Abstract
Sequencing cancer genomes may enable tailoring of therapeutics to the underlying biological abnormalities driving a particular patient’s tumor. However, sequencing-based strategies rely heavily on representative sampling of tumors. To understand the subclonal structure of primary breast cancer, we applied whole genome and targeted sequencing to multiple samples from each of 50 patients’ tumors (total 303). The extent of subclonal diversification varied among cases and followed spatial patterns. No strict temporal order was evident, with point mutations and rearrangements affecting the most common breast cancer genes, including PIK3CA, TP53, PTEN, BRCA2 and MYC, occurring early in some tumors and late in others. In 13/50 cancers, potentially targetable mutations were subclonal. Landmarks of disease progression, such as resisting chemotherapy and acquiring invasive or metastatic potential, arose within detectable subclones of antecedent lesions. These findings highlight the importance of including analyses of subclonal structure and tumor evolution in clinical trials of primary breast cancer.
Introduction
Driver mutations occur in single cells, and are associated with subsequent clonal expansions. Consequently, a given patient’s breast tumor comprises a complex patchwork of genetically related, competing clones1-3. Genome sequencing has enabled analysis of clonal evolution in breast cancer through sequencing of primary tumor and metastasis pairs in a few cases4,5, sequencing of single cells2,6 and xenograft models7 as well as deep sequencing for subclonal mutations1,3. These studies have revealed that subclonal evolution is found in breast cancer, albeit based on relatively small sample sizes.
Most breast cancers are localized at first presentation and managed with curative intent using surgery often combined with radiotherapy and systemic therapies. Therapies targeted against the estrogen and HER2 receptors improve survival and benefit may extend to cases where the targetable alteration is subclonal8,9. Therapies directed against a wider range of biological targets are currently in early phase trials but heterogeneity could complicate study design and confound analysis10,11. The optimal therapy may be directed against mutations shared by all cells in a cancer but later, subclonal mutations may be important if they enable subclones to resist treatment or confer metastatic capacity. In colon, pancreatic and hematological cancers, preferred temporal orders of somatic mutation accumulation may predominate12-15, but whether this exists in breast cancer has not been evaluated. In renal, pancreatic, colon and prostate tumors, geographical stratification of clonal structure is common, with subclones containing driver mutations expanding locally16-21. Whether early breast cancers show similar patterns is unknown.
Results
Multiregion sequencing of breast cancer
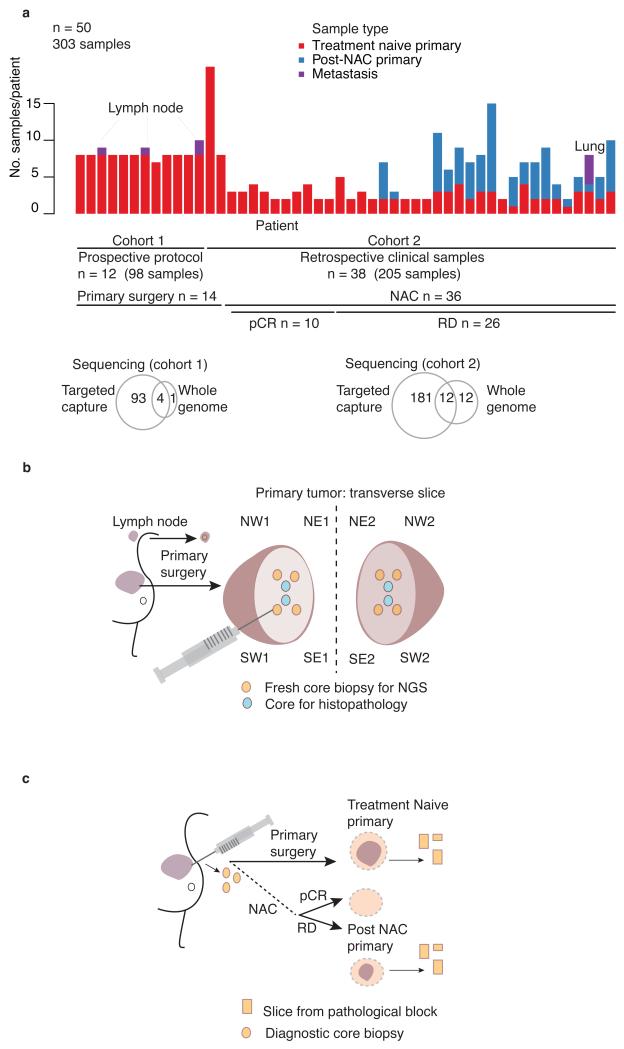
To determine the patterns of spatial evolution in primary breast cancer, we undertook sequencing of multiregion samples from 50 invasive cancers (27 ER+/HER2−, 3 ER+HER2+, and 20 triple negative (ER−PgR−HER2−); Supplementary Table 1). We sequenced the cancers in two cohorts. In cohort 1, we performed prospective, systematic needle biopsy sampling of 12 primary, treatment-naïve, surgically excised cancers (Fig. 1a–b). In cohort 2, we studied multiple treatment-naïve needle biopsy or tissue block samples from 38 cancers (Fig. 1a and c). All but 2 cases in cohort 2 underwent