Summary
Ebolavirus NP oligomerizes into helical filaments found at the core of the virion, encapsidates the viral RNA genome, and serves as a scaffold for additional viral proteins within the viral nucleocapsid. We identified a portion of the phosphoprotein homologue VP35 that binds with high affinity to nascent NP and regulates NP assembly and viral genome binding. Removal of the VP35 peptide leads to NP self-assembly via its N-terminal oligomerization arm. NP oligomerization likely causes a conformational change between the NP N- and C-terminal domains, facilitating RNA binding. These functional data are complemented by a crystal structure of the NP°-VP35 complex at 2.4 Å resolution. The interactions between NP and VP35 illuminated by these structures are conserved among filoviruses and provide key targets for therapeutic intervention.
INTRODUCTION
Ebola virus causes severe hemorrhagic fever and is responsible for a significant and sustained outbreak in Western Africa 2013–2015 (Agua-Agum et al., 2015; Burke et al., 1978). Ebola virus belongs to the family filoviridae, which also contains four other members of the ebolavirus genus, the pathogenic Marburg virus (MARV) and the recently discovered Lloviu virus (LLOV) (Negredo et al., 2011). Filoviruses belong to the order Mononegavirales, which also contains the pathogenic respiratory syncytial virus (RSV), measles, mumps, Nipah, rabies and other viruses. All members of Mononegavirales bear a nonsegmented, negative-sense RNA genome (NNS) that is encapsidated by a viral nucleoprotein, termed N or NP (Knipe and Howley, 2001). The viral polymerase (L) and a phosphoprotein polymerase cofactor (typically called P) direct viral transcription and replication. In filoviruses, the phosphoprotein homolog is VP35 (Muhlberger et al., 1998), though this protein is not strongly phosphorylated (Elliott et al., 1985). Filoviruses contain an additional polymerase co-factor, VP30, which facilitates read-through of RNA secondary structure in the ebolavirus NP transcript (Weik et al., 2002), and contributes to the continuation of transcription at gene junctions (Martinez et al., 2008). Filovirus NP binds both VP35 and VP30 (Hartlieb et al., 2007; Huang et al., 2002), which in turn, both interact with the viral polymerase to assemble the viral replication complex (Groseth et al., 2009; Trunschke et al., 2013).
The nucleoproteins of NNS viruses also protect viral RNA genomes from recognition by the cellular innate immune response and render the genome resistant to ribonucleases (Hornung et al., 2006; Knipe and Howley, 2001). Recombinant expression of ebolavirus NP in the absence of viral infection produces long filament-like, helical NP coils bound to cellular RNA suggesting that the NP does not specifically recognize viral RNA (Bharat et al., 2012; Mavrakis et al., 2002). The long helical NP filament serves as a scaffold for the assembly of the filovirus nucleocapsid which includes VP35, VP30, VP24 and L (Elliott et al., 1985; Huang et al., 2002). NP oligomers further interact with the matrix protein, VP40, allowing recruitment of the nucleocapsid into progeny virions (Noda et al., 2006). The multiple functions of Ebola virus NP make NP an essential piece of machinery for viral RNA synthesis and virus assembly alike.
Despite the importance of the NP protein to the ebolavirus life cycle, the structure of its RNA-binding and self-assembling domains as well as the mechanisms for its oligomerization and RNA binding have remained elusive. However, nucleoprotein structures for several other NNS viruses have been determined (Alayyoubi et al., 2015; Albertini et al., 2006; Green et al., 2006; Rudolph et al., 2003; Tawar et al., 2009; Yabukarski et al., 2014). These structures reveal a shared general fold in which N- and C-terminal domains form a bi-lobed structure with RNA binding in a cleft between the two lobes. In the oligomeric nucleoprotein structures available, N- and C-terminal arms or loops extend from each of these lobes to contact adjacent nucleoproteins in the oligomer. In many NNS viruses, the viral phosphoprotein (P) contains a peptide in its N-terminal region which chaperones the viral nucleoprotein (N) in a monomeric, RNA-free state called N°-P (Curran et al., 1995; Leyrat et al., 2011; Majumdar et al., 2004; Yabukarski et al., 2014). This P peptide prevents premature nucleoprotein oligomerization and aberrant, nonproductive interactions of nascent nucleoprotein with cellular RNA. Two crystal structures of N°-P have so far been described. For vesicular stomatitis virus (VSV), P directly blocks both the nucleoprotein RNA-binding site and the binding site of the nucleoprotein N-terminal arm domain, preventing RNA binding and nucleoprotein oligomerization (Leyrat et al., 2011). In the Nipah virus N°-P structure, the P peptide does not occlude the RNA-binding site and instead likely interferes with the binding of both the N- and C-terminal arms of adjacent nucleoproteins (Yabukarski et al., 2014). Additionally, the Nipah virus N° structure is in an open conformation, while the oligomeric, RNA-bound nucleoprotein of RSV is in a closed conformation (Tawar et al., 2009). The open conformation of Nipah N°-P is believed to be incapable of binding to RNA. Despite the commonality of N°-P complexes amongst NNS viruses, filovirus VP35 were previously hypothesized to lack this chaperoning activity (Noda et al., 2011).
In this work, we identify an N-terminal binding site on filovirus VP35 for NP and demonstrate that this VP35-NP interaction chaperones the NP in a monomeric, RNA-free state. We term this complex NP°-VP35. Characterization of this complex has allowed us to determine crystal structures of the Ebola virus NP° core domain bound to a VP35 peptide to 2.4 Å resolution in multiple space groups. The interactions observed in these structures are conserved across the filovirus family. Accompanying biophysical analyses demonstrate that the VP35 Nterminal peptide binds NP with high affinity and that binding is coupled to conformational changes in one or both proteins. We further identify the N- and C-terminal oligomerization arms of the Ebola virus NP and show their importance in the formation of large NP polymers. Further data also demonstrate that the binding of RNA to NP is dependent upon NP oligomerization, rather than the absence of the VP35 N-terminal peptide chaperone. Based on these results, we propose a structure-based model for the assembly of the oligomeric Ebola virus NP from newly synthesized NP°.
RESULTS
Identification of a second NP binding site on VP35
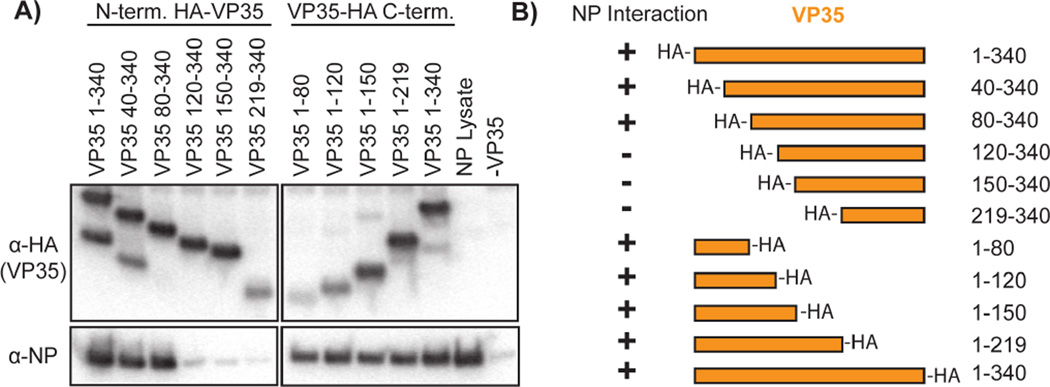
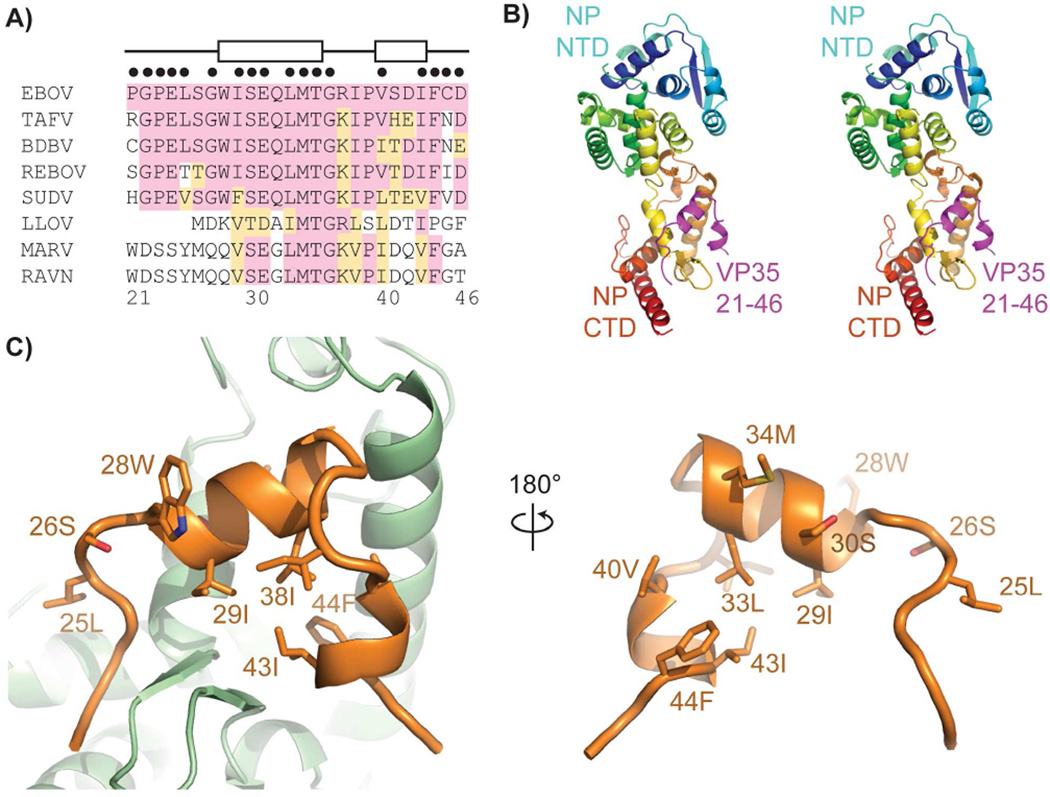
To examine the interactions between Ebola virus (EBOV formerly known as Zaire ebolavirus, ZEBOV) VP35 and NP, we prepared a series of N- and C-terminal truncations of VP35, each containing an HA-epitope tag for purification and detection. Each truncation construct was designed to incorporate the HA-tag such that one native terminus is preserved in each construct (i.e. N-terminal truncations contain N-terminal HA tags). We co-transfected each of these VP35 constructs with NP into HEK 293T cells. Western blotting of anti-HA pull downs for VP35 (anti-HA) and NP (Fig. 1) clearly shows that either VP35 1-80 or VP35 80-340 are sufficient to pull-down NP. Hence, VP35 has two unique and independent binding sites for the viral NP: residues 1–80 and 80–340. The VP35 80-340 region contains a putative coiled-coil oligomerization domain (residues 80–120) (Reid et al., 2005) as well as a folded C-terminal domain (residues 219–340) (Kimberlin et al., 2010; Leung et al., 2010). A previous study (Prins et al., 2010) showed that the C-terminal domain of VP35 binds to NP. Our identification of a stronger interaction by residues 80–340 than 219–340 suggests a role for VP35 oligomerization in enhancing the avidity of the VP35 C-terminal domain interaction. Comparison of the N-terminal sequence (residues 1–80) of EBOV VP35 to that of the other ebolaviruses, and more broadly to other filoviruses, highlights a region of high sequence conservation (Fig. 2A) (Karlin and Belshaw, 2012).
Figure 1. Pull-down of NP with VP35 truncations.
HA-tagged EBOV VP35 constructs were co-expressed with NP and pulled down on anti-HA beads. A) The Western blotting results probing for VP35 (anti-HA) or NP. The ‘NP lysate’ positive control is cleared cell lysate from cells transfected with NP alone. The ‘-VP35’ negative control is an anti-HA pull-down of cells transfected with NP alone. B) Results summary of the pull-down experiment.
Figure 2. Structure of the NP°-VP35 complex.
A) Sequence alignment of the conserved VP35 N-terminal peptide. Amino acids sharing identity with EBOV are shaded pink, similar amino acids are shaded orange. Amino acids participating in helices as observed in the crystal structure are indicated at top by boxes. Amino acid positions within 4 Å of NP are indicated with black dots above the alignment. B) Stereoview of the NP°-VP35 crystal structure. NP is colored from the N terminus (blue) to the C terminus (red) with the VP35 peptide in magenta. C) The VP35 peptide (orange) uses a hydrophobic face and conserved amino acids to contact the NP (green). See also Figure S2 and S3 and Table S1.
The VP35 peptide chaperones NP in a monomeric form
The N-terminal 450 residues of NP were previously shown to be sufficient for RNA-binding and oligomerization of NP (Bharat et al., 2012; Watanabe et al., 2006). We hypothesized that the VP35 N-terminal 1–80 peptide identified by the pull-down may play a role in chaperoning the NP, analogous to the P proteins of other nonsegmented negative-sense RNA viruses. To test this hypothesis we co-expressed the VP35 1-80 peptide and NP amino acids 1–450 in E. coli. We observe that NP 1-450 co-purifies with His-tagged VP35 1-80. The copurification of these regions of VP35 and NP confirms the results of the pull down and indicates that the VP35 N-terminal peptide interacts with the N-terminal 450 amino acids of NP. Size exclusion chromatography with in-line multi-angle light scattering (SEC-MALS), indicates this sample to be a monomer in solution (Table 1, sample 1). However, over time, this sample gradually shifts to larger molecular weight species.
Table 1.
EBOV NP oligomerization assessed by multi-angle light scattering.
| # | Sample | VP35 1- 80 Added |
Monomeric NP (kDa) Calculated |
NP MALS |
|---|---|---|---|---|
| Monomeric NP°-VP35 Co-expression | ||||
| 1 | EBOV VP35 1-80 + NP 1-450 | 50 | 72 | |
| 2 | EBOV NP 1-450, AEX | 50 | >1000 | |
| Monomeric NP°-VP35 fusions | ||||
| 3 | EBOV VP35 1-80-NP 1-450 | 61 | 58 | |
| 4 | EBOV VP35 1-80-NP 1-739 | 94 | 110 | |
| 5 | SUDV VP35 1-49-NP 1-450 | 58 | 52 | |
| 6 | MARV VP35 1-41-NP 1-430 | 54 | 51 | |
| 7 | EBOV VP35 15-60-NP 34-367 | 42 | 40 | |
| RNA-independent NP Oligomerization and Reversibility | ||||
| 8 | EBOV VP35 1-80-TEV-NP 1-450 | 60 | 74 | |
| 9 | EBOV VP35 1-80 + NP 1-450, TEV cleaved | 50 | 70 | |
| 10 | EBOV NP 1-450, TEV cleaved, AEX | 50 | >1000 | |
| 11 | EBOV NP 1-450 | + | 50 | 74 |
| Importance of Terminal Arm Domains in NP Oligomerization | ||||
| 12 | EBOV NP 34-367 | 40 | 38 | |
| 13 | EBOV NP 34-367 | + | 40 | 51 |
| 14 | MBP-EBOV NP 34-367 | 81 | 78 | |
| 15 | MBP-EBOV NP 34-367 | + | 81 | 85 |
| 16 | MBP-EBOV NP 34-450 | 90 | 88 | |
| 17 | MBP-EBOV NP 1-367 | 85 | >1000 | |
| 18 | MBP-EBOV NP 19-389 | 85 | >1000 | |
| Oligomeric NP-RNA | ||||
| 19 | MBP-EBOV NP 1-450 | 94 | >1000 | |
| 20 | MBP-EBOV NP 1-450 | + | 94 | >1000 |
| 21 | EBOV NP 1-450 | 50 | >1000 | |
| 22 | EBOV NP 1-450 | + | 50 | >1000 |
| 23 | MBP-EBOV NP 1-739 | 127 | >1000 | |
| 24 | MBP-EBOV NP 1-739 | + | 127 | >1000 |
Calculated molecular weights are based on the expected mass of a single NP. Multi-angle light scattering data was collected with in-line size exclusion chromatography (SEC-MALS). In the Sample column hyphens between two protein domains represent protein fusions whereas plus signs indicate two distinct protein chains whether the result of co-expression or the result of a TEV protease cleavage. The data representing the co-expression of NP with VP35 (samples 1–2) or the RNA-independent NP oligomerization and reversibility of this oligomerization (samples 8–11) each represent a single protein preparation that was subsequently processed with additional steps such as cleavage by TEV protease, purification by anion exchange chromatography (AEX), or addition of purified VP35 1-80 peptide. See also Figure S1 for imaging of selected samples with negative stain electron microscopy.
To stabilize the VP35 1-80–NP 1-450 complex, the two proteins were engineered as a fusion. The VP35 1-80–NP 1-450 fusion protein was similarly monomeric in solution (Table 1, sample 3, Fig. S1) and purified completely free of E. coli nucleic acids. In addition, the corresponding fusion proteins for Sudan ebolavirus (SUDV) and Marburg virus (MARV) and the full-length EBOV NP are also monomeric (Table 1, samples 4–6) and RNA-free as determined by the ratio of their UV absorbance at 260 and 280 nm. To characterize the EBOV VP35 1-80– NP 1-450 fusion protein, we performed hydrogen/deuterium exchange mass spectrometry (H/DXMS). This analysis illustrates that VP35 residues 18–47 are protected from solvent exchange in the presence of NP 1-450 (Fig. S2). These solvent-protected VP35 residues correspond closely to those conserved across ebolaviruses. H/DXMS also shows that NP residues 39–356 exchange more slowly with solvent than the terminal regions and suggests that NP 39-356 forms a folded core.
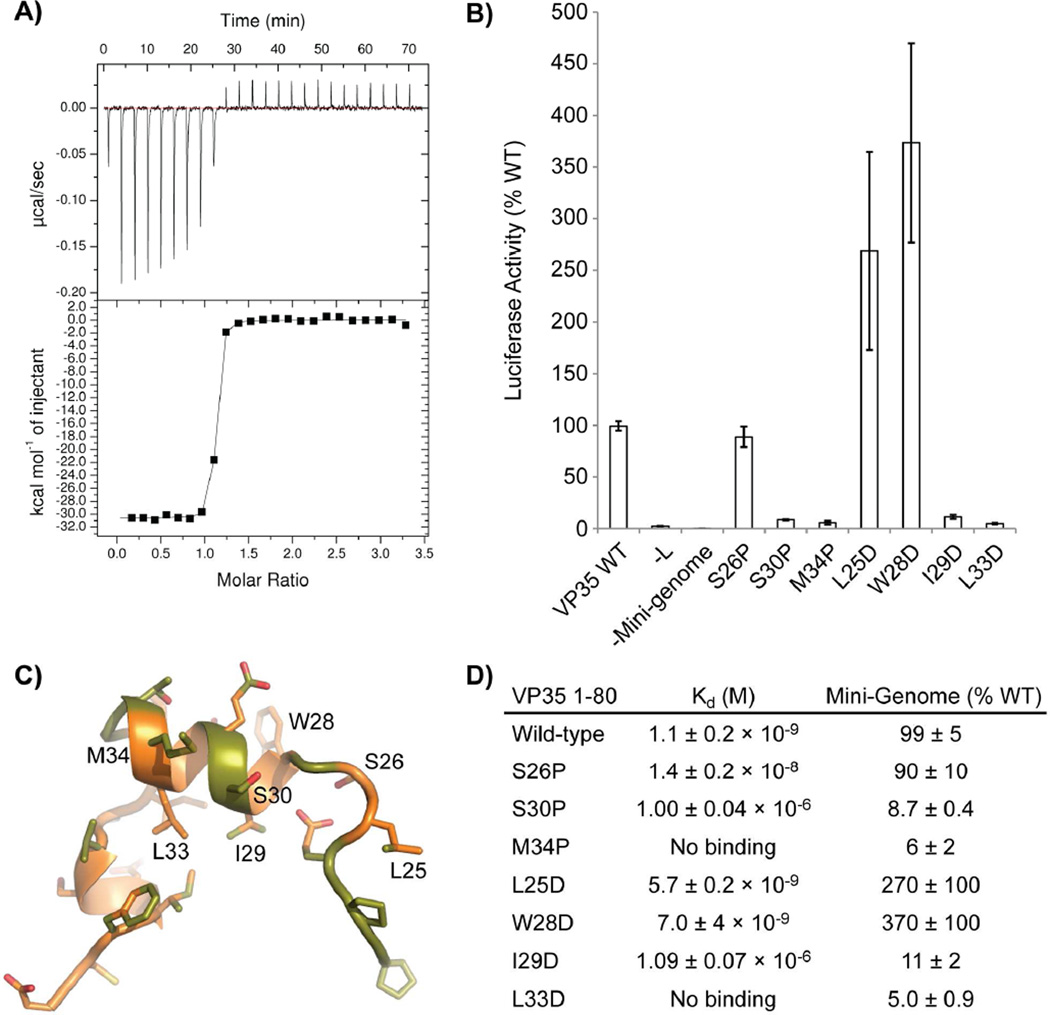
We hypothesized that the folded core of EBOV NP corresponds to the bi-lobed fold of other NNS virus nucleoproteins (Tawar et al., 2009). We further hypothesized the presence of additional N- and C-terminal arms of the EBOV NP, which appear free to solvent exchange in the chaperoned monomeric fusion protein and would ordinarily facilitate NP oligomerization in the absence of VP35. Using SEC-MALS and isothermal titration calorimetry (ITC), we found that a protein construct containing the folded NP core, but excluding the terminal regions (NP 34-367) is monomeric and capable of binding the VP35 1-80 peptide (Table 1 samples 12–13, and Fig. 3). Correspondence between these solution binding data for the NP°-VP35 interaction and the solvent protection observed by H/DXMS for the VP35 1-80-NP 1-450 fusion protein demonstrates the VP35-NP fusion protein is an accurate model of NP°-VP35. These data support the conclusion that the EBOV VP35 N-terminal peptide chaperones NP as a monomeric, RNA-free NP°-VP35 complex.
Figure 3. VP35 N-terminal peptide and NP° core interactions.
A) Isothermal titration calorimetry (ITC) was used to assess the binding of the VP35 1-80 peptide to the folded NP° core (NP 34-367). A representative trace is presented for the binding reaction using wild-type VP35 1-80. B) Activity of VP35 mutants in the mini-genome assay presented as a percent of wild-type VP35 activity. C) VP35 21-46 peptide from the NP°-VP35 complex. Mutated positions are labeled. Carbon atoms within 4 Å of NP° are colored green. Leucine 33 is buried by other VP35 residues. It does not directly contact NP°. D) Calculated molecular dissociation constants for the wild-type interaction and various VP35 1-80 mutants with corresponding activities in the mini-genome assay. The reported error represents the standard deviation of three replicate experiments. For the M35P and L33D mutants, there was no observed binding in the ITC experiment using 30 µM NP° core and 300µM mutant VP35 1-80. See also Figure S5.
VP35 chaperoning of NP mechanism: Structure and high-affinity binding
In order to use crystallography to understand the mechanisms of interaction in the NP°-VP35 complex, we constructed a fusion protein lacking extraneous regions of VP35 and NP that freely exchange with solvent (Table 1, sample 7). This fusion protein (VP35 15-60–NP 34-367) crystallizes and diffracts to 2.4 Å. The structure could not be solved by molecular replacement using the known NNS virus nucleoproteins or nucleoprotein domain structures as search models. Instead, we determined the structure of the complex using X-ray crystallography with Sulfur-SAD phasing from endogenous protein sulfur atoms. The experimentally phased structure was then used as a search model for molecular replacement to solve the structure of VP35 15-60–NP 34-367 in two additional space groups (Table S1).
The structure of the EBOV NP RNA-binding domain is primarily alpha-helical and is composed of N- and C-terminal domains that together form a bi-lobed structure (Fig 2B). Nearly all residues were resolved, with the exception of a handful of disordered amino acids at the N and C termini of NP and VP35, as well as sparse electron density in a region spanning residues 121–143 of NP. This region of NP has high B-factors for the residues modeled in each space group and corresponds to a region of high solvent exchange in H/DXMS (Fig. S2) reflecting the inherent flexibility of these amino acids. These residues are best ordered in the P22121 space group.
The VP35 N-terminal peptide binds to one side of the C-terminal domain of the NP° core (Fig. 2C) and adopts an unusual straddling conformation as compared to other NNS virus P homologs. The P peptides observed to chaperone VSV or Nipah virus nucleoproteins are primarily alpha-helical (Leyrat et al., 2011; Yabukarski et al., 2014). However, the 26 amino acids of the EBOV VP35 peptide ordered in the structure are mostly lacking secondary structure and contain only 12 amino acids in a helical conformation. The N-terminus of the VP35 peptide lies near the C-terminus of the NP core and the VP35 chain progresses towards the junction of the NP N- and C-terminal domains. However, before reaching this junction, the VP35 peptide chain abruptly turns and progresses towards the rear of the C-terminal domain. The path of the VP35 N-terminal peptide allows the VP35 peptide to straddle a β-hairpin in the NP C-terminal domain. This β-hairpin is unique to Ebola virus NP and is not observed in known nucleoprotein structures of other NNS viruses.
The chaperoning VP35 peptide buries 1,260 Å2 of molecular surface on NP. The interacting region of VP35 contains both hydrophilic and hydrophobic regions. However, the buried surface of NP is primarily hydrophobic and involves both the body of the C-terminal domain as well as the ebolavirus-specific, NP β-hairpin. Many of the interacting residues on VP35 and NP are highly conserved across the ebolavirus genus and to a lesser extent across the filoviruses family (Fig. 2A and Fig. S3).
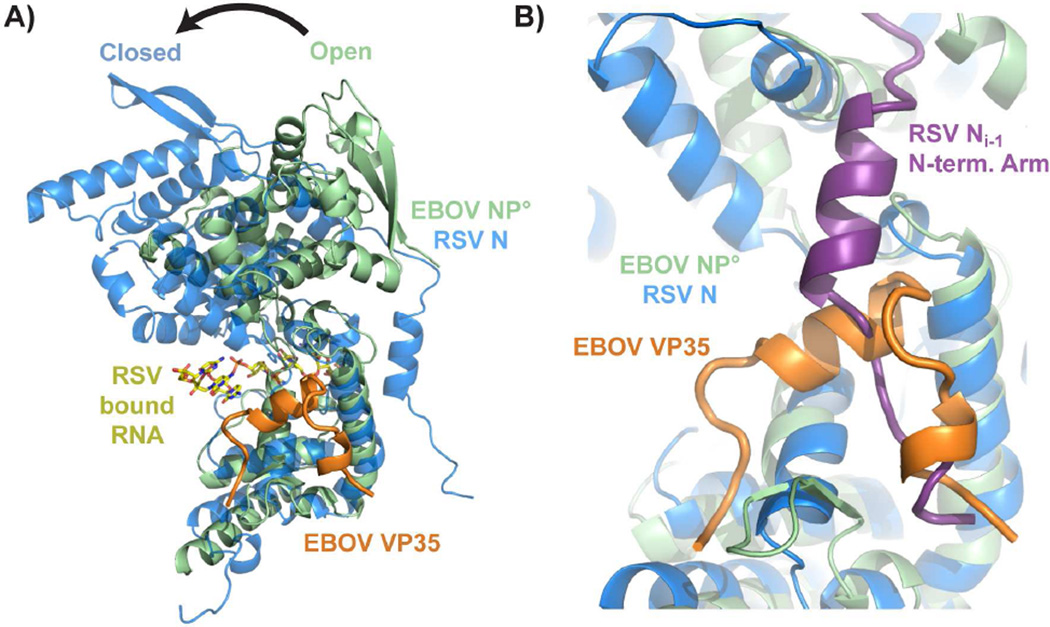
The bi-lobed fold of the EBOV NP is most similar to that of the closely related RSV (Tawar et al., 2009), parainfluenza virus 5 (PIV5) (Alayyoubi et al., 2015) and Nipah virus (Yabukarski et al., 2014) nucleoproteins. The C-terminal domains of the EBOV and RSV nucleoproteins structurally align more closely while the N-terminal domains are more divergent (Fig. S4). Structural alignment of the oligomeric RSV N and the monomeric EBOV NP° core-VP35 peptide complex shows the EBOV NP° core to be in an open conformation (Fig. 4) similar to Nipah virus N°-P. Homology with the RNA-bound RSV nucleoprotein suggests the RNA-binding site of EBOV NP runs between the N- and C-terminal domains. The RNA-binding site of RSV nucleoprotein is composed of regions from both the N- and C-terminal domains. In contrast to RSV nucleoprotein, the EBOV NP°-VP35 structure is in an open conformation with the two domains splayed apart. Hence, this monomeric complex is unlikely to bind RNA.
Figure 4. Comparison of the Ebola virus NP°-VP35 complex with the respiratory syncytial virus N-RNA complex.
A) Chain A of PDB 2WJ8 was superimposed onto the Ebola virus NP° core domain-VP35 N-terminal peptide complex using the nucleoprotein C-terminal domains. A transition from the open state (monomeric EBOV NP) to the closed state (oligomeric RSV N) is likely necessary to form the RNA-binding site, which is not occluded by the EBOV VP35 peptide. B) The EBOV VP35 N-terminal peptide occupies the site on EBOV NP° core that is bound by the N-terminal oligomerization arm of an adjacent nucleoprotein (NPi−1) in the oligomeric RSV N structure. For a structural alignment of the individual N- and C-terminal domains of RSV and EBOV NP, see figure S4.
To examine the binding of VP35 to NP, we performed isothermal titration calorimetry (ITC) using the VP35 N-terminal peptide (1–80) and the NP° core domain (34–367). The interaction between these two partners is remarkably strong, with an equilibrium dissociation constant (Kd) of 1.1 ± 0.2 nM (Fig. 3). We mutated the VP35 N-terminal peptide N-terminal to, and within the first helical region. Mutated VP35 1-80 was used in ITC analysis, while fulllength VP35 mutants were used in Ebola virus mini-genome assays. While the ITC assays directly measure the strength of the interaction between the NP° core and VP35 N-terminal peptide, the mini-genome assay assesses the abilities of these mutants in the background of full-length viral proteins in mammalian cells to support replication/transcription of a reporter gene whose expression is controlled by viral sequences (Jasenosky et al., 2010). Selected VP35 hydrophobic amino acids were mutated to aspartate to disrupt hydrophobic interactions. Additional amino acid positions were mutated to proline to disrupt the peptide backbone conformation. All of the VP35 mutants negatively affected protein binding in ITC although to greatly differing extents (Fig. 3). Aspartate mutations to hydrophobic residues with solvent-exposed side chains or a proline mutation within a non-helical region had minimal effects on VP35 binding; these mutants maintain high affinity binding with Kds of less than 20 nM. These same mutations maintain at least wild-type activity to support mini-genome replication/transcription with two of the mutants showing enhancements in reporter expression over the wild-type VP35. While these positions are highly conserved in ebolaviruses, they are polymorphic across the filoviruses reflecting their lessor importance in maintaining the NP°-VP35 interaction. In contrast, aspartate mutations to VP35 hydrophobic residues buried in the NP°-VP35 complex or proline mutations targeted to helical regions had low affinities for NP, having Kds of 1 µM or greater. The mutations with detrimental effects on VP35 binding to NP in ITC further have minimal activity in the replication/transcription of the EBOV mini-genome. VP35 positions, which have detrimental effects in either the ITC binding assay or the mini-genome assay, are much more highly conserved across the filovirues family. This indicates the importance of the first helical region and buried hydrophobic amino acids in maintaining the interaction. The experimentally measured effects of each of these mutations accurately correspond to predictions made from the NP° core-VP35 peptide complex crystal structure and the VP35 sequence alignment across the filovirus family.
Because the crystal structures reveal such a large hydrophobic binding surface between VP35 and NP, we characterized the burial of non-polar surface area upon binding in solution using ITC. By measuring the change in enthalpy (ΔH) upon VP35 1-80 binding to the NP° core domain at different temperatures, we calculated a heat capacity change upon binding (ΔCp) of −946 cal mol−1 K−1 (Fig. S5). This ΔCp indicates that approximately 3,400 Å2 of nonpolar surface is buried upon VP35 peptide binding to the NP° core (Ha et al., 1989). However, this large buried non-polar surface area is far greater than the 2,520 Å2 total area buried as observed in our NP° core-VP35 N-terminal peptide crystal structure (Krissinel and Henrick, 2007) and is strongly suggestive of a significant conformational change upon binding in either or both VP35 and NP proteins to bury additional non-polar surface. Using trends in the entropy change upon binding (ΔS) with temperature and the ΔCp, we estimate that upon VP35 peptide binding to NP° core, approximately 43 amino acids are rigidified (Spolar and Record, 1994). As ITC measures the observed change in the thermodynamic parameters between the starting components and final complex, it is difficult to assign these changes to either NP° core or VP35 peptide. The loss of flexibility in the VP35 N-terminal peptide by binding the NP° core is likely to contribute substantially to the rigidification. However, we observe only 26 amino acids of VP35 consistently in the crystal structures and only 30 amino acids of VP35 are protected from solvent exchange in H/DXMS. The difference between these figures and the likely ~43 amino acids in total that are rigidified, demonstrates additional rigidification occurs beyond that accounted for by the VP35 peptide. Therefore, even if the VP35 N-terminal peptide rigidifies from a completely flexible peptide to the bound form observed in the crystal structure, these data predict a decrease in the number of conformational states within NP° core upon VP35 peptide binding.
Removal of the peptide causes NP oligomerization in vitro
While NP 1-450 co-expressed with VP35 1-80 initially purifies as a monomer (Table 1, sample 1), prolonged storage leads to the gradual formation of large oligomers. We exploited this phenomenon to explore the oligomerization dynamics of NP starting with NP°-VP35 monomers. When the VP35 1-80 peptide is purified away from NP 1-450 using anion exchange chromatography, the NP forms oligomers (Table 1 sample 2). The chromatographic separation of the NP°-VP35 complex into its constituents is only possible when using NP constructs containing regions sufficient for NP oligomerization (for example NP 1-450 and not for the shorter NP core, 34–367). To obtain better control of oligomerization and to produce preparations of NP free from any contaminating cellular RNA, we constructed a TEV proteasecleavable VP35 1-80–NP 1-450 fusion protein. This construct expresses as an RNA-free monomer (Table 1, sample 8). After TEV cleavage, the VP35-bound NP 1-450 remains mostly monomeric (Table 1, sample 9) with some of the NP losing its VP35 1-80 and shifting to higher oligomers. The monomeric nature of the fusion protein cleaved with TEV protease mirrors that of the co-expressed VP35 1-80 and NP 1-450 supporting the use of this system to examine NP oligomerization.
Anion exchange chromatography completely removes the TEV protease-cleaved VP35 peptide from NP 1-450, shifting the entire NP sample to large oligomers (Table 1, sample 10). To eliminate the possibility that the observed large oligomers may be soluble aggregates, we imaged this sample using negative-stain electron microscopy. The RNA-free, VP35-free, oligomeric NP forms large ring-like structures approximately 50 nm in diameter (Fig. S1). This is analogous to the ring formation of VSV, Rabies virus and RSV nucleoproteins using heterologous expression systems (Albertini et al., 2006; Green et al., 2006; Tawar et al., 2009) although the rings formed by EBOV NP are considerably larger. The assembly of NP into large oligomers indicates that removal of the VP35 N-terminal peptide allows the EBOV NP to oligomerize in vitro and that NP oligomerization is independent of RNA binding. Furthermore, the oligomerization of the RNA-free EBOV NP 1-450 is reversible. Incubating oligomeric NP 1-450 with an excess of VP35 1-80 peptide overnight reverts the sample entirely to a monomeric form (Table 1, sample 11). Our data demonstrate that competition exists between the binding of the VP35 N-terminal peptide and NP oligomerization.
We aligned the crystal structures of the monomeric EBOV NP° core-VP35 peptide complex with the oligomeric RSV nucleoprotein (2WJ8.pdb). This alignment shows an overlap between the binding sites of the EBOV VP35 21-46 peptide and the RSV nucleoprotein N-terminal arm from an adjacent protomer on the nucleoproteins’ core domains (Fig 4B). The adjacent RSV protomer is in the 3’ direction along the bound RNA strand (Ni−1). The structural and functional homology between EBOV and RSV nucleoproteins leads us to hypothesize that EBOV VP35 directly competes with the N-terminal arm of other NP protomers to prevent NP oligomerization and maintain NP as monomeric NP°.
NP oligomerization domains
We performed H/DXMS analysis of the oligomeric, RNA-free EBOV NP 1-450 to examine NP oligomerization (Fig. S2). Comparison of the oligomeric NP with the monomeric NP°-VP35 indicates that a shared folded core (NP 39-356) is maintained in both samples, though the C-terminal region of the NP core is better protected from solvent exchange in the oligomeric sample than the monomeric sample. Additionally, NP regions 20–29 and 357–381 show protection from solvent exchange uniquely in the oligomeric NP sample, suggesting these are the N- and C-terminal oligomerization arms of NP. To test this hypothesis, we produced a series of constructs lacking one or both terminal regions, as well as a protein construct truncated to contain the putative minimal oligomerization arms. These protein constructs were expressed as MBP-fusion proteins to facilitate their purification by affinity chromatography and the NP oligomeric state was assessed by SEC-MALS. The fusion of MBP was found to not impair NP oligomerization (Table 1, samples 19 and 21 and Fig S1). Deletion of the C-terminal arm domain had no effect on protein oligomerization (Table 1, sample 17). However, deletion of the N-terminal arm domain rendered the protein monomeric (Table 1, sample 16). Deletion of both arm domains also yields monomeric protein, as also observed with non-MBP tagged NP core (Table 1, samples 12–15). Finally, the NP core containing both of the putative minimal N- and C-terminal arms (NP 19-389) was oligomeric (Table 1, sample 18) in agreement with the oligomeric NP H/DXMS results. NP 19-389 with the minimal oligomerization arms was confirmed to form oligomeric NP rings by electron microscopy (Fig S1).
Our crystal structure of the EBOV NP° core-VP35 N-terminal peptide complex and structural homology with the RSV nucleoprotein (Tawar et al., 2009) suggests that the EBOV VP35 N-terminal peptide directly competes for NP binding with the N-terminal oligomerization arm of the NPi−1. However, VP35 is not expected to interfere with the binding of the C-terminal oligomerization arm of the NPi+1 protomer. Our SEC-MALS analyses demonstrate that only the N-terminal arm domain is required for NP oligomerization and that the C-terminal arm domain is non-essential. These oligomerization and the structural data indicate that it is sufficient for VP35 to block the binding of the NP N-terminal arm in order to maintain the EBOV NP in a monomeric state.
NP Oligomerization allows RNA binding
We determined that all of the monomeric NP constructs could be purified free of E. coli RNA regardless of whether the VP35 1-80 peptide was present to chaperone NP°. However, oligomeric NP 1-450 co-purifies with RNA which could not be dissociated. These anecdotal data suggest that there is a connection between RNA binding and the oligomeric state of the NP. As noted above, the EBOV NP° core-VP35 peptide complex structure is in an open conformation similar to Nipah virus N°-P (Yabukarski et al., 2014). As the NP RNA-binding site is likely a composite of amino acids contributed by the N- and C-terminal domains, we predict that the EBOV NP open conformation with these domains splayed apart is incapable of presenting a fully formed RNA-binding site as observed in the RSV nucleoprotein closed conformation (Tawar et al., 2009). The VP35 N-terminal peptide binds to the side of the C-terminal NP core domain, and does not occlude the RNA binding site, in contrast to the VSV N°-P complex (Leyrat et al., 2011). The VP35 peptide also makes only limited contact with the N-terminal domain and is not expected to clash with a closed conformation of EBOV NP. Thus, VP35 is unlikely to directly block such a conformational transition.
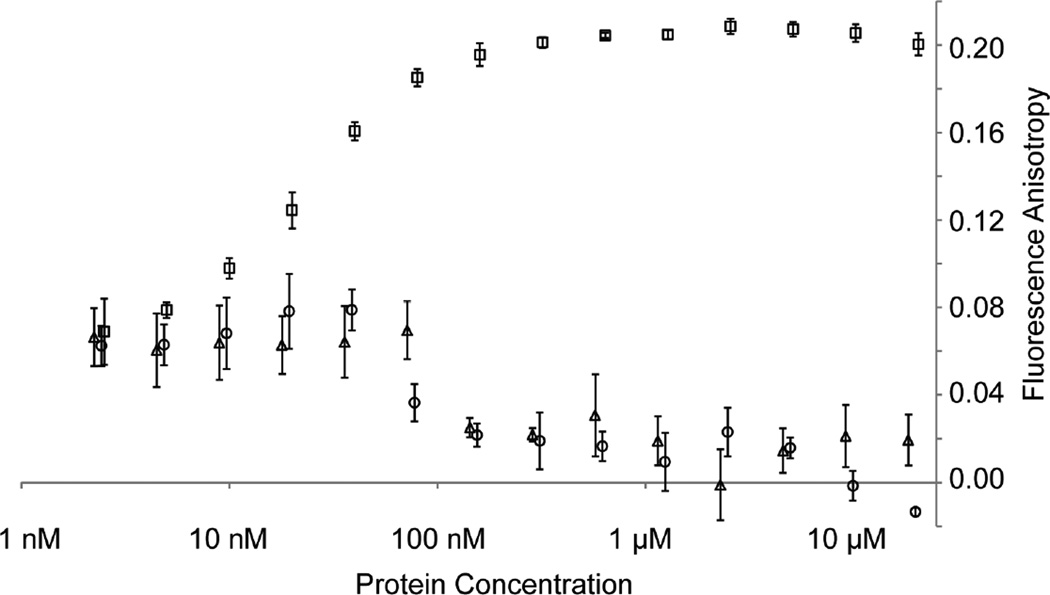
We hypothesized that rather than directly preventing the binding of NP to RNA or modulating the conformational transition from open to closed states, the VP35 chaperone functions to maintain NP in a monomeric form and that this monomeric form does not have an appreciable affinity for RNA. We tested this hypothesis by measuring the binding of RNA to monomeric or oligomeric NPs using fluorescence anisotropy. The oligomeric NP 1-450 readily binds to an 18 nucleotide RNA with low nanomolar affinity (~20 nM) (Fig. 5). However, neither of the monomeric NPs: chaperoned NP° core-VP35 peptide complex and unchaperoned NP° core (NP 34-367), show any significant binding to RNA with protein concentrations up to 20 µM. These data suggest that even without the VP35 peptide, monomeric NP° remains in an open conformation incapable of binding RNA.
Figure 5. Ebolavirus nucleoprotein RNA binding.
The potential of different NP N-terminal domain constructs to bind a FITC-labeled 18 nucleotide RNA was assessed. Binding was measured by changes in the fluorescence anisotropy of the FITC-RNA with increasing protein concentrations. The oligomeric NP 1-450 (squares) shows 50% binding at a protein concentration of ~20 nM. No appreciable binding is observed for the monomeric NP°-VP35 complex (circles, VP35 15-60-NP34-367 protein fusion) or for the monomeric NP° core (triangles, NP 34-367).
The VP35 peptide does not dissociate large NP-RNA oligomers
Above, we determined that NP oligomerization was not dependent on bound RNA and that RNA-free NP 1-450 oligomers could be reverted to a monomeric NP° by incubating the oligomers with the VP35 N-terminal peptide. However, NP purified directly from E. coli in an oligomeric form contains bound cellular RNA as assessed by the ratio of UV absorbance at 260 and 280 nm. Incubation of either the RNA-bound NP 1-450 or RNA-bound full-length NP with the VP35 N-terminal peptide did not convert oligomeric NP to a monomeric form (Table 1, samples 19–24). This suggests that while not required for oligomerization, RNA stabilizes the oligomeric NP complex. This result indicates that the VP35 N-terminal peptide is not capable of disassembling NP oligomer-RNA complexes into NP°-VP35 during viral replication to expose the RNA genome for RNA synthesis by the viral polymerase.
DISCUSSION
Our work has demonstrated that the EBOV VP35 contains a conserved peptide near its N terminus which is used to chaperone viral NP in a high affinity NP°-VP35 complex. Our structures of the NP° core-VP35 N-terminal peptide complex demonstrate that the VP35 N-terminal peptide binds a hydrophobic site on the side of the NP C-terminal domain. Further, the VP35 peptide adopts a unique conformation to straddle a β-hairpin so far only observed in filovirus NP. The EBOV VP35 N-terminal peptide binds to the NP° core with very high affinity and the binding is coupled to conformational changes in both NP and VP35. We have also identified the EBOV NP oligomerization arms and shown that NP oligomerization is dependent on the N-terminal arm which likely competes directly with the VP35 N-terminal peptide. The RNA-binding affinity of NP is not directly modulated by the presence of the VP35 N-terminal peptide, but rather indirectly by preventing NP oligomerization: neither the monomeric NP° core nor NP° core-VP35 peptide complex bind to RNA with any appreciable affinity. Finally, we have shown that the VP35 N-terminal peptide can reverse the oligomerization of RNA-free NP oligomers, but has no effect on RNA-bound NP oligomers. The structural and biochemical analysis performed here allows us to present a working model for ebolavirus NP assembly into a nucleocapsid-like oligomer (Fig. 6) and provide insights into NP’s role as a part of the viral RNA synthesis machinery.
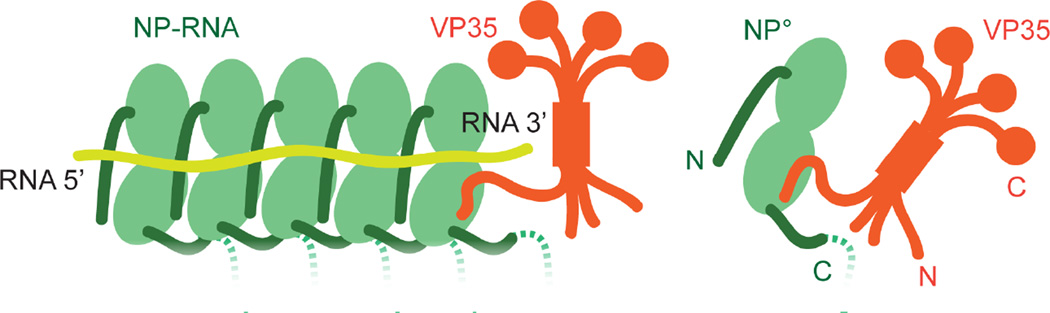
Figure 6. Ebolavirus VP35-NP° and assembly of NP-RNA.
This cartoon model of the assembly of NP-RNA oligomers from VP35-NP° monomers depicts the interactions between VP35 and NP as well as adjacent NPs. The NP core is shown in light green with terminal arms in dark green. The C-terminal tails are depicted as dashed lines. RNA is colored yellow and VP35 is in orange. Termini of proteins are labeled with N or C. The incoming VP35-NP° complex must displace the VP35 bound to the NP-RNA in order to continue the encapsidation of the viral RNA in the 5’ to 3’ direction. Once the VP35 has been displaced, its former binding site on NP is occupied by the N-terminal arm of the incoming NP-VP35 complex and the incoming NP undergoes a transition from an open to a closed conformation to bind RNA.
In our model for NP assembly into nucleocapsids, newly synthesized NP is chaperoned by high-affinity binding to the N-terminal region of VP35 creating a NP°-VP35 chaperone complex. The chaperoned NP must then find its way to an actively replicating nucleocapsid. As VP35 contains two independent binding sites for NP, the VP35 C-terminal domain may associate with nascent nucleocapsids thus guiding NP° to the RNA synthesis machinery. Alternatively, this may also be accomplished by VP35 binding to the viral polymerase, L.
The extremely high affinity binding of the VP35 N-terminal peptide to NP° would suggest that VP35 does not simply fall off of the NP°. Rather, this suggests that a process must occur in order to displace the VP35 peptide from NP°. As the VP35 peptide is able to dissociate NP oligomers in the absence of RNA in vitro, NP oligomerization alone is unlikely to be sufficient to displace the VP35 peptide from NP°. Because the NP° core domain, with or without bound VP35 N-terminal peptide, does not have appreciable affinity for RNA, RNA binding alone is also not likely to be the driving force behind VP35 peptide removal. It is possible that the peptide is actively removed by a protein component of the viral RNA replication machinery. Alternatively, the coupled process of NP oligomerization and concurrent RNA binding at the viral replication complex may provide the necessary force to displace the VP35 N-terminal peptide and provide the specificity for encapsidation of viral genomes.
Removal of the VP35 N-terminal peptide would reveal the hydrophobic surface on the C-terminal domain of NP°. Once this surface has been exposed, NP rapidly oligomerizes using its N- and C-terminal arms, spanning amino acids 20–38 and 356–381 respectively. Based on homology with RSV nucleoprotein, it is likely that the N-terminal oligomerization arm of an incoming NP occupies the hydrophobic surface formerly occupied by the VP35 N-terminal peptide to become the NPi−1 protomer (Fig 6.). We predict that the oligomerization of NP will trigger a conformational change between the N- and C-terminal domains transitioning the protein from an open to a closed form capable of efficiently binding RNA. It has been demonstrated for vesicular stomatitis virus (VSV) that phosphoprotein induces nucleoprotein to bind specifically to viral RNAs (Chen et al., 2013). As the RNA-binding by EBOV NP is believed to be nonspecific, but is nonetheless a high-affinity interaction (Fig. 5), RNA-binding by NP is likely to occur concurrently with or in very rapid succession to the removal of the VP35 N-terminal peptide and oligomerization of NP in order to ensure that only viral RNA and not cellular RNA is encapsidated.
In the available RNA-bound NNS virus nucleoprotein structures, the encapsidated RNA is hypothesized to be inaccessible for RNA synthesis and must first be displaced from the nucleoprotein prior to its use as a template by the viral polymerase (Green et al., 2006). Given the EBOV VP35 N-terminal peptide’s ability to dissociate EBOV NP oligomers, it is tempting to conclude that this peptide plays a role in disassembling NP oligomers during RNA synthesis to allow the viral polymerase to gain access to the genome. However, our in vitro data show that oligomeric NP-RNA complexes cannot be dissociated by the VP35 N-terminal peptide. It is possible though, that the VP35 N-terminal peptide may play a role in chaperoning the viral NP after RNA has been released during the transient disassociation of the RNA from the nucleocapsid that is believed to take place during NNS viral RNA synthesis (Green et al., 2006). It is also possible that the VP35 N-terminal peptide binds to the viral NP at the RNA genome 3’ end as this NP would not have an adjacent NPi−1 protomer N-terminal arm occupying the VP35 peptide-binding site (Leyrat et al., 2011). A bound VP35 at the 3’ end of the viral nucleocapsid could serve to recruit the viral polymerase, L, to the RNA genome transcription start site for viral transcription and replication.
During the preparation of this work, a second group working independently determined a crystal structure of an EBOV NP fragment bound to a VP35 peptide to 3.7 Å resolution (Leung et al., 2015). This structure and most of the accompanying data are consistent with the findings presented here. However, we do note several differences and our additional results lead us to present differing conclusions. In this work, we are able to confirm that EBOV VP35 possesses two unique and independent binding sites for NP interaction. We advance this finding to show that VP35 plays a similar role across the filoviruses in chaperoning NP as an RNA-free monomer. Our thermodynamic analysis of the NP°-VP35 binding event suggests that a large conformational change occurs upon binding, supporting the hypothesis that the VP35 N-terminal peptide is disordered in solution. The H/DXMS data and extensive truncation studies presented here clearly delineate the NP core domain and identify boundaries for the terminal oligomerization arms. Our smaller NP core (NP 34-367) is sufficient for the VP35 N-terminal peptide interaction and interacts with higher affinity (1 nM) than that observed by Leung et al. (18–29 nM). This likely owes to the inclusion in the Leung et al. construct (NP 25-457) of a portion of the N-terminal arm domain. Our data suggest the N-terminal arm domain (NP 20-29) competes with VP35 N-terminal peptide binding to the NP core and this competition may decrease the previously observed affinity from the affinity observed here. Similarly, Leung et al. observe the NP affinity for RNA to be 620 nM whereas we observe a much higher affinity of 20 nM. Again, this difference likely arises from the different NP constructs used for these determinations. Leung et al. use NP 25-457 for their RNA binding assays whereas here we use NP 1-450, the previously recognized region sufficient for NP RNA binding and oligomerization (Noda et al., 2011; Watanabe et al., 2006). Our data show the N-terminal arm is essential for NP oligomerization and that NP oligomerization is important for RNA binding. The partial deletion of the N-terminal arm in NP 25-457 is likely to destabilize the oligomeric NP. According to our model, in which a conformational change upon NP oligomerization forms the RNA binding site, deletion of half the N-terminal oligomerization arm could destabilize the NP oligomer and impair the affinity of NP for RNA. These affinity differences highlight the importance of experimental identification of the domain boundaries of the folded NP core (NP 34-367) and the terminal arm domains (NP 20-29 and 357-381) prior to considering models for VP35 and RNA binding to NP. While Leung et al. and the data presented here both show that NP can be dissociated into monomers by the VP35 peptide, we find that this dissociation can only be accomplished when the NP 1-450 is purified free from RNA. The RNA content of the Leung et al. NP 1-457 sample is unclear. However, the previous conclusion that the VP35 peptide dissociates NP-RNA oligomers is inconsistent with the observations that oligomeric NP-RNA in virions (Bharat et al., 2012), cells (Noda et al., 2011) or in vitro (as presented here) are not dissociated into monomers when bound by VP35.
The structure by Leung et al. differs from the structures presented here in the inclusion of a greater extent of the NP C-terminal region. This NP C-terminal region forms an additional helix which mediates crystal contacts and may represent a physiological oliogomerization interaction between NP C-terminal domains. These C-terminal helix interactions are not unique to EBOV, but were also observed to occur in the oligomeric, RNA-bound PIV5 nucleoprotein (Alayyoubi et al., 2015). Leung et al.’s proposed function of this region is in agreement with our H/DXMS data and accompanying NP truncation analysis using MALS and electron microscopy as well as our proposed model for NP oligomerization.
Finally, Leung et al. propose a process of NP assembly where the VP35 N-terminal peptide is first removed from the NP, then NP binds the RNA and finally NP oligomerizes. The results presented here revise this model. We show that the monomeric NP core has high affinity for the VP35 N-terminal peptide and no appreciable affinity for RNA. Further, our data show that NP oligomerization can occur independently of RNA binding. Based on these results we instead propose the displacement of the VP35 peptide from NP by an incoming NPi−1 with subsequent RNA binding by the oligomeric NP.
In this work, we have shown that the ebolavirus VP35 contains a chaperoning activity in its N-terminal region. This VP35 peptide is responsible for preventing premature NP oligomerization and RNA binding by maintaining the protein in a NP°-VP35 complex. This work has also yielded a higher-resolution view of the chaperoned filovirus NP core domain structure and delineated the regions of NP that likely correspond to the oligomerization-facilitating arm domains observed in other NNS virus nucleoproteins (Ruigrok et al., 2011). The scope of data presented here has granted a working model of NP assembly and RNA binding. Given the high level of sequence conservation across the filoviruses of both the VP35 N-terminal peptide and the NP RNA-binding domain, these results and conclusions are likely to be applicable across this family of viruses. Indeed, our data show the VP35 chaperoning activity to follow suit for both SUDV and MARV. These studies provide mechanistic insight into the workings of the complex RNA replication machinery belonging to this pathogenic family of viruses and yield a starting point for targeting these weaknesses with antiviral drugs.
EXPERIMENTAL PROCEDURES
Pull-downs
293T cells were transfected with HA-tagged full-length VP35 or VP35 truncation mutants in the vector pDisplay with an equal amount of NP-pcDNA. Cells were lysed after two days and HA-tagged VP35 was bound to anti-HA agarose beads (Roche) and washed with lysis buffer. Beads were resuspended and boiled in SDS-PAGE sample buffer prior to loading SDS-PAGE gels for Western blotting with anti-HA (VP35) or anti-NP antibodies.
Recombinant Protein Expression and Purification
Proteins were expressed in Escherichia coli using the pET46 vector (Novagen). For VP35 + NP co-expressions, VP35 was cloned in frame with upstream His-tags and the two proteins were expressed from a bicistronic mRNA. For preparation of oligomeric NP, an N-terminal MBP fusion was used as a spacer between the His-tag and the NP. In all cases, Rosetta2 pLysS E. coli (Novagen) were transformed with the expression vectors and induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) at an optical density at 600 nm of 0.4. Following overnight expression at 25°C, bacterial cultures were harvested and lysed using a microfluidizer. Cleared lysates were applied to Ni-NTA Agarose and eluted with lysis buffer containing 300 mM imidazole. For removal of purification tags, proteins were incubated with 0.5% (w/w) TEV protease overnight at 4°C. In cases where the protein was subsequently purified by anion exchange (MonoQ), the proteins were dialyzed beforehand into a low salt buffer. All proteins were finally purified by size exclusion chromatography (Superdex200). Nucleic acid content was assessed by measuring the A260/A280 ratios of the purified proteins. A260/A280 ratios less than 0.7 were considered to be RNA-free.
Hydrogen/Deuterium Exchange Mass Spectrometry
The monomeric, chaperoned, fusion protein, VP35 1-80–NP 1-450 or in vitro oligomerized, RNA-free NP 1-450 was diluted into D2O buffer for 10, 100, or 1000 s. Reactions were quenched by the addition of guanidine hydrochloride, glycerol and formic acid. Proteins were digested with pepsin and subjected to mass spectroscopic analysis.
Crystallization and Structure Solution
The EBOV VP35 15-60–NP 34-367 fusion protein was crystallized using sitting drop vapor diffusion techniques at 4°C. Crystals were cryoprotected in glycerol prior to cryo-cooling in liquid nitrogen. Data were collected at the Advanced Photon Source at Argonne National Labs, beamlines 23ID-B and 23ID-D. Data were reduced with XDS (Kabsch, 2010) and merged with AIMLESS (Evans, 2011) or XSCALE (Kabsch, 2010). Structure solution was performed in Phenix (Adams et al., 2010). Solutions were rebuilt in Coot (Emsley et al., 2010) and refined with Phenix (Adams et al., 2010) and BUSTER-TNT (Blanc et al., 2004) with final rounds of refinement in Refmac (Murshudov et al., 2011).
Multi-angle Light Scattering
Approximately 100 µg of purified NP protein was used for light scattering experiments. In cases where VP35 1-80 peptide was mixed with NP, a 50-fold molar excess of peptide was used and the samples were incubated overnight at 4°C. Light scattering data were collected on a Dawn MiniTreos (Wyatt Technologies) in line with a Superdex200 column and analyzed with ASTRA (Wyatt Technologies).
Negative Stain Electron Microscopy
Copper grids (carbon coated, 400 mesh, Electron Microscopy Sciences, Hatfield PA) were glow discharged. Samples were applied to grids and then stained with uranyl acetate. Dried grids were examined on a Philips CM100 electron microscope (FEI) at 80 kv.
Isothermal Titration Calorimetry
Concentrated proteins were dialyzed overnight into the matching buffers. NP samples were loaded into the ITC cell and VP35 peptide samples were loaded into the syringe. Reactions were run by performing 20 injections of VP35 into the cell at indicated temperatures. Data were processed using Origin (OriginLab).
RNA-Binding Assay
Two-fold serial dilutions of EBOV NP proteins were mixed with carboxyfluoroscein-labeled 18 nucleotide RNA (final concentration 4 nM, Integrated DNA Technologies). Fluorescence anisotropy was read on a Tecan Infinite 200 Pro (Rossi and Taylor, 2011).
Mini-genome Assay
Screening of VP35 mutants was done as in (Jasenosky et al., 2010). HEK 293T cells were transfected with plasmids expressing NP, VP35, VP30, L, T7 RNA polymerase, Renilla luciferase proteins and the gene for firefly luciferase flanked by viral sequences whose expression is driven by a T7 RNA polymerase promoter. VP35 was either wild-type or mutated in the N-terminal conserved region. Dual luciferase assays were performed according to manufacturer’s instructions (Promega).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yoshihiro Kawaoka for providing us with plasmids for the EBOV mini-genome assay. This work was funded by the Skaggs Institute of Chemical Biology and the National Institutes of Health 1R56 AI118016-01. R.N.K. was funded by a National Institutes of Health training grant 5T32 AI007354-25. E.O.S was funded by an Investigators in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Fund. GM/CA at the Advanced Photon Source has been funded in whole or in part with Federal funds from the National Cancer Institute ACB-12002 and the National Institute of General Medical Sciences AGM-12006. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (D.O.E.) Office of Science User Facility operated for the D.O.E. Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This is TSRI manuscript number 29051
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Coordinates of the EBOV NP° core bound to the VP35 N-terminal peptide in three different space groups have been deposited in the Protein Data Bank with accession numbers 4ZTA, 4ZTI and 4ZTG.
Supplemental information accompanying this work includes Supplemental figures S1–S5 and Extended Experimental Procedures.
AUTHOR CONTRIBUTIONS
R.N.K designed constructs and carried out protein characterizations. D.M.A produced plasmid DNA constructs. S.L. performed the H/DXMS analysis. M.R.W. imaged samples with negative-stain electron microscopy. R.N.K. and E.O.S designed experiments and wrote the manuscript.
The authors declare no conflicts of interest.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agua-Agum J, Ariyarajah A, Aylward B, Blake IM, Brennan R, Cori A, Donnelly CA, Dorigatti I, Dye C, Eckmanns T, et al. West African Ebola epidemic after one year-- slowing but not yet under control. N. Engl. J. Med. 2015;372:584–587. doi: 10.1056/NEJMc1414992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alayyoubi M, Leser GP, Kors CA, Lamb RA. Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein-RNA complex. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E1792–E1799. doi: 10.1073/pnas.1503941112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313:360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- Bharat TA, Noda T, Riches JD, Kraehling V, Kolesnikova L, Becker S, Kawaoka Y, Briggs JA. Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4275–4280. doi: 10.1073/pnas.1120453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- Burke J, Declerq R, Ghysebrechts G, Pattyn SR, Piot P, Ruppol JF, Thonon D, Van Der Groen G, Van Nieuwenhov S, Witvrouwen M, et al. Ebola haemorrhagic fever in Zaire, 1976. Bull. World Health Organ. 1978;56:271–293. [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang S, Banerjee AK, Chen M. N-terminal phosphorylation of phosphoprotein of vesicular stomatitis virus is required for preventing nucleoprotein from binding to cellular RNAs and for functional template formation. J. Virol. 2013;87:3177–3186. doi: 10.1128/JVI.02761-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J, Marq JB, Kolakofsky D. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J. Virol. 1995;69:849–855. doi: 10.1128/jvi.69.2.849-855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott LH, Kiley MP, McCormick JB. Descriptive analysis of Ebola virus proteins. Virology. 1985;147:169–176. doi: 10.1016/0042-6822(85)90236-3. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TJ, Zhang X, Wertz GW, Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- Groseth A, Charton JE, Sauerborn M, Feldmann F, Jones SM, Hoenen T, Feldmann H. The Ebola virus ribonucleoprotein complex: a novel VP30-L interaction identified. Virus Res. 2009;140:8–14. doi: 10.1016/j.virusres.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha JH, Spolar RS, Record MT., Jr Role of the hydrophobic effect in stability of site-specific protein-DNA complexes. J. Mol. Biol. 1989;209:801–816. doi: 10.1016/0022-2836(89)90608-6. [DOI] [PubMed] [Google Scholar]
- Hartlieb B, Muziol T, Weissenhorn W, Becker S. Crystal structure of the C-terminal domain of Ebola virus VP30 reveals a role in transcription and nucleocapsid association. Proc. Natl. Acad. Sci. U. S. A. 2007;104:624–629. doi: 10.1073/pnas.0606730104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Huang Y, Xu L, Sun Y, Nabel GJ. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol. Cell. 2002;10:307–316. doi: 10.1016/s1097-2765(02)00588-9. [DOI] [PubMed] [Google Scholar]
- Jasenosky LD, Neumann G, Kawaoka Y. Minigenome-based reporter system suitable for high-throughput screening of compounds able to inhibit Ebolavirus replication and/or transcription. Antimicrob. Agents Chemother. 2010;54:3007–3010. doi: 10.1128/AAC.00138-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin D, Belshaw R. Detecting remote sequence homology in disordered proteins: discovery of conserved motifs in the N-termini of Mononegavirales phosphoproteins. PLoS One. 2012;7:e31719. doi: 10.1371/journal.pone.0031719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin CR, Bornholdt ZA, Li S, Woods VL, Jr, MacRae IJ, Saphire EO. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc. Natl. Acad. Sci. U. S. A. 2010;107:314–319. doi: 10.1073/pnas.0910547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe DM, Howley PM. Field's virology. 4th edn. Philadelphia, PA: Lippincott Williams and Wilkins; 2001. [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Leung DW, Borek D, Luthra P, Binning JM, Anantpadma M, Liu G, Harvey IB, Su Z, Endlich-Frazier A, Pan J, et al. An Intrinsically Disordered Peptide from Ebola Virus VP35 Controls Viral RNA Synthesis by Modulating Nucleoprotein-RNA Interactions. Cell Rep. 2015;11:376–389. doi: 10.1016/j.celrep.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Prins KC, Borek DM, Farahbakhsh M, Tufariello JM, Ramanan P, Nix JC, Helgeson LA, Otwinowski Z, Honzatko RB, et al. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat. Struct. Mol. Biol. 2010;17:165–172. doi: 10.1038/nsmb.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyrat C, Yabukarski F, Tarbouriech N, Ribeiro EA, Jr, Jensen MR, Blackledge M, Ruigrok RW, Jamin M. Structure of the vesicular stomatitis virus N(0)-P complex. PLoS Pathog. 2011;7:e1002248. doi: 10.1371/journal.ppat.1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A, Bhattacharya R, Basak S, Shaila MS, Chattopadhyay D, Roy S. P-protein of Chandipura virus is an N-protein-specific chaperone that acts at the nucleation stage. Biochemistry. 2004;43:2863–2870. doi: 10.1021/bi035793r. [DOI] [PubMed] [Google Scholar]
- Martinez MJ, Biedenkopf N, Volchkova V, Hartlieb B, Alazard-Dany N, Reynard O, Becker S, Volchkov V. Role of Ebola virus VP30 in transcription reinitiation. J. Virol. 2008;82:12569–12573. doi: 10.1128/JVI.01395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis M, Kolesnikova L, Schoehn G, Becker S, Ruigrok RW. Morphology of Marburg virus NP-RNA. Virology. 2002;296:300–307. doi: 10.1006/viro.2002.1433. [DOI] [PubMed] [Google Scholar]
- Muhlberger E, Lotfering B, Klenk HD, Becker S. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J. Virol. 1998;72:8756–8764. doi: 10.1128/jvi.72.11.8756-8764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negredo A, Palacios G, Vazquez-Moron S, Gonzalez F, Dopazo H, Molero F, Juste J, Quetglas J, Savji N, de la Cruz Martinez M, et al. Discovery of an ebolavirus-like filovirus in europe. PLoS Pathog. 2011;7:e1002304. doi: 10.1371/journal.ppat.1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Ebihara H, Muramoto Y, Fujii K, Takada A, Sagara H, Kim JH, Kida H, Feldmann H, Kawaoka Y. Assembly and budding of Ebolavirus. PLoS Pathog. 2006;2:e99. doi: 10.1371/journal.ppat.0020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Kolesnikova L, Becker S, Kawaoka Y. The importance of the NP: VP35 ratio in Ebola virus nucleocapsid formation. J. Infect. Dis. 2011;204(Suppl 3):S878–S883. doi: 10.1093/infdis/jir310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins KC, Binning JM, Shabman RS, Leung DW, Amarasinghe GK, Basler CF. Basic residues within the ebolavirus VP35 protein are required for its viral polymerase cofactor function. J. Virol. 2010;84:10581–10591. doi: 10.1128/JVI.00925-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP, Cardenas WB, Basler CF. Homo-oligomerization facilitates the interferon-antagonist activity of the ebolavirus VP35 protein. Virology. 2005;341:179–189. doi: 10.1016/j.virol.2005.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi AM, Taylor CW. Analysis of protein-ligand interactions by fluorescence polarization. Nat. Protoc. 2011;6:365–387. doi: 10.1038/nprot.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MG, Kraus I, Dickmanns A, Eickmann M, Garten W, Ficner R. Crystal structure of the borna disease virus nucleoprotein. Structure. 2003;11:1219–1226. doi: 10.1016/j.str.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Ruigrok RW, Crepin T, Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr. Opin. Microbiol. 2011;14:504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- Tawar RG, Duquerroy S, Vonrhein C, Varela PF, Damier-Piolle L, Castagne N, MacLellan K, Bedouelle H, Bricogne G, Bhella D, et al. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009;326:1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- Trunschke M, Conrad D, Enterlein S, Olejnik J, Brauburger K, Muhlberger E. The L-VP35 and L-L interaction domains reside in the amino terminus of the Ebola virus L protein and are potential targets for antivirals. Virology. 2013;441:135–145. doi: 10.1016/j.virol.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Noda T, Kawaoka Y. Functional mapping of the nucleoprotein of Ebola virus. J. Virol. 2006;80:3743–3751. doi: 10.1128/JVI.80.8.3743-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weik M, Modrof J, Klenk HD, Becker S, Muhlberger E. Ebola virus VP30-mediated transcription is regulated by RNA secondary structure formation. J. Virol. 2002;76:8532–8539. doi: 10.1128/JVI.76.17.8532-8539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabukarski F, Lawrence P, Tarbouriech N, Bourhis JM, Delaforge E, Jensen MR, Ruigrok RW, Blackledge M, Volchkov V, Jamin M. Structure of Nipah virus unassembled nucleoprotein in complex with its viral chaperone. Nat. Struct. Mol. Biol. 2014;21:754–759. doi: 10.1038/nsmb.2868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.