Abstract
Background
Phosphoinositides, lipid-signaling molecules, participate in diverse brain processes within a wide metabolic cascade.
Hypothesis
Gene transcriptional networks coordinately regulate the phosphoinositide cascade during human brain Development and Aging.
Methods
We used the public BrainCloud database for human dorsolateral prefrontal cortex to examine age-related expression levels of 49 phosphoinositide metabolic genes during Development (0 to 20+ years) and Aging (21+ years).
Results
We identified three groups of partially overlapping genes in each of the two intervals, with similar intergroup correlations despite marked phenotypic differences between Aging and Development. In each interval, ITPKB, PLCD1, PIK3R3, ISYNA1, IMPA2, INPPL1, PI4KB, and AKT1 are in Group 1, PIK3CB, PTEN, PIK3CA, and IMPA1 in Group 2, and SACM1L, PI3KR4, INPP5A, SYNJ1, and PLCB1 in Group 3. Ten of the genes change expression nonlinearly during Development, suggesting involvement in rapidly changing neuronal, glial and myelination events. Correlated transcription for some gene pairs likely is facilitated by colocalization on the same chromosome band.
Conclusions
Stable coordinated gene transcriptional networks regulate brain phosphoinositide metabolic pathways during human Development and Aging.
Introduction
Phosphoinositides, inositol-containing derivatives of phosphatidic acid that lack nitrogen, participate in neurotransmission, autophagy, apoptosis, neuronal and glial growth, myelination, and membrane trafficking in brain [1–3]. Their participation is highly energy dependent [1–3]. Phosphoinositide metabolism is disturbed in many human brain diseases [1, 4–7] and in animal models for some of these diseases [8–10]. Changes in phosphoinositide metabolites and enzymes also accompany normal human brain development and aging [4–7, 11, 12].
The complexity of brain phosphoinositide metabolism limits our understanding the roles of phosphoinositides in Development and Aging and our ability to design therapeutic interventions in disease states [10, 13–17]. One way to address these limitations may be to analyze age-related transcription of phosphoinositide genes in brain over the lifespan. During Development (0 to ~20 years), the human brain undergoes marked nonlinear changes in synaptic and dendritic growth and pruning, neuronal loss, glial elaboration and myelination, in arachidonic and docosahexaenoic acid concentrations, and it shifts from ketone body to glucose consumption for ATP synthesis [18–24]. During later Aging (21+ years), brain function and metabolism are maintained in a more homeostatic range, although risk for neurodegeneration increases [25].
Several databases are available to examine age changes in gene expression in the human brain, including the publically accessible BrainCloud for the dorsolateral prefrontal cortex (http://braincloud.jhmi.edu) [26–28]. We recently used BrainCloud to demonstrate age-related coordinated expression patterns during Development and Aging of genes of phospholipase A2 (PLA2)-initiated arachidonic acid (AA, 20:4n-6) and docosahexaenoic acid (22:6n-3) metabolic cascades [29] and of genes for cytokines, chemokines, and other inflammatory proteins [30].
In the present study, we used BrainCloud to compare age-related expression in human dorsolateral prefrontal cortex of 49 genes involved in phosphoinositide synthesis, degradation, and signaling [1, 2]. Based on our prior studies [29, 30], we hypothesized that we could identify coordinated expression of these genes during the Development and Aging intervals. Such changes might correspond to changes in biochemical reactions involving the gene products and be facilitated by colocalization on a chromosomal band [29–34].
Methods
We selected 49 genes involved in phosphoinositide metabolism, based on canonical pathways reported in Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, Redwood City, CA, http://www.ingenuity.com) and other sources [1, 2]. Expression data for each gene were exported from the BrainCloud database from 231 males and females ranging in age from birth to 78 years [26]. No subject had a history of significant psychiatric, neurological disorder, or drug abuse, or postmortem evidence of neuropathology.
As described in our prior studies, we separated the samples into Development (0 to 20.95 years, 87 subjects) and Aging (21 to 78.2 years, 144 subjects) intervals [29, 30]. Gender and race breakdowns, as well as a description of the data in BrainCloud, have been reported earlier [29, 30].
Twenty-two of the 49 genes chosen were detected by more than one probe in the BrainCloud database. When possible (18 of these 22 genes), the probe covering all possible alternate transcripts of the gene was chosen using the Gene View tab on BrainCloud. The probe that covered all transcripts also was the highest intensity probe for all but one gene (PIP5K1A), for which we used the probe covering all the transcripts. If one probe did not cover all possible transcripts, we chose the highest intensity probe (SLC5A3, PTGS2, and ITPK1).
Statistical tests were performed using Partek Genomics Suite (Version 6.6, Partek, St. Louis, MO, USA) and GraphPad Prism 5 (Version 5.02, GraphPad Software, La Jolla, CA, USA). First, a t-test was performed in Partek to determine whether mean expression levels differed significantly between the Aging and Development intervals for each gene. Pearson’s r correlations were performed in GraphPad Prism 5 (Graph Pad Software, La Jolla, CA) for each gene, to determine correlation with age in each interval. Visual observation during Development suggested nonlinear expression for some genes. We therefore compared goodness of fit of the data with a nonlinear equation, Y = (Y0 - Plateau)*exp(-K*A) + Plateau (where Y = expression level at age A, and Y0 expression level at A = 0 years), to goodness of fit with a linear regression between 0 and 20 years [29, 30].
Data also were analyzed with Cluster 3.0 software [35], without filtering or adjustment. As described earlier [29, 30], we calculated distance between probes using the Euclidean distance calculation and clustering using the centroid linkage method expression [36]. The output.cdt file was loaded into TreeView program [37] to generate figures showing relatedness among genes of interest.
Similarity matrices using Pearson’s r correlations and hierarchical clustering also were created using Partek, to make correlation (heat) maps showing correlation coefficients and gene clusters in expression between pairs of genes during Development and Aging [29, 30]. Correlation data from the heat maps were used to construct corresponding statistical significance matrices. GraphPad Prism was used to calculate Pearson’s r correlations for pairs of genes closely located on the same chromosome band.
Ethics Statement. This research was supported entirely by the Intramural Programs of the National Institute on Aging, the National Institute of Alcohol Abuse and Alcoholism, and the National Institute of Mental Health, National Institutes of Health. No author has a conflict of interest. Samples were collected under NIH protocol number NCT00001260, 900142, which include written informed consent from next-of-kin including consent for clinical records to be used. Every brain is consented.
Results
Construction of phosphoinositide metabolic pathways
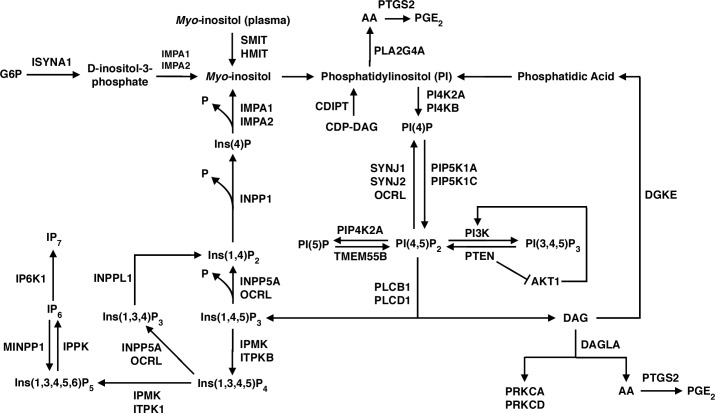
We constructed pathways of phosphoinositide metabolism within a large cascade, as illustrated in Fig 1, when using ingenuity analysis (http://www.ingenuity.com) and other literature references [1, 2]. In the upper left of Fig 1, inositol-3-phosphate synthase (ISYNA1) is shown to catalyze synthesis of D-inositol-3-phosphate from glucose-6-phosphate (G6P). Then, inositol monophosphatase (IMPA1, IMPA2) hydrolyzes D-inositol-3-phosphate to form myo-inositol, which also can be transported into brain from blood by the sodium/myo-inositol transporter (SMIT, SLC5A3) or the H+/myo-inositol symporter (HMIT, SLC2A13) [1, 38]. CDP (cytidine 5'-diphospho)-diacylglycerol-inositol-3-phosphatidyltransferase (CDIPT) phosphorylates myo-inositol to phosphatidyl-1D-myo-inositol (phosphatidylinositol, PI) using CDP-diacylglycerol. PI constitutes the largest phosphoinositide pool in brain [2].
Fig 1. Pathways of phosphoinositide metabolism.
See Results section for details.
Phosphatidylinositol is phosphorylated by PI 4-kinase (PI4KB, PI4K2A) to PI(4)P, which in turn is phosphorylated to PI(4,5)P2 by PI(4)P 5-kinase (PIP5K1A, PIP5K1C) [2]. These reactions occur primarily in the endoplasmic reticulum, from where phosphoinositides can be brought to the plasma membrane via membrane trafficking, but some reactions also take place at the plasma membrane itself [2]. Reconversion (dephosphorylation) of PI(4,5)P2 to PI(4)P can be catalyzed by synaptojanin (SYNJ1, SYNJ2) and Oculocerebrorenal Syndrome Of Lowe (OCRL) [2, 39].
Phospholipase C (PLCB1, PLCD1) is a critical enzyme that integrates phosphoinositide metabolism with neurotransmission, hormonal, and other signaling processes. PLC can be activated by G-protein mechanisms coupled to muscarinic cholinergic M1,3,5, dopaminergic D2,3,4, serotonergic 5-HT2A,C, and other post-synaptic neuroreceptors [40], or to hormone receptors [2]. PLC cleaves membrane PI(4,5)P2 to form two important second messengers, cytosol-soluble inositol 1,4,5-trisphosphate (Ins(1,4,5)P3, or IP3) and membrane-bound diacylglycerol (DAG) [1, 14].
Newly formed IP3 moves through the cytosol and binds to IP3 receptors on the endoplasmic reticulum, thereby stimulating calcium release [41, 42]. Released calcium creates a positive feedback loop to PLC and to store-operated plasma membrane calcium channels [41, 42]. IP3 also can be dephosphorylated by Type I inositol 1,4,5-trisphosphate 5-phosphatase (INPP5A) to Ins(1,4)P2. Ins(1,4)P2 in turn can be dephosphorylated to Ins(4)P and thence by IMPA1 or IMPA2 to myo-inositol. Released myo-inositol becomes available for synthesis of PI and the resynthesis of PI (4,5)P2 to complete the “PI cycle” (Fig 1) [1, 2, 43].
Additionally, IP3 can be converted to highly phosphorylated inositol pyrophosphates like IP7, which participate in cell growth and apoptosis [44] (Fig 1, lower left). IP4 and IP5 are formed primarily by inositol polyphosphate mutikinase (IPMK); deleting IPMK in mice prevents formation of IP5, IP6, and IP7 [44, 45].
IPMK also can act as a PI3 kinase (PI3K), which has implications in the mTOR (mammalian target of rapamycin)/protein kinase B (AKT) signaling pathway that participates in growth processes [44]. Inhibition of this pathway, either directly by rapamycin or indirectly by caloric restriction, has been implicated in increased longevity in animal models [46]. Also, apoptosis and autophagy are mediated by phosphatidylinositol 4,5-bisphosphate 3-kinase (PIK3CB, PIK3CA) and AKT [47]. AKT interacts with phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase (PTEN) (Fig 1, center).
The other second messenger released by PLC mediated hydrolysis is membrane-bound DAG, which binds to and activates protein kinase C (PKC) and other kinases that phosphorylate intracellular proteins [2]. DAG can be lost by phosphorylation by diacylglycerol kinase (DGKE) and recycled into phosphatidylinositol (PI), effectively terminating activation of PKC, or it can be hydrolyzed by diacylglycerol lipase (DAGLA) to arachidonic acid (AA). Arachidonic acid in turn can be oxidized by COX-2 (PTGS2) to produce pro-inflammatory metabolites like prostaglandin E2 (PGE2) (Fig 1, lower right) [29, 48]. Arachidonic acid also may be released from PI by calcium-dependent cPLA2 type IVA (PLA2G4A), then converted to PGE2 by COX-2 (Fig 1, top) [29, 31].
Age correlations in expression of genes and gene pairs
Table 1 lists the 49 genes in this analysis, their corresponding protein names, chromosomal locations, probes used, and results of t-tests comparing mean expression levels between Aging and Development. All fold changes are less than |2|, suggesting relatively stable expression throughout the lifespan. SYNJ2 has the highest fold increase (1.81, p = 10−15) and SLC2A13 has the largest fold decrease (-1.67, p = 10−9) in Aging compared with Development. Correcting for 49 comparisons, at p < 0.001, 14 genes are higher and of 6 genes are lower in mean expression during Aging than Development.
Table 1. Gene list with protein name, chromosomal location, and ANOVA comparing mean expression levels between Aging and Development.
| Gene Name | Protein Name | Chromosome | Probe | Fold change | p-value |
|---|---|---|---|---|---|
| CYTH3 | General receptor for phosphoinositides | 7p22.1 | 1 | -1.16 | 0.0001 |
| IMPA1 | Inositol monophosphatase 1 | 8q21.1-q21.3 | 1 | 1.21 | 3.27E-07 |
| INPP5A | Type I inositol 1,4,5-trisphosphate 5-phosphatase | 10q26.3 | 1 | 1.23 | 9.44E-07 |
| IP6K1 | Inositol hexakisphosphate kinase 1 | 3p21.31 | 1 | 1.2 | 5.19E-07 |
| ITPKB | Inositol-trisphosphate 3-kinase B | 1q42.12 | 2 | 1.62 | 2.16E-13 |
| MTM1 | Myotubularin | Xq27.3-q28 | 2 | 1.26 | 1.21E-06 |
| MTMR14 | Myotubularin-related protein 14 | 3p26 | 2 | 1.14 | 1.31E-08 |
| PIK3R4 | Phosphoinositide 3-kinase regulatory subunit 4 | 3q22.1 | 1 | 1.27 | 8.08E-10 |
| PIP5K1C | Phosphatidylinositol 4-phosphate 5-kinase type-1 gamma | 19p13.3 | 2 | -1.22 | 4.64E-07 |
| PRKCD | Protein kinase C, delta | 3p21.31 | 1 | -1.27 | 2.99E-06 |
| SACM1L | Phosphatidylinositide phosphatase SAC1 | 3p21.3 | 1 | 1.27 | 1.16E-11 |
| SLC2A13 | Solute carrier family 2 (facilitated glucose transporter), member 13 (HMIT) | 12q12 | 1 | -1.67 | 1.29E-09 |
| SYNJ1 | Synaptojanin-1 | 21q22.2 | 2 | 1.32 | 9.25E-06 |
| SYNJ2 | Synaptojanin-2 | 6q25.3 | 1 | 1.81 | 1.85E-15 |
| TMEM55B | Type 1 phosphatidylinositol 4,5-bisphosphate 4-phosphatase | 14q11.1 | 1 | 1.2 | 9.55E-13 |
| PIP4K2A | Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | 10p12.2 | 2 | 1.27 | 0.0005 |
| PTEN | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase | 10q23 | 1 | -1.27 | 0.0004 |
| PIK3R2 | Phosphatidylinositol 3-kinase regulatory subunit beta | 19p13.11 | 1 | -1.12 | 0.0005 |
| PRKCA | Protein kinase C, alpha | 17q22-q24 | 2 | 1.14 | 0.0006 |
| PIK3R1 | Phosphatidylinositol 3-kinase regulatory subunit alpha | 5q13.1 | 2 | 1.16 | 0.00071 |
| ISYNA1 | Inositol-3-phosphate synthase 1 | 19p13.11 | 1 | ns | |
| MINPP1 | Multiple inositol polyphosphate phosphatase 1 | 10q23 | 2 | ns | |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | 1q25.2-q25.3 | 4 | ns | |
| CDIPT | CDP-diacylglycerol—inositol 3-phosphatidyltransferase | 16p11.2 | 2 | ns | |
| GRASP | General receptor for phosphoinositides associated scaffold protein | 12q13.13 | 1 | ns | |
| PI4KB | Phosphatidylinositol 4-kinase beta | 1q21 | 2 | ns | |
| INPP1 | Inositol polyphosphate 1-phosphatase | 2q32 | 1 | ns | |
| PIK3C3 | Phosphatidylinositol 3-kinase catalytic subunit type 3 | 18q12.3 | 1 | ns | |
| IPMK | Inositol polyphosphate multikinase | 10q21.1 | 1 | ns | |
| INPP4A | Type I inositol 3,4-bisphosphate 4-phosphatase | 2q11.2 | 2 | ns | |
| PIK3C2B | Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit beta | 1q32 | 1 | ns | |
| DAGLA | Diacylglycerol lipase, alpha | 11q12.3 | 1 | ns | |
| AKT1 | Protein kinase B | 14q32.32-q32.33 | 2 | ns | |
| PI4K2A | Phosphatidylinositol 4-kinase type 2-alpha | 10q24 | 1 | ns | |
| PIK3R3 | Phosphatidylinositol 3-kinase regulatory subunit gamma | 1p34.1 | 2 | ns | |
| OCRL | Inositol polyphosphate 5-phosphatase OCRL-1 | Xq25 | 2 | ns | |
| INPPL1 | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | 11q23 | 2 | ns | |
| PIK3CB | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform | 3q22.3 | 1 | ns | |
| PIP5K1A | Phosphatidylinositol 4-phosphate 5-kinase type-1 alpha | 1q21.3 | 3 | ns | |
| SLC5A3 | Solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 (SMIT) | 21q22.11 | 4 | ns | |
| PLA2G4A | Phospholipase A2, group IVA (cytosolic, calcium-dependent) | 1q25 | 1 | ns | |
| PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | 3q26.3 | 1 | ns | |
| PLCB1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | 20p12 | 2 | ns | |
| DGKE | diacylglycerol kinase, epsilon | 17q22 | 2 | ns | |
| IPPK | Inositol-pentakisphosphate 2-kinase | 9q22.31 | 1 | ns | |
| PLCD1 | 1-Phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1 | 3p22-p21.3 | 1 | ns | |
| ITPK1 | Inositol-tetrakisphosphate 1-kinase | 14q32.12 | 4 | ns | |
| IMPA2 | Inositol monophosphatase 2 | 18p11.2 | 1 | ns | |
| PIK3C2A | Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha | 11p15.5-p14 | 1 | ns |
The probe column indicates type of probe used: 1, only one probe in database; 2, highest intensity probe covering all transcripts; 3, probe used covers all transcripts but is not highest intensity probe; 4, highest intensity probe as none of the probes covered all transcripts. The ANOVA column shows whether levels in Aging are higher or lower than in Development, after correcting for 49 comparisons (p < 0.001). ns, not significant.
Table 2 shows statistically significant correlations between gene expression levels and age in the two intervals. Correcting for 98 comparisons, at p < 0.0005 fewer age correlations are evident during Aging than Development. Only expression of ITPKB (r = 0.34) and of GRASP (r = - 0.38) correlates significantly with age during the Aging interval. During Development, on the other hand, expression of PIP42K2A, SYNJ2, SACM1L, IMPA1, PLA2G4A, SYNJ1, PIK3CB, PIK3R4, INPP1, PRKCD, PIK3CD, PIK3C3, and ITPKB increase significantly, while expression of AKT1, PIK3R2, IPMK, CYTH3, PIP5K1C, and PIK3C2B decrease significantly.
Table 2. Statistically significant correlations of gene expression with age during Development and Aging intervals.
| Gene name | Development | Aging | ||
|---|---|---|---|---|
| Pearson r | p value | Pearson r | p value | |
| PIP4K2A | 0.66 | < 0.0001 | ||
| AKT1 | -0.59 | < 0.0001 | ||
| SYNJ2 | 0.81 | < 0.0001 | ||
| SACM1L | 0.60 | < 0.0001 | ||
| IMPA1 | 0.60 | < 0.0001 | ||
| PLA2G4A | 0.55 | < 0.0001 | ||
| SYNJ1 | 0.52 | < 0.0001 | ||
| PIK3CB | 0.53 | < 0.0001 | ||
| PIK3R2 | -0.58 | < 0.0001 | ||
| PIK3R4 | 0.46 | < 0.0001 | ||
| INPP1 | 0.45 | < 0.0001 | ||
| IPMK | -0.43 | < 0.0001 | ||
| CYTH3 | -0.45 | < 0.0001 | ||
| PIP5K1C | -0.46 | < 0.0001 | ||
| PIK3C2B | -0.40 | 0.0001 | ||
| PRKCD | 0.39 | 0.0002 | ||
| PIK3C3 | 0.37 | 0.0004 | ||
| ITPKB | 0.37 | 0.0004 | 0.34 | < 0.0001 |
| GRASP | -0.38 | < 0.0001 | ||
Statistical significance, corrected for 98 comparisons, taken as p < 0.0005. Nonsignificant values not shown.
Visual observation of expression levels during Development suggested nonlinear changes for some genes. We tested this by comparing nonlinear to linear goodness of fits for each of the 49 genes. For 10 of them, as illustrated in Fig 2, expression of ITBKB, PIP4K2A, SYNJ2, PRKCD, GRASP, PLA2G4A and PTGS2 increase non-linearly in the first years of life before reaching a plateau, whereas expression of PIK3C2B, CYTH3, and DGKE decline before reaching a plateau.
Fig 2. Nonlinear gene expression changes in relation to age during Development, for 10 genes.
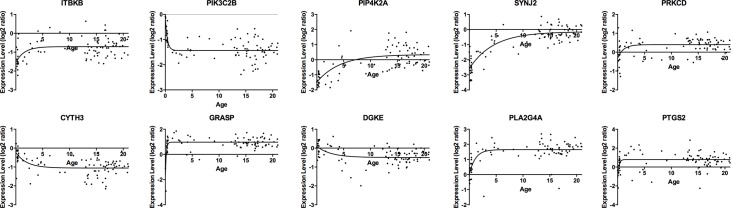
Data were fit by following equation, Y = (Y0 - Plateau)*exp(-K*A) + Plateau (where Y = expression level at age A, and Y0 expression level at A = 0 years).
Colocalization on a chromosome may facilitate transcription of genes whose protein products participate in tightly connected metabolic pathways [29, 32]. To consider this mechanism for our genes, we list in Table 3 statistically significant (p < 0.0001) correlations between genes on the same chromosomal band. On 1q25, expression of PLA2G4A (cPLA2 Type IVA) correlates with expression of PTGS2 (COX-2) during both Development and Aging. Genes located on 3p21-23, PLCD1, SACMIL, IP6K1, and PRKCD, are significantly correlated with each other during Development and/or Aging. Significant correlations during both intervals occur between PIK3R4 and PIK3CB on 3q22 and between ITPK1 and AKT1 on 14q32. During Development DGKE and PRKCA are significantly correlated on 17q22.
Table 3. Significant correlations between pairs of genes located on the same chromosomal band during the Development and Aging intervals.
| Development | Aging | |||||
|---|---|---|---|---|---|---|
| Chromosomal Location | Gene | Gene | Pearson's r | p-value | Pearson's r | p-value |
| 1q25 | PLA2G4A | PTGS2 | 0.58 | < 0.0001 | 0.54 | < 0.0001 |
| 3p21-23 | SACM1L | PLCD1 | -0.41 | < 0.0001 | ||
| 3p21-23 | IP6K1 | PLCD1 | -0.43 | < 0.0001 | -0.49 | < 0.0001 |
| 3p21-23 | PRKCD | PLCD1 | -0.52 | < 0.0001 | ||
| 3q22 | PIK3R4 | PIK3CB | 0.48 | < 0.0001 | 0.38 | < 0.0001 |
| 14q32 | ITPK1 | AKT1 | 0.46 | < 0.0001 | 0.48 | < 0.0001 |
| 17q22 | DGKE | PRKCA | 0.49 | < 0.0001 | ||
Nonsignificant correlations not shown.
Cooperative clustered transcription correlations within extended groups
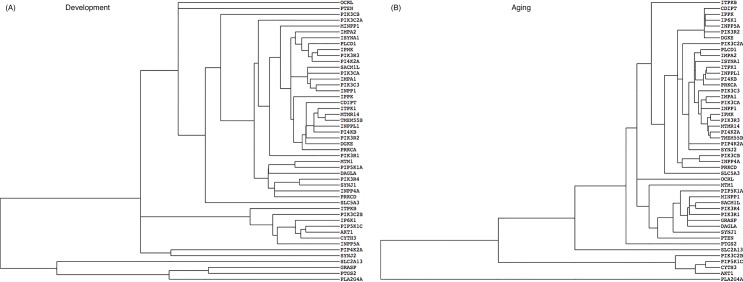
TreeView dendrograms can identify genes whose transcription is coordinated or clustered in a hierarchical cascade, indicating relatedness and common cellular processes [29, 30, 49]. For example, the Development dendrogram (Fig 3A) shows hierarchical interactions of IMPA2 and ISYNA1 (involved in myo-inositol synthesis); of PLCD1, IPMK, PIK3R3, and PI4K2A; and of SACMIL, PIK3CA, IMPA1, PIK3C3, and INPP1. PLA2G4A, PTGS2 GRASP, and SLC2A13 are distant (more dissimilar expression patterns) from the other genes. The Aging dendrogram (Fig 3B) shows that IMPA2 is closely tied to PLCD1 and that PLA2G4A also is distant from the other genes.
Fig 3. Hierarchical clustering of gene expression during Development (A) and Aging (B) intervals.
Horizontal length indicates relative relatedness of age-dependent gene expression levels based on cluster calculations.
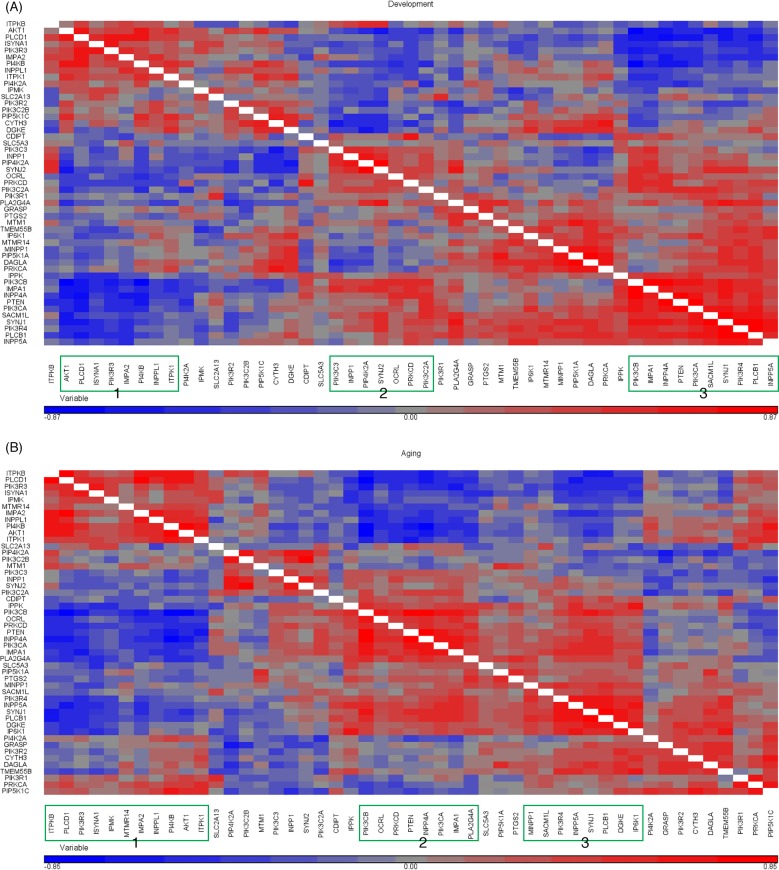
Pearson’s correlation matrices (heat maps) of pairwise correlations among the 49 genes were created using unsupervised hierarchical clustering within the Development (Fig 4A) and Aging (Fig 4B) intervals. Hierarchical clustering row and column titles are not conserved between heat maps in the two intervals, as they represent the highest probability of correctly clustering genes based on Pearson’s r correlation in each interval. In the figures, genes that are positively intercorrelated within a cluster are highlighted in red, while those that are negatively intercorrelated are shown in blue.
Fig 4. Heat maps for pairwise correlations of genes in Development (A) and Aging (B) intervals.
Correlations or p-values are hierarchically clustered using the centroid linkage method.
Three distinct clusters of genes with highly positively intercorrelated expression levels are identified in both the Development and Aging intervals (green outlined boxes on x-axis). During Development (Fig 4A), the three groups are: AKT1, PLCD1, ISYNA1, PIK3R3, IMPA2, PI4KB, INPPL1, and ITPK1 (Group 1); PIK3C3, INPP1, PIP4K2A, SYNJ2, OCRL, PRKCD, and PIK3C2A (Group 2); and PIK3CB, IMPA1, INPP4A, PTEN, PIK3CA, SACM1L, SYNJ1, PIK3R4, PLCB1, and INPP5A (Group 3).
S1A Fig presents correlation values corresponding to Fig 4A, and identifies correlations significant at p < 0.001 with green highlighting. It shows that genes within each of the three separate groups of Fig 4A are significantly intercorrelated. Genes in Group 1 are highly and negatively correlated with genes in Group 2. Genes in Group 1 and Group 3 also are highly and negatively correlated across groups, while genes in Group 2 and Group 3 are positively correlated across groups.
The heat map for Aging (Fig 4B) also identifies three distinct groups of intercorrelated genes, with many similarities (bolded) to the respective Development groups in Fig 4A: ITPKB, PLCD1, PIK3R3, ISYNA 1, IPMK, MTMR14, IMPA2, INPPL1, PI4KB, and AKT1 (Group 1); PIK3CB, OCRL, PRKCD, PTEN, INPPLA, PIK3CA, IMPA1, and PLA2G4A (Group 2); and MINPP1, SACM1L, PI3KR4, INPP5A, SYNJ1, PLCB1, DGKE, and IP6K1 (Group 3). The matrix of corresponding exact correlation values at p < 0.001 (S1B Fig) shows similar patterns to what is found during Development. Thus, genes within each of the three groups are significantly intercorrelated. Genes in Group 1 are negatively correlated with genes in Group 2, and genes in Group 3 are negatively correlated with ITPKB, PLCD1, PIK3R3, ISYNA1, and IPMK in Group 1. Genes in Groups 2 and 3 are positively correlated across groups.
Discussion
Based on the literature, we constructed a phenotypic cascade containing known pathways of brain phosphoinositide metabolism and identified 49 genes whose protein products participate in this cascade (Fig 1). Among the 49 genes, we identified three groups in both Development and Aging and showed that the groups have similar intercorrelations and partially overlapping composition in the two intervals. In both intervals, ITPKB, PLCD1, PIK3R3, ISYNA1, IMPA2, INPPL1, PI4KB, and AKT1 are Group 1, PIK3CB, PTEN, PIK3CA, and IMPA1 are in Group 2, and SACM1L, PI3KR4, INPP5A, SYNJ1, and PLCB1 are in Group 3. Genes in Group 1 are negatively correlated with genes in Groups 2 and 3. Genes in Groups 2 and 3 are positively correlated across groups.
The similar gene groups and their interrelations in Development and Aging suggest that stable transcriptional networks underlie brain phosphoinositide metabolism throughout the entire lifespan, despite marked phenotypic differences between the two intervals [18–24]. Such stable networks likely arose through evolutionary constraints that maximized functional efficiency and minimized energy requirements of the metabolic pathways that were regulated [3, 50, 51]. A number of mechanisms may have contributed to their elaboration, such required coupling of reaction products and enzymes, enzyme colocalization at a cellular cite, organization of the nucleosome to bring genes and promoter regions together for co-transcription, and gene colocalization on common chromosomal bands [29, 32, 52].
We considered gene colocalization on the same chromosomal band in Table 3. Thus, expression levels correlate during Development and Aging for PLA2G4A and PTGS2 colocalized on 1q25, consistent with evidence that arachidonic acid may have to be liberated from phospholipid by PLA2 before it can be oxidized to PGE2 by COX-2 [53, 54]. Levels for PIK3R4 and PIK3CB on 3q22 and for ITPK1 and AKT1 on 14q32 also correlate significantly in the two age intervals. Levels for PLCD1, SACMIL, IP6K1, and for PRKCD on 3p21-23 correlate during Development and/or Aging, and levels for DGKE and PRKCA on 17q22 correlated during Development.
The dendrograms of Fig 4 identify certain gene expression hierarchies in the two intervals. For example, the Development dendrogram (Fig 4A) shows hierarchical relations among IMPA2, ISYNA1, PLCD1, IPMK, PIK3R3, and PI43K, which are distant from PLA2G4A and PTGS2 within the arachidonic acid cascade [29]. PLA2G4A also is separated from the phosphoinositide hierarchy in the Aging dendrogram (Fig 3B).
Our separating Aging and Development in this study is consistent with our prior studies using BrainCloud [29, 30], and with phenotypic differences between the two intervals [18–24]. Of the 49 genes studied, 15 have a higher and 5 genes a lower mean expression level during Aging than Development (Table 1). Significant age correlations also are fewer during Aging than Development (Table 2), but lesser variation during Aging may have reduced our power to determine statistically significant correlations [55].
Nonlinear increases in ITBKB, PIP4K2A, SYNJ2, PRKCD, GRASP, PLA2G4A and PTGS2 expression and decreases in PIK3C2B, CYTH3, and DGKE expression during Development likely correspond to the many nonlinear phenotypic changes that have ben described, suggesting a role for phosphoinositides in them. There is rapid neuronal loss in the first year of life, dendritic growth followed by pruning over a 15 year period, declining myelination, changing arachidonic and docosahexaenoic acid concentrations throughout the entire interval, and conversion from ketone body to glucose use for oxidative metabolism in the first year [18–24].
Some of our correlations are consistent with reported enzyme coupling in phosphoinositide enzyme pathways (Fig 1). For example, PTEN (phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase) dephosphorylates PI(3,4,5)P3 to PI(4,5)P2 and negatively influences both AKT (AKT1) and PI(3,4,5)P3 signaling [56]. PTEN and AKT1 are in separate groups that are correlated negatively in the Development and Aging heat maps (Fig 4A and 4B). Aberrant activation of neuronal PI3K/AKT/mTOR and PTEN signaling may be an early prelude of Alzheimer disease [57, 58].
The association between PLCD1 and IMPA2 in Group 1 in each age interval also is of interest because IMPA2, which dephosphorylates myo-inositol monophosphate within the “PI cycle”, has been proposed as the preferred target of lithium in the treatment bipolar disorder [59–61]. The PI cycle is initiated by PLC-hydrolysis of PI(4,5)P2, followed by formation Ins(1,4,5)P3, Ins(1,4)P2, Ins(4)P, myo-inositol, phosphatidylinositol (PI), PI(4)P and finally resynthesis of PI(4,5)P2 [43, 60].
During Development, the positive age correlations of SYNJ1 and SYNJ2 expression (Table 2), whose protein products modify clathrin-mediated synaptic endocytosis [2, 39], correspond to a reported increase in SYNJ immunoreactivity and dendritic spine density in brain [19, 62]. SYNJ2 expression is 1.8 fold higher in Aging than Development, suggesting late stage synaptic changes [63]. Finally, SYNJ1, SYNJ2, and INPP1 are in Group 3 with PTEN in both intervals. It is reasonable that genes whose proteins degrade PI(3,4,5)P3 should be positively correlated with PTEN, because if PTEN negatively regulates PI(3,4,5)P3 signaling it could do so by interacting with other enzymes that degrade PI(3,4,5)P3.
The 1.67 fold decrement in SLC2A13 (HMIT) expression in Aging compared with Development does not correspond to a decreased brain myo-inositol concentration [64, 65]. However, the negative age correlation in GRASP expression (Table 2) during Aging suggests glutamatergic alterations, since GRASP links Group 1 metabotropic glutamate receptors to neuronal proteins [66]. The highly positive correlation of ITPKB expression during Aging differs from a report that ITPKB mRNA was not increased with age in postmortem brain [67].
Class IA PI3K dimers evolved from a single enzyme in unicellular eukaryotes [68]. They consist of a p110 catalytic and a p85 regulatory subunit, each of which has three isoforms, PIK3CA, PIK3CB, and PIK3CD, and PIK3R1, PIK3R2 and PIK3R3, respectively [68]. Expression of PIK3R2 decreases while that of PIK3R4 increases during Development (Table 2), demonstrating the principle of divergent expression after gene duplication [69]. The regulatory subunit PIK3R3 is in Group 1 while the catalytic subunits PIK3CA and PIK3CB are in Group 3 in both the Development and Aging heat maps and the genes in the two groups are negatively correlated.
Although BrainCloud is a powerful tool for examining gene expression changes with age, it has limitations. It only contains data for the dorsolateral prefrontal cortex [26], but expression patterns differ between brain regions [70]. It also does not distinguish between cell types, which can also have distinct transcriptional trajectories [27, 28, 70]. However, BrainCloud does have a large number of samples, which increases its statistical power. The Allen Human Brain Atlas contains data only from 3 men, while the Loerch et al. study contains 28 brain samples [26–28].
In the future, it would be of interest to investigate mechanisms underlying coordinated transcription in relation to changing levels of the transcribed proteins. Methylation of gene promoters, histone acetylation and methylation state, transcription factors, miRNAs, DNA sequences of cis-elements (transcription factor binding sites), and feedback regulation by metabolites can influence expression [71–73]. Genes whose expression decreases with age appear to have higher promoter GC content than the other genes [71], suggesting differences in methylation state [74].
In summary, we hade described coordinated changes during Development and Aging in transcription of genes coding for multiple aspects of brain phosphoinositide metabolism, suggesting important roles for these genes. Three somewhat similar groups of genes with distinct expression intercorrelations were identified in each of the two intervals, and some pairwise correlations could be related to colocalization on the same chromosomal band. Nonlinear changes during Development likely participate in concurrent nonlinear phenotypic changes within this period. Mechanisms of coordinated transcription in normal as well as pathological human brain deserve to be explored further.
Supporting Information
Correlations highlighted in green are significant at p < 0.05 and p < 0.001 shown on separate tabs.
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files. All BrainCloud files are available from the GEO Accession database (accession number: GSE30272).
Funding Statement
The Intramural Research Programs of the National Institute on Aging, National Institute on Alcohol Abuse and Alcoholism, National Institute of Mental Health supported this research.
References
- 1. Fisher SK, Novak JE, Agranoff BW. Inositol and higher inositol phosphates in neural tissues: homeostasis, metabolism and functional significance. Journal of neurochemistry. 2002;82(4):736–54. Epub 2002/10/03. . [DOI] [PubMed] [Google Scholar]
- 2. Falkenburger BH, Jensen JB, Dickson EJ, Suh BC, Hille B. Phosphoinositides: lipid regulators of membrane proteins. The Journal of physiology. 2010;588(Pt 17):3179–85. Epub 2010/06/04. 10.1113/jphysiol.2010.192153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Purdon AD, Rapoport SI. Energy consumption by phospholipid metabolism in mammalian brain In: Gibson G, Dienel G, editors. Neural Energy Utilization: Handbook of Neurochemistry and Molecular Biology. Handbook of Neurochemistry and Molecular Neurobiology. 3rd ed. New York: Springer; 2007. p. 401–27. [Google Scholar]
- 4. Huang W, Alexander GE, Daly EM, Shetty HU, Krasuski JS, Rapoport SI, et al. High brain myo-inositol levels in the predementia phase of Alzheimer's disease in adults with Down's syndrome: a 1H MRS study. Am J Psychiatry. 1999;156(12):1879–86. . [DOI] [PubMed] [Google Scholar]
- 5. Kantarci K, Jack CR Jr, Xu YC, Campeau NG, O'Brien PC, Smith GE, et al. Regional metabolic patterns in mild cognitive impairment and Alzheimer's disease: A 1H MRS study. Neurology. 2000;55(2):210–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jolles J, Bothmer J, Markerink M, Ravid R. Reduced phosphatidylinositol kinase activity in Alzheimer's disease: effects of age and onset. Dementia. 1993;4(2):81–6. Epub 1993/03/01. . [DOI] [PubMed] [Google Scholar]
- 7. Pacheco MA, Jope RS. Phosphoinositide signaling in human brain. Progress in neurobiology. 1996;50(2–3):255–73. Epub 1996/10/01. . [DOI] [PubMed] [Google Scholar]
- 8. Voronov SV, Frere SG, Giovedi S, Pollina EA, Borel C, Zhang H, et al. Synaptojanin 1-linked phosphoinositide dyshomeostasis and cognitive deficits in mouse models of Down's syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(27):9415–20. Epub 2008/07/02. 10.1073/pnas.0803756105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shetty HU, Holloway HW, Acevedo LD, Galdzicki Z. Brain accumulation of myo-inositol in the trisomy 16 mouse, an animal model of Down syndrome. Biochem J. 1996;313:31–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Law AJ, Wang Y, Sei Y, O'Donnell P, Piantadosi P, Papaleo F, et al. Neuregulin 1-ErbB4-PI3K signaling in schizophrenia and phosphoinositide 3-kinase-p110delta inhibition as a potential therapeutic strategy. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(30):12165–70. Epub 2012/06/13. 10.1073/pnas.1206118109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Greenwood AF, Powers RE, Jope RS. Phosphoinositide hydrolysis, G alpha q, phospholipase C, and protein kinase C in post mortem human brain: effects of post mortem interval, subject age, and Alzheimer's disease. Neuroscience. 1995;69(1):125–38. Epub 1995/11/01. . [DOI] [PubMed] [Google Scholar]
- 12. Shetty HU, Schapiro MB, Holloway HW, Rapoport SI. Polyol profiles in Down syndrome. myo-Inositol, specifically, is elevated in the cerebrospinal fluid. J Clin Invest. 1995;95(2):542–6. Epub 1995/02/01. 10.1172/JCI117696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benjamin J, Agam G, Levine J, Bersudsky Y, Kofman O, Belmaker RH. Inositol treatment in psychiatry. Psychopharmacology bulletin. 1995;31(1):167–75. Epub 1995/01/01. . [PubMed] [Google Scholar]
- 14. Irvine RF. 20 years of Ins(1,4,5)P3, and 40 years before. Nature reviews Molecular cell biology. 2003;4(7):586–90. Epub 2003/07/03. 10.1038/nrm1152 . [DOI] [PubMed] [Google Scholar]
- 15. Heffron TP, Salphati L, Alicke B, Cheong J, Dotson J, Edgar K, et al. The design and identification of brain penetrant inhibitors of phosphoinositide 3-kinase alpha. Journal of medicinal chemistry. 2012;55(18):8007–20. Epub 2012/09/06. 10.1021/jm300867c . [DOI] [PubMed] [Google Scholar]
- 16. Ishrat T, Sayeed I, Atif F, Hua F, Stein DG. Progesterone is neuroprotective against ischemic brain injury through its effects on the phosphoinositide 3-kinase/protein kinase B signaling pathway. Neuroscience. 2012;210:442–50. Epub 2012/03/28. 10.1016/j.neuroscience.2012.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Delgado-Esteban M, Martin-Zanca D, Andres-Martin L, Almeida A, Bolanos JP. Inhibition of PTEN by peroxynitrite activates the phosphoinositide-3-kinase/Akt neuroprotective signaling pathway. Journal of neurochemistry. 2007;102(1):194–205. Epub 2007/02/17. 10.1111/j.1471-4159.2007.04450.x . [DOI] [PubMed] [Google Scholar]
- 18. Huttenlocher PR. Synapse elimination and plasticity in developing human cerebral cortex. American journal of mental deficiency. 1984;88(5):488–96. Epub 1984/03/01. . [PubMed] [Google Scholar]
- 19. Petanjek Z, Judas M, Simic G, Rasin MR, Uylings HB, Rakic P, et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(32):13281–6. Epub 2011/07/27. 10.1073/pnas.1105108108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yakovlev PI, Lecours AR. The myelogenetic cycles of regional maturation of the brain In: Minkowski A, editor. Regional Development of the Brain in Early Life. Philadelphia: F. A. Davis; 1967. p. 3–70. [Google Scholar]
- 21. Sowell ER, Thompson PM, Toga AW. Mapping changes in the human cortex throughout the span of life. The Neuroscientist: a review journal bringing neurobiology, neurology and psychiatry. 2004;10(4):372–92. 10.1177/1073858404263960 . [DOI] [PubMed] [Google Scholar]
- 22. Rapoport SI. Integrated phylogeny of the primate brain, with special reference to humans and their diseases. Brain research Brain research reviews. 1990;15(3):267–94. Epub 1990/09/01. . [DOI] [PubMed] [Google Scholar]
- 23. Carver JD, Benford VJ, Han B, Cantor AB. The relationship between age and the fatty acid composition of cerebral cortex and erythrocytes in human subjects. Brain Res Bull. 2001;56(2):79–85. . [DOI] [PubMed] [Google Scholar]
- 24. Chugani HT, Phelps ME. Imaging human brain development with positron emission tomography. Journal of nuclear medicine: official publication, Society of Nuclear Medicine. 1991;32(1):23–6. Epub 1991/01/01. . [PubMed] [Google Scholar]
- 25. Shankar SK. Biology of aging brain. Indian journal of pathology & microbiology. 2010;53(4):595–604. Epub 2010/11/04. 10.4103/0377-4929.71995 . [DOI] [PubMed] [Google Scholar]
- 26. Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478(7370):519–23. Epub 2011/10/28. 10.1038/nature10524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–76. Epub 2006/12/08. 10.1038/nature05453 . [DOI] [PubMed] [Google Scholar]
- 28. Loerch PM, Lu T, Dakin KA, Vann JM, Isaacs A, Geula C, et al. Evolution of the aging brain transcriptome and synaptic regulation. PloS one. 2008;3(10):e3329 Epub 2008/10/03. 10.1371/journal.pone.0003329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ryan VH, Primiani CT, Rao JS, Ahn K, Rapoport SI, Blanchard H. Coordination of gene expression of arachidonic and docosahexaenoic acid cascade enzymes during human brain development and aging. PloS One. 2014;9(6):e100858 Epub 2014/06/26. 10.1371/journal.pone.0100858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Primiani CT, Ryan VH, Rao JS, Cam MC, Ahn K, Modi HR, et al. Coordinated Gene Expression of Neuroinflammatory and Cell Signaling Markers in Dorsolateral Prefrontal Cortex during Human Brain Development and Aging. PloS One. 2014;9(10):e110972 Epub 2014/10/21. 10.1371/journal.pone.0110972 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laposata M, Majerus PW. Measurement of icosanoid precursor uptake and release by intact cells. Methods Enzymol. 1987;141:350–5. . [DOI] [PubMed] [Google Scholar]
- 32. Dai Z, Dai X. Nuclear colocalization of transcription factor target genes strengthens coregulation in yeast. Nucleic acids research. 2012;40(1):27–36. Epub 2011/09/02. 10.1093/nar/gkr689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hu L, Grosberg AY, Bruinsma R. Are DNA transcription factor proteins maxwellian demons? Biophysical journal. 2008;95(3):1151–6. Epub 2008/05/06. 10.1529/biophysj.108.129825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tamarkin-Ben-Harush A, Schechtman E, Dikstein R. Co-occurrence of transcription and translation gene regulatory features underlies coordinated mRNA and protein synthesis. BMC genomics. 2014;15:688 Epub 2014/08/20. 10.1186/1471-2164-15-688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–4. Epub 2004/02/12. 10.1093/bioinformatics/bth078 . [DOI] [PubMed] [Google Scholar]
- 36. Eisen M. Cluster 3.0 Manual In: Hoon Md, editor. University of Tokyo: Human Genome Center; 1998. [Google Scholar]
- 37. Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–8. Epub 2004/06/08. 10.1093/bioinformatics/bth349 . [DOI] [PubMed] [Google Scholar]
- 38. Hauser G, Finelli VN. The Biosynthesis of Free and Phosphatide Myo-Inositol from Glucose by Mammalian Tissue Slices. The Journal of biological chemistry. 1963;238:3224–8. Epub 1963/10/01. . [PubMed] [Google Scholar]
- 39. Wenk MR, De Camilli P. Protein-lipid interactions and phosphoinositide metabolism in membrane traffic: insights from vesicle recycling in nerve terminals. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(22):8262–9. Epub 2004/05/18. 10.1073/pnas.0401874101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cooper JR, Bloom FE, Roth RH. The Biochemical Basis of Neuropharmacology. 8th ed. Oxford: Oxford University Press; 2003. 404 p. [Google Scholar]
- 41. Miyazaki S. Inositol trisphosphate receptor mediated spatiotemporal calcium signalling. Current opinion in cell biology. 1995;7(2):190–6. Epub 1995/04/01. . [DOI] [PubMed] [Google Scholar]
- 42. Putney JW. Capacitative calcium entry: from concept to molecules. Immunological reviews. 2009;231(1):10–22. Epub 2009/09/17. 10.1111/j.1600-065X.2009.00810.x . [DOI] [PubMed] [Google Scholar]
- 43. Dawson AP, Lea EJ, Irvine RF. Kinetic model of the inositol trisphosphate receptor that shows both steady-state and quantal patterns of Ca2+ release from intracellular stores. The Biochemical journal. 2003;370(Pt 2):621–9. Epub 2002/12/14. 10.1042/BJ20021289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee JY, Kim YR, Park J, Kim S. Inositol polyphosphate multikinase signaling in the regulation of metabolism. Annals of the New York Academy of Sciences. 2012;1271:68–74. Epub 2012/10/12. 10.1111/j.1749-6632.2012.06725.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim S, Kim SF, Maag D, Maxwell MJ, Resnick AC, Juluri KR, et al. Amino acid signaling to mTOR mediated by inositol polyphosphate multikinase. Cell metabolism. 2011;13(2):215–21. Epub 2011/02/03. 10.1016/j.cmet.2011.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, et al. Global epigenomic reconfiguration during Mammalian brain development. Science. 2013;341(6146):1237905 Epub 2013/07/06. 10.1126/science.1237905 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dall'Armi C, Devereaux KA, Di Paolo G. The role of lipids in the control of autophagy. Current biology: CB. 2013;23(1):R33–45. Epub 2013/01/12. 10.1016/j.cub.2012.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rapoport SI. Brain arachidonic and docosahexaenoic acid cascades are selectively altered by drugs, diet and disease. Prostaglandins, leukotrienes, and essential fatty acids. 2008;79(3–5):153–6. Epub 2008/11/01. 10.1016/j.plefa.2008.09.010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(25):14863–8. Epub 1998/12/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ma K, Deutsch J, Villacreses NE, Rosenberger TA, Rapoport SI, Shetty HU. Measuring brain uptake and incorporation into brain phosphatidylinositol of plasma myo-[2H6]inositol in unanesthetized rats: an approach to estimate in vivo brain phosphatidylinositol turnover. Neurochem Res. 2006;31(6):759–65. . [DOI] [PubMed] [Google Scholar]
- 51.Cunnane SC, Crawford MA. Energetic and nutritional constraints on infant brain development: Implications for brain expansion during human evolution. Journal of human evolution. 2014. Epub 2014/06/15. 10.1016/j.jhevol.2014.05.001 . [DOI] [PubMed]
- 52.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nature reviews Molecular cell biology. 2015. Epub 2015/02/05. 10.1038/nrm3941 . [DOI] [PubMed]
- 53. Neufeld EJ, Majerus PW, Krueger CM, Saffitz JE. Uptake and subcellular distribution of [3H]arachidonic acid in murine fibrosarcoma cells measured by electron microscope autoradiography. J Cell Biol. 1985;101(2):573–81. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Furth EE, Laposata M. Mass quantitation of agonist-induced arachidonate release and icosanoid production in a fibrosarcoma cell line. Effect of time of agonist stimulation, amount of cellular arachidonate, and type of agonist. J Biol Chem. 1988;263(31):15951–6. . [PubMed] [Google Scholar]
- 55. Sullivan GM, Feinn R. Using Effect Size-or Why the P Value Is Not Enough. Journal of graduate medical education. 2012;4(3):279–82. Epub 2013/09/03. 10.4300/JGME-D-12-00156.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Downward J. PI 3-kinase, Akt and cell survival. Seminars in cell & developmental biology. 2004;15(2):177–82. Epub 2004/06/24. . [DOI] [PubMed] [Google Scholar]
- 57. O' Neill C. PI3-kinase/Akt/mTOR signaling: impaired on/off switches in aging, cognitive decline and Alzheimer's disease. Experimental gerontology. 2013;48(7):647–53. Epub 2013/03/09. 10.1016/j.exger.2013.02.025 . [DOI] [PubMed] [Google Scholar]
- 58. Griffin RJ, Moloney A, Kelliher M, Johnston JA, Ravid R, Dockery P, et al. Activation of Akt/PKB, increased phosphorylation of Akt substrates and loss and altered distribution of Akt and PTEN are features of Alzheimer's disease pathology. Journal of neurochemistry. 2005;93(1):105–17. Epub 2005/03/19. 10.1111/j.1471-4159.2004.02949.x . [DOI] [PubMed] [Google Scholar]
- 59. Sjoholt G, Ebstein RP, Lie RT, Berle JO, Mallet J, Deleuze JF, et al. Examination of IMPA1 and IMPA2 genes in manic-depressive patients: association between IMPA2 promoter polymorphisms and bipolar disorder. Molecular psychiatry. 2004;9(6):621–9. Epub 2003/12/31. 10.1038/sj.mp.4001460 . [DOI] [PubMed] [Google Scholar]
- 60. Berridge MJ. The Albert Lasker Medical Awards. Inositol trisphosphate, calcium, lithium, and cell signaling. Jama. 1989;262(13):1834–41. . [PubMed] [Google Scholar]
- 61. Serretti A, Drago A, De Ronchi D. Lithium pharmacodynamics and pharmacogenetics: focus on inositol mono phosphatase (IMPase), inositol poliphosphatase (IPPase) and glycogen sinthase kinase 3 beta (GSK-3 beta). Current medicinal chemistry. 2009;16(15):1917–48. Epub 2009/05/16. . [DOI] [PubMed] [Google Scholar]
- 62. Arai Y, Ijuin T, Itoh M, Takenawa T, Takashima S, Becker LE. Developmental changes of synaptojanin expression in the human cerebrum and cerebellum. Brain research Developmental brain research. 2001;129(1):1–9. Epub 2001/07/17. . [DOI] [PubMed] [Google Scholar]
- 63. George AA, Hayden S, Holzhausen LC, Ma EY, Suzuki SC, Brockerhoff SE. Synaptojanin 1 is required for endolysosomal trafficking of synaptic proteins in cone photoreceptor inner segments. PloS one. 2014;9(1):e84394 Epub 2014/01/07. 10.1371/journal.pone.0084394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pouwels PJ, Brockmann K, Kruse B, Wilken B, Wick M, Hanefeld F, et al. Regional age dependence of human brain metabolites from infancy to adulthood as detected by quantitative localized proton MRS. Pediatric research. 1999;46(4):474–85. Epub 1999/10/06. 10.1203/00006450-199910000-00019 . [DOI] [PubMed] [Google Scholar]
- 65. Grachev ID, Swarnkar A, Szeverenyi NM, Ramachandran TS, Apkarian AV. Aging alters the multichemical networking profile of the human brain: an in vivo (1)H-MRS study of young versus middle-aged subjects. Journal of neurochemistry. 2001;77(1):292–303. Epub 2001/03/30. . [DOI] [PubMed] [Google Scholar]
- 66. Kitano J, Kimura K, Yamazaki Y, Soda T, Shigemoto R, Nakajima Y, et al. Tamalin, a PDZ domain-containing protein, links a protein complex formation of group 1 metabotropic glutamate receptors and the guanine nucleotide exchange factor cytohesins. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2002;22(4):1280–9. Epub 2002/02/19. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Saetre P, Jazin E, Emilsson L. Age-related changes in gene expression are accelerated in Alzheimer's disease. Synapse. 2011;65(9):971–4. Epub 2011/03/23. 10.1002/syn.20933 . [DOI] [PubMed] [Google Scholar]
- 68. Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nature reviews Genetics. 2006;7(8):606–19. Epub 2006/07/19. 10.1038/nrg1879 . [DOI] [PubMed] [Google Scholar]
- 69. Ohno S. Evolution by Gene Duplication. Berlin: Springer Verlag; 1970. 160 p. [Google Scholar]
- 70. Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489(7416):391–9. Epub 2012/09/22. 10.1038/nature11405 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Somel M, Guo S, Fu N, Yan Z, Hu HY, Xu Y, et al. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome research. 2010;20(9):1207–18. Epub 2010/07/22. 10.1101/gr.106849.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang D, Dubois RN. The Role of Anti-Inflammatory Drugs in Colorectal Cancer. Annual review of medicine. 2012. Epub 2012/10/02. 10.1146/annurev-med-112211-154330 . [DOI] [PubMed]
- 73. Persengiev S, Kondova I, Bontrop R. Insights on the functional interactions between miRNAs and copy number variations in the aging brain. Frontiers in molecular neuroscience. 2013;6:32 10.3389/fnmol.2013.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. American journal of human genetics. 2012;90(2):260–72. Epub 2012/02/07. 10.1016/j.ajhg.2011.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlations highlighted in green are significant at p < 0.05 and p < 0.001 shown on separate tabs.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All BrainCloud files are available from the GEO Accession database (accession number: GSE30272).