neo-adjuvant chemotherapy with 10 demonstrating a complete pathological response and 26 having histopathologically confirmed residual disease. For 18 of these 26 cancers, we sequenced samples from both pre-treatment and post-treatment residual, invasive disease. For 290 samples from the 50 cancers, we sequenced to high coverage (mean = 166x) (Supplementary Table 2) 360 known cancer genes, chosen from review of published literature22-24 and including over 40 genes recurrently mutated in breast cancer3,25-30 (Supplementary Table 3). For 13 of these cancers we sequenced selected tumor samples (n = 29) and a matched constitutional DNA sample in each case to whole genome level with an average depth of 40-fold (Supplementary Table 2).
Figure 1. Study design.
(a) Summary of samples within cohorts 1 and 2. n, number of subjects. (b) Geographical sampling approach: NW, northwest; NE, northeast; SW, southwest; SE, southeast within tumor hemisphere 1 and 2, plus 1 or 2 involved lymph nodes in 3 cases. For multifocal cancers all samples are taken from the single largest focus. (c) Source of retrospective clinical samples in relation to primary tumor management. RD, residual disease; NAC, Neo-adjuvant chemotherapy; pCR, pathological complete response.
We identified driver mutations as recurrent mutations in oncogenes or truncating mutations and recurrent missense substitutions in tumor suppressor genes3,14,25,27,28,30,31 (details of driver mutation annotation in Online Methods). Copy number analysis focused on the 5 most frequent arm level copy number changes32 and 12 frequently amplified genes in breast cancer33,34 (Supplementary Table 3b).
False positive and false negative mutation calls in multi-sample studies can lead to the appearance of subclonal heterogeneity that is in fact artifactual. To validate our pipeline, we repeated the targeted capture experiment using independent libraries for 38 needle biopsy samples from 5 cancers. Positive predictive values and, critically, negative predictive values were on average > 99%, confirming our ability to call both the presence and the absence of individual mutations across multiple samples from a single cancer (Supplementary Table 4). From whole genome sequencing data, we successfully verified 2,217 of 2,235 (99%) substitutions and 18 of 19 (95%) indels (Supplementary Table 4). We confirmed 1,567 of 1,778 (88%) structural variants using PCR or breakpoint-associated copy number changes (Supplementary Table 4). We achieved 97% concordance between copy number amplifications called by targeted gene sequencing and by multiplex-ligation dependent probe amplification (Supplementary Table 4). We validated phylogenetic trees reconstructed from whole genome data (Supplementary Fig. 1, Supplementary Table 5) by targeted deep sequencing of mutations on each proposed branch. Cohort 2 contained fresh frozen and FFPE samples with no systematic differences in mutation calls (Supplementary Fig. 2).
Geographical patterns of subclonal growth
To assess the spatial distribution of subclones for 12 cancers, we sliced the tumour in half immediately after surgical resection and obtained six needle biopsy samples from the cut face of each half (Fig. 1b). We performed targeted gene sequencing from 8 biopsies from each primary tumor and an associated lymph node metastasis in 3 cases (Fig. 1a). We evaluated the remaining four biopsies from each primary tumour by histopathology to confirm the presence of invasive cancer and assess Ki67 levels (Supplementary Table 1).
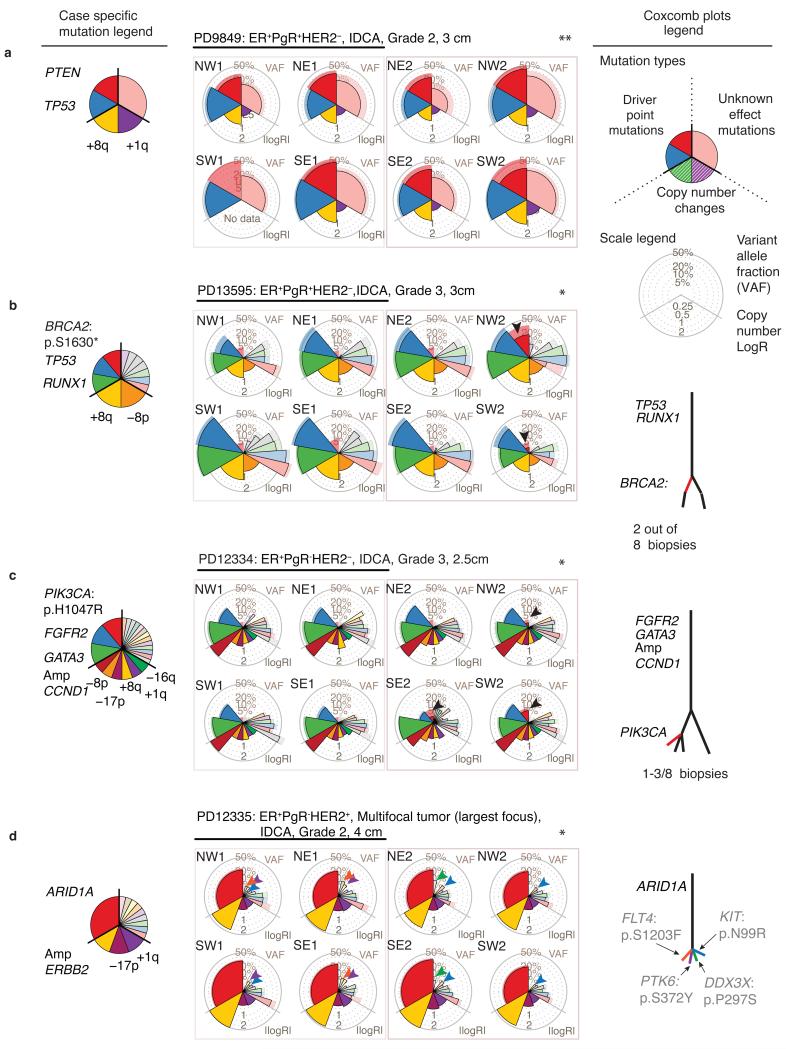
Eight of twelve tumors demonstrated statistically significant spatial heterogeneity of point mutations (q < 0.05) and a further 2 samples displayed heterogeneity of copy number changes alone (Fig. 2a–d, Supplementary Table 6). Layering mutational data onto the spatial arrangement of biopsies demonstrated that local, geographically constrained expansion is the predominant pattern of heterogeneity with 10 out of 12 cancers having at least one mutation confined to 1–3 adjacent regions.
Figure 2. Systematic sampling reveals spatial and temporal tumor evolution.
(a–d) Somatic mutation genotypes, presented as coxcomb plots, overlaid on the sample schema described in Figure 1b. Point estimates of the variant allele fraction (VAF) or copy number (LogR) is represented by the lateral extension of the outlined wedge. Pale wedges lacking an outline represent the 95% confidence interval – if coverage is low the confidence of the VAF is reduced and the pale wedge appears beyond the point estimate. ER, Estrogen receptor; PgR, Progesterone receptor; IDCA, invasive ductal carcinoma. Driver mutations and arm level copy number gains (+) and losses (−) detected in each cancer are annotated in the case-specific mutation legend. Significant heterogeneity amongst point mutations in individual cancers is determined using generalized linear models (glm) and Benjamini-Hochberg correction: *, q < 0.05 indicates significant point mutational heterogeneity; **, non-significance. (a) No detected intra-tumoral heterogeneity (q=0.8). (b–c) Local expansion of subclones (red arrow heads). (d) Complex intermixing of subclones: Individual mutations (each highlighted with a different colored arrowhead) appear in different combinations of samples. Mock phylogenetic trees are also shown: The presence and absence of mutations across related samples indicate distinct subclones and dictate the branching structure, the number of mutations in each subclone determine branch lengths. See ftp://ftp.sanger.ac.uk/pub/cancer/YatesEtAl/ for coxcomb and heatmap plots for every cancer in the cohort.
Localized confinement of subclones was not always the case. In four cancers, we found evidence of admixture of clones (Fig. 2d, Supplementary Fig. 3–4). The subclonal mutations often had low, but variably distributed, allele fractions in the samples where they were detected, a pattern suggestive of extensive intermingling of subclones across wide geographical ranges. This pattern was only common amongst larger tumors (4 of 5 tumors > 3cