1. Assembling a regulatory network in the lens
Previously, we had proposed that integrative analysis of diverse systems-level information and traditional molecular experimental data can be used to facilitate the assembly of regulatory networks in eye development [21]. Here, using the Mafg −/−:Mafk +/− study [1] as an example, we outline in detail the various analytical methods that can be applied to identify promising targets and derive a regulatory sub-network for the lens. This analysis can be used to identify and validate direct targets of MafG using functional assays and also to test how these genes are involved mechanistically in the ocular pathology of Mafg −/−:Mafk +/− mutants. These targets include promising candidates such as Hspb1, which is significantly downregulated in Mafg −/−:Mafk +/− lens microarrays, is lens-enriched according to iSyTE, harbors a bZIP binding motif and is identified as a target of the small Maf binding partner Nrf2 in ChIP analysis. It is evident from this example that a gene expression resource for a specific cell or tissue type, such as the iSyTE database, is essential for effectively integrating these different datasets. To begin with, iSyTE led to the identification of the small Maf proteins Mafg and Mafk as potential new regulators in lens cells, a hypothesis that was proved to be correct by the analysis of Mafg −/−:Mafk +/− mouse compound mutants. Further, overlay of lens gene expression information present in iSyTE served to effectively and selectively provide lens relevance to the seemingly discreet information present in various publically available regulatory and molecular interaction datasets. For example, non-lens derived ChIP data could be utilized to predict putative direct targets of small Mafs in the lens because iSyTE could screen for only those lens-expressed candidates within these datasets that were also found to be differentially expressed in Mafg −/−:Mafk +/− mutant lenses. Similarly, iSyTE could select for lens-expressed protein–protein or other relevant interactions within the Mafg −/−:Mafk +/− differentially expressed gene dataset. These “lens-filtered” data, along with cis-regulatory motif identification and gene ontology-based functional classification allowed for the prediction of a small Maf regulatory network in the lens that indicates defects in sterol synthesis or elevated oxidative stress to contribute to the phenotype in Mafg −/−:Mafk +/− mutants. Future experiments can now be directed to test the robustness of this predicted network and to further understand how its specific perturbation may result in lens defects such as cataracts.
In sum, we outline in detail a strategy that utilized iSyTE to screen for lens relevant information within publically available datasets and led to the effective prioritization of Mafg −/−:Mafk +/− differentially expressed genes. It is expected that development of resources similar to iSyTE will allow the extension of this integrative approach to mutant microarray data in other cell or tissue types.
The following are the supplementary data related to this article.
List of microarray identified differentially expressed genes in mutant lens.
Functional enrichment analysis of DEGs with the tool DAVID.
Conflict of interest
The authors have no conflicts of interest to declare.
Acknowledgements
The authors thank Mr. Atul Kakrana for helpful discussions. This work was supported by the National Institutes of Health/National Eye Institute grant R01 EY021505 and a Fight For Sight Grant-in-Aid award to S.A.L. S.A.L. is a Pew Scholar in Biomedical Sciences.
References
- 1.Agrawal S.A., Anand D., Siddam A.D., Kakrana A., Dash S., Scheiblin D.A. Compound mouse mutants of bZIP transcription factors Mafg and Mafk reveal a regulatory network of non-crystallin genes associated with cataract. Hum. Genet. 2015 doi: 10.1007/s00439-015-1554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lachke S.A., Ho J.W.K., Kryukov G.V., O'Connell D.J., Aboukhalil A., Bulyk M.L. iSyTE: integrated Systems Tool for Eye gene discovery. Invest. Ophthalmol. Vis. Sci. 2012;53:1617–1627. doi: 10.1167/iovs.11-8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du P., Kibbe W.A., Lin S.M. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 4.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- 5.Huang D.W., Sherman B.T., Zheng X., Yang J., Imamichi T., Stephens R. Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinformatics. 2009 doi: 10.1002/0471250953.bi1311s27. (Unit 13.11. Chapter 13) [DOI] [PubMed] [Google Scholar]
- 6.Lachke S.A., Alkuraya F.S., Kneeland S.C., Ohn T., Aboukhalil A., Howell G.R. Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science. 2011;331:1571–1576. doi: 10.1126/science.1195970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasaikina M.V., Fomenko D.E., Labunskyy V.M., Lachke S.A., Qiu W., Moncaster J.A. Roles of the 15-kDa selenoprotein (Sep15) in redox homeostasis and cataract development revealed by the analysis of Sep 15 knockout mice. J. Biol. Chem. 2011;286:33203–33212. doi: 10.1074/jbc.M111.259218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lachke S.A., Higgins A.W., Inagaki M., Saadi I., Xi Q., Long M. The cell adhesion gene PVRL3 is associated with congenital ocular defects. Hum. Genet. 2012;131:235–250. doi: 10.1007/s00439-011-1064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf L., Harrison W., Huang J., Xie Q., Xiao N., Sun J. Histone posttranslational modifications and cell fate determination: lens induction requires the lysine acetyltransferases CBP and p300. Nucleic Acids Res. 2013;41:10199–10214. doi: 10.1093/nar/gkt824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manthey A.L., Lachke S.A., FitzGerald P.G., Mason R.W., Scheiblin D.A., McDonald J.H. Loss of Sip1 leads to migration defects and retention of ectodermal markers during lens development. Mech. Dev. 2014;131:86–110. doi: 10.1016/j.mod.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manthey A.L., Terrell A.M., Lachke S.A., Polson S.W., Duncan M.K. Development of novel filtering criteria to analyze RNA-sequencing data obtained from the murine ocular lens during embryogenesis. Genomics Data. 2014;2:369–374. doi: 10.1016/j.gdata.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anchan R.M., Lachke S.A., Gerami-Naini B., Lindsey J., Ng N., Naber C. Pax6- and Six3-mediated induction of lens cell fate in mouse and human ES cells. PLoS ONE. 2014;9:e115106. doi: 10.1371/journal.pone.0115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terrell A.M., Anand D., Smith S.F., Dang C.A., Waters S.M., Pathania M. Molecular characterization of mouse lens epithelial cell lines and their suitability to study RNA granules and cataract associated genes. Exp. Eye Res. 2015;131:42–55. doi: 10.1016/j.exer.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motohashi H., O'Connor T., Katsuoka F., Engel J.D., Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 15.Malhotra D., Portales-Casamar E., Singh A., Srivastava S., Arenillas D., Happel C. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chorley B.N., Campbell M.R., Wang X., Karaca M., Sambandan D., Bangura F. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirotsu Y., Katsuoka F., Funayama R., Nagashima T., Nishida Y., Nakayama K. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res. 2012;40:10228–10239. doi: 10.1093/nar/gks827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wasserman W.W., Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 19.Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A. STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lachke S.A., Maas R.L. Building the developmental oculome: systems biology in vertebrate eye development and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010;2:305–323. doi: 10.1002/wsbm.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of microarray identified differentially expressed genes in mutant lens.
Functional enrichment analysis of DEGs with the tool DAVID.




