Abstract
Objectives
The objective of this study was to explore the activity of ceftazidime and ceftazidime/avibactam against a collection of isogenic strains of Escherichia coli DH10B possessing SHV and KPC β-lactamases containing single amino acid substitutions in the Ω-loop (residues 164–179).
Methods
Ceftazidime and ceftazidime/avibactam MICs were determined by the agar dilution method for a panel of isogenic E. coli strains expressing SHV-1 and KPC-2 with amino acid substitutions at positions 164, 167, 169 or 179. Two KPC-2 β-lactamase variants that possessed elevated MICs of ceftazidime/avibactam were selected for further biochemical analyses.
Results
Avibactam restored susceptibility to ceftazidime for all Ω-loop variants of SHV-1 with MICs <8 mg/L. In contrast, several of the Arg164 and Asp179 variants of KPC-2 demonstrated MICs of ceftazidime/avibactam >8 mg/L. β-Lactamase kinetics showed that the Asp179Asn variant of KPC-2 demonstrated enhanced kinetic properties against ceftazidime. The Ki app, k2/K and koff of the Arg164Ala and Asp179Asn variant KPC-2 β-lactamases indicated that avibactam effectively inhibited these enzymes.
Conclusions
Several KPC-2 variants demonstrating ceftazidime resistance as a result of single amino acid substitutions in the Ω-loop were not susceptible to ceftazidime/avibactam (MICs >8 mg/L). We hypothesize that this observation is due to the stabilizing interactions (e.g. hydrogen bonds) of ceftazidime within the active site of variant β-lactamases that prevent avibactam from binding to and inhibiting the β-lactamase. As ceftazidime/avibactam is introduced into the clinic, monitoring for new KPC-2 variants that may exhibit increased ceftazidime kinetics as well as resistance to this novel antibiotic combination will be important.
Keywords: β-lactamase inhibitors, extended-spectrum β-lactamases, ESBLs, antibiotic resistance
Introduction
β-Lactam antibiotics are highly effective inhibitors of the PBPs in bacteria. The binding of β-lactams to PBPs prevents peptidoglycan cross-linking and eventually leads to cell lysis and death. Bacteria continually evolve in response to antibiotic threats and many Gram-negative bacteria produce β-lactamase enzymes that are able to cleave the β-lactam bond of the antibiotics and render them ineffective.
β-Lactamases are grouped into four classes according to the Ambler system; class A, C and D β-lactamases utilize an active-site serine residue, while class B metallo-β-lactamases possess Zn2+ ions as a nucleophile.1 β-Lactamases are also phenotypically or functionally classified into three groups according to the Bush–Jacoby classification system. The group 1 β-lactamases (class C) hydrolyse cephalosporins more than penicillins as their defining characteristic. The group 2 enzymes (class A and D) hydrolyse penicillins and possess variants with extended-spectrum properties that also hydrolyse oxyimino-cephalosporins. In addition, other variants are present in group 2 that exhibit resistance to β-lactamase inhibitors. The group 3 β-lactamases (class B) confer broad-spectrum hydrolysis with the exception of monobactams.2,3
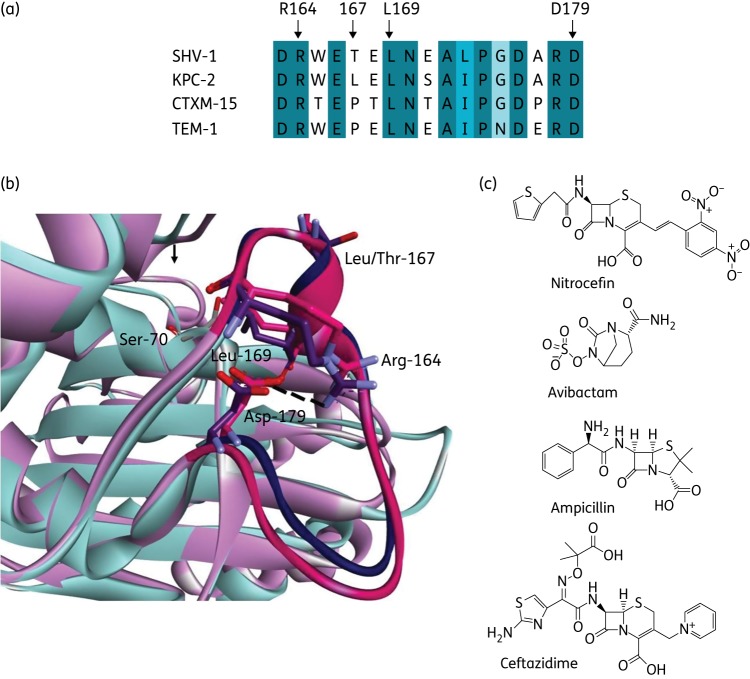
Several conserved motifs are present in class A β-lactamases including the Ω-loop, which encompasses amino acid residues Arg164 to Asp179.4–10 The Ω-loop is particularly important as it contains two amino acids involved in the acylation and deacylation of substrates by β-lactamases, Glu166 and Asn170 (Figure 1a and b).5,6,10–12 Single amino acid substitutions, particularly at positions 164, 167, 169 and 179 within the Ω-loop of class A β-lactamases, lead to reduced susceptibility to ceftazidime (Figure 1b).5–9,12–19 Enhanced kinetics towards ceftazidime is responsible for the extension of the substrate profile of some TEM-1 and SHV-1 variants at these positions.12,14,20–28
Figure 1.
(a) Amino acid sequence alignment of the Ω-loop in four class A β-lactamases. The black arrows mark the sites of focus in this paper (Arg164, Leu/Pro167, Leu169 and Asp179). (b) Overlay of the X-ray crystallographic protein structure of SHV-1 (PDB ID: 1SHV, pink/magenta) and KPC-2 (PDB ID: 2OV5, cyan/purple) showing the Ω-loop in deeper colours with the catalytic Ser70, Arg164, Leu/Thr167, Leu169 and Asp179 labelled and the Arg164–Asp179 salt bridge drawn as a broken black line. (c) Chemical structures of the compounds tested in this paper. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Avibactam is a novel non-β-lactam β-lactamase inhibitor that is being developed with ceftazidime for the treatment of Gram-negative bacterial infections. Ceftazidime/avibactam demonstrates potent activity against Enterobacteriaceae possessing extended-spectrum β-lactamases (ESBLs).29–33 Single amino acid substitutions in the Ω-loop that confer the ESBL phenotype are commonly encountered in clinical isolates (Table 1) and the inhibition of these Ω-loop variants by avibactam has not previously been explored. Thus, we tested to see if avibactam could restore the activity of ceftazidime against several SHV-1 and KPC-2 class A β-lactamase variants possessing single amino acid substitutions in the Ω-loop. Little is known about the interplay of cephalosporins in the active site of ESBLs when coupled with a novel β-lactamase inhibitor from the diazabicyclooctanone class like avibactam. However, other traditional β-lactamase inhibitors (i.e. clavulanic acid, tazobactam, sulbactam) have shown activity in partnership with a penicillin or cephalosporin in clinical and non-clinical settings against Ω-loop ESBL SHV variants.18,34,35
Table 1.
Clinical Ω-loop variants of the SHV and KPC β-lactamasesa
| Enzyme | Amino acid substitution |
|---|---|
| SHV-6 | D179A |
| SHV-8 | D179N |
| SHV-16 | 5 amino acid duplication at E166 |
| SHV-19, -20, -21 | L173F |
| SHV-24 | D179G |
| SHV-51 | G175A |
| SHV-57 | L169R |
| SHV-79 | A172V |
| SHV-111 | P174S |
| SHV-143 | R164L |
| KPC-12 | L169M |
aObtained from the Lahey website: http://www.lahey.org/Studies.
We discovered that avibactam is able to inhibit the Ω-loop variant β-lactamases, as evidenced by comparable inhibitory kinetic parameters for selected KPC variants. Yet ceftazidime/avibactam MICs were still higher for certain KPC variants with Ω-loop amino acid substitutions. We advance that resistance to ceftazidime/avibactam may be due to enhanced kinetics against ceftazidime (e.g. lower Km), as previously described for the R164S variant of KPC-2, thus preventing avibactam from inhibiting the enzyme.15
Materials and methods
Site-directed mutagenesis
The Agilent site-directed mutagenesis XL kitTM was used to create codon changes corresponding to particular amino acid substitutions at positions 164, 167, 169 and 179 in the blaSHV-1 gene in the pBC SK(−) and the blaKPC-2 gene in the pBR322-catI plasmid. The cloning of these β-lactamase genes into their respective plasmids was previously described.36,37 McLab (http://www.mclab.com/) was used to sequence each plasmid-encoded β-lactamase gene to verify the success of the mutagenesis reaction.
MIC measurement
Agar-dilution MICs were determined according to the CLSI protocol.38 Briefly, Mueller–Hinton (M–H) agar was used to pour plates with doubling dilutions of antibiotics. Bacteria were grown overnight in M–H broth and then diluted and stamped onto the plates with a SteersTM replicator to deliver 10 μL of a 104 bacterial load per spot. The following day, the plates were read and the MIC was defined as the antibiotic concentration at which bacterial growth was no longer observed. We also performed a second set of MICs using Escherichia coli clones containing blaSHV-1 and blaKPC-2 on LB agar in order to compare values obtained on these two different agar compositions. In addition to agar-dilution MICs, ceftazidime Etest (bioMérieux Diagnostics) MIC assays were determined using the manufacturer's instructions. Broth microdilution using frozen panels (ThermoFisher Scientific, Cleveland, OH, USA) with ceftazidime according to CLSI guidelines and MicroScan panels were also conducted.
Compounds
Ceftazidime was purchased from Sigma-Aldrich, Research Products International Corp. and its commercial source. Avibactam was acquired through a research contract with AstraZeneca Pharmaceuticals. Nitrocefin was purchased from Becton-Dickinson.
β-Lactamase preparation
Four litres of super-optimal broth supplemented with chloramphenicol at 20 mg/L for plasmid maintenance were grown overnight at 37°C with E. coli DH10B cells containing pBR322-catI-blaKPC-2, pBR322-catI-blaKPCArg164Ala or pBR322-catI-blaKPCAsp179Asn. The cells were pelleted and the supernatant was discarded. The cell pellets were frozen at −20°C. The pellets were thawed, resuspended in Tris–HCl, pH 7.4 with lysozyme, benzonuclease, MgSO4 and EDTA added as previously described.39 The cellular debris was then pelleted and the supernatant was filtered twice through a Corning 28 mm 0.20 μm syringe filter and these crude periplasmic extracts were used for enzyme kinetic analysis. We note that, during attempted procedures to obtain pure enzyme, these Ω-loop variants were unstable. In addition, the variants were not expressed equally at steady-state; therefore, the amount of extract used for kinetic analysis was normalized to the level of nitrocefin hydrolysis.15
Steady-state enzyme kinetic measurements
All experiments were performed at room temperature (25°C) in 10 mM PBS pH 7.4 using crude extracts under steady-state conditions on an Agilent 8453 spectrophotometer as previously described with all values having an error of 10%.40 Due to the use of the periplasmic extracts, all values are reported as the apparent kinetic constants. For nitrocefin, the apparent Km and Vmax were determined by measuring initial velocities at a variety of nitrocefin concentrations. Then, the data were fitted to the Michaelis–Menten equation (non-linear least-square fit) using EnzFitter to obtain the apparent Vmax and Km.
For avibactam, Ki apparent or Ki app was determined by a competition assay with nitrocefin as previously reported.41 Briefly, increasing concentrations of avibactam (from 2.5 to 20 μM) were added to extract/nitrocefin mixtures and the initial velocities (vi) were measured. Excel was used to plot 1/vi versus the concentration of avibactam and the data were fitted to a linear equation, where the y-intercept divided by the slope of the line was defined as the Ki app observed. This value was corrected for the use of nitrocefin according to equation 1 and this corrected value was defined as Ki app.
| (1) |
The apparent onset of acylation (k2/K) was approximated for avibactam as previously described.41,42 Briefly, the apparent k2/K values were determined by measuring timed inactivation of the periplasmic extracts using 100 μM of nitrocefin as a reporter substrate and increasing concentrations of avibactam over a 400 s time course. Origin 8.1 was used to fit each time course to equation 2 to obtain a kobs value for each avibactam concentration. Here, vf is final velocity and Ai is the initial absorbance at λ = 482 nm.
| (2) |
Then, the kobs values were plotted against [avibactam] and fitted to a linear equation using Excel. The k2/K observed was derived using equation 3. The k2/K observed was then corrected for the use of nitrocefin to obtain the apparent k2/K value according to equation 4.
| (3) |
| (4) |
The apparent koff was determined as previously described.41,42 Periplasmic extracts were incubated with an excess of avibactam (as determined by full enzyme inhibition using nitrocefin as a reporter substrate) for 5 min. Serial dilutions of the enzyme/inhibitor mixes were performed to eventually dilute the mixture 1: 1000 with 100 μM nitrocefin as a reporter substrate and the hydrolysis was measured for 3600 s. The apparent koff was calculated by fitting the resulting curve to a single exponential equation using Excel.
Ceftazidime (50 μM) hydrolysis curves were measured at λ = 260 nm using periplasmic extracts of KPC-2 and the KPC-2 Asp179Asn variant at room temperature (25°C) in PBS during a 100 s time course. The lines were compared with background spontaneous hydrolysis of ceftazidime alone and fitted to a simple linear equation in Excel to obtain the rate of hydrolysis as the slope of the line.
Rapid-mixing stopped-flow kinetics
Pre-steady-state kinetics were measured on an Applied Photophysics SX-20 Stopped-Flow spectrophotometer using similar conditions as for the ceftazidime hydrolysis curves described above. However, for stopped-flow kinetics, a cuvette with a 0.2 cm pathlength was used and data were collected on a logarithmic scale measuring 1000 points over 1 s.
Results
Susceptibility to ceftazidime/avibactam combinations
The MIC measurements for the Ω-loop variants of SHV-1 and KPC-2 expressed in E. coli DH10B are shown in Table 2. Many of the SHV-1 Ω-loop variants raised the ceftazidime MIC from 4–8 mg/L for WT SHV-1 to 32–128 mg/L. Similarly, several of the KPC-2 Ω-loop variants increased the ceftazidime MIC from 64 mg/L for WT KPC-2 to 256 and >512 mg/L. When avibactam was added to ceftazidime, the MICs were lowered for all of the bacterial strains. Avibactam decreased the ceftazidime MICs to 0.5 mg/L or lower for all of the SHV-1 variants. However, ceftazidime/avibactam MICs remained >8 mg/L for five of the KPC-2 variants—Arg164Ala (16 mg/L), Arg164Pro (64 mg/L), Asp179Ala (64 mg/L), Asp179Gln (32 mg/L) and Asp179Asn (64 mg/L)—while the ceftazidime/avibactam MIC for WT KPC-2 was lowered to 1 mg/L.
Table 2.
MICs in mg/L for various Ω-loop mutants of KPC and SHV tested with ceftazidime and ceftazidime/avibactam on M–H agara,b
| Strain | Ceftazidime | Ceftazidime/avibactam |
|---|---|---|
| E. coli DH10B | 0.25 | 0.25 |
| E. coli DH10B pBC SK(−) emptyc | 0.5 | 0.25 |
| E. coli DH10B pBC SK(−) blaSHV-1 | 4–8 | 0.5 |
| E. coli DH10B pBC SK(−) blaSHV Arg164Ala | 128 | 0.5 |
| E. coli DH10B pBC SK(−) blaSHV Arg164His | 64 | 0.5 |
| E. coli DH10B pBC SK(−) blaSHV Arg164Pro | 32 | 0.5 |
| E. coli DH10B pBC SK(−) blaSHV Thr167Ala | 8 | 0.5 |
| E. coli DH10B pBC SK(−) blaSHV Thr167Leu | 8 | 0.5 |
| E. coli DH10B pBC SK(−) blaSHV Thr167Pro | 0.5 | 0.5 |
| E. coli DH10B pBC SK(−) blaSHV Leu169Ala | 4 | 0.25 |
| E. coli DH10B pBC SK(−) blaSHV Leu169Arg | 64 | 0.5 |
| E. coli DH10B pBC SK(−) blaSHV Asp179Ala | 64 | 0.5 |
| E. coli DH10B pBC SK(−) blaSHV Asp179Gln | 32 | 0.5 |
| E. coli DH10B pBC SK(−) blaSHV Asp179Asn | 32 | 0.5 |
| E. coli DH10B pBR322-catI-blaKPC-2 | 64 | 1 |
| E. coli DH10B pBR322-catI-blaKPC Arg164Ala | 256 | 16 |
| E. coli DH10B pBR322-catI-blaKPC Arg164His | 256 | 8 |
| E. coli DH10B pBR322-catI-blaKPC Arg164Pro | 256 | 64 |
| E. coli DH10B pBR322-catI- blaKPC Arg164Ser | 256 | 8 |
| E. coli DH10B pBR322-catI-blaKPC Leu167Ala | 32 | 1 |
| E. coli DH10B pBR322-catI-blaKPC Leu167Pro | 16 | 1 |
| E. coli DH10B pBR322-catI-blaKPC Leu167Thr | 32 | 1 |
| E. coli DH10B pBR322-catI-blaKPC Leu169Ala | 128 | 2 |
| E. coli DH10B pBR322-catI-blaKPC Leu169Arg | 256 | 0.5 |
| E. coli DH10B pBR322-catI-blaKPC Asp179Ala | 512 | 64 |
| E. coli DH10B pBR322-catI-blaKPC Asp179Gln | 512 | 32 |
| E. coli DH10B pBR322-catI-blaKPC Asp179Asn | >512 | 32 |
Potency of ceftazidime preparations and MIC creep
The ceftazidime MIC measurements that we obtained for E. coli DH10B expressing pBC SK(−) with blaSHV-1 (4–8 mg/L) and pBR322-catI- with blaKPC-2 (64 mg/L) are higher than those previously reported by our laboratory: 1–2 mg/L for SHV-1 and 32 mg/L for KPC-2.15,43 To address these higher ceftazidime MICs, we repeated ceftazidime MICs using three different frozen stocks of E. coli DH10B pBC SK(−) blaSHV-1 with three different formulations of ceftazidime. We obtained MICs between 4 and 8 mg/L for all measurements. Ceftazidime Etest MICs were also conducted and an MIC of 4 mg/L was observed (see the Acknowledgements section). Broth microdilution MICs using frozen panels from Thermofisher Scientific and MicroScan also produced results of 4–8 mg/L for ceftazidime.
However, the previously reported MICs used LB agar for determination of the agar dilution MICs whereas we used M–H agar for our studies. Therefore, we performed a comparison of LB and M–H agar for MIC determination. When agar dilution MICs were performed with the LB agar, we determined MICs of 2–4 mg/L for the SHV-1 clone and 32 mg/L for the KPC-2 clone. Therefore, we believe that our higher MICs are due to the characteristics of the M–H agar. The MIC comparison of the different variant enzymes is valid as all MICs were performed using M–H agar.
Biochemical assays for avibactam inhibition and ceftazidime hydrolysis by KPC-2 and selected variants
To begin to understand the mechanistic details behind the elevated ceftazidime/avibactam MICs in the five KPC Ω-loop variants, we selected the Arg164Ala and Asp179Asn variants for further biochemical assays with avibactam and ceftazidime. We used crude β-lactamase extracts to perform inhibitory enzyme kinetics on these β-lactamase variants as the purified proteins were unstable under our purification conditions.
Nitrocefin kinetic assays were performed initially to assess the catalytic activity of these variants. The apparent Km for nitrocefin for the Asp179Asn variant was similar to WT KPC-2, but the Arg164Ala variant had a 10-fold higher Km for nitrocefin (Table 3). Additionally, the Vmax/Km ratios were similar for the Asp179Asn variant and KPC-2, but 10-fold lower for the Arg164Ala variant.
Table 3.
Enzyme kinetics of periplasmic extracts of KPC variants
| KPC-2 | KPC-2 Arg164Ala | KPC-2 Asp179Asn | |
|---|---|---|---|
| Nitrocefin | |||
| Km (μM) | 15 ± 4 | 118 ± 50 | 5 ± 1 |
| Vmax/Km (s−1) | 0.06 | 0.005 | 0.09 |
| Avibactam | |||
| Ki app (μM) | 1.2 ± 0.1 | 2.2 ± 0.2 | 0.4 ± 0.04 |
| k2/K (M−1 s−1) | 17 000 | 7000 | 38 000 |
| koff | 1.1 × 10−4 | 4.9 × 10−5 | 4.9 × 10−5 |
To assess the inhibitory capacity of avibactam against the Arg164Ala and Asp179Asn variants, the Ki app and k2/K values were determined. Similar concentrations of avibactam were necessary to obtain full inhibition of each enzyme variant (Figure 2). The variants were rapidly acylated with a k2/K value lowest for the Arg164Ala variant and highest for the Asp179Asn variant (Table 3). The Ki app was similar among the three enzymes (Table 3). The koff was also slow for all three variant enzymes (Table 3).
Figure 2.
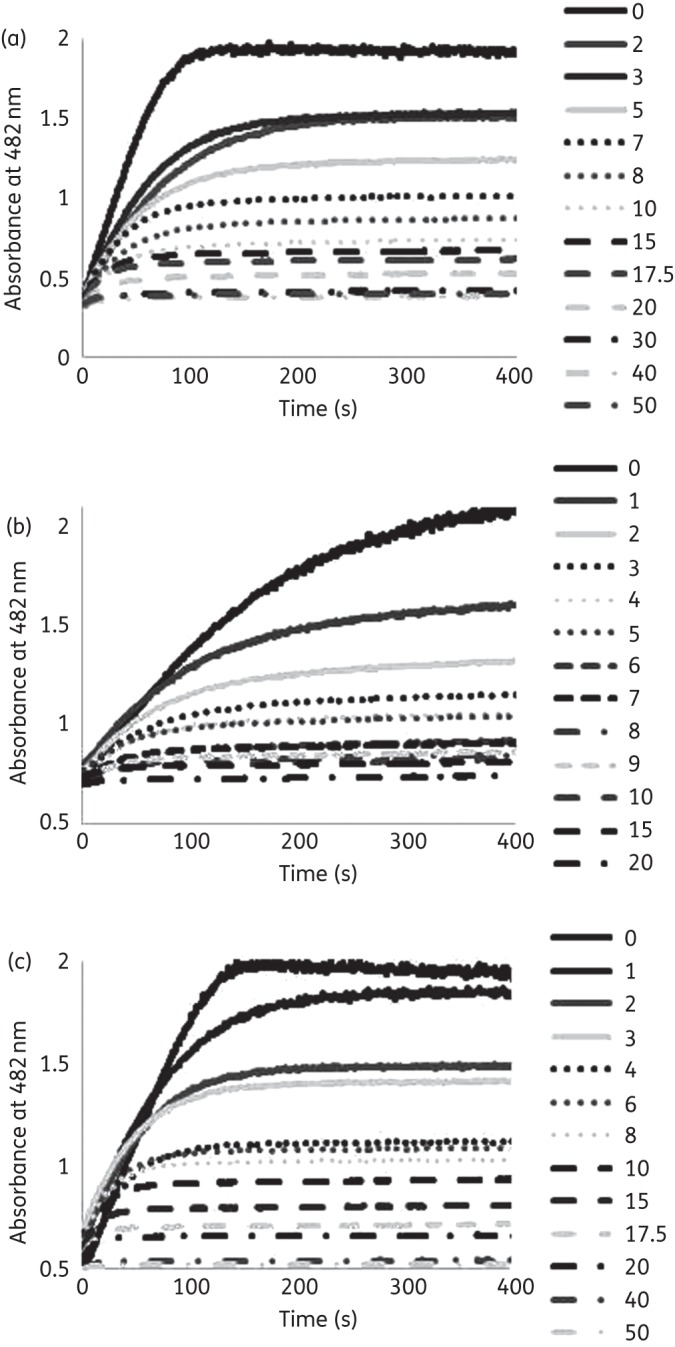
(a) Inhibition of nitrocefin (100 μM) hydrolysis by KPC-2 with increasing concentrations of avibactam (2–50 μM). (b) Inhibition of nitrocefin (100 μM) hydrolysis by KPC Arg164Ala with increasing concentrations of avibactam (1–20 μM). (c) Inhibition of nitrocefin (100 μM) hydrolysis by KPC Asp179Asn with increasing concentrations of avibactam (1–50 μM).
To evaluate the impact of the single amino acid substitutions in the Ω-loop on ceftazidime hydrolysis, extracts of KPC-2 and the Asp179Asn variant were incubated with ceftazidime during two different time courses (Figure 3). During a 100 s time course, the ceftazidime hydrolysis rates were similar between the two enzymes. However, using rapid-mixing stopped-flow spectroscopy during a 1.5 ms time course, a ‘burst’ in ceftazidime hydrolysis was observed with the Asp179Asn variant, but not with KPC-2. The level of ceftazidime hydrolysis by KPC-2 was similar to the background level of spontaneous ceftazidime hydrolysis under these pre-steady-state conditions.
Figure 3.
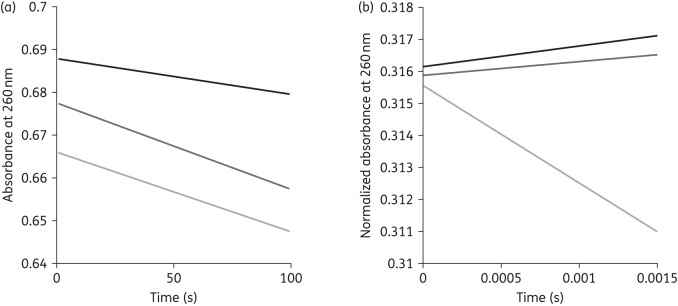
(a) Hydrolysis of ceftazidime (50 μM) by periplasmic extracts of KPC-2 (dark grey) and Asp179Asn (light grey); ceftazidime alone (black) during a 100 s time course. (b) Hydrolysis of ceftazidime (50 μM) by periplasmic extracts of KPC-2 (dark grey) and Asp179Asn (light grey); ceftazidime alone (black) during a 1.5 ms time course.
Discussion
Variant class A β-lactamases with single amino acid substitutions in the Ω-loop pose a significant threat against expanded-spectrum cephalosporins as these enzymes hydrolyse ceftazidime and other oxyimino-cephalosporins at a greater rate than their parent enzyme. We showed that single amino acid substitutions at positions 164, 169 and 179 of SHV-1 and KPC-2 raised the ceftazidime MIC for isogenic strains of E. coli containing these variant enzymes. Notably, we did not observe elevated ceftazidime MICs for strains carrying 167 variants. Conversely, substitutions at position 167 in the CTX-M or TEM class A β-lactamases were shown to convey increased ceftazidime MICs.6,17,19 The addition of avibactam to ceftazidime was able to reduce ceftazidime MICs for all of the variants with single amino acid substitutions in the Ω-loops of SHV-1 and KPC-2. However, five of the KPC-2 variants demonstrated ceftazidime/avibactam MICs >8 mg/L.
Notably, ceftazidime MICs were higher for E. coli DH10B pBC SK(−) expressing SHV-1 and pBR322-catI- producing KPC-2 than previously reported by our group.15,43 After testing multiple formulations of ceftazidime using three different frozen stocks of E. coli DH10B pBC SK(−) expressing SHV-1, we concluded that the difference can be attributed to the use of M–H agar in this study versus LB agar in previous work.15,43 As three different frozen stocks of bacteria were used for this analysis, we did not believe that this was an issue with our clones. However, we also verified each clone by DNA sequencing and found the bla genes and promoters to be identical. Further studies will be completed to evaluate the effect of the different agar formulations on MIC determination.
Selected variants of KPC-2 with elevated MICs to ceftazidime/avibactam were assayed kinetically with avibactam and ceftazidime. The inhibitory avibactam kinetics were similar between KPC-2 and the Arg164Ala and Asp179Asn variants, indicating that avibactam was able to encounter and inactivate these β-lactamases with similar potency. The ceftazidime kinetics revealed that the KPC-2 and Asp179Asn variant hydrolysed ceftazidime at a similar rate during a 100 s study period. Conversely, rapid-mixing stopped-flow kinetics of KPC-2 and the Asp179Asn variant on a 1.5 ms timescale showed a phenomenon that was previously observed with the Arg164Ser variant of KPC-2 and ceftazidime.15 The Arg164Ser variant showed a ‘burst’, where ceftazidime was rapidly bound to the variant enzyme and unable to interact with its PBP target. This ‘burst’ leads to ceftazidime resistance despite comparable hydrolysis rates among the enzymes in steady-state experiments due to an increased affinity of ceftazidime for the enzyme. We qualitatively observed a ‘burst’ with the Asp179Asn variant. Thus, we postulate that increased affinity for ceftazidime may prevent binding of avibactam to our enzymes with Ω-loop substitutions, resulting in elevated ceftazidime/avibactam MICs despite potent avibactam inhibition of these enzymes. Since avibactam has not been shown to have activity on its own, the PBP targets are not inactivated. As a result, elevated ceftazidime/avibactam MICs were observed. Further biochemical analyses of Ω-loop variants of KPC-2 are currently underway to dissect their mechanisms of ceftazidime resistance and will be the focus of separate studies.
One KPC-2 variant with an amino acid substitution in the Ω-loop has emerged clinically (KPC-12; Table 1). Thus, the appearance of bacteria containing Ω-loop variants of KPC-2 may threaten the efficacy of ceftazidime/avibactam combination therapy. However, since there appears to be a severe evolutionary cost of Ω-loop amino acid substitutions that results in impaired stability, we hypothesize that our tested variants may not emerge clinically without a secondary stabilizing substitution (i.e. a ‘global suppressor’ substitution).4–7,44,45 Evidence of global suppressor mutations in the class A TEM-1 β-lactamase has been well established; their role in KPC-2 is not yet defined.44,45 As ceftazidime/avibactam is introduced into the clinic, being aware of their limitations of efficacy against ESBL enzyme variants with high ceftazidime resistance is essential.
Conclusions
Here, we revealed that resistance to ceftazidime/avibactam might occur via mutations to KPC enzymes that result in enhanced ceftazidime kinetics rather than reduced avibactam inhibition. The likelihood of this as a clinical risk remains to be ascertained. This is in contrast to typical inhibitor resistant class A β-lactamases, which commonly manifest this phenotype as a result of resistance to the inhibitor mediated by changes in the amino acid sequence.46,47 All class A β-lactamase variants resistant to the commercially available combinations of amoxicillin/clavulanic acid, ampicillin/sulbactam and piperacillin/tazobactam are resistant due to inhibitor-resistant substitutions (e.g. substitutions at positions 69, 130 and 234), not due to increased resistance to the β-lactam partner.46–48 Here, Ω-loop variants are more resistant to ceftazidime/avibactam, due to ESBL substitutions (e.g. Ω-loop) that alter ceftazidime kinetics. This lowers the effectiveness of the ceftazidime/avibactam combination but does not lead to inhibitor resistance due to an effect on avibactam kinetics. Clinicians will be administering ceftazidime/avibactam to patients who possess infections with ceftazidime-resistant strains; if these ceftazidime-resistant strains also contain certain Ω-loop substitutions, the efficacy of the combination may be in question. Being aware of the emergence of variants with enhanced ceftazidime kinetic properties as the ceftazidime/avibactam combination completes Phase III clinical trials and begins to be introduced for the treatment of complicated clinical infections is important. These data also stress the importance of ceftazidime in this novel β-lactam/β-lactamase inhibitor combination.
Funding
This work was supported by the National Institutes of Health and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health, the Cleveland Department of Veterans Affairs and AstraZeneca. In addition, this work was supported in part by the National Institutes of Health (Grant T32-GM-7250 to Case Western Reserve University, M. L. W.), the Veterans Affairs Career Development Award (K. M. P.-W.), the Veterans Affairs Merit Review Program (R. A. B.), the National Institutes of Health (Grant AI072219-05 and Grant AI063517-07 to R. A. B.) and the Geriatric Research Education and Clinical Center VISN 10 (R. A. B.).
Transparency declarations
K. M. P.-W. has received research funding from AstraZeneca. R. A. B. has received research funding from AstraZeneca, Merck and Checkpoints, and has served on a Tetraphase drug safety monitoring board. M. L. W.: none to declare.
Acknowledgements
We thank AstraZeneca Pharmaceuticals for providing the avibactam powder. We thank Dr Michael R. Jacobs for conducting broth microdilution and MicroScan experiments for ceftazidime susceptibility assessment. We would like to thank Scott A. Becka and Steven M. Marshall for testing various formulations of ceftazidime for susceptibility testing. We would also like to thank Dr Laurent Poirel at the University of Fribourg for the independent testing of ceftazidime susceptibility of these strains.
References
- 1.Ambler RP, Coulson AF, Frere JM, et al. A standard numbering scheme for the class A β-lactamases. Biochem J 1991; 276: 269–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother 1995; 39: 1211–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 2010; 54: 969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vakulenko SB, Toth M, Taibi P, et al. Effects of Asp-179 mutations in TEMpUC19 β-lactamase on susceptibility to β-lactams. Antimicrob Agents Chemother 1995; 39: 1878–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palzkill T, Le QQ, Venkatachalam KV, et al. Evolution of antibiotic resistance: several different amino acid substitutions in an active site loop alter the substrate profile of β-lactamase. Mol Microbiol 1994; 12: 217–29. [DOI] [PubMed] [Google Scholar]
- 6.Petrosino JF, Palzkill T. Systematic mutagenesis of the active site omega loop of TEM-1 β-lactamase. J Bacteriol 1996; 178: 1821–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vakulenko SB, Taibi-Tronche P, Toth M, et al. Effects on substrate profile by mutational substitutions at positions 164 and 179 of the class A TEMpUC19 β-lactamase from Escherichia coli. J Biol Chem 1999; 274: 23052–60. [DOI] [PubMed] [Google Scholar]
- 8.Arpin C, Labia R, Andre C, et al. SHV-16, a β-lactamase with a pentapeptide duplication in the omega loop. Antimicrob Agents Chemother 2001; 45: 2480–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yi H, Cho KH, Cho YS, et al. Twelve positions in a β-lactamase that can expand its substrate spectrum with a single amino acid substitution. PLoS One 2012; 7: e37585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerjee S, Pieper U, Kapadia G, et al. Role of the Ω-loop in the activity, substrate specificity, and structure of class A β-lactamase. Biochemistry 1998; 37: 3286–96. [DOI] [PubMed] [Google Scholar]
- 11.Guillaume G, Vanhove M, Lamotte-Brasseur J, et al. Site-directed mutagenesis of glutamate 166 in two β-lactamases. Kinetic and molecular modeling studies. J Biol Chem 1997; 272: 5438–44. [DOI] [PubMed] [Google Scholar]
- 12.Delmas J, Robin F, Bittar F, et al. Unexpected enzyme TEM-126: role of mutation Asp179Glu. Antimicrob Agents Chemother 2005; 49: 4280–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirel L, Naas T, Le Thomas I, et al. CTX-M-type extended-spectrum β-lactamase that hydrolyzes ceftazidime through a single amino acid substitution in the omega loop. Antimicrob Agents Chemother 2001; 45: 3355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurokawa H, Yagi T, Shibata N, et al. A new SHV-derived extended-spectrum β-lactamase (SHV-24) that hydrolyzes ceftazidime through a single-amino-acid substitution (D179G) in the Ω-loop. Antimicrob Agents Chemother 2000; 44: 1725–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levitt PS, Papp-Wallace KM, Taracila MA, et al. Exploring the role of a conserved class A residue in the Ω-loop of KPC-2 β-lactamase: a mechanism for ceftazidime hydrolysis. J Biol Chem 2012; 287: 31783–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sampson JM, Ke W, Bethel CR, et al. Ligand-dependent disorder of the Ω loop observed in extended-spectrum SHV-type β-lactamase. Antimicrob Agents Chemother 2011; 55: 2303–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bae IK, Lee BH, Hwang HY, et al. A novel ceftazidime-hydrolysing extended-spectrum β-lactamase, CTX-M-54, with a single amino acid substitution at position 167 in the omega loop. J Antimicrob Chemother 2006; 58: 315–9. [DOI] [PubMed] [Google Scholar]
- 18.Ma L, Alba J, Chang FY, et al. Novel SHV-derived extended-spectrum β-lactamase, SHV-57, that confers resistance to ceftazidime but not cefazolin. Antimicrob Agents Chemother 2005; 49: 600–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturenburg E, Kuhn A, Mack D, et al. A novel extended-spectrum β-lactamase CTX-M-23 with a P167 T substitution in the active-site omega loop associated with ceftazidime resistance. J Antimicrob Chemother 2004; 54: 406–9. [DOI] [PubMed] [Google Scholar]
- 20.Chanal C, Poupart MC, Sirot D, et al. Nucleotide sequences of CAZ-2, CAZ-6, and CAZ-7 β-lactamase genes. Antimicrob Agents Chemother 1992; 36: 1817–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutmann L, Kitzis MD, Billot-Klein D, et al. Plasmid-mediated β-lactamase (TEM-7) involved in resistance to ceftazidime and aztreonam. Rev Infect Dis 1988; 10: 860–6. [DOI] [PubMed] [Google Scholar]
- 22.Kurokawa H, Shibata N, Doi Y, et al. A new TEM-derived extended-spectrum β-lactamase (TEM-91) with an R164C substitution at the Ω-loop confers ceftazidime resistance. Antimicrob Agents Chemother 2003; 47: 2981–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul GC, Gerbaud G, Bure A, et al. TEM-4, a new plasmid-mediated β-lactamase that hydrolyzes broad-spectrum cephalosporins in a clinical isolate of Escherichia coli. Antimicrob Agents Chemother 1989; 33: 1958–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petit A, Sirot DL, Chanal CM, et al. Novel plasmid-mediated β-lactamase in clinical isolates of Klebsiella pneumoniae more resistant to ceftazidime than to other broad-spectrum cephalosporins. Antimicrob Agents Chemother 1988; 32: 626–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinn JP, Miyashiro D, Sahm D, et al. Novel plasmid-mediated β-lactamase (TEM-10) conferring selective resistance to ceftazidime and aztreonam in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother 1989; 33: 1451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sougakoff W, Petit A, Goussard S, et al. Characterization of the plasmid genes blaT-4 and blaT-5 which encode the broad-spectrum β-lactamases TEM-4 and TEM-5 in Enterobacteriaceae. Gene 1989; 78: 339–48. [DOI] [PubMed] [Google Scholar]
- 27.Rasheed JK, Jay C, Metchock B, et al. Evolution of extended-spectrum β-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob Agents Chemother 1997; 41: 647–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arlet G, Rouveau M, Philippon A. Substitution of alanine for aspartate at position 179 in the SHV-6 extended-spectrum β-lactamase. FEMS Microbiol Lett 1997; 152: 163–7. [DOI] [PubMed] [Google Scholar]
- 29.Lahiri SD, Johnstone MR, Ross PL, et al. Avibactam and class C β-lactamases: mechanism of inhibition, conservation of the binding pocket, and implications for resistance. Antimicrob Agents Chemother 2014; 58: 5704–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keepers TR, Gomez M, Celeri C, et al. Bactericidal activity, absence of serum effect, and time–kill kinetics of ceftazidime-avibactam against β-lactamase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 2014; 58: 5297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sader HS, Castanheira M, Flamm RK, et al. Antimicrobial activity of ceftazidime-avibactam against Gram-negative organisms collected from U.S. medical centers in 2012. Antimicrob Agents Chemother 2014; 58: 1684–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Zhang F, Zhao C, et al. In vitro activities of ceftazidime-avibactam and aztreonam-avibactam against 372 Gram-negative bacilli collected in 2011 and 2012 from 11 teaching hospitals in China. Antimicrob Agents Chemother 2014; 58: 1774–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porres-Osante N, Dupont H, Torres C, et al. Avibactam activity against extended-spectrum AmpC β-lactamases. J Antimicrob Chemother 2014; 69: 1715–6. [DOI] [PubMed] [Google Scholar]
- 34.Falagas ME, Karageorgopoulos DE. Extended-spectrum β-lactamase-producing organisms. J Hosp Infect 2009; 73: 345–54. [DOI] [PubMed] [Google Scholar]
- 35.Afridi FI, Farooqi BJ. Activity of β-lactam β-lactamase inhibitor combinations against extended spectrum β-lactamase producing Enterobacteriaceae in urinary isolates. J Coll Physicians Surg Pak 2012; 22: 358–62. [PubMed] [Google Scholar]
- 36.Papp-Wallace KM, Taracila M, Hornick JM, et al. Substrate selectivity and a novel role in inhibitor discrimination by residue 237 in the KPC-2 β-lactamase. Antimicrob Agents Chemother 2010; 54: 2867–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuzin AP, Nukaga M, Nukaga Y, et al. Structure of the SHV-1 β-lactamase. Biochemistry 1999; 38: 5720–7. [DOI] [PubMed] [Google Scholar]
- 38. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-fourth Informational Supplement M100-S24. CLSI, Wayne, PA, USA, 2014. [Google Scholar]
- 39.Papp-Wallace KM, Bethel CR, Distler AM, et al. Inhibitor resistance in the KPC-2 β-lactamase, a preeminent property of this class A β-lactamase. Antimicrob Agents Chemother 2010; 54: 890–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Padayatti PS, Sheri A, Totir MA, et al. Rational design of a β-lactamase inhibitor achieved via stabilization of the trans-enamine intermediate: 1.28Å crystal structure of wt SHV-1 complex with a penam sulfone. J Am Chem Soc 2006; 128: 13235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papp-Wallace KM, Winkler ML, Gatta JA, et al. Reclaiming the efficacy of β-lactam-β-lactamase inhibitor combinations: avibactam restores the susceptibility of CMY-2-producing Escherichia coli to ceftazidime. Antimicrob Agents Chemother 2014; 58: 4290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ehmann DE, Jahic H, Ross PL, et al. Kinetics of avibactam inhibition against class A, C, and D β-lactamases. J Biol Chem 2013; 288: 27960–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bethel CR, Hujer AM, Hujer KM, et al. Role of Asp104 in the SHV β-lactamase. Antimicrob Agents Chemother 2006; 50: 4124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang W, Palzkill T. A natural polymorphism in β-lactamase is a global suppressor. Proc Natl Acad Sci USA 1997; 94: 8801–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marciano DC, Pennington JM, Wang X, et al. Genetic and structural characterization of an L201P global suppressor substitution in TEM-1 β-lactamase. J Mol Biol 2008; 384: 151–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun T, Bethel CR, Bonomo RA, et al. Inhibitor-resistant class A β-lactamases: consequences of the Ser130-to-Gly mutation seen in Apo and tazobactam structures of the SHV-1 variant. Biochemistry 2004; 43: 14111–7. [DOI] [PubMed] [Google Scholar]
- 47.Winkler ML, Rodkey EA, Taracila MA, et al. Design and exploration of novel boronic acid inhibitors reveals important interactions with a clavulanic acid-resistant sulfhydryl-variable (SHV) β-lactamase. J Med Chem 2013; 56: 1084–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Helfand MS, Hujer AM, Sonnichsen FD, et al. Unexpected advanced generation cephalosporinase activity of the M69F variant of SHV β-lactamase. J Biol Chem 2002; 277: 47719–23. [DOI] [PubMed] [Google Scholar]