Abstract
Viruses manipulate cellular machinery and functions to subvert intracellular environment conducive for viral proliferation. They strategically alter functions of the multitasking mitochondria to influence energy production, metabolism, survival, and immune signaling. Mitochondria either occur as heterogeneous population of individual organelles or large interconnected tubular network. The mitochondrial network is highly susceptible to physiological and environmental insults, including viral infections, and is dynamically maintained by mitochondrial fission and fusion. Mitochondrial dynamics in tandem with mitochondria-selective autophagy ‘mitophagy’ coordinates mitochondrial quality control and homeostasis. Mitochondrial dynamics impacts cellular homeostasis, metabolism, and innate-immune signaling, and thus can be major determinant of the outcome of viral infections. Herein, we review how mitochondrial dynamics is affected during viral infections and how this complex interplay benefits the viral infectious process and associated diseases. This article is part of a Special Issue entitled: Mitophagy.
Keywords: Mitophagy, Mitochondrial fission and fusion, Innate immunity, Virus, Hepatitis virus
Highlights
-
•
Mitochondrial dynamics influences mitochondrial and cellular functions.
-
•
Mitochondrial dynamics is affected during viral infections.
-
•
Viruses exploit mitochondrial dynamics and mitophagy to benefit infectious process.
-
•
Virus-altered mitochondrial dynamics determines the outcome of infection.
-
•
Disruption of mitochondrial dynamics promotes viral pathogenesis.
1. Introduction
Viruses are obligate parasites completely reliant on host cell energy and molecular machinery to multiply. In order to attain a cellular environment conducive for proliferation, the viruses strategically modulate host cells' metabolism and physiology, resulting in a dramatic alteration of cellular and sub-cellular architecture and functions [1].
Mitochondria have emerged as one of the key organelles in the maintenance of cellular homeostasis, metabolism, aging, innate immunity, apoptosis and other signaling pathways [2], [3]. In the last decade, work on the mitochondria has expanded our knowledge of its roles in cellular homeostasis in many parallel ways. At subcellular level, the size, shape and motility of the mitochondria are governed by mitochondrial dynamics that has emerged as a key event which influences many cellular processes [4]. Once believed to be a solitary and rigid organelle, the mitochondria constitute a population of organelles that continuously elongate (by fusion), divide (by fission) and undergo a controlled turnover (by mitophagy) [4]. The processes of fusion, fission and mitophagy set a fundamental framework of mitochondrial dynamics. The mitochondrial dynamics (fusion and fission) in concert with mitophagy sustains mitochondrial homeostasis and constitutes an important arm of mitochondrial quality control. This dynamic process is sensitive to the changes in the cellular physiological or metabolic conditions and guards against detrimental stresses that thwart cellular homeostasis [4].
The mitochondrial dynamics and mitophagy play crucial roles in metabolism, cellular differentiation, cancer, neurodegeneration and numerous pathologies [4]. In the past, the aspects of mitochondrial dynamics were broadly studied in the context of neurodegenerative disorders like Parkinson's, Alzheimer's and Huntington's diseases [5], [6]. The role of mitochondrial dynamics in viral infections is scant and described only for few viruses. Currently, our understanding of mitochondrial dynamics and mitophagy during viral infections is still in its infancy, but certainly represents an emerging avenue of molecular investigations in virology. The integral roles of the mitochondria as a hub of innate immune signaling and metabolism implicate its role in viral pathogenesis. The mitochondria are either directly targeted by viral proteins or influenced by the physiological alterations to cellular environment during viral pathogenesis like deregulated calcium homeostasis, endoplasmic reticulum stress, oxidative stress and hypoxia. Recent work by Kim et al., revealed intriguing aspects of how hepatitis B virus (HBV) and hepatitis C virus (HCV) utilize the alterations in mitochondrial dynamics for the maintenance of persistent infection [7], [8], [9]. Further investigation of the effect of mitochondrial dynamics in viral infections will enhance our understanding of virus–host interactions and their role in pathogenesis. Such an understanding will be useful in the therapeutic design of new antiviral strategies. In this review article, we discuss the events of virus-regulated mitochondrial dynamics and their implications in viral pathogenesis. The impact of viral infection on mitochondrial functions has been described elsewhere [10], [11].
2. Mitochondrial dynamics and mitophagy
Mitochondrial homeostasis is broadly maintained by the two closely interlinked and crucial processes, i.e. mitochondrial dynamics and mitophagy [12]. Molecular and morphological analysis of mitochondrial network provides evidence that a single mitochondrion, in its life cycle, undergoes fission and fusion several times and when the cell's machinery senses any impaired mitochondria, they are eliminated by a process known as mitophagy [13]. A series of key events associated with this process is described in Fig. 1 . Although mitophagy is broadly considered to be a central route for the elimination of dysfunctional mitochondria, in many biological events mitophagy plays an active role in eliminating the fully functional mitochondria such as during starvation, fertilization and erythroid differentiation [14], [15], [16]. The fission, fusion and mitophagy are interlinked but distinct processes that together facilitate segregation of impaired mitochondria and subsequent delivery to the lysosomes for complete elimination [13]. Fission has been implicated in the sorting of the mitochondria with mutant mtDNA copies [17]. Unlike fission, the fusion is a route by which the mitochondria quickly exchange and equilibrate matrix metabolites, intact mtDNA copies, and mitochondrial membrane components. Therefore, this process serves to alleviate the damage by fusing a partially impaired mitochondrion to a fully healthy, tubular mitochondrial network by simple exchange of matrix proteins. At first, based on the above explanation, it is easy to make out the inverse correlation between the mitochondrial fusion and mitophagy, however in cellular context, both processes are complementary and together they aid in the maintenance of homeostasis [12].
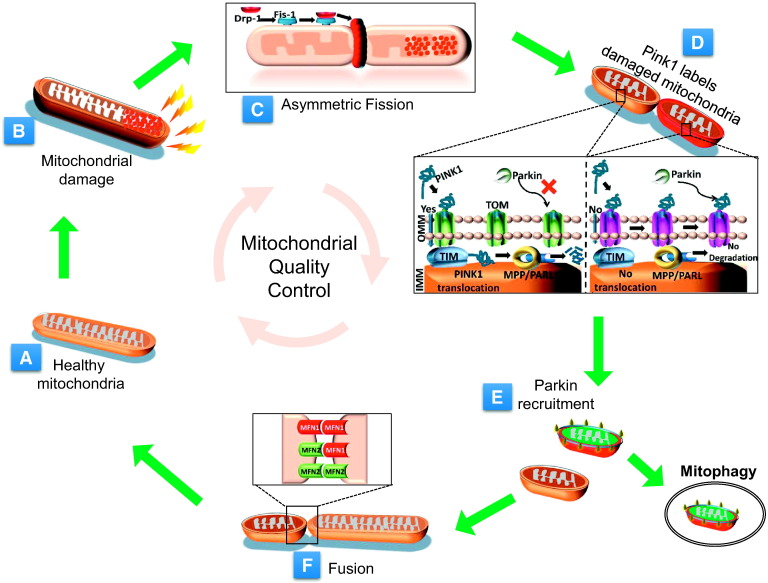
Fig. 1.
Mitochondrial quality control. The key events associated with mitochondrial dynamics and mitophagy affecting the mitochondrial quality control are depicted here. Under normal physiological conditions, the mitochondria are usually tubular (A). Physiological stress induces mitochondrial injury and causes mitochondrial impairment (B). The impaired part of the mitochondria is segregated by the fission process (C). The impaired mitochondria are selectively flagged by the PINK1 (D) that facilitates Parkin recruitment and mitophagy (E). All the repaired mitochondria are recruited to the functional mitochondrial network by fusion with other mitochondria (F). The factors that orchestrate the fission, fusion and mitophagy activities are shown in the inset.
The mitochondria that are highly depolarized and irreversibly damaged are destined to form a pre-mitophagic pool and are eliminated selectively by mitophagy. The mitochondria that undergo fusion are selectively rescued from this pre-mitophagic pool and incorporated back into the functional mitochondrial population. The mitochondrial membrane potential (∆Ψm) plays an important role in the process of sorting out and segregation of defective mitochondria [18]. Depending on the extent of damage, the impaired and segregated daughter mitochondria are either restored, undergo repair and incorporated back or are completely eliminated by mitophagy, if the ∆Ψm is not repairable [18], [19].
The events of fusion and fission are highly dynamic. In a cellular cycle, the mitochondria follow a kiss-and-run pattern, which entails a brief fusion event (for seconds) followed by fission [20]. In eukaryotic cells, mitochondrial life cycle can be seen in two distinct periods i.e. pre-fusion period and post-fusion period [21]. In pre-fusion stage, the mitochondria are solitary, while post-fusion stage structures them in a complex network [21]. Sometimes, in the solitary stage of the mitochondria, even asymmetric fission can induce remarkable alteration in the ∆Ψm because of the unequal distribution of the membrane surface and nucleoid structure between the two daughter mitochondria. Thus, it is presumed that the depolarized mitochondria can also result from the spontaneous depolarization during solitary stage [12]. Whether it is spontaneous or stress-induced event, such depolarized mitochondria are destined for either repair or clearance. Studies that correlate the size of the mitochondria with the rate of mitophagy show that the elongated mitochondria exhibit reduced rate of mitophagy and the mitochondria inside the autophagosomes undergoing mitophagy are smaller in size (< 1 μM) [22], [23], [24]. Mitophagy of the larger elongated mitochondria appears less likely. Fission facilitates the sorting of impaired mitochondria from the healthy mitochondrial pool. If the damage in a sorted mitochondrion is repairable, it may follow the fusion phase but those in which the damage is irreversible are eliminated by mitophagy [19]. In this manner, the mitochondrial dynamics and mitophagic machinery work in concert to prevent circulation of severely damaged mitochondria into the active and healthy pool (Fig. 1C–E).
3. Mitochondrial fusion and fission machinery and their regulation
Mitochondrial dynamics i.e. fission, and fusion, is coordinated by dynamin-related proteins. Dynamin has the capacity to oligomerize into spirals and mechanically remodel membranes by utilizing energy from GTP hydrolysis. They commonly promote membrane scission however their role in mitochondrial fusion is quite unique. Mitochondrial function and quality are determined by the subtle balance between continuous fission and fusion. Below, we discuss the central players of these processes.
3.1. Fusion proteins
The Fuzzy onion (Fzo) was the first fusion regulating protein identified in Drosophila. Fzo regulates mitochondrial fusion and its deletion results in fragmented mitochondria [25]. In the Fzo mutant flies, the mitochondria aggregate and fail to fuse leading to the accumulation of mitochondria in spermatid [26]. The molecules commandeering fusion pathway in mammals are Mitofusins (Mfn1 and Mfn2) and optic atrophy 1 protein (OPA1). Knockdown of any of these proteins leads to embryonic lethality and mitochondrial dysfunction [27]. Mfn1 and Mfn2 are homologs of Fzo and have related but discrete roles in controlling mitochondrial morphology. Mitofusins are the members of the membrane anchored dynamin family expressed in a wide variety of mammalian cells and uniformly localize to the outer mitochondrial membrane (OMM) [28]. Mitochondrial fusion involves fusion of the outer and inner mitochondrial membranes resulting in the merger of two individual mitochondria and concomitant mixing of the mitochondrial contents [28]. The OMM fusion is coordinated by Mfn1 and Mfn2. Mutations in either of the Mfns lead to abnormalities in mitochondrial morphology associated with dramatic reduction in mitochondrial fusion. Depletion of both Mfn1 and 2 in cells results in poor cell growth, decreased cellular respiration associated with the heterogeneity of mitochondrial membrane potential [29]. Inner mitochondrial membrane (IMM) fusion is orchestrated by OPA1. OPA1 is a dynamin-related GTPase expressed in many mammalian cell types and localizes on IMM [30]. OPA1 is considered a multifunctional protein with pivotal roles in mitochondrial cristae architecture, mitochondrial bioenergetics and apoptosis [13], [31].
Overall the mitochondrial fusion is a multistep process with Mfns mediating the outer membrane fusions followed subsequently by inner membrane fusions by OPA1 [13]. The presence of Mfns on both the fusing mitochondria is a prerequisite for outer membrane fusion. The Mfns on adjacent mitochondria oligomerize bringing the opposing membranes within close proximity and eventually promoting fusion by membrane merger [27]. Both Mfn1 and Mfn2 physically interact homotypically and heterotypically resulting in Mfn1 homotypic oligomers, Mfn2 homotypic oligomers and Mfn1–Mfn2 heterotypic oligomers (Fig. 1F). Interestingly, inner membrane fusion by OPA1 does not require its presence on adjacent fusing membranes [32].
Both Mfns and OPA1 undergo post-translational modifications and proteolysis. Due to differential RNA splicing and precursor protein processing, OPA1 exists as distinct isoforms. The long isoform has membrane anchor but the short isoform lacks membrane anchor but does interact with membranes [33]. Usually both long and short isoforms drive membrane fusion but under conditions of stress, the long isoform solely promotes mitochondrial fusion. The activity of OPA1 is regulated by SIRT3-mediated deacetylation [34]. Moreover, it has recently been shown that decrease in mitochondrial membrane potential triggers the proteolysis of OPA1 by the isoenzymes of the adenosine triphosphate (ATP)-dependent matrix AAA (ATPase associated with diverse cellular activities [m-AAA]) protease and OMA1 [35], [36]. OMA1 is constitutively active under normal cellular conditions. However, various stress stimuli can enhance its catalytic activity several fold leading to enhanced turnover. In a recent study, Baker et al., have identified a stress sensor N-terminal domain of OMA1 that is responsible for stress-induced activation of OMA1 [37]. This mechanism probably drives selective fusion of normal mitochondria resulting in isolation of defective/dysfunctional mitochondria. JNK-mediated phosphorylation of Mfn2 during genotoxic stress leads to interaction with Huwe1, an E3 ubiquitin ligase, resulting in proteasomal degradation of Mfn2, mitochondrial fragmentation, and apoptotic cell death [38]. Similarly recruitment of Parkin to damaged mitochondria also results in the ubiquitination and degradation of Mfns, which prevents the fusion of damaged mitochondria with the healthy mitochondrial pool [39].
3.2. Fission proteins
The central player in mitochondrial fission is a cytosolic dynamin-related GTPase, dynamin-related protein 1 (Drp1). Drp1 is localized primarily in the cytosol and in mammalian cells the accessory proteins Mid49, Mid51, and mitochondrial fission factor (Mff) mediate its recruitment to the mitochondria by acting as ligands for Drp1 on the mitochondria. Once recruited to the mitochondria, Drp1 oligomerizes as rings on mitochondrial tubules and causes scission of both outer and inner mitochondrial membranes [40], [41]. Our current understanding of the mitochondrial fission mechanism is incomplete, however recent studies suggest that ER tubules wrap around and constrict the mitochondria, marking them for subsequent scission by Drp1 [42]. These sites of ER–mitochondrial contact are enriched with Drp1 suggesting that ER contact with mitochondria directs mitochondrial division. Interestingly, mitochondrial constriction at ER contact sites was also apparent in cells depleted of Drp1 and Mff suggesting that the ER-mediated flagging of mitochondrial division sites occurs prior to the mitochondrial recruitment of Drp1 [42]. The mechanical force required for the constriction of mitochondrial tubules by the ER seems to be generated by actin assembly at the ER–mitochondria contact sites via interaction with ER-associated formin INF2 [43]. Genetic or chemical inhibition of Drp1 leads to elongated mitochondria. The first molecule to be identified as Drp1 receptor was the Fission1 (Fis1) in yeast. Fis1, is a small C-tail protein anchored on OMM and displays uniform distribution [44]. However, the role of mammalian Fis1 in mitochondrial fission is enigmatic and mitochondrial morphology or mitochondrial recruitment of Drp1 was unaffected in Fis1 knockout cells, raising the concern about the role of mammalian Fis1 in mitochondrial fission [45]. Mitochondrial fission factor Mff is also a C-tail anchored protein on OMM and a well-recognized receptor of Drp1. It transiently interacts with Drp1 through its N-terminal cytoplasmic region. In contrast to uniform localization of Fis1 on OMM, Mff mostly colocalizes with Drp1 foci on the OMM. Overexpression of Mff stimulates mitochondrial recruitment of Drp1 and mitochondrial fragmentation and silencing of Mff results in mitochondrial elongation [46], [47]. The MiDs can mediate mitochondrial fission in the absence of Fis1 and Mff. Knockdown of either gene results in the increment of mitochondrial length and interconnectivity suggesting that these proteins positively regulate mitochondrial fission. Interestingly, overexpression of MiDs also results in the elongation of the mitochondria. The mitochondrial elongation caused by MiD overexpression is associated with enhanced phosphorylation of Drp1 at S637 site, which negatively regulates the Drp1 function [40], [41]. Although the cells mutated for Drp1 resist mitochondrial fragmentation induced by depolarization, they exhibit substantial mitochondrial fragmentation when treated with apoptotic stimuli such as actinomycin D and etoposide, suggesting that Drp1-independent mechanisms of mitochondrial fission also occur. The mitochondrial fragmentation induced by the pore-forming toxin listeriolysin O, secreted by the bacterium Listeria monocytogenes, is independent of traditional fission protein Drp1 but dependent on actin cytoskeleton [48].
Several posttranslational modifications of Drp1 regulate mitochondrial fission dictated by various physiological cues. In normal cells, Drp1 is mostly cytoplasmic with about ~ 3% of total Drp1 associated with the mitochondria. Overexpression of Drp1 does not usually lead to enhanced fission but stimulation of Drp1 mitochondrial recruitment, GTPase activity, and spiral assembly directly influences mitochondrial fission (Fig. 1A) [49]. During mitosis Cdk1/cyclin B phosphorylates Drp1 at S616 residue (amino acid numbering corresponds to human Drp1), resulting in the upregulation of mitochondrial recruitment of Drp1 and fission activity. This regulation is essential in cells undergoing mitosis to ensure equal distribution of the mitochondria to daughter cells [50]. During mitosis, Aurora A kinase also promotes mitochondrial fission by phosphorylating small Ras-like GTPase, RalA leading to its relocation to mitochondria, followed by the recruitment of its effector RalBP1 to the mitochondria. RalBP1 then facilitates CDK1-mediated Drp1 S616 phosphorylation and recruitment to the mitochondria [51]. Interestingly, a study indicates that high-glucose stimulation of liver-derived cells induces Ca2 +-mediated MAP kinase signaling leading to ERK1/2-dependent Drp1 S616 phosphorylation and enhanced mitochondrial fission [52]. Phosphorylation of Drp1 at S637 residue leads to inhibition of fission activity. Protein kinase A (PKA) or cAMP-dependent protein kinase phosphorylates Drp1 on S637 located in the GTPase effector domain (GED) and impairs intramolecular interaction of GED and GTPase domains [53]. During hypoxia, PKA-mediated phosphorylation of Drp1 is affected due to the proteasomal degradation of mitochondrial A-kinase anchoring protein 121 (Akap1) that anchors PKA to mitochondria [54], [55]. Calcium-dependent phosphatase calcineurin dephosphorylates Drp1 at S637 and facilitates mitochondrial recruitment of Drp1 followed by fission [56]. The same residue is also dephosphorylated by the phosphatase, PP2A/Bβ 2 [55]. In contrast, phosphorylation of same residue in response to rise in intracellular calcium by Ca2 +/calmodulin-dependent protein kinase α (CaMKIα) in neuronal cells or by ROCK1, results in enhanced fission [57], [58]. Surprisingly, recent reports reveal the importance of upstream kinases, rather than the site of phosphorylation as an important factor that determines the activity of Drp1. For example, the S616-Drp1 phosphorylation by CDK1 or PKCΔ, is reported to enhance fission by activating Drp1 but if the same site is phosphorylated by CDK5, it was explained to enhance the dissociation of Drp1 from its oligomeric to monomeric form and severely reduce its fission-prompting activity [59]. Calcineurin mediated dephosphorylation of S637 as well as phosphorylation of S637 by CaMKIα also enigmatically leads enhanced fission suggesting that different cellular contexts may have different effects of Drp1 phosphorylation on same serine residue. However, further studies are needed to clarify how phosphorylation at the same site, by different kinases can modulate Drp1 activity with different consequences. In addition, ubiquitination, sumoylation, and s-nitrosylation have also been shown to modulate Drp1 activity [60], [61], [62]. Parkin and mitochondria ubiquitin ligase (MITOL) facilitate Drp1 degradation via proteasomal pathways [63], [64]. All the post-translational modifications of the key molecules involved in the processes of fission and fusion are outlined in Table 1 .
Table 1.
Post-translational modifications of the protein involved in mitochondrial dynamics. List of important post-translational modifications of the key molecules of mitochondrial dynamics and their upstream modulators with its final consequences on cell physiology.
| Protein | Modification | Induced by | Effect | Ref |
|---|---|---|---|---|
| Drp-1 | Phosphorylation (S616) | CDK1 | Enhanced mitochondrial fission | [50] |
| Erk1/2 | Enhanced mitochondrial fission | [52] | ||
| PKCδ | Enhanced mitochondrial fission | [142] | ||
| CDK5 | Reduced mitochondrial fission | [59] | ||
| Phosphorylation S637 | PKA, | Reduced mitochondrial fission | [53] | |
| CaMK1α | Enhanced mitochondrial fission | [57] | ||
| ROCK1 | Enhanced mitochondrial fission | [58] | ||
| Dephosphorylation S637 | Calcineurin | Enhanced mitochondrial fission | [56] | |
| S-Nitrosylation C644 | Nitric oxide | Enhanced mitochondrial fission | [60] | |
| Sumoylation | MAPL | Enhanced mitochondrial fission | [61] | |
| Desumoylation | SENP5 | Reduced mitochondrial fission | [62] | |
| Ubiquitination | MITOL | Degradation | [64] | |
| Parkin | Degradation | [79] | ||
| Fis-1 | Ubiquitination | MITOL | Degradation | [64] |
| Mfn-1 | Ubiquitination | Parkin | Degradation | [39] |
| Ubiquitination | MARCH5 | Degradation | [143] | |
| Mfn-2 | Ubiquitination | Parkin | Degradation | [39] |
| Phosphorylation T111, S442 | PINK1 | Parkin recruitment | [74] | |
| Opa-1 | Acetylation K926/931 | Stress | Reduced GTPase activity | [34] |
| De-acetylation K926/931 | SIRT3 | High GTPase activity | [34] |
4. Mitophagy machinery and its regulation
Autophagy is a catabolic process, in which a destined cargo is actively engulfed by phagophore and subsequently followed by fusion of autophagosome with lysosomes to deliver the cargo to the lysosome for degradation [65]. Autophagy is a nonselective process, but recent advancements in this field have established the presence of selective autophagy in which specific cargo or damaged organelles are selectively eliminated. Here, we discuss the selective autophagy of the mitochondria, termed mitophagy. Mammalian mitophagy can be broadly classified in two distinct groups; Parkin-dependent, and independent forms of mitophagy. Parkin-dependent mitophagy involves selective tagging of damaged mitochondria by PTEN-induced putative kinase 1 (PINK1) followed by Parkin recruitment to the impaired mitochondria and subsequent mitophagy [66]. The serine/threonine kinase, PINK1 contains a mitochondrial targeting sequence (MTS) and is sequentially imported into the healthy mitochondria via the translocase of outer mitochondrial membrane (TOM) and translocase of inner mitochondrial membrane (TIM). At the IMM, PINK1 is processed by the mitochondrial processing protease (MPP) to excise the MTS, and further cleaved by the presenilin-associated rhomboid-like protease (PARL) to generate a 52 kDa protein, which is then rapidly degraded. However in the impaired mitochondria, the loss of membrane potential (∆Ψm) rapidly perturbs the import of PINK1 to the IMM and prevents its degradation by MPP, PARL and other proteases (Fig. 1D). This leads to the display of PINK1 on the surface of the mitochondria, which thereby marks the damaged mitochondria for Parkin recruitment [66], [67] to initiate the process of mitophagy. Several studies have explained how PINK1 recruits Parkin to the mitochondria. PINK1 auto-phosphorylation on depolarized mitochondria is required for Parkin recruitment and direct phosphorylation of Parkin by PINK1 at S65 residue has been shown to stimulate Parkin's E3 ligase activity [68]. PINK1 also phosphorylates S65 residue of ubiquitin that subsequently unlocks Parkin's autoinhibition at the active cysteine site [69]. Ubiquitin mutant S65A inhibited Parkin translocation to impaired mitochondria [70]. Based on these premise small molecules that mimic the S65 phospho-ubiquitin can be used as potential therapeutic tool against the Parkinson's disease [71]. Together these studies propose a feed-forward mechanism for Parkin recruitment to the mitochondria involving PINK1 mediated phosphorylation of both Parkin and poly-UB chains synthesized by Parkin to the inner mitochondrial membrane [70], [71], [72]. Some studies suggest a direct physical interaction between Parkin and PINK1 [66]. It is likely that other substrates of PINK1 kinase activity are involved in Parkin recruitment to the mitochondria. Although PINK1 facilitates Parkin recruitment to the mitochondria, in flies it is not the only means for Parkin recruitment. Mitochondrial morphology in PINK-1 deficient flies can be rescued by Parkin overexpression and Parkin deficient flies had more severe phenotype than the later, suggestive of PINK1 independent functions of Parkin in flies [73]. Can Parkin be recruited to the mitochondria independent of PINK1 in mammalian cells remains to be determined. However, in one study, PINK1 mediated phosphorylation of Mfn2 was shown to promote Parkin recruitment on depolarized mitochondria [74].
Parkin ubiquitinates and facilitates degradation OMM proteins such as Mfn1, Mfn2 and TOM complex which seems to be essential for subsequent autophagic removal of the damaged mitochondria [39]. In agreement with this notion, inhibition of proteasomal activity impairs Parkin-dependent mitophagy. The accumulation of proteasomal subunits on OMM following mitochondrial recruitment of Parkin, suggests that the proteasomal machinery is directly recruited to the mitochondria to facilitate degradation. The AAA+ATPase p97, which structurally remodels or unfolds ubiquitinated client proteins destined for degradation is shown to be recruited to the mitochondria along with Parkin and regulate protein degradation at OMM [39]. Currently, it is unclear how proteins are targeted by Parkin and which of these targets are critical for initiation of mitophagy. Parkin not only promotes K-48 linked polyubiquitination which leads to proteasomal degradation but can also induce the formation of various other lysine-linked polyubiquitin chains conjugated to its target proteins [72]. Considering Parkin-mediated K-63 linked ubiquitination, it is likely that p62 adaptor protein that binds to K-63 linked polyubiquitin conjugated proteins, plays a role in recruiting mitochondria to autophagosomes [75]. However, mitochondrial elimination has been reported even in the absence of p62 clustering at the mitochondria [76]. The histone deacetylase 6 (HDAC6), also involved in binding ubiquitinated proteins, is recruited to the mitochondria in Parkin-dependent manner. HDAC6 has been shown to play a role in autophagosome maturation [77]. PINK1 and Parkin have also been shown to directly interact with Beclin-1 and PI3K (phosphatidylinositol-4,5-bisphosphate 3-kinase) complex to promote autophagy. Parkin also facilitates the recruitment of activating molecule in Beclin-1 regulated autophagy (Ambra-1) to the mitochondria, which could activate Beclin1 and promote the nucleation of isolation membranes (phagophore) around the mitochondria [78].
Mutations in both Parkin and PINK1 are the most prevalent cause of autosomal recessive genetic disorder Parkinsonism, which is usually associated with accumulation of impaired mitochondria leading to neurodegeneration in Parkinson's disease (PD). This lends support to the view that PINK1/Parkin-mediated mitophagy is important for mitochondrial quality control [5]. PINK1/Parkin pathway, not only initiates the mitophagic removal of damaged mitochondria, but also facilitates the segregation of the damaged mitochondria by preventing their fusion with healthy functional mitochondria. This segregation is achieved in two different ways; making the impaired mitochondria fusion defective by Parkin-mediated degradation of Mfns and by restricting cytosolic motility of the damaged mitochondria by targeting of Miro (Rho-GTPase), a mitochondrial adapter molecule that anchors kinesin motor complex to the outer surface of the mitochondria. PINK1 phosphorylates Miro, which is recognized and degraded by the Parkin. Miro degradation releases the kinesin motor from the mitochondria, which hinders their motility [39], [79]. These events clearly illustrate that mitochondrial dynamics and mitophagy are interlinked and regulated in a coordinated manner.
Several deviations to the PINK1–Parkin pathway support the existence of PINK1–Parkin-independent pathways of mitophagy. PINK1–Parkin pathway may not represent the only pathway for steady state turnover of the mitochondria, because Parkin deficiency does not lead to dramatic loss of mitochondrial mass and mice deficient in Parkin are normal unless exposed to stress [80]. Allen et al., observed that iron chelation induces mitophagy independent of PINK1 stabilization and Parkin activation in primary human fibroblasts as well as those isolated from PD patients with Parkin mutations [81]. AMBRA-1 is reported to induce Parkin-independent mitophagy by directly interacting with LC3 via its LC3-interacting region (LIR). AMBRA-1 can induce mitophagy in Parkin-deficient cells as well as potentiate the Parkin-mediated mitophagy [82]. In response to energy depletion, AMPK stimulates autophagy by phosphorylating and activating ULK1 [83]. Loss of AMPK or ULK1 in mammalian cells leads to accumulation of the autophagy adaptor p62 and deficient mitophagy suggesting that ULK1 is required for mitochondrial and cellular homeostasis during starvation [83]. A recent study suggests that ULK1 translocates to the mitochondria and phosphorylates the FUN14 domain-containing protein (FUNDC1), which directly binds to LC3 via its LIR motif to regulate mitophagy [84]. Similarly, BCL2/adenovirus E1B 19 kDa protein-interacting protein 3-like (BNIP3L) or Nix facilitates the elimination of the mitochondria from reticulocytes during erythrocyte maturation by directly interacting with LC3 via its LIR motif [85]. Another study implicates FUNDC1, an OMM protein as mediator of hypoxia-induced mitophagy [86]. In neurons, cardiolipin externalization to the outer mitochondrial membrane serves as a signal for elimination of the damaged mitochondria. The phospholipid cardiolipin, present in the IMM of the mitochondria when externalized to the OMM, interacts with LC3 to facilitate mitochondrial degradation [87], [88]. It is still unknown whether the parkin-dependent and -independent pathways function in parallel during depolarization-induced mitophagy.
5. Role of mitochondrial dynamics and mitophagy in cell physiology
Over the past decade, it has become evident that mitochondrial dynamics and mitophagy constitute the two main pathways required to maintain mitochondrial quality control and cellular homeostasis. Perturbations in these pathways have physiological consequences and are among the most common reason underlying the pathogenesis of many neurodegenerative disorders, cancer, inflammation, metabolic syndrome and cardiac dysfunctions. We discuss, below, the roles of mitochondrial dynamics and mitophagy in cellular physiology.
5.1. Mitochondrial quality control; cell survival and apoptosis
Perturbation in mitochondrial dynamics and mitophagy usually generates an unhealthy pool of the mitochondria that create an energy-deprived condition in the cytosol that can eventually lead to apoptosis. Mitochondrial fragmentation has been implicated in apoptosis. Enhanced fragmentation increases the sensitivity of the cells for apoptotic stimuli while enhanced fusion has been associated with delay in Bax activation, cytochrome C release and cell death [89]. Drp1-mediated fission has been shown to facilitate cytochrome C release from the mitochondria [90]. The Drp-1-dependent remodeling of the OMM facilitates the oligomerization and insertion of Bax into the mitochondria [91]. Conversely, both Bax/Bak facilitate Drp-1 sumoylation and stabilize Drp1 on the mitochondria thereby promoting fission [92]. In the neuronal cell model of nitric oxide-induced oxidative stress, mitochondrial fission preceded apoptosis, and inhibition of fission conferred protection [93]. Enhanced mitochondrial fusion has been shown to protect cells from mitochondria-triggered apoptosis but not the mitochondria-independent apoptotic stimuli [94]. Contradicting the common notion, cytochrome C release was also observed in Drp1 defective cells upon induction of apoptosis, suggesting that mitochondrial fission and mitochondrial outer membrane permeabilization are independent of each other. Currently, the precise role of mitochondrial fission in apoptosis is not clearly defined, however the anti-apoptotic Bcl2 family members in their pursuit to promote cellular adaptability to various stressful conditions may modulate mitochondrial dynamics.
Defective mitochondria are a potential source of reactive oxygen species (ROS), which can also lead to damage of the healthy mitochondria. Hence, perturbation in the rapid clearance of damaged mitochondria will initiate a vicious cycle of ROS generation and mitochondrial damage, which eventually deteriorates other cellular components and promotes cell death [95], [96], [97]. Defective mitochondrial quality has been associated with induction of inflammatory responses via the activation of inflammasome [98]. Pre-conditioning the cells to upregulate mitochondrial quality control pathways has been shown to enhance resistance to stressful stimuli. Interorganelle contacts are essential for mitochondrial functions and are implicated in mitochondrial dynamics. One such example is the involvement of the ER–mitochondria encountered structure (ERMES) in maintenance of mitochondrial morphology in budding yeast [99]. In mammals, mitochondrial-associated membranes (MAMs) represent the ER–mitochondria contact sites [89]. Mfn2 is enriched in MAMs and it is proposed that Mfn2 on the ER is required to stabilize interaction between ER and mitochondria by engaging in homotypic and heterotypic complexes with Mfn1/2 on the surface of the mitochondria. Alterations in Mfn2 expression affect ER–mitochondria contact followed by hindrance to lipid exchange and calcium signaling between ER and mitochondria [100]. In mammals many other interactions between ER and mitochondrial proteins have been reported, however it remains to be determined if these interactions are an integral part of a physical complex [101].
5.2. Innate immunity
The role of mitochondrial dynamics in innate immune signaling is an upcoming field. Pattern recognition receptors (PRRs), the components of the innate immune system, recognize pathogen associated molecular patterns (PAMPs) and activate a cascade of complex pathways to eliminate infection, sometimes involving the apoptosis of infected cells [1]. Many PRRs have been characterized and each of them has a specific ligand. Many viral PAMPs are recognized by the retinoic acid-inducible gene I (RIG-I) and RIG-I like receptors (RLRs), which then activate the mitochondria associated antiviral signaling protein (MAVS) via binding facilitated by the CARD domains of each protein [102]. Recent studies implicate the role of mitochondrial dynamics and mitophagy in modulation of this intrinsic defense strategy [103]. Mitochondrial fusion and fission play a central role in amplifying or dampening the RLR signaling. Mitochondrial fusion serves to increase MAVS interactions with downstream signaling molecules resulting in enhanced interferon (IFN) synthesis upon polyI:C stimulation or Sendai virus infection [103]. In contrast, the mitochondrial fission serves to block MAVS downstream signaling resulting in reduced IFN synthesis [103]. In agreement with this notion, fibroblasts lacking both Mfn1 and 2 were impaired in interferon and pro-inflammatory cytokine synthesis in response to viral infection [104]. Absence of fusion leads to the loss of mitochondrial membrane potential in these double-knockout cells, which correlated with reduced MAVS signaling suggesting that mitochondrial membrane potential may be required for MAVS signaling [104]. It is also envisaged that mitochondrial fusion and the resultant tubular mitochondrial network formation around viral replication sites assist in concentrating the MAVS signalosome around sites of viral replication [103], [105]. A recent report claims that Mfn1 promotes dramatic redistribution of MAVS on the mitochondria after RLR activation and that Mfn1 is required for recruitment of MAVS-enriched mitochondria to the RIG-I enriched sites of viral replication [106]. The effect of mitochondrial morphology is also implicated in regulating the signaling of ER-associated innate immunity molecule STING (stimulator of interferon genes). Tubular elongated mitochondria tend to interact more with the ER and therefore assist in STING's downstream interaction with MAVS [103], [105]. We observed that inhibition of mitochondrial fission by silencing Drp1 during HCV infection enhanced IFN production indicating that HCV-mediated altered mitochondrial dynamics serves to perturb the antiviral defense [8]. This may operate in addition to the HCV NS3/4a-orchestrated proteolytic cleavage of MAVS to cripple host innate immunity [107].
6. Mitochondrial dynamics during viral infection
Viruses interfere with the mitochondrial pathways and distort mitochondrial functions to facilitate their proliferation. Viral strategy to impede mitochondria-associated antiviral signaling mechanism is one such paradigm of the virus–mitochondria interactions [102]. The functional implication of mitochondrial dynamics in various mitochondrial and cellular functions suggests that alteration of mitochondria dynamics can serve as an efficient viral strategy to promote interference of cellular signaling pathways [103], [105]. In addition, the sensitivity of mitochondrial dynamics to subtle physiological perturbations in cellular environment makes the effect of viral infections on mitochondrial dynamics a likely inevitable consequence. The viral infection causes many physiological alterations in host cell and many of those alterations can directly affect the mitochondrial dynamics and mitophagy. The overall effects of viral infection on mitochondrial dynamics and its consequences on viral pathogenesis are illustrated in Fig. 2 . We discuss, below, few of the viruses, which have been characterized for their effect on mitochondrial dynamics and highlighting the significance of these alterations in the viral infectious process. A brief description of few viruses and their effects on mitochondrial dynamics and mitophagy are summarized in Table 2 .
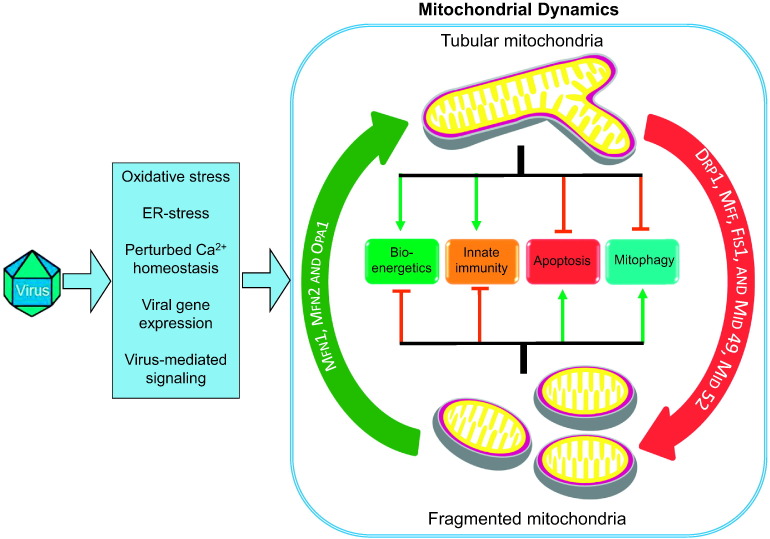
Fig. 2.
Viruses and mitochondrial dynamics. Model of mitochondrial dynamics during virus infection explaining the upstream causes of virus induced disruption in mitochondrial dynamics along with their possible downstream consequences.
Table 2.
Alteration of mitochondrial dynamics by viruses and its physiological significance. List of viruses known to disrupt mitochondrial dynamics and the consequences on cell physiology and viral pathogenesis.
| Virus [Ref] | Effect on mitochondrial dynamics | Viral protein involved | Affected protein(s) | Consequences on cell physiology |
|---|---|---|---|---|
| Hepatitis C virus [8], [9] | Enhanced fission and mitophagy | Core, E1–E2 | Activation of Drp-1, and mitochondrial translocation of Parkin | Inhibition of apoptosis and innate immune response, facilitates persistent infection |
| Pseudorabies virus [136] | Fragmented mitochondria | Glycoprotein B (GB) | Altered functioning of Miro protein | Affects intracellular calcium signaling and mitochondrial motility |
| Human cytomegalovirus [130] | Enhanced fission | vMIA | Affects mechanism of Bax | Inhibition of apoptosis |
| Epstein–Barr virus [128] | Enhanced fission | LMP2A | Up regulation of Drp-1 | Cell migration and apoptosis |
| Hepatitis B virus [7] | Enhanced fission and mitophagy | HBx | Parkin and PINK1 up-regulation and Drp-1 phosphorylation | Inhibition of apoptosis and innate immune response, facilitates persistent infection |
| Influenza A virus [98] | Induction of mitophagy | Unknown | The NOD2 and RIPK2 promote ULK1 phosphorylation to induce mitophagy | Inhibits inflammasome activation and reduces disease severity |
| Influenza A virus [137], [138] | Induction of mitochondrial fragmentation | PB1-F2 | Inhibit MAVS and NLRP3 inflammasome activation | Inhibit the antiviral response via mitochondrial pathways |
| Measles virus and Newcastle disease virus [139], [140], [144] | Induction of mitophagy | Unknown | Reduces the level of MAVS via p62 dependent mitophagy | Cripples innate immune signaling |
| SARS coronavirus [141] | Enhanced fusion | ORF-9b | Reduces the level of Drp1 and MAVS | Cripples innate immune signaling |
6.1. Hepatitis viruses
Chronic infection with hepatitis C virus (HCV) and hepatitis B virus (HBV) has long been associated with prominent mitochondrial injury of the liver [108]. Although HBV and HCV differ in their genomic organization and life cycle, they share similar pathologies, and promote chronic hepatitis, which subsequently progresses into liver fibrosis, cirrhosis and hepatocellular carcinoma [108], [109]. Histopathological features commonly observed in HBV/HCV infection include mitochondrial swelling and loss of mitochondrial cristae and number [108]. HBV and HCV infection-associated ER-stress triggers leakage of ER-calcium stores and their subsequent uptake by the adjacent mitochondria promotes mitochondrial depolarization and dysfunction, associated with mitochondrial ROS generation [108]. We have recently investigated the effect of HBV and HCV infections on host cell mitochondrial dynamics and established a direct relationship between viral infections, mitochondrial dynamics, mitochondrial homeostasis, and viral persistence.
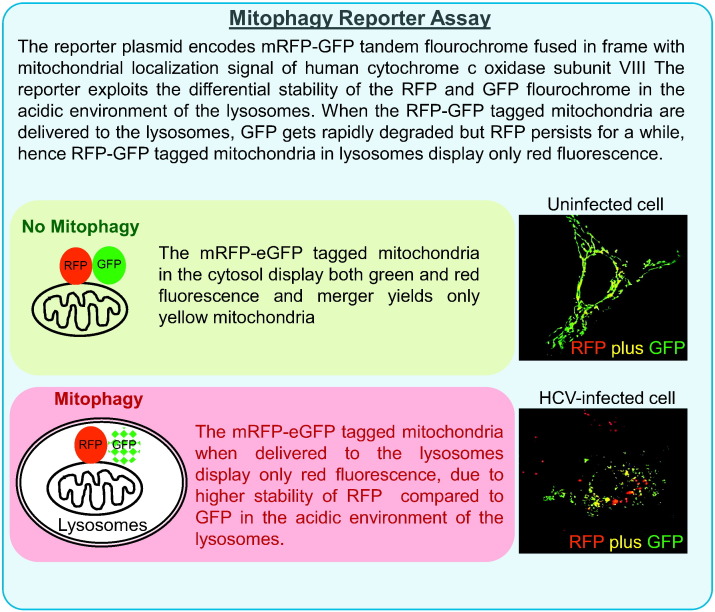
HCV: HCV is a positive strand RNA virus belonging to the genus Hepacivirus in the Flaviviridae family. The 9.5 kb RNA genome is translated via an IRES element located in the 5′ untranslated region (UTR) producing a ~ 3000 amino acid precursor polyprotein, which is processed by cellular and virus-encoded proteases to produce 3 structural and 7 non-structural viral proteins with various functions [110]. Most of the viral proteins remain associated with the ER membrane and promote ER stress. HCV induces ER stress and promotes autophagy which plays a crucial role in HCV RNA translation and HCV replication [111], [112]. HCV infection promotes mitochondrial abnormalities, and it is assumed that the ER-stress mediated depletion of ER Ca2 + stores and uptake by mitochondria paves the way for mitochondrial dysfunctions [113], [114]. HCV proteins have also been shown to directly associate with the mitochondria and localize to the OMM and MAM [107]. HCV core and NS5a proteins have been shown to perturb complex 1 activity and promote mitochondrial Ca2 + uptake, ROS production, and mitochondrial permeability transition [115], [116]. HCV NS3/4a protease localizes to MAM and cleaves MAM-associated MAVS to facilitate evasion from innate immune response [117]. HCV infection leads to elevation of ROS in the mitochondria that is induced by calcium signaling [113]. Mitochondrial damage during HCV infection is implicated in the inhibition of mitochondrial β-oxidation of fatty acids [109]. This is in conjunction with enhanced lipogenesis and lipid uptake during HCV infection ultimately leads to intracellular accumulation of lipid droplets. Hepatosteatosis is characterized by the accumulation of LDs which accelerate progression into end stage liver diseases in chronic hepatitis C [118]. Other effects on mitochondrial functions during HCV infection include reduced oxidative phosphorylation and ATP generation [119]. HCV infection induces perinuclear clustering of the mitochondria, a notable phenotype in cells exposed to oxidative stress [9]. Our studies have shown that HCV promotes mitochondrial fragmentation by inducing Drp-1 S616 phosphorylation. Drp1 phosphorylation at S616 promotes its mitochondrial translocation and subsequent fission/fragmentation of the mitochondria. HCV-infected cells also displayed enhanced expression of Parkin and PINK1 and mitochondrial recruitment of Parkin. Mitochondrial recruitment of Parkin in HCV-infected cells was associated with concomitant increase in autophagic removal of the damaged mitochondria by Parkin-dependent mitophagy which was elegantly demonstrated using the mitophagy reporter assay (Box-1 ) [8], [9]. Depletion of Drp-1 and Parkin in HCV-infected cells hindered HCV-induced mitochondrial fission and mitophagy respectively, but led to a marked increase in mitochondrial apoptotic signaling as evidenced by robust cytochrome C release and increase in caspase-3 activity. Our study suggested that HCV-induced mitochondrial fission followed by complete mitophagy is required to sustain mitochondrial homeostasis during the infection and promote viability of infected cells. The upregulation of mitochondrial quality control pathway likely attenuates the impending cell death due to the mitochondrial injury incurred during infection. In addition to promoting viability of infected cells, HCV-induced alteration of mitochondrial dynamics also subverted the mitochondria-associated antiviral signaling which was evident by a marked increase in interferon (IFN) production by silencing Drp-1 expression in HCV-infected cell [8]. HCV cripples the innate immune response by NS3/4a protease-mediated cleavage of MAVS protein suggesting that the effect of altered mitochondrial dynamics on MAVS signaling, further complements this effective viral strategy [120]. These studies clearly show that HCV-mediated alteration of mitochondrial dynamics and mitophagy is a major determinant of viral persistence and sheds light on the novel aspect of viruses exploiting the mitochondrial dynamics and quality control mechanism to favor viral propagation. Inhibition of mitochondrial fission affects HCV secretion that presumably occurs due to altered rate of glycolysis and ATP synthesis. Alternatively, reactive oxygen species emanating from accumulated dysfunctional mitochondria can damage lipids and proteins that could play a crucial role in HCV secretion [8].
Box 1.
Mitophagy reporter assay.
HBV: Unlike HCV, the genetic makeup of hepatitis B virus (HBV) consists of DNA. HBV replicates via an RNA intermediate and amplifies its genome by reverse transcription, a strategy similar to retroviruses [121], [122]. A successful recombinant vaccine against HBV is available with significant level of protection (about 98%) against infection. HBV encodes a regulatory protein, termed HBx or HBV X protein. HBx is localized to the mitochondria via its association with VDAC, alters membrane potential, and elevates the levels of calcium and ROS, ultimately causing damage to the mitochondria [108], [123], [124]. HBx-regulated calcium signaling and ROS are involved in activation of latent transcription factors such as STAT-3, NF-kB and NFAT [125]. HBV also induces early autophagic pathways that subsequently help the viral DNA replication [126].
HBV, like HCV also induced Drp1-dependent mitochondrial fission, and Parkin-dependent mitophagy [7]. Disruption of mitochondrial dynamics was observed in cell expressing full-length HBV genome or only HBx but HBx-defective (HBV-∆X) genome had no effect on mitochondrial dynamics reinforcing the issue that HBx is sufficient to promote mitochondrial damage and alter mitochondrial dynamics [7]. HBV-altered mitochondrial dynamics and mitophagy effectively contribute to mitochondrial quality control resulting in mitochondrial homeostasis. This promotes the maintenance of persistent infection by inhibition of apoptosis of HBV infected cells. In Parkin-silenced cells HBV infection promoted apoptosis as evidenced by the burst of cytochrome C release, caspase-3 activation and poly (ADP-ribose) polymerase (PARP) cleavage [7]. These observations collectively implicate mitochondrial dynamics in the maintenance of persistent phenotype in HBV infected hepatocytes [7]. Depletion of Parkin also modestly affected HBV DNA synthesis, presumably due to the impaired mitochondrial functions or enhanced innate immunity [7].
6.2. Epstein–Barr virus (EBV)
EBV is an oncogenic virus belonging to the herpes virus family and is implicated in various lymphoid and epithelial cancers [127]. A recent study has shown that the viral latent membrane protein 2A (LMP2A) causes elevated mitochondrial fission in gastric and breast cancer cells. Their observations revealed that LMP2A-triggered Notch pathway leads to induction in Drp1 expression and manifests in enhanced mitochondrial fission [128]. However, it has to be noted that Drp1 overexpression per se did not lead to enhance fission, but the mitochondrial recruitment of Drp1 and trigger of its GTPase activity is essential to promote fission. Drp1 has been implicated in the invasive ability and enhanced epithelial–mesenchymal transition of breast cancer cells. Concurrently, this study establishes that altered mitochondrial dynamics plays a critical role in the metastatic behavior of EBV-associated gastric and breast carcinomas.
6.3. Human cytomegalovirus (HCMV)
Cytomegaloviruses are ubiquitous viruses and pose an important health problem because of the high frequency of congenital infections [129]. Severe infections are associated with immunosuppression [129]. In an interesting report by McCormick et al., it has been observed that fibroblast infected with human cytomegalovirus exhibits a peculiar characteristic of punctate and dispersed mitochondria reminiscent of mitochondrial fission at 24 h post infection. The factor responsible for this alteration is the product of UL37x1 gene, which is an early anti-apoptotic gene and also referred to as viral mitochondrion localized inhibitor of apoptosis (vMIA) [130]. vMIA contains two domains. The first domain is located between amino acids 5 and 34 (30 amino acids), overlaps with the mitochondrial signal sequence that targets vMIA to the mitochondria [131]. The second domain located between amino acids 118 and 147 displays anti-apoptotic properties [131]. A recombinant fusion protein derived from vMIA consisting of these two domains is fully functional. vMIA is conserved in all human CMV strains as well as in other primate CMVs that have been examined [132]. The vMIA-mediated inhibition of apoptosis is dependent on its interaction with the pro-apoptotic Bax and Bak proteins [133], [134]. But the primary sequence of vMIA has very low similarity with BCL-xl (anti-apoptotic). The ectopic Bax is able to rescue the vMIA-mediated fragmented phenotype, whereas the BH3 mutant Bax did not show any rescuing effect [135]. This study indicates involvement of the functions of the Bax and Bak BH3 domains in both promoting apoptosis as well as regulating mitochondrial morphology in healthy cells.
6.4. Pseudorabies virus (alphaherpesvirus)
Pseudorabies virus (PRV) and herpes simplex virus type 1 (HSV-1) infection disrupts mitochondrial motility and morphology in the superior cervical ganglion (SCG) neurons of rodents [136]. During PRV infection the fusion events mediated by glycoprotein B (gB) resulted in electrical coupling of neurons and increased action potential firing rates that were causes of intracellular Ca2 + increase and altered mitochondrial dynamics [136]. This mechanism is governed by the Ca2 + sensitive cellular protein, Miro, which inhibits the recruitment of kinesin-1 HC to the mitochondria. PRV-mediated disruption in mitochondrial dynamics is required for efficient growth and spread of PRV, indicating that altered mitochondrial transport enhances alphaherpesvirus pathogenesis and infection. This study has opened the possibility that kinesin-1 HC is hijacked and recruited for the movement of viral particles or protein cargo during PRV assembly and/or egress [136].
6.5. Influenza virus
Infections by influenza viruses account for millions of deaths worldwide. An interesting study demonstrates that NOD2 and receptor interacting protein kinase 2 (RIPK2) respond to influenza A virus (IAV) infection by promoting ULK1 phosphorylation and inducing mitophagy [98]. Ripk2−/− cells exhibited defective mitophagy and triggered greater inflammasome activation because of the accumulation of the damaged mitochondria. This suggests that upregulation of mitochondrial quality control during IAV infection dampens inflammasome activation and IL-18 production [98]. This induction in mitophagy was independent of Parkin but was associated with ULK1 phosphorylation at serine 555. However, it is not clear if RIPK2 directly phosphorylates ULK1 or whether other intermediates are involved. Overall this study suggests that NOD2–RIPK2 signaling protects against virally triggered immunopathology by upregulating ULK1-dependent mitophagy [98]. The influenza A viral protein PB1-F2 binds to MAVS and affects IFN synthesis [137]. PB1-F2 also induces mitochondrial fragmentation, modulates innate immune response and NLRP3 inflammasome activation [138]. PB1-F2 translocates to mitochondrial inner membrane space via TOM40 channel and leads to the reduction of mitochondrial membrane potential. The PB1-F2 variant lacking C-terminal domain does not affect mitochondrial function [138].
6.6. Measles virus and Newcastle disease virus (NDV)
Measles is an acute, highly infectious disease. The Edmonston strain of Measles virus (MV-Edm) triggers p62-mediated mitophagy in non-small cell lung cancer (NSCLC) cells [139]. Defect in autophagy decreases viral titers and cell death induced by MV-Edm in NSCLC cells. MV-Edm triggered mitophagy results in the reduction of MAVS protein levels and subsequent dampening of the innate immune response. Silencing p62 expression inhibited mitophagy and resulted in the preservation of mitochondrial mass in MV-Edm-infected cells. Overall this study suggests that MV usurps mitophagy to mitigate the innate immune response mediated by RIG-I/MAVS signaling [139]. Interestingly, a similar study by the same group suggests that MV-Edm triggered p62-mediated mitophagy that subverts apoptosis of infected NSCLC cells by preventing cytochrome C release and leads to enhanced viral replication [140]. Autophagy-sustained persistent viral replication and eventually promotes necrotic cell death due to ATP exhaustion and inhibition of autophagy resulted in marked decline in MV-Edm associated oncolytic activity. Though both the studies agree that MV-Edm induces p62-dependent mitophagy, and that this induction is required to promote viral replication, both differ on the physiological outcome of mitophagy. The same group also showed that the Newcastle disease virus (NDV) uses a similar strategy and exploits p62-mediated mitophagy to promote viral propagation. Induction of mitophagy attenuated cytochrome C release and intrinsic apoptotic signaling in NDV-infected NSCLC cells and inhibition of mitophagy enhanced the oncolytic effect of NDV [140]. It is also likely that inhibition of mitophagy promotes innate immune signaling leading to cell death or both mitochondrial – apoptosis and – innate immune signaling contribute to cell death/oncolysis.
6.7. Severe acute respiratory syndrome-coronavirus
Severe acute respiratory syndrome-coronavirus (SARS-CoV) is a novel coronavirus, which has emerged as a lethal pathogen and threat to human health. How SARS-CoV escapes the innate immune signaling is poorly understood. A recent study revealed that SARS-CoV protein, open reading frame-9b (ORF-9b) localizes to the mitochondria and promotes proteasomal degradation of Drp1 leading to mitochondrial fusion [141]. Expression of 9b in HEK 293 cells led to ~ 70% reduction in Drp1 levels and this reduction was sensitive to proteasome inhibition but unaffected by inhibition of autophagy [141]. Interestingly, in this study, lowering Drp1 expression was associated with impaired MAVS signaling, although this reduction was not as effective in comparison to exogenous expression of ORF-9b protein suggesting that other mechanisms may be operative. The notion that reduced Drp1 expression or enhanced mitochondrial fusion hampers MAVS signaling is in sharp contrast to the common belief that MAVS signaling and IFN production are enhanced and dampened by mitochondrial fusion and fission respectively [141]. It is further shown that ORF-9b also promotes degradation of MAVS and its interacting partners by targeting poly(rC) binding protein 2 (PCBP2) and the HECT domain E3 ligase AIP4 to trigger the degradation of MAVS, TRAF3, and TRAF6 leading to disruption of MAVS signaling and IFN production. The localization of ORF-9b to the mitochondria led to the translocation of PCBP2 from the nucleus to the mitochondria, which subsequently interacts with AIP4 and targets it to the mitochondria. It remains to be clarified if TRAF3 and -6, are direct targets of AIP4. Silencing PCBP2 or AIP4 expression restored MAVS signaling and IFN production [141]. These results indicate that SARS-CoV ORF-9b manipulates mitochondrial dynamics and targets mitochondrial innate immune signaling to evade host innate immunity. However, more studies will be required to elucidate the physiological significance of enhanced mitochondrial fusion in the viral infectious processes or disease pathogenesis.
7. Conclusions
Despite the efforts made in the last few decades in research focusing on the role of the mitochondria and viral infections, many issues remain unresolved. Recently, the mitochondrial dynamics and mitophagy have gained substantial attention as these events modulate mitochondrial functions during viral infections. Current findings indeed have placed the mitochondrial dynamics and viral infections at the crossroad. Undoubtedly, there is a close relationship between the mitochondrial dynamics and viral infections but in-depth characterizations and their relevance to pathogenesis may help in understanding the pathogenic processes. It is important to determine how the players of mitochondrial dynamics and mitophagy are differentially regulated during viral infections and elucidate the different ways viruses exploit these alterations to their own benefits. Elucidation of the functional relevance of mitochondrial dynamics and viral pathogenesis will open new avenues for therapeutic design of strategies to combat viral infections and associated diseases. Greater understanding of the critical mitochondrial functions like bioenergetics, apoptosis, innate anti-viral signaling and inter-organelle cross talk needs to be pursued in future investigations to elucidate the impact of viral infections on mitochondria and mitochondrial dynamics.
Acknowledgement
Research in the laboratory (A.S.) is supported by grants from NIH (DK077704, DK08379 and AI085087) and Michael J Fox Foundation.
Footnotes
This article is part of a Special Issue entitled: Mitophagy.
References
- 1.Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.McBride H.M., Neuspiel M., Wasiak S. Mitochondria: more than just a powerhouse. Curr. Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 3.Ernster L., Schatz G. Mitochondria: a historical review. J. Cell Biol. 1981;91:227s–255s. doi: 10.1083/jcb.91.3.227s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan D.C. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Detmer S.A., Chan D.C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 6.Su B., Wang X., Zheng L., Perry G., Smith M.A., Zhu X. Abnormal mitochondrial dynamics and neurodegenerative diseases. Biochim. Biophys. Acta. 2010;1802:135–142. doi: 10.1016/j.bbadis.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S.J., Khan M., Quan J., Till A., Subramani S., Siddiqui A. Hepatitis B virus disrupts mitochondrial dynamics: induces fission and mitophagy to attenuate apoptosis. PLoS Pathog. 2013;9:e1003722. doi: 10.1371/journal.ppat.1003722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S.J., Syed G.H., Khan M., Chiu W.W., Sohail M.A., Gish R.G., Siddiqui A. Hepatitis C virus triggers mitochondrial fission and attenuates apoptosis to promote viral persistence. Proc. Natl. Acad. Sci. U. S. A. 2014;111:6413–6418. doi: 10.1073/pnas.1321114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S.J., Syed G.H., Siddiqui A. Hepatitis C virus induces the mitochondrial translocation of Parkin and subsequent mitophagy. PLoS Pathog. 2013;9:e1003285. doi: 10.1371/journal.ppat.1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anand S.K., Tikoo S.K. Viruses as modulators of mitochondrial functions. Adv. Virol. 2013;2013:738794. doi: 10.1155/2013/738794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claus C., Liebert U.G. A renewed focus on the interplay between viruses and mitochondrial metabolism. Arch. Virol. 2014;159:1267–1277. doi: 10.1007/s00705-013-1841-1. [DOI] [PubMed] [Google Scholar]
- 12.Twig G., Shirihai O.S. The interplay between mitochondrial dynamics and mitophagy. Antioxid. Redox Signal. 2011;14:1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H., Chan D.C. Mitochondrial dynamics—fusion, fission, movement, and mitophagy—in neurodegenerative diseases. Hum. Mol. Genet. 2009;18:R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebato C., Uchida T., Arakawa M., Komatsu M., Ueno T., Komiya K., Azuma K., Hirose T., Tanaka K., Kominami E., Kawamori R., Fujitani Y., Watada H. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Shitara H., Kaneda H., Sato A., Inoue K., Ogura A., Yonekawa H., Hayashi J.I. Selective and continuous elimination of mitochondria microinjected into mouse eggs from spermatids, but not from liver cells, occurs throughout embryogenesis. Genetics. 2000;156:1277–1284. doi: 10.1093/genetics/156.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takano-Ohmuro H., Mukaida M., Kominami E., Morioka K. Autophagy in embryonic erythroid cells: its role in maturation. Eur. J. Cell Biol. 2000;79:759–764. doi: 10.1078/0171-9335-00096. [DOI] [PubMed] [Google Scholar]
- 17.Suen D.F., Narendra D.P., Tanaka A., Manfredi G., Youle R.J. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattenberger Y., James D.I., Martinou J.C. Fusion of mitochondria in mammalian cells is dependent on the mitochondrial inner membrane potential and independent of microtubules or actin. FEBS Lett. 2003;538:53–59. doi: 10.1016/s0014-5793(03)00124-8. [DOI] [PubMed] [Google Scholar]
- 19.Kim I., Lemasters J.J. Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antioxid. Redox Signal. 2011;14:1919–1928. doi: 10.1089/ars.2010.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., Alroy J., Wu M., Py B.F., Yuan J., Deeney J.T., Corkey B.E., Shirihai O.S. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Twig G., Hyde B., Shirihai O.S. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim. Biophys. Acta. 2008;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elmore S.P., Qian T., Grissom S.F., Lemasters J.J. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 23.Gomes L.C., Scorrano L. High levels of Fis1, a pro-fission mitochondrial protein, trigger autophagy. Biochim. Biophys. Acta. 2008;1777:860–866. doi: 10.1016/j.bbabio.2008.05.442. [DOI] [PubMed] [Google Scholar]
- 24.Navratil M., Terman A., Arriaga E.A. Giant mitochondria do not fuse and exchange their contents with normal mitochondria. Exp. Cell Res. 2008;314:164–172. doi: 10.1016/j.yexcr.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Hermann G.J., Thatcher J.W., Mills J.P., Hales K.G., Fuller M.T., Nunnari J., Shaw J.M. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hales K.G., Fuller M.T. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 27.Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., Chan D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santel A., Fuller M.T. Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- 29.Chen H., Chomyn A., Chan D.C. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 30.Misaka T., Miyashita T., Kubo Y. Primary structure of a dynamin-related mouse mitochondrial GTPase and its distribution in brain, subcellular localization, and effect on mitochondrial morphology. J. Biol. Chem. 2002;277:15834–15842. doi: 10.1074/jbc.M109260200. [DOI] [PubMed] [Google Scholar]
- 31.Chan D.C. Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 32.Song Z., Ghochani M., McCaffery J.M., Frey T.G., Chan D.C. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol. Biol. Cell. 2009;20:3525–3532. doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song Z., Chen H., Fiket M., Alexander C., Chan D.C. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samant S.A., Zhang H.J., Hong Z., Pillai V.B., Sundaresan N.R., Wolfgeher D., Archer S.L., Chan D.C., Gupta M.P. SIRT3 deacetylates and activates OPA1 to regulate mitochondrial dynamics during stress. Mol. Cell. Biol. 2014;34:807–819. doi: 10.1128/MCB.01483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Head B., Griparic L., Amiri M., Gandre-Babbe S., van der Bliek A.M. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 2009;187:959–966. doi: 10.1083/jcb.200906083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehses S., Raschke I., Mancuso G., Bernacchia A., Geimer S., Tondera D., Martinou J.C., Westermann B., Rugarli E.I., Langer T. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 2009;187:1023–1036. doi: 10.1083/jcb.200906084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker M.J., Lampe P.A., Stojanovski D., Korwitz A., Anand R., Tatsuta T., Langer T. Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J. 2014;33:578–593. doi: 10.1002/embj.201386474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leboucher G.P., Tsai Y.C., Yang M., Shaw K.C., Zhou M., Veenstra T.D., Glickman M.H., Weissman A.M. Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol. Cell. 2012;47:547–557. doi: 10.1016/j.molcel.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.F., Karbowski M., Youle R.J. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loson O.C., Song Z., Chen H., Chan D.C. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell. 2013;24:659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer C.S., Osellame L.D., Laine D., Koutsopoulos O.S., Frazier A.E., Ryan M.T. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., Voeltz G.K. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korobova F., Ramabhadran V., Higgs H.N. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoon Y., Krueger E.W., Oswald B.J., McNiven M.A. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee Y.J., Jeong S.Y., Karbowski M., Smith C.L., Youle R.J. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol. Biol. Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gandre-Babbe S., van der Bliek A.M. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otera H., Wang C., Cleland M.M., Setoguchi K., Yokota S., Youle R.J., Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stavru F., Palmer A.E., Wang C., Youle R.J., Cossart P. Atypical mitochondrial fission upon bacterial infection. Proc. Natl. Acad. Sci. U. S. A. 2013;110:16003–16008. doi: 10.1073/pnas.1315784110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smirnova E., Griparic L., Shurland D.L., van der Bliek A.M. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taguchi N., Ishihara N., Jofuku A., Oka T., Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 51.Kashatus D.F., Lim K.H., Brady D.C., Pershing N.L., Cox A.D., Counter C.M. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat. Cell Biol. 2011;13:1108–1115. doi: 10.1038/ncb2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu T., Jhun B.S., Yoon Y. High-glucose stimulation increases reactive oxygen species production through the calcium and mitogen-activated protein kinase-mediated activation of mitochondrial fission. Antioxid. Redox Signal. 2011;14:425–437. doi: 10.1089/ars.2010.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang C.R., Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J. Biol. Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- 54.Merrill R.A., Dagda R.K., Dickey A.S., Cribbs J.T., Green S.H., Usachev Y.M., Strack S. Mechanism of neuroprotective mitochondrial remodeling by PKA/AKAP1. PLoS Biol. 2011;9:e1000612. doi: 10.1371/journal.pbio.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dickey A.S., Strack S. PKA/AKAP1 and PP2A/Bbeta2 regulate neuronal morphogenesis via Drp1 phosphorylation and mitochondrial bioenergetics. J. Neurosci. Off. J. Soc. Neurosci. 2011;31:15716–15726. doi: 10.1523/JNEUROSCI.3159-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cereghetti G.M., Stangherlin A., Martins de Brito O., Chang C.R., Blackstone C., Bernardi P., Scorrano L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han X.J., Lu Y.F., Li S.A., Kaitsuka T., Sato Y., Tomizawa K., Nairn A.C., Takei K., Matsui H., Matsushita M. CaM kinase I alpha-induced phosphorylation of Drp1 regulates mitochondrial morphology. J. Cell Biol. 2008;182:573–585. doi: 10.1083/jcb.200802164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang W., Wang Y., Long J., Wang J., Haudek S.B., Overbeek P., Chang B.H., Schumacker P.T., Danesh F.R. Mitochondrial fission triggered by hyperglycemia is mediated by ROCK1 activation in podocytes and endothelial cells. Cell Metab. 2012;15:186–200. doi: 10.1016/j.cmet.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho B., Cho H.M., Kim H.J., Jeong J., Park S.K., Hwang E.M., Park J.Y., Kim W.R., Kim H., Sun W. CDK5-dependent inhibitory phosphorylation of Drp1 during neuronal maturation. Exp. Mol. Med. 2014;46:e105. doi: 10.1038/emm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cho D.H., Nakamura T., Fang J., Cieplak P., Godzik A., Gu Z., Lipton S.A. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Braschi E., Zunino R., McBride H.M. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10:748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zunino R., Schauss A., Rippstein P., Andrade-Navarro M., McBride H.M. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J. Cell Sci. 2007;120:1178–1188. doi: 10.1242/jcs.03418. [DOI] [PubMed] [Google Scholar]
- 63.Wang H., Song P., Du L., Tian W., Yue W., Liu M., Li D., Wang B., Zhu Y., Cao C., Zhou J., Chen Q. Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson disease. J. Biol. Chem. 2011;286:11649–11658. doi: 10.1074/jbc.M110.144238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yonashiro R., Ishido S., Kyo S., Fukuda T., Goto E., Matsuki Y., Ohmura-Hoshino M., Sada K., Hotta H., Yamamura H., Inatome R., Yanagi S. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 66.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meissner C., Lorenz H., Weihofen A., Selkoe D.J., Lemberg M.K. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J. Neurochem. 2011;117:856–867. doi: 10.1111/j.1471-4159.2011.07253.x. [DOI] [PubMed] [Google Scholar]
- 68.Kondapalli C., Kazlauskaite A., Zhang N., Woodroof H.I., Campbell D.G., Gourlay R., Burchell L., Walden H., Macartney T.J., Deak M., Knebel A., Alessi D.R., Muqit M.M. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., Endo T., Fon E.A., Trempe J.F., Saeki Y., Tanaka K., Matsuda N. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 70.Kane L.A., Lazarou M., Fogel A.I., Li Y., Yamano K., Sarraf S.A., Banerjee S., Youle R.J. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kazlauskaite A., Kondapalli C., Gourlay R., Campbell D.G., Ritorto M.S., Hofmann K., Alessi D.R., Knebel A., Trost M., Muqit M.M. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ordureau A., Sarraf S.A., Duda D.M., Heo J.M., Jedrychowski M.P., Sviderskiy V.O., Olszewski J.L., Koerber J.T., Xie T., Beausoleil S.A., Wells J.A., Gygi S.P., Schulman B.A., Harper J.W. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park J., Lee S.B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J.M., Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 74.Chen Y., Dorn G.W., II PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chan N.C., Salazar A.M., Pham A.H., Sweredoski M.J., Kolawa N.J., Graham R.L., Hess S., Chan D.C. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Narendra D., Kane L.A., Hauser D.N., Fearnley I.M., Youle R.J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee J.Y., Nagano Y., Taylor J.P., Lim K.L., Yao T.P. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J. Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ashrafi G., Schwarz T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang X., Winter D., Ashrafi G., Schlehe J., Wong Y.L., Selkoe D., Rice S., Steen J., LaVoie M.J., Schwarz T.L. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kubli D.A., Zhang X., Lee Y., Hanna R.A., Quinsay M.N., Nguyen C.K., Jimenez R., Petrosyan S., Murphy A.N., Gustafsson A.B. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J. Biol. Chem. 2013;288:915–926. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Allen G.F., Toth R., James J., Ganley I.G. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–1135. doi: 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Strappazzon F., Nazio F., Corrado M., Cianfanelli V., Romagnoli A., Fimia G.M., Campello S., Nardacci R., Piacentini M., Campanella M., Cecconi F. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2014 doi: 10.1038/cdd.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim J., Kundu M., Viollet B., Guan K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu W., Tian W., Hu Z., Chen G., Huang L., Li W., Zhang X., Xue P., Zhou C., Liu L., Zhu Y., Zhang X., Li L., Zhang L., Sui S., Zhao B., Feng D. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014;15:566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang J., Ney P.A. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu L., Feng D., Chen G., Chen M., Zheng Q., Song P., Ma Q., Zhu C., Wang R., Qi W., Huang L., Xue P., Li B., Wang X., Jin H., Wang J., Yang F., Liu P., Zhu Y., Sui S., Chen Q. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat. Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 87.Chu C.T., Bayir H., Kagan V.E. LC3 binds externalized cardiolipin on injured mitochondria to signal mitophagy in neurons: implications for Parkinson disease. Autophagy. 2014;10:376–378. doi: 10.4161/auto.27191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chu C.T., Ji J., Dagda R.K., Jiang J.F., Tyurina Y.Y., Kapralov A.A., Tyurin V.A., Yanamala N., Shrivastava I.H., Mohammadyani D., Qiang Wang K.Z., Zhu J., Klein-Seetharaman J., Balasubramanian K., Amoscato A.A., Borisenko G., Huang Z., Gusdon A.M., Cheikhi A., Steer E.K., Wang R., Baty C., Watkins S., Bahar I., Bayir H., Kagan V.E. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frank S., Gaume B., Bergmann-Leitner E.S., Leitner W.W., Robert E.G., Catez F., Smith C.L., Youle R.J. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 90.Estaquier J., Arnoult D. Inhibiting Drp1-mediated mitochondrial fission selectively prevents the release of cytochrome c during apoptosis. Cell Death Differ. 2007;14:1086–1094. doi: 10.1038/sj.cdd.4402107. [DOI] [PubMed] [Google Scholar]
- 91.Montessuit S., Somasekharan S.P., Terrones O., Lucken-Ardjomande S., Herzig S., Schwarzenbacher R., Manstein D.J., Bossy-Wetzel E., Basanez G., Meda P., Martinou J.C. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell. 2010;142:889–901. doi: 10.1016/j.cell.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wasiak S., Zunino R., McBride H.M. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J. Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barsoum M.J., Yuan H., Gerencser A.A., Liot G., Kushnareva Y., Graber S., Kovacs I., Lee W.D., Waggoner J., Cui J., White A.D., Bossy B., Martinou J.C., Youle R.J., Lipton S.A., Ellisman M.H., Perkins G.A., Bossy-Wetzel E. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J. 2006;25:3900–3911. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G.V., Rudka T., Bartoli D., Polishuck R.S., Danial N.N., De Strooper B., Scorrano L. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 95.Frank M., Duvezin-Caubet S., Koob S., Occhipinti A., Jagasia R., Petcherski A., Ruonala M.O., Priault M., Salin B., Reichert A.S. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim. Biophys. Acta. 2012;1823:2297–2310. doi: 10.1016/j.bbamcr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 96.Murphy M.P. Mitochondrial dysfunction indirectly elevates ROS production by the endoplasmic reticulum. Cell Metab. 2013;18:145–146. doi: 10.1016/j.cmet.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 97.Sena L.A., Chandel N.S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lupfer C., Thomas P.G., Anand P.K., Vogel P., Milasta S., Martinez J., Huang G., Green M., Kundu M., Chi H., Xavier R.J., Green D.R., Lamkanfi M., Dinarello C.A., Doherty P.C., Kanneganti T.D. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat. Immunol. 2013;14:480–488. doi: 10.1038/ni.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Murley A., Lackner L.L., Osman C., West M., Voeltz G.K., Walter P., Nunnari J. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2013;2:e00422. [Google Scholar]
- 100.de Brito O.M., Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 101.Lee H., Yoon Y. Mitochondrial fission: regulation and ER connection. Mol. Cells. 2014;37:89–94. doi: 10.14348/molcells.2014.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 103.Castanier C., Garcin D., Vazquez A., Arnoult D. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 2010;11:133–138. doi: 10.1038/embor.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koshiba T., Yasukawa K., Yanagi Y., Kawabata S. Mitochondrial membrane potential is required for MAVS-mediated antiviral signaling. Sci. Signal. 2011;4:ra7. doi: 10.1126/scisignal.2001147. [DOI] [PubMed] [Google Scholar]
- 105.West A.P., Shadel G.S., Ghosh S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 2011;11:389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Onoguchi K., Onomoto K., Takamatsu S., Jogi M., Takemura A., Morimoto S., Julkunen I., Namiki H., Yoneyama M., Fujita T. Virus-infection or 5′ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS Pathog. 2010;6:e1001012. doi: 10.1371/journal.ppat.1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Horner S.M., Liu H.M., Park H.S., Briley J., Gale M., Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bouchard M.J., Navas-Martin S. Hepatitis B and C virus hepatocarcinogenesis: lessons learned and future challenges. Cancer Lett. 2011;305:123–143. doi: 10.1016/j.canlet.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Syed G.H., Amako Y., Siddiqui A. Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol. Metab. 2010;21:33–40. doi: 10.1016/j.tem.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lindenbach B.D., Rice C.M. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 111.Dreux M., Gastaminza P., Wieland S.F., Chisari F.V. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sir D., Chen W.L., Choi J., Wakita T., Yen T.S., Ou J.H. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tardif K.D., Waris G., Siddiqui A. Hepatitis C virus, ER stress, and oxidative stress. Trends Microbiol. 2005;13:159–163. doi: 10.1016/j.tim.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 114.Waris G., Tardif K.D., Siddiqui A. Endoplasmic reticulum (ER) stress: hepatitis C virus induces an ER-nucleus signal transduction pathway and activates NF-kappaB and STAT-3. Biochem. Pharmacol. 2002;64:1425–1430. doi: 10.1016/s0006-2952(02)01300-x. [DOI] [PubMed] [Google Scholar]
- 115.Lai M.M. Hepatitis C virus proteins: direct link to hepatic oxidative stress, steatosis, carcinogenesis and more. Gastroenterology. 2002;122:568–571. doi: 10.1053/gast.2002.31474. [DOI] [PubMed] [Google Scholar]
- 116.Gong G., Waris G., Tanveer R., Siddiqui A. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kappa B. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9599–9604. doi: 10.1073/pnas.171311298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li X.D., Sun L., Seth R.B., Pineda G., Chen Z.J. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Negro F. Abnormalities of lipid metabolism in hepatitis C virus infection. Gut. 2010;59:1279–1287. doi: 10.1136/gut.2009.192732. [DOI] [PubMed] [Google Scholar]
- 119.Piccoli C., Scrima R., Quarato G., D'Aprile A., Ripoli M., Lecce L., Boffoli D., Moradpour D., Capitanio N. Hepatitis C virus protein expression causes calcium-mediated mitochondrial bioenergetic dysfunction and nitro-oxidative stress. Hepatology. 2007;46:58–65. doi: 10.1002/hep.21679. [DOI] [PubMed] [Google Scholar]
- 120.Loo Y.M., Owen D.M., Li K., Erickson A.K., Johnson C.L., Fish P.M., Carney D.S., Wang T., Ishida H., Yoneyama M., Fujita T., Saito T., Lee W.M., Hagedorn C.H., Lau D.T., Weinman S.A., Lemon S.M., Gale M., Jr. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Koziel M.J., Siddiqui A. 6th ed. Elsevier; 2005. Hepatitis B Virus and Hepatitis Delta Virus. (Place Published) [Google Scholar]
- 122.Seeger C., Mason W.S. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rahmani Z., Huh K.W., Lasher R., Siddiqui A. Hepatitis B virus X protein colocalizes to mitochondria with a human voltage-dependent anion channel, HVDAC3, and alters its transmembrane potential. J. Virol. 2000;74:2840–2846. doi: 10.1128/jvi.74.6.2840-2846.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bouchard M.J., Wang L.H., Schneider R.J. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2001;294:2376–2378. doi: 10.1126/science.294.5550.2376. [DOI] [PubMed] [Google Scholar]
- 125.Waris G., Huh K.W., Siddiqui A. Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT-3 and NF-kappa B via oxidative stress. Mol. Cell. Biol. 2001;21:7721–7730. doi: 10.1128/MCB.21.22.7721-7730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sir D., Tian Y., Chen W.L., Ann D.K., Yen T.S., Ou J.H. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4383–4388. doi: 10.1073/pnas.0911373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Young L.S., Rickinson A.B. Epstein–Barr virus: 40 years on. Nat. Rev. Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 128.Pal A.D., Basak N.P., Banerjee A.S., Banerjee S. Epstein–Barr virus latent membrane protein-2A alters mitochondrial dynamics promoting cellular migration mediated by Notch signaling pathway. Carcinogenesis. 2014;35:1592–1601. doi: 10.1093/carcin/bgu069. [DOI] [PubMed] [Google Scholar]
- 129.Sinzger C., Jahn G. Human cytomegalovirus cell tropism and pathogenesis. Intervirology. 1996;39:302–319. doi: 10.1159/000150502. [DOI] [PubMed] [Google Scholar]
- 130.McCormick A.L., Smith V.L., Chow D., Mocarski E.S. Disruption of mitochondrial networks by the human cytomegalovirus UL37 gene product viral mitochondrion-localized inhibitor of apoptosis. J. Virol. 2003;77:631–641. doi: 10.1128/JVI.77.1.631-641.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hayajneh W.A., Colberg-Poley A.M., Skaletskaya A., Bartle L.M., Lesperance M.M., Contopoulos-Ioannidis D.G., Kedersha N.L., Goldmacher V.S. The sequence and antiapoptotic functional domains of the human cytomegalovirus UL37 exon 1 immediate early protein are conserved in multiple primary strains. Virology. 2001;279:233–240. doi: 10.1006/viro.2000.0726. [DOI] [PubMed] [Google Scholar]
- 132.Goldmacher V.S. vMIA, a viral inhibitor of apoptosis targeting mitochondria. Biochimie. 2002;84:177–185. doi: 10.1016/s0300-9084(02)01367-6. [DOI] [PubMed] [Google Scholar]
- 133.Arnoult D., Bartle L.M., Skaletskaya A., Poncet D., Zamzami N., Park P.U., Sharpe J., Youle R.J., Goldmacher V.S. Cytomegalovirus cell death suppressor vMIA blocks Bax- but not Bak-mediated apoptosis by binding and sequestering Bax at mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7988–7993. doi: 10.1073/pnas.0401897101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Poncet D., Pauleau A.L., Szabadkai G., Vozza A., Scholz S.R., Le Bras M., Briere J.J., Jalil A., Le Moigne R., Brenner C., Hahn G., Wittig I., Schagger H., Lemaire C., Bianchi K., Souquere S., Pierron G., Rustin P., Goldmacher V.S., Rizzuto R., Palmieri F., Kroemer G. Cytopathic effects of the cytomegalovirus-encoded apoptosis inhibitory protein vMIA. J. Cell Biol. 2006;174:985–996. doi: 10.1083/jcb.200604069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Karbowski M., Norris K.L., Cleland M.M., Jeong S.Y., Youle R.J. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- 136.Kramer T., Enquist L.W. Alphaherpesvirus infection disrupts mitochondrial transport in neurons. Cell Host Microbe. 2012;11:504–514. doi: 10.1016/j.chom.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Varga Z.T., Grant A., Manicassamy B., Palese P. Influenza virus protein PB1-F2 inhibits the induction of type I interferon by binding to MAVS and decreasing mitochondrial membrane potential. J. Virol. 2012;86:8359–8366. doi: 10.1128/JVI.01122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yoshizumi T., Ichinohe T., Sasaki O., Otera H., Kawabata S., Mihara K., Koshiba T. Influenza A virus protein PB1-F2 translocates into mitochondria via Tom40 channels and impairs innate immunity. Nat. Commun. 2014;5:4713. doi: 10.1038/ncomms5713. [DOI] [PubMed] [Google Scholar]
- 139.Xia M., Gonzalez P., Li C., Meng G., Jiang A., Wang H., Gao Q., Debatin K.M., Beltinger C., Wei J. Mitophagy enhances oncolytic measles virus replication by mitigating DDX58/RIG-I-like receptor signaling. J. Virol. 2014;88:5152–5164. doi: 10.1128/JVI.03851-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xia M., Meng G., Jiang A., Chen A., Dahlhaus M., Gonzalez P., Beltinger C., Wei J. Mitophagy switches cell death from apoptosis to necrosis in NSCLC cells treated with oncolytic measles virus. Oncotarget. 2014;5:3907–3918. doi: 10.18632/oncotarget.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shi C.S., Qi H.Y., Boularan C., Huang N.N., Abu-Asab M., Shelhamer J.H., Kehrl J.H. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Qi X., Disatnik M.H., Shen N., Sobel R.A., Mochly-Rosen D. Aberrant mitochondrial fission in neurons induced by protein kinase C{delta} under oxidative stress conditions in vivo. Mol. Biol. Cell. 2011;22:256–265. doi: 10.1091/mbc.E10-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Park Y.Y., Nguyen O.T., Kang H., Cho H. MARCH5-mediated quality control on acetylated Mfn1 facilitates mitochondrial homeostasis and cell survival. Cell Death Dis. 2014;5:e1172. doi: 10.1038/cddis.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Meng G., Xia M., Wang D., Chen A., Wang Y., Wang H., Yu D., Wei J. Mitophagy promotes replication of oncolytic Newcastle disease virus by blocking intrinsic apoptosis in lung cancer cells. Oncotarget. 2014;5:6365–6374. doi: 10.18632/oncotarget.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]