Abstract
Somatic cells can be transdifferentiated to other cell types without passing through a pluripotent state by ectopic expression of appropriate transcription factors1,2. Recent reports have proposed an alternative transdifferentiation method in which fibroblasts are directly converted to various mature somatic cell types by brief expression of the induced pluripotent stem cell (iPSC) reprogramming factors Oct4, Sox2, Klf4 and c-Myc (OSKM) followed by cell expansion in media that promote lineage differentiation3–6. Here we test this method using genetic lineage tracing for expression of endogenous Nanog and Oct4 and for X chromosome reactivation, as these events mark acquisition of pluripotency. We show that the vast majority of reprogrammed cardiomyocytes or neural stem cells obtained from mouse fibroblasts by OSKM-induced transdifferentiation pass through a transient pluripotent state, and that their derivation is molecularly coupled to iPSC formation mechanisms. Our findings underscore the importance of defining trajectories during cell reprogramming by different methods.
Somatic cell transdifferentiation involves ectopic expression of lineage master regulators that induce transformation into a different somatic cell type without going through a pluripotent configuration. For example, expression of C/EBPα converts Pro-B cells into macrophage-like cells7. Recently, a new approach to somatic transdifferentiation, called OSKM-mediated transdifferentiation (OSKM-TD), has been described in which Yamanaka’s four original pluripotency reprogramming factors2 are briefly expressed for periods as short as 3-10 days to induce an intermediate, partially reprogrammed and presumably ‘plastic’ state3-6. Next, lineage-specifying media that lack conventional pluripotency-promoting cytokines, such as Leukemia Inhibitory Factor (LIF), are provided to shift these intermediate cells toward a desired somatic cell fate without their ever becoming pluripotent3-6. The conclusion that the method circumvents pluripotency was supported by the experimental protocol and results3–5. Brief OSKM induction of <10 days was deemed insufficient to yield iPSCs. Culture conditions, particularly the absence of LIF and the presence of JAK1 small-molecule inhibitors (J1i) to block Stat3 signaling, were designed to prevent acquisition of pluripotency. However, lineage-tracing tools that could unequivocally determine whether the cells attained pluripotency were not used. Thus, it remains unclear whether somatic cells produced by this technique transdifferentiate or, alternatively, go through a transient state of induced pluripotency and then differentiate to a somatic lineage according to the media conditions applied. Addressing the latter question is fundamental to understating mechanisms of cellular reprogramming, and relevant to evaluating the safety and quality of cells reprogrammed via this approach.
Our interest in this question arose from the observation that Nanog-GFP+ iPSCs appear at low efficiency during reprogramming with different Doxycycline (Dox)-inducible OSKM transgenic systems10,29–30 after as few as 3 days of Dox induction in conditions of 15% FBS, 5% KSR and LIF (Fig. 1a). Furthermore, when we induced OSKM10,29–30 with Dox in Oct4-GFP secondary reporter fibroblast cells using cardiogenic or neural stem cell (NSC) growth conditions instead of conventional LIF-containing pluripotency conditions, we obtained GFP+ embryonic stem cell (ESC)-like colonies during the OSKM induction phase, and observed ‘hybrid’ colonies with Oct4-GFP+ cells in the center of the colony, while their edges showed clear signs of neuronal differentiation lack of Oct4-GFP (Supplementary Fig. 1a-c). These results emphasized the need to exclude the possibility that iPSCs may form rapidly under suboptimal reprogramming conditions and may be a source of ‘trans-differentiated’ cells generated by OSKM-TD approaches3,4.
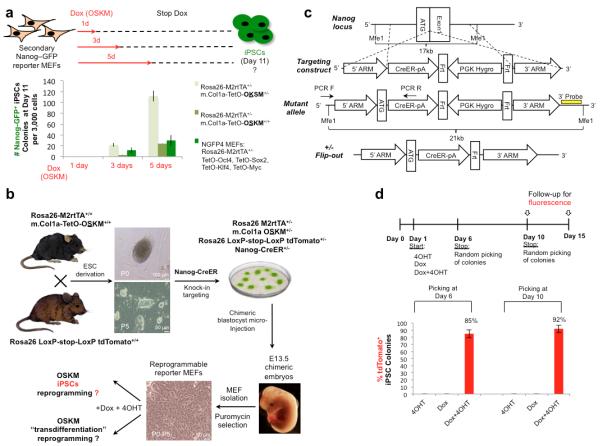
Figure 1. ineage tracing for endogenous Nanog reactivation during reprogramming.
a. MEFs from three indicated different secondary reprogramming systems, all carrying Nanog-GFP knock-in reporter for pluripotency, were subjected to Dox induced reprogramming. Dox was applied for the indicated time points, and then withdrawn. iPSCs formation was evaluated at day 11 without passaging. Error bars indicate s.e.m of biological triplicates (1 out of 2 representative experiments is shown). b. Scheme illustrating generation of quadruple knock-in-allele-reporter in reprogrammable MEFs, utilized for either OSKM-iPSCs or OSKM trans-differentiation (OSKM-TD) reprogramming. c. Nanog-CreER knock-in targeting strategy. d. Reprogrammable Nanog-CreER MEFs were subjected to iPSCs reprogramming protocol in the presence of Dox, 4OHT or both, which were withdrawn at day 6 or 10 as indicated. 48 Colonies were randomly subcloned and validated as iPSCs (Nanog, SSEA1 and AP staining) from each condition and time point, and then scored for the presence of tdTomato+ signal. Bar plot showing percentage of tdTomato+ iPSCs clones obtained, indicates the sensitivity of the system. Error bars indicate s.e.m of biological duplicates (1 out of 3 representative experiments is shown).
We next engineered a system to track transient acquisition of pluripotency during in vitro reprogramming. Previous OSKM-TD studies argued that the lack of Nanog reactivation in the bulk of reprogramming cultures3,4 proved that the cells did not become pluripotent. Thus, we designed a genetic tracing system for reactivation of endogenous Nanog, which occurs at the late stages of iPSC formation3,4,8,9. In our reprogramming system, transient activation of Nanog is monitored by permanent tdTomato fluorescence, indicating that pluripotency was achieved even if only temporarily. For this purpose we first mated the Rosa26 M2rtTA/m.Col1a OSKM+/+ reprogrammable mouse strain10 with Rosa26 LoxP-stop-LoxP tdTomato+/+ reporter mice, and derived and validated mESC lines from day E3.5 blastocysts that are heterozygous for the triple knock-in alleles: Rosa26 M2rtTA+/−, Rosa26 LoxP-Stop-LoxP tdTomato+/− and m.Col1a-TetO-OSKM+/− (Fig. 1b). Next, we generated a BAC recombineered Nanog-CreER targeting construct, which introduces a knock-in Tamoxifen-inducible CreER cassette under the control of the endogenous Nanog promoter (Fig. 1c, Supplementary Fig. 2a). The quadruple knock-in mESCs were microinjected into host blastocysts to generate chimeric animals from which transgenic fibroblasts were extracted for experimental analysis (Fig. 1b).
We validated the sensitivity and specificity of this system. Mouse ESCs reactivated tdTomato rapidly after 48 hours of 4OHT induction (95-100% of ESC colonies), and no fluorescence was detected in mESCs passaged over extended periods of time in the absence of 4OHT (Supplementary Fig. 2b). Nanog-CreER was not active in nascent neuroectoderm or mesoderm, as NCAM+ sorted cells from early embryoid bodies (EBs), did not activate the tdTomato reporter after 4OHT treatment (Supplementary Fig. 3). MEFs expanded with or without Dox in the absence of 4OHT did not show any reactivation of tdTomato, further indicating specificity of the system and excluding leakiness during expansion or reprogramming. The fidelity of the reporter was evaluated by reprogramming the transgenic MEFs to pluripotency, by adding Dox and 4OHT to the medium and withdrawing both molecules at days 6 and 10. Adding 4OHT alone did not produce any colonies whereas adding Dox alone produced mES-like colonies, all of which were tdTomato negative (Fig. 1d, Supplementary Fig. 2c). mES-like colonies formed in each of the conditions at days 6 and 10 (48 colonies for each group) were randomly subcloned and grown for another 5-6 days, and the presence of tdTomato+ clones was assessed as a percentage of the total number of developed and validated Dox-independent iPSC clones. We found the fidelity of our system to be ~85% and ~92% for iPSCs obtained after 6 and 10 days of Dox+4OHT induction, respectively (Fig. 1d). Lack of 100% sensitivity might be explained by limitations of the Nanog knock-in allele activation or a reflection of a true biological outcome, given that Nanog activity can be dispensable during iPSC generation9. Nevertheless, these results validate the ability of the system to specifically document acquisition of pluripotency during iPSC reprogramming with high specificity.
Next, we applied the Nanog tracing system and reporter-engineered MEFs to previously reported OSKM-TD experiments on converting mouse fibroblasts into neural stem cells (NSCs)3 and cardiomyocytes (CMs)4. The published protocols were meticulously followed (Fig. 2a and Methods Section), including the use of conditions without LIF or feeder cells.
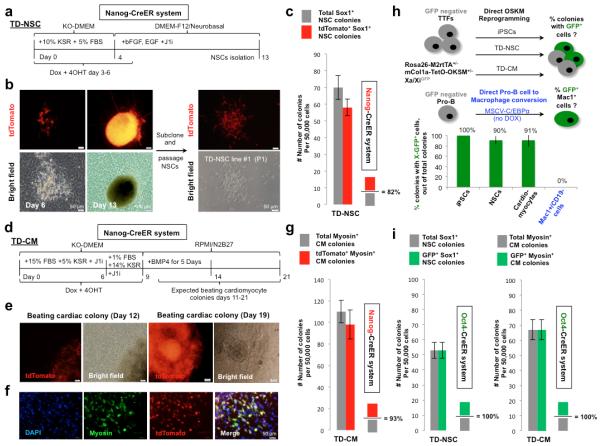
Figure 2. Frequent and transient acquisition of pluripotency during OSKM-TD.
a. Schematic illustration of NSCs trans-differentiation with indicated time points for medium switch, small molecules and growth factors additions. b. Left - Representative images of NSC tdTomato+ colonies on the primary plate generated by the above mentioned protocol (a), at the start and end points of neural reprogramming. Right -Passage of subcloned NSC colony grown in NSC medium, showing typical NSCs morphology and tdTomato+ fluorescence. c. Bar plot of immunostained Sox1+ TD-NSC colonies, showing the number of total and of tdTomato+ colonies obtained. Error bars indicate s.e.m of biological triplicate wells (1 out of 2 representative experiments is shown). d. Schematic illustration of TD-CM with indicated time points for medium switch, small molecules and growth factors additions. e. Representative images of cardiomyocytes colonies reprogrammed by the above-mentioned protocol (d) with tdTomato+ fluorescence, and repeated rhythmic beating motion after 12 or 19 days of reprogramming. f. Representative immunostaining for Myosin in TD-CM with overlay of tdTomato fluorescence. g. Bar plot quantifying validated immunostained Myosin+ TD-CM colonies, showing total number of tdTomato+ colonies obtained. Error bars indicate s.e.m of biological triplicate wells (1 out of 2 representative experiments is shown). h. Tail tip fibroblasts (TTFs) were established from the indicated adult transgenic reprogrammable female mice. GFP negative populations were sorted, purified and subjected to OSKM iPSCs or TD protocols. Pro-B cells were purified form the same mice, and subjected to C/EBPα mediated conversion to macrophage like cells (without OSKM induction). Percentage of GFP+ cells or GFP+ cell containing colonies, detected at day 12-13 (for OSKM reprogramming) and day 8 (for Pro-B to macrophage conversion), is indicated. Error bars indicate s.e.m of biological duplicates (1 out of 3 representative experiments is shown). i. Bar plot of immunostained Sox1+ TD-NSC colonies and Myosin+ TD-CM colonies derived from Oct4-CreER mT/mG reporter MEFs, showing the number of total colonies and of GFP+ colonies obtained. Error bars indicate s.e.m of biological triplicate wells (1 out of 2 representative experiments is shown).
The TD-NSC protocol3 generated Sox1+ NSC colonies that could be subcloned and expanded as stable NSC lines (Fig. 2b, Supplementary Fig. 4) as previously shown. RNA-Seq analysis confirmed the transcriptional similarity of TD-NSCs to NSCs derived from primary embryos, and their difference from mouse fibroblasts and ESCs/iPSCs (Supplementary Fig. 5a). Notably, ~82% of Sox1+ TD-NSC colonies were tdTomato+, indicating acquisition of pluripotency during their conversion from MEFs (Fig. 2c, Supplementary Fig. 5b, 6, 7a,b).
Similarly, the TD-CM protocol (Fig. 2d and Methods) yielded TD-CMs that expressed early and late cardiomyocytes markers (Myosin, cardiac Troponin T), many of which showed beating rhythmicity (Fig. 2e-f, Supplementary Fig. 5b-c, Supplementary Videos 1-2). Approximately 93% of Myosin+ TD-CM colonies, and within each colony the majority of cells, were tdTomato+, indicating transient acquisition of pluripotency (Fig. 2e-g, Supplementary Fig. 5b-c, 7b-c).
It is well established that an additional late and specific hallmark of acquisition of naïve pluripotency during reprogramming to iPSCs is reactivation of silenced X chromosome. Subsequent differentiation of iPSCs involves random X inactivation11. To determine whether X chromosome reactivation occurs in OSKM-TD, we crossed female Rosa26 M2rtTA+/+/m.Col1a-TetO-OSKM+/+ mice10 with male mice carrying an X-linked GFP reporter transgene12, and purified GFP− tail tip fibroblasts from female offspring; all GFP− reprogrammable cells carry the GFP transgene on the inactive X chromosome (Fig. 2h). Notably, GFP fluorescence was identified in >90% of TD-NSC and TD-CM colonies (Fig. 2h), and in 100% of iPSC colonies obtained. Reprogramming of female-derived XGFP− Pro-B lymphocytes into macrophage-like cells by C/EBPα overexpression13 did not yield any GFP+/Mac1+ colonies (Fig. 2h), thus excluding promiscuous, non-specific X chromosome reactivation in the transdifferentiation protocols applied.
Finally, we used MEFs derived from mice carrying a Oct4-CreER knock-in reporter and a Rosa26 mTomato/mGFP (mT/mG) reporter cassette; in these cells mTomato expression is switched to EGFP expression upon Oct4 locus reactivation in the presence of 4OHT (Supplementary Fig. 8a)14. Repeating OSKM-TD protocols with either polycistronic lentiviral vectors or with individual moloney viruses encoding OSKM (Fig. 2i) showed that 100% of Sox1+ TD-NSC and Myosin+ TD-CM colonies lost mTomato expression and became GFP+, marking endogenous Oct4 reactivation during their reprogramming process (Fig. 2i, Supplementary Fig. 8b-d).
The higher efficiency of Oct4 reactivation (100%) compared with Nanog and X reactivation that we detected may be explained by the fact that Oct4 is expressed in all states of pluripotency (primed, metastable and naïve), whereas Nanog and X reactivation are more stringent markers observed only in naïve pluripotency8,15. Alternatively, endogenous Oct4 reactivation in isolation may not unequivocally indicate the reacquisition of pluripotency16. Nevertheless, the Oct4, Nanog and X chromosome lineage tracing results collectively corroborate the conclusion that the vast majority of mouse OSKM-TD-derived cells pass through a pluripotent state before transitioning towards overt somatic differentiation. Acquisition of pluripotency in these secondary Nanog-CreER cells did not was not the result of higher levels of OSKM compared with those in ESCs or during reprogramming to iPSCs, and was reproduced with multiple different OSKM delivery methods and reporters (Supplementary Fig. 9a,b).
The above results indicate that the majority of OSKM-TD cells pass through a transient Nanog+ pluripotent state before differentiating into various somatic linages according to the culture conditions applied. To validate and evaluate features of this transient pluripotent state, we sorted Nanog-CreER reprogramed cells at the early end point of OSKM induction of the TD protocol (day 6 for TD-NSC and day 9 for TD-CM protocol) (Fig. 3a, Supplementary Fig. 10a-b). Sorted tdTomato+ cells were immediately subjected to various functional and molecular tests of pluripotency.
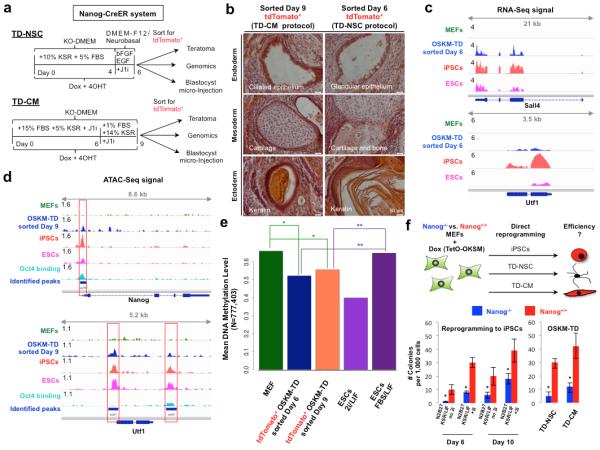
Figure 3. Molecular and functional evidence for transient acquisition of pluripotency during OSKM-TD.
a. Schematic representation of OSKM-TD “intermediate” Nanog-CreER reporter cells sorted for tdTomato+ at the end of OSKM induction step and utilized for teratoma assay, blastocyst microinjection or high-throughput genomics. OSKM-TD protocols were applied on Nanog-CreER reporter MEFs, and stopped at the end of OSKM induction step, at day 6 and 9, respectively. b. Representative images of Hematoxylin & Eosin stain of teratoma sections with representation of all three germ layers (endoderm, mesoderm and ectoderm). c. Transcriptional profile of Sall4 and Utf1 measured by poly-A RNA sequencing, in four samples: MEFs (green), sorted tdTomato+ OSKM-TD day6 in NSC conditions (blue), iPSCs (red) and ESCs (magenta). Data range is indicated at the left side of each panel. d. Chromatin accessibility signal measured by ATAC-Seq of Nanog and Utf1 in MEF (green), sorted tdTomato+ OSKM-TD day 9 in CM conditions (blue), iPSCs (red) and ESCs (magenta), in addition to Oct4 binding measured by ChIP-Seq in iPSCs (light blue). All peaks identified by MACS are indicated by blue lines. Peaks which fall in promoters or in potential enhancers are indicated by red squares. e. Mean DNA methylation level measured in MEFs, OSKM-TD day 6 and day 9 tdTomato+ sorted cells, and ES cells (in ground state naïve 2i/LIF conditions, or metastable naïve FBS/LIF conditions). Mean methylation was measured using whole-genome bisulfite sequencing (WGBS, see Methods) and calculated across all CpGs that were covered in all samples. *indicates p-value<10−15, **indicates p-value<2.2×10−16, both in Wilcoxon signed-rank test. f. Reprogramming efficiencies of Nanog−/− and Nanog+/+ MEFs to either iPSCs, TD-NSC or TD-CM, evaluated by AP, Sox1 or Myosin colony staining, respectively. Reprogramming was conducted following transduction with FUW-M2rtTA and FUWTetO-OKSM lentiviral vectors. Error bars indicate s.e.m of biological triplicates (1 out of 2 representative experiments is shown). *Indicates Student t-test p-value <0.05 (in comparison to matched Nanog+/+ sample).
Both day 6 and day 9 sorted tdTomato+ cells obtained early during TD-NSC and TD-CM protocols, respectively, generated teratomas in vivo, with mature differentiation towards all three germ layers (Fig. 3b, Supplementary Fig. 10c). tdTomato+ sorted cells at both time points, but not pluripotent primed EpiSCs or somatic MEFs, contributed to mouse chimeric embryos after microinjection into host E3.5 blastocysts (Supplementary Fig. 10d,e).
RNA-Seq analysis of sorted day 6 tdTomato+ TD-NSC derived cells validated reactivation of bona fide pluripotency genes, including Nanog, Sall4, Nr5A2, Cdh1, Utf1 and Epcam (Fig 3c, Supplementary Fig. 11). qPCR and western blot analysis for endogenous reactivation of several pluripotency factors (Supplementary Fig. 12a), further validated these results. The transcriptional profiles of these cells consistently clustered with those of iPSCs and ESCs and apart from those of established primary NSCs, TD-NSCs and MEFs (Supplementary Fig. 12b,c). As expected, they were not fully identical to those of established ESCs and iPSCs as these early isolated tdTomato+ cells were reprogrammed in sub-optimal conditions and were not allowed to consolidate their pluripotency program under optimal naïve pluripotency conditions and long-term passaging.
Global chromatin accessibility measured by assay for transposase-accessible chromatin followed by sequencing (ATAC-Seq)17 showed reactivation of enhancer and promoter regions of key bona fide pluripotency genes in day 9 tdTomato+ sorted cells from the TD-CM protocol, similar to what is observed in ESC and iPSC lines (Fig. 3d, Supplementary Fig. 13-14). Although the OSKM-TD protocol does not use 2i/LIF conditions, genome-wide DNA methylation assessment via Whole Genome Bisulfite Sequencing (WGBS) showed significant global reduction in DNA methylation levels in tdTomato+ sorted cells toward what is typically measured in ground-state naïve 2i/LIF murine pluripotent cells (Fig. 3e). The latter result documents transient global DNA demethylation during reprogramming by OSKM without applying 2i, and may be worth further investigation.
We next set out to explore the functional role of endogenous Nanog reactivation in OSKM-TD. Endogenous Nanog reactivation has been shown to synergistically boost iPSC reprogramming by OSKM18. We used Nanog−/− knock out and Nanog+/+ mESCs18. Nanog−/− mESCs can be expanded in N2B27 2i/LIF/15%KSR conditions while maintaining their pluripotency19,20 (Supplementary Fig. 15a). Reprogramming of Nanog−/− and Nanog+/+ MEFs to pluripotency was done following OSKM transduction, and yielded validated pluripotent Nanog−/− iPSCs (Supplementary Fig. 15b-d). Reprogramming efficiency to pluripotency of Nanog−/− MEFs as compared to Nanog wild type MEFs, showed a 90% and 55% reduction in iPSC reprogramming efficiency when evaluated at days 6 and 10 after OSKM induction, respectively (Fig. 3f, Supplementary Fig. 15e, 16). Next, Nanog−/− and Nanog+/+ MEFs were used in OSKM-TD protocols for TD-NSC and TD-CM. Transient reactivation of pluripotency markers like Utf1 or SSEA1 was evident in Nanog−/− MEFs in iPSC and OSKM-TD protocols, by the end of OSKM induction steps (Supplementary Fig. 17-18). Upon completion of the reprogramming process, validated TD-NSCs and TD-CMs were obtained from Nanog−/− cells (Supplementary Fig. 19). This revealed a reduction of OSKM-TD efficiency by 80-85% in the Nanog−/− MEFs as compared to Nanog+/+ cells (Fig. 3f and Supplementary Fig. 15e and 16a). Collectively, these results show that endogenous Nanog reactivation is functionally coupled to OSKM-TD efficiency, and at an equivalent robustness to that observed during OSKM-iPSC formation.
Finally, OSKM-TD protocols make use of media devoid of LIF or with added JAK1-Inhibitor (J1i), which blocks Stat3 pathway signaling, under the assumption that these conditions completely prevent the formation of pluripotent iPSCs3,4. Given that OSKM transgenes have been found sufficient to support induction and expansion of pluripotent cells from non-permissive mouse strains like NOD in LIF-only conditions21, we wondered whether they could completely substitute for exogenous LIF/Stat3 signaling. To address this issue, we reprogrammed Nanog-CreER reporter MEFs to pluripotency with added J1i and LIF-free N2B27 medium (Supplementary Fig. 20a-b). Transgene-dependent iPSCs were derived, expanded and shown to be pluripotent by positive staining for alkaline phosphatase, SSEA1 and Nanog and by the potential to form mature teratomas (Supplementary Fig. 20c-d). These results corroborate previous findings that Stat3 can be dispensable for re-establishing pluripotency22, and that exclusion of LIF/Stat3 does not constitute an unequivocal proof for lack of acquisition of pluripotency when providing exogenous pluripotency factor encoding transgenes, as the latter can substitute for the requirement for exogenous LIF addition to induce and maintain pluripotency21.
Conversion of cell fate in vitro and in vivo holds great potential for regenerative medicine. There may be different routes of reprogramming to achieve a given cell type23, and it is important to understand their technical and functional advantages and disadvantages and the molecular pathways and trajectories of each method24,25. Although our study of OSKM-mediated ‘transdifferentiation’ shows that the vast majority of the cells are reprogrammed to pluripotency and then rapidly differentiate according to the media used, we cannot exclude that some cells do transdifferentiate without passing through a pluripotent state. From the perspective of potential applications of OSKM-TD, both the advantages and disadvantages associated with iPSC generation are likely to be relevant. It will also be important to conduct similar studies on OSKM-TD of human cells5,6,26. Finally, considering the continued discovery of factors, inhibitors and cytokines that can induce and maintain pluripotency16,27,28, our findings underscore the importance of using unbiased molecular lineage tracing to track cell trajectories in different reprogramming protocols.
ONLINE METHODS
Generation of Reprogrammable Nanog-CreER Reporter Mice
Homozygote Rosa26 LoxP-stop-LoxP tdTomato+/+ mice were cross bred with the reprogrammable double homozygote Rosa26-M2rtTA and m.Col1a OSKM+/+ mice10. At day E3.5 blastocysts were extracted and individually seeded in a flat bottom 96 well with plated gamma-irradiated feeder cells, in mESCs growth medium: DMEM (Gibco 41965-039) supplemented with human LIF (20ng/ml prepared in house), 1% L-Glutamine, 1% Non-essential amino acids, 1% penicillin/streptomycin, 0.1 mM β-mercaptoethanol and 15% FBS (Biological Industries). Developed ESCs colonies were expanded as single clonal lines. In order to make a targeting construct for inserting CreERT2 gene in frame with mouse Nanog we used Red/ET based recombineering technique (GeneBridges). Briefly, CreERT-pgk-gb2-Kanamycin cassette was inserted into a BAC covering murine Nanog gene region in a way that starting codon of Nanog now became starting codon of CreERT2. Next, BAC fragment containing CreERT insert and appropriate homology arms were cloned into pDTA plasmid encoding Diphtheria Toxin gene cassette used for negative selection in mESCs. The Kanamycin selection cassette was replaced with PGK-Hygromycin cassette by ligation. The resulting recombineered Nanog-CreER pgk-Hygro construct (Deposited by our group on Addgene: #59721) was linearized with Sal1 and electroporated into the triple allele knock-in ESCs. Hygromycin resistant clones (10 days selection) were subcloned and screened by southern blot using a 3′ external probe (Deposited by our group on Addgene: #60037) and a PCR creating a 2kb amplification of the 5′ arm, with upstream external forward primer and reverse primer from the Nanog CreER knock-in. Correctly targeted clones were transfected with Flippase encoding construct to remove the inverted Hygromycin resistance cassette, while no difference in tracing results were observed following conducting this optional step. Properly targeted ESCs were microinjected into blastocysts to create chimeric animals. At day E12.5-E13.5, embryos were extracted and used to derive MEFs containing the reporter system with all four knock-in alleles. V6.5 ESCs were also similarly targeted with Nanog-CreER reporter construct and used for some of the analysis. Karyotype analysis was conducted to validated chromosomal stability of all lines used, and genotyping to validate their alleles. Mycoplasma testing is conducted routinely bi-weekly in our lab (MycoAlert Kit, Lonza), and all reported results were conducted on non-contaminated cells.
Southern blot analysis
Genomic DNA was extracted from each Hygromycin resistant targeted subclone. 10-15ϻg of genomic DNA was digested with Mfe1 restriction enzyme for 5 hours and separated by gel electrophoresis. The DNA was transferred to a nitrocellulose membrane that was next hybridized with a radioactive labeled probe and developed using ECL (Thermo Scientific).
iPSC reprogramming by OSKM
For mouse iPSC reprogramming experiments, secondary OSKM transgenic reporter MEFs were plated at day 0 in MEF medium: DMEM supplemented with 1% L-Glutamine, 1% Non-essential amino acids, 1% penicillin/streptomycin and 10% FBS, and the following day reprogramming was initiated by changing medium to N2B27 2i/LIF: DMEM-F12 (BI 01-170-1a) and Neurobasal medium (Gibco 21103-049) mixed by 1:1 ratio and supplemented with N2 (Insulin sigma I-1882, Apo-Transferrin sigma T-1147, Progesterone sigma P8783, Putrescine sigma P5780, sodium selenite sigma S5261), B27 (Gibco 17504-044) and 2i: CHIR (Axon 99021, 3ϻM) PD0325901 (Tocris 4192, 1ϻM), LIF and adding Doxycycline hyclate (sigma D9891, 4ϻg/mL), with 4-Hydroxytamoxifen (sigma H7904, 1ϻM) for activation of the CreER reporter system. Media was replaced every 48 hours, and colonies were analyzed at the indicated time points. Additional FUW-M2RtTA and/or TetO-STEMCCA lentiviral induction were optionally used to enhance reprogramming efficiency. Nanog+/+ and Nanog−/− ESCs18 were rendered transgenic for a constitutively expressed Puromycin resistance cassette29, and were subsequently microinjected into host blastocysts. Chimeric embryos were used to derive MEFs, which were selected by Puromycin (2ϻg/mL) resistance for at least 3 days then used for transduction and reprogramming in 2i/LIF/15% KSR (Invitrogen) enriched conditions. In Fig. 1a and Supplementary Fig. 1, MEFs were obtained from Rosa26-m2rtTA(+/+ or +/−); m.Col1a-OSKM(+/+) mice (Jackson Laboratories #011004), Rosa26-m2rtTA(+/−); m.Col1a STEMCCA-OKSM(+/−) mice (Jackson Laboratories #011001) that were bred with Oct4-GFP or Nanog-GFP knock-in reporter mice30. NGFP4 iPSC line was generated as previously described30.
Trans-differentiation Reprogramming with OSKM
Reprogramming of MEFs to cardiomyocytes was done in media containing: knock-out DMEM (Invitrogen) with supplemented 15% FBS and 5% KSR with added JAK1 inhibitor (Calbiochem 420099, 0.5ϻM) for 6 days, followed by switching to 1% FBS and 14% KSR and JAK1 inhibitor for 3 days. The medium was supplemented with 1% L-Glutamine, 1% Non-essential amino acids, 1% penicillin/streptomycin and 0.1 mM β-mercaptoethanol. Doxycycline hyclate (Dox) and 4-Hydroxytamoxifen (4OHT) were added to the medium at day 1 and continuously kept in the medium until day 6-9. From day 9 onwards, cells were cultured in chemically defined RPMI (Gibco 21875-034)/N2B27 medium (0.5XN2, 1XB27) with the cardio-inductive growth factor BMP4 (Peprotech 120-05ET) for the first 5 days, and supplemented with BSA fraction V 0.05% (Gibco 15260-037) 1% L-Glutamine, 1% Non-essential amino acids, 1% penicillin/streptomycin and 0.1 mM β-mercaptoethanol. Colonies were allowed to develop for 21 days or until beating cardiomyocytes (CMs) were noticed among the plated cells.
Reprogramming of MEFs to NSCs was carried out in knock-out DMEM medium (Gibco 10829-018) with 10% KSR, 5% FBS, 1% L-Glutamine, 1% Non-essential amino acids, 1% penicillin/streptomycin, 0.05 mM β-mercaptoethanol and with Dox+4OHT for the first 3-6 days. Thereafter, Dox and 4OHT were withdrawn from the medium, and cells were grown in neural reprogramming medium: DMEM-F12 and Neurobasal medium mixed by 1:1 ratio and supplemented with BSA fraction V 0.05%, 1XN2, 1XB27, 1% L-Glutamine, 1% Non-essential amino acids, 1% penicillin/streptomycin and 0.1 mM β-mercaptoethanol with bFGF (Peprotech 450-33, 20ng/mL) and EGF (Peprotech 315-09,20ng/mL), until NSCs colonies were formed by day 13. From day 4 onwards JAK1 inhibitor was added to the culture medium. At day 13, formed colonies were subcloned and grown in NSCs medium on Poly-D-Lysine and Laminin coated plates. We noted that OSKM activation by Dox for 3 days was sufficient for OSKM-TD, though substantially fewer colonies developed at these minimally required activation periods, as reported in the original papers3,4. When applying OSKM-TD protocols without 4OHT or with ethanol (ETOH) as a control replacement, all CMs and NSCs colonies were negative for tdTomato signal, excluding leakiness of this system under OSKM-TD conditions as similarly observed with iPSCs (Fig. 1d).
Genetic tracing for X chromosome reactivation in reporter murine female cells
Female Rosa26 M2rtTA+/+/m.Col1a-TetO-OSKM+/+ mice10 were bred with male mice carrying a CMV-GFP knock-in to the X chromosome. Adult Female F1 mice were used as a source for adult tail tip fibroblast or Pro-B somatic cells. Additional FUW-M2RtTA and TetO-STEMCCA lentiviral induction were used to enhance reprogramming efficiency. GFP negative population were sorted on a FACSAria III five-laser equipped cell sorter (BD Biosciences) and further expanded for OSKM-iPSCs and OSKM-TD reprogramming experiments. Colonies were scored for GFP signal on Zeiss Axioscope D1 microscope, and as expected, iPSCs were homogenously GFP positive (because they maintain both X chromosomes active), while OSKM-TD colonies had patchy GFP expression consistent with random inactivation of the X alleles. Yet, the presence of GFP positive cells in TD colonies indicates reactivation of the X chromosome in the clonal population, followed by random inactivation during differentiation.
Genetic tracing with Oct4 CreER mT/mG reporter MEFs
The Oct4-CreER reporter system has been previously reported and validated elsewhere14. Briefly, Tamoxifen inducible Cre recombinase gene was inserted into the 3′-untranslated region (UTR) of Oct4. The knock-in allele was maintained on a C57BL/6 mice strain background. Oct4-MerCreMer mice are deposited with The Jackson Laboratory (Stock No. 016829). For lineage tracing, MEFs were derived from E12.5 embryos obtained following of Oct4-MerCreMer mice mating with homozygous double-fluorescent Rosa26 LoxP-mTomato-stop-LoxP-mEGFP reporter mice (The Jackson Laboratory, Stock No. 007576). The latter system allows marking tamoxifen dependent Cre recombination by replacement of membrane targeted tdTomato with membrane targeted enhanced green fluorescent protein (EGFP) expression.
Mouse embryo micromanipulation
Pluripotent ESCs, day-9 iPSCs, EpiSCs or Nanog-CreER (day 6 or day 9) tdTomato+ cells were injected into BDF2 diploid blastocysts (10 cells per embryo, unless specified otherwise), harvested from hormone primed BDF1 6-week-old females. MEF cells constitutively labeled with tdTomato were used as negative control (3 MEF cells were injected per blastocyst). All cells were subjected to the same sorting conditions, and were immediately used for microinjections. Microinjection into BDF2 E3.5 blastocysts placed in M2 medium under mineral oil was done by a flat-tip microinjection pipette. After microinjection, blastocysts were returned to KSOM media (Zenith) and placed at 37°C until transferred to recipient females. Ten to fifteen injected blastocysts were transferred to each uterine horn of 2.5 days post coitum pseudo-pregnant females. 129 EpiSCs, constitutively labeled with tdTomato exogenous transgene, were treated with ROCK inhibitor 48h before harvesting with Accutase (Sigma Aldrich), to increased survival yield. The latter experiments were approved by Weizmann Institute IACUC (00330111-2). Animals were not randomized or blinded throughout this study. No animals were excluded from any analysis throughout this study. Accurate mouse embryos/animal group size is indicated in for Supplementary Fig. 10c,e.
Viral production
For primary cell reprogramming, ~3×106 293T cells in a 10cm culture dish were transfected with a solution made of 770μL DMEM (Invitrogen) together with 50μl of TransIT®-LT1, pPAX (3.5 μg), pMDG (1.5 μg) and 5μg of the lentiviral target plasmid (FUW-M2rtTA, FUW-TetO-STEMCCA-OKSM, FUW-TetO-OSKM). Viral supernatant was harvested 48 and 72 hours post transfection, filtered through 0.45micron sterile filters (Nalgene) and added freshly to the reprogrammed cells. MSCV-C/EBPα retrovirus stocks were prepared by transient transfection of Phoenix-Eco cells using Fugene (Roche), and supernatants were harvested 48 hours later and filtered. Pro-B cell expansion on OP9 cells and transduction with C/EBPα was conducted as previously described30.
Immunostaining
Nanog-CreER iPSCs in LIF-free N2B27 medium with supplemented JAK1-Inhibitor and Dox, and Nanog−/− iPSCs were cultured on glass cover slips (13 mm 1.5H; Marienfeld, 0117530), washed three times with PBS and fixed with 4% paraformaldehyde for 10 minutes at room temperature. Cells were then permeabilized and blocked in 0.1% Triton, 0.1% Tween, and 5% FBS in PBS for 15 min at room temperature. Primary antibodies were incubated for two hours at room temperature and then washed with 0.1% Tween and 1% FBS in PBS three times. Next, cells were incubated with secondary antibody for one hour at room temperature, washed and counterstained with DAPI, mounted with Shandon Immu-Mount (Thermo Scientific) and imaged. All secondary antibodies were diluted 1:200.
For staining of trans-differentiated NSCs, CMs and intermediate cells, all cells were reprogrammed in 6 well tissue culture plates, fixed and stained in the tissue culture wells. The following antibodies were used: polyclonal goat Gata4 antibody (Santa Cruz SC-1237, 1:200), mouse monoclonal Myosin antibody (Hybridoma MF-20, 1:200), goat polyclonal Sox1 antibody (R&D AF-3369, 1:200), mouse monoclonal Tuj1 antibody (Covance MMS-435P, 1:500), mouse monoclonal Nestin antibody (Hybridoma Rat-401, 1:20), rabbit polyclonal Sox2 antibody (Millipore AB5603, 1:200), Chicken polyclonal MAP2 antibody (abcam ab5392, 1:5,000), rabbit polyclonal O2 antibody (Millipore AB9610, 1:1,000), mouse monoclonal IgM O4 antibody (R&D MAB1326, 1:500), mouse monoclonal Oct4 antibody clone C10 (Santa Cruz SC5279, 1:200), rabbit polyclonal Nanog antibody (Bethyl A300-397A, 1:200), mouse monoclonal IgM SSEA1antibody (Hybridoma MC-480 clone, 1:20) and rabbit polyclonal Utf1 antibody (abcam ab24273 1:1,000). All antibodies in this study have been validated in the literature and by our internal tests on primary cell lines.
Microscopy and image analysis
Images were acquired with A1 Axioscope microscope (Carl Zeiss) equipped with DP73 camera (Olympus) or with Z1 Axioscope microscope (Carl Zeiss), using a 20X Plan-Apochromat objective (numerical aperture 0.4). All images were acquired in sequential mode. Images were processed with Zeiss Zenblue 2011 software (Carl Zeiss) and Adobe Photoshop CS4. In house algorithm was developed and implemented in Matlab to process the images and analyze the percentage of tdTomato+/ green-fluorescence+ overlay area out of total green-fluorescence+ area (Supplementary Fig. 5c and 7). The algorithm consists of the following steps: 1) Noise reduction using sliding median filter. 2) Creating a binary mask image per channel using a detection threshold defined as the overall image median value plus bias (defined as 10% of the dynamic range). All pixels with values above the threshold are “detected” 3) Overlaying tdTomato+ and green-fluorescence+ mask images and calculating the percentage of tdTomato+ & green-fluorescence+ pixels out of all green-fluorescence+ pixels.
Western blot analysis
Whole-cell protein extracts were isolated from wild type Nanog+/+, Nanog−/− mESCs, Nanog-CreER reporter MEFs and sorted day 6 tdTomato+ intermediate cells. Blots were incubated with the following antibodies in 5% BSA/TBST: rabbit polyclonal Hsp90β antibody (Calbiochem CA1016, 1:5,000), rabbit monoclonal Gapdh antibody (Epitomics 2251-1, 1:5000), rabbit polyclonal Oct4 antibody clone H-134 (Santa Cruz SC9081, 1:1,000), rabbit polyclonal Sall4 antibody (abcam ab29112 1:1000) and rabbit polyclonal Nanog antibody (Bethyl A300-397A, 1:1,000). Secondary antibodies were horseradish peroxidase-linked goat anti-rabbit (1:10,000; Jackson). Blots were developed using ECL (Thermo Scientific).
Teratoma assay
In Supplementary Figure 10c, sorted Nanog-CreER tdTomato+ cells at day 6 and 9 of OSKM-TD protocols were injected subcutaneously to the flanks of immune deficient NSG mice. Sorted ESCs were used as a positive control. For each time point, 2×105 sorted tdTomato+ cells were collected in 25x diluted Matrigel (BD Bioscience), and subsequently injected subcutaneously in NSG mice. Three animals were injected (on one side) per each group. After 4 weeks, all injected mice were sacrificed and the tumor mass extracted, measured and fixed in 4% paraformaldehyde over-night. In Supplementary Fig. 20, Nanog-CreER iPSCs expanded in LIF-free N2B27 medium with supplemented JAK1-Inhibitor and doxycycline, and Nanog−/− iPSCs grown in N2B27 2i/LIF cell lines were expended for over 8 passages and injected (2×106 cells) subcutaneously to the flanks of immune deficient NSG mice and dissected 4-6 weeks afterwards. Slides were prepared from the paraffin embedded fixed tissue, which were next Hematoxylin & Eosin stained and inspected for representation of all three germ layers. Differentiation results were confirmed by a board-certified histopathologist.
Alkaline Phosphatase staining
Cells were grown on 6 well tissue culture plates and were washed three times with PBS and fixed with 4% paraformaldehyde for 2 minutes at room temperature. Alkaline phosphatase staining was performed according to the manufacturer protocol (Millipore SCR004).
RNA sequencing and gene expression profiling analysis
RNA was extracted from Trizol pellets, and utilized for RNA-Seq by TruSeq RNA Sample Preparation Kit v2 (Illumina) according to manufacturer’s instruction. DNA sequencing was conducted on Illumina Hiseq1500. Tophat software version 2.0.10 was used to align reads to mouse mm10 reference genome (UCSC, December 2011). Read counts per exon were calculated over all 628,052 exons in mm10 ensemble GTF (UCSC, December 2011), using bedtools coverage command (version 2.16.2). Exons annotated as protein coding, pseudogene or lincRNA (n=459,556) were selected for further analysis. Exon counts were normalized by the exon length in Kbp and by million number of aligned reads per sample, to give RPKM (Read-Per-Kilobase-per-Million-reads) values. Gene expression was defined by the average expression level (RPKM) of all exons associated with a certain gene. Sample hierarchical clustering was done over primary brain derived NSCs, MEFs, mESCs and two subclones of TD-NSCs (Supplementary Fig. 5a), only exons with at least one RPKM call > 2 were selected, resulting in 371,807 exons corresponding to 19,755 genes. Additional hierarchical clustering was done over all samples (Supplementary Fig. 12c), only exons with at least one RPKM call > 2 were selected, resulting in 379,435 exons corresponding to 20,419 genes. Hierarchical clustering was carried out in both figures, using Matlab (version R2011b) clustergram command, with Spearman correlation as a distance metric, average linkage, and per-row standardization, such that the mean of each row is 0 and its std is 1. Gene profiles (Fig. 3c Supplementary Fig. 11 and 12b) were generated using IGV version 2.3.26.
Whole-genome bisulphite sequencing for DNA methylation
Nanog-CreER reporter MEFs were reprogrammed in TD-NSC or TD-CM protocols, and sorted for tdTomato+ the end of OSKM induction step, at day 6 and 9 respectively. Similarly, cells were collected form MEF cultures, as well as V6.5 mouse ESCs grown in FBS/LIF (taken from GSE41923) or in naïve ground state KSR/2i/LIF conditions. DNA was isolated from snap-frozen intermediate sorted cells using the Quick-gDNA mini prep kit (Zymo). DNA was then bisulfite converted using the EZ DNA Methylation-Gold kit (Zymo). Sequencing libraries were created using the EpiGnome Methyl-Seq (Epicentre) and sequenced on an Illumina HiSeq 1500 system. The sequencing reads were aligned to the Mouse Genome Build 37 (mm9) using a proprietary script based on Bowtie 2. Due to the low sequencing depth of some of the samples, the methylation calls were down-sampled so that each CpG was covered by a single call (being either ‘methylated’ or ‘non-methylated’). Mean methylation was then calculated over all CpGs that had calls in all samples (either methylated “1” or non-methylated “0”). WGBS data is deposited under GEO submission: GSE67299.
Assay for Transposase-Accessible Chromatin using sequencing (ATAC-Seq)
Nanog-CreER reporter MEFs were reprogrammed using OSKM-TD protocol until the end of OSKM induction step, at which point tdTomato+ intermediate cells were sorted and applied for ATAC sequencing. Briefly, 50,000 sorted cells were centrifuged at 500g for 3 minutes, followed by a wash using 50 μL of cold PBS and centrifugation at 500g for 3 min. Cells were lysed using cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2 and 0.1% IGEPAL CA-630). Immediately after lysis, nuclei were spun at 500g for 10 min using a refrigerated centrifuge. Next, the pellet was re-suspended in the transposase reaction mix (25 μL 2× TD buffer, 2.5 μL transposase (Illumina) and 22.5 μL nuclease-free water). The transposition reaction was carried out for 30 minutes at 37 °C and immediately put on ice. Directly afterwards, the sample was purified using a Qiagen MinElute kit. Following purification, the library fragments were amplified using custom Nextera PCR primers 1 and 2 for a total of 12 cycles. Following PCR amplification the libraries were purified using a Qiagen MinElute Kit.
ATAC-Seq analysis
Reads were aligned to mm10 mouse genome using Bowtie2 with the parameter -X2000 (allowing fragments up to 2 kb to align). Duplicated aligned reads were removed using Picard MarkDuplicates tool with the command REMOVE_DUPLICATES=true. To identify chromatin accessibility signal we considered only short reads (≤ 100bp) that correspond to nucleosome free region17. To detect and separate accessible loci in each sample, we used MACS version 1.4.2-1 with --call-subpeaks flag (PeakSplitter version 1.0). Next, summits in previously annotated spurious regions were filtered out using a custom blacklist targeted at mitochondrial homologues. To develop this blacklist, we generated 10,000,000 synthetic 34mer reads derived from the mitochondrial genome. After mapping and peak calling of these synthetic reads we found 28 high-signal peaks for the mm10 genome. For all subsequent analysis we discarded peaks falling within these regions. Each peak in each sample was represented by a 300bp region around the summit center. The peaks from all samples were unified and merged (using bedtools unionbedg and merge commands), to create a single list of 94,855 identified accessible loci. Accessibility signal and peaks, alongside previously published Oct4 binding signal (GSE49767) were visualized using IGV version 2.3.26 (Fig. 3d and Supplementary Fig. 13 and 14a).
To quantify the change in accessibility between samples, we estimated read coverage in all accessible loci (n=94,855) using bedtools coverageBed command (version 2.16.2). Read coverage was normalized by peak length in Kbp and by million number of aligned reads per sample, to give RPKM values. Further analysis was done using Matlab version R2011b. Differential peaks were defined by 4-fold change difference between MEF and ES samples, resulting in 20,540 peaks. Correlation matrix was calculated using Spearman correlation over all 20,540 differential peaks, followed by hierarchical clustering performed using Spearman correlation as a distance metric and average linkage (Supplementary Fig. 14b). ATAC-Seq data is deposited under GEO submission: GSE67299.
PCR analysis
Total RNA was isolated using the RNeasy kit (Qiagen). 3 μg of total RNA was treated with DNase I to remove potential contamination of genomic DNA using a DNA Free RNA kit (Zymo Research). 1 μg of DNase-I-treated RNA was reverse transcribed using a First Strand Synthesis kit (Invitrogen) and ultimately re-suspended in 100 μl of water. Quantitative PCR analysis was performed in triplicate using 1/50 of the reverse transcription reaction on Viia7 platform (Applied Biosystems). Error bars indicate standard deviation of technical triplicate for each measurement. For mouse pluripotency factors relative expression, the following validated mouse specific primer sequences were used: Oct4 forward primer: 5′ ACA TCG CCA ATC AGC TTG G 3′ reverse primer: 5′ AGA ACC ATA CTC GAA CCA CAT CC 3′. Sox2 forward primer: 5′ ACA GAT GCA ACC GAT GCA CC 3′ reverse primer: 5′ TGG AGT TGT ACT GCA GGG CG 3′. Klf4 forward primer: 5′ GCA CAC CTG CGA ACT CAC AC 3′ reverse primer: 5′ CCG TCC CAG TCA CAG TGG TAA 3′. For genotyping the following primers were used: M2rtTA genotyping: Forward primer 5′ AAA GTC GCT CTG AGT TGT TAT 3′; Reverse primer 5′ GCG AAG AGT TTG TCC TCA ACC 3′. Col1a knock-in allele genotyping: Forward primer 5′ GCA CAG CAT TGC GGA CAT G 3′; Reverse primer 5′ TTG CTC AGC GGT GCT GTC CA 3′. tdTomato genotyping: Forward primer 5′ CTG TTC CTG TAC GGC ATG G 3′; Reverse primer 5′ GGC ATT AAA GCA GCG TAT CC 3′. CreER genotyping: Forward primer 5′ TCC ATT TGT CAC GTC CTG CAC G 3′; Reverse primer 5′ GAA GGA ACC TGG CTT TGC CCT G 3′.
Supplementary Material
Supplementary Video 1: Representative phase-contrast live imaging of a contracting OSKM-TD cardiomyocyte colony.
Supplementary Video 2: Fluorescent live imaging of tdTomato+ beating cardiomyocyte colony shown in Supplementary Video 1. Please note that the filming is not synchronized, but was conducted 1 minute later on the same shown beating colony.
Acknowledgements
J.H.H is supported by a generous gift from Ilana and Pascal Mantoux; the New York Stem Cell Foundation (NYSCF), FAMRI, the Kimmel Innovator Research Award, the ERC (StG-2011-281906), the Leona M. and Harry B. Helmsley Charitable Trust, Moross Cancer Institute, the Israel Science Foundation Regular research program, the ICRF Foundation, Helen and Martin Kimmel Institute for Stem Cell research (HMKISCR), the Benoziyo Endowment fund. J.H.H and W.G. are supported by an HFSPO research grant. J.H.H. is a New York Stem Cell Foundation - Robertson Investigator. We thank K. Hochedlinger for mutual exchange of results and discussions prior to publication. We thank Weizmann Institute management for providing critical financial and infrastructural support. Genomic sequencing data is deposited under GEO submission: GSE67299.
Footnotes
The authors have no competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this paper
References
- 1.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Kim J, et al. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. USA. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Efe JA, et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 5.Zhu S, et al. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature. 2014;508:93–97. doi: 10.1038/nature13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurian L, et al. Conversion of human fibroblasts to angioblast-like progenitor cells. Nat. Methods. 2013;10:77–83. doi: 10.1038/nmeth.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silva J, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis-Dusenbery BN, Koszka K, Ichida JK, Eggan K. Nanog-Independent Reprogramming to iPSCs with Canonical Factors. Stem Cell Reports. 2014;2:119–126. doi: 10.1016/j.stemcr.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carey BW, Markoulaki S, Beard C, Hanna J, Jaenisch R. Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nat. Methods. 2010;7:56–59. doi: 10.1038/nmeth.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Hadjantonakis AK, Cox LL, Tam PP, Nagy A. An X-linked GFP transgene reveals unexpected paternal X-chromosome activity in trophoblastic giant cells of the mouse placenta. Genesis. 2001;29:133–140. doi: 10.1002/gene.1016. [DOI] [PubMed] [Google Scholar]
- 13.Rodríguez-Ubreva J, et al. Pre-B cell to macrophage transdifferentiation without significant promoter DNA methylation changes. Nucleic Acids Res. 2012;40:1954–1968. doi: 10.1093/nar/gkr1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greder LV, et al. Brief Report: Analysis of Endogenous Oct4 Activation during Induced Pluripotent Stem Cell Reprogramming Using an Inducible Oct4 lineage label. Stem Cells. 2012;30:2596–2601. doi: 10.1002/stem.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chambers I, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 16.Buganim Y, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 19.Festuccia N, et al. Esrrb Is a Direct Nanog Target Gene that Can Substitute for Nanog Function in Pluripotent Cells. Cell Stem Cell. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarz BA, Bar-Nur O, Silva J, Hochedlinger K. Nanog Is Dispensable for the Generation of Induced Pluripotent Stem Cells. Curr. Biol. 2014;24:347–350. doi: 10.1016/j.cub.2013.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanna J, et al. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell. 2009;4:513–524. doi: 10.1016/j.stem.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, et al. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell. 2010;7:319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papp B, Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marro S, et al. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell. 2011;9:374–382. doi: 10.1016/j.stem.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko K, Araúzo-Bravo MJ, Kim J, Stehling M, Schöler HR. Conversion of adult mouse unipotent germline stem cells into pluripotent stem cells. Nat. Protoc. 2010;5:921–928. doi: 10.1038/nprot.2010.44. [DOI] [PubMed] [Google Scholar]
- 26.Szabo E, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 27.Yuan X, et al. Combined Chemical Treatment Enables Oct4-Induced Reprogramming from Mouse Embryonic Fibroblasts. Stem Cells. 2011;29:549–553. doi: 10.1002/stem.594. [DOI] [PubMed] [Google Scholar]
- 28.Gafni O, et al. Derivation of novel human ground state naïve pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 29.Rais Y, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- 30.Hanna J, et al. Direct Reprogramming of Terminally Differentiated Mature B Lymphocytes to Pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1: Representative phase-contrast live imaging of a contracting OSKM-TD cardiomyocyte colony.
Supplementary Video 2: Fluorescent live imaging of tdTomato+ beating cardiomyocyte colony shown in Supplementary Video 1. Please note that the filming is not synchronized, but was conducted 1 minute later on the same shown beating colony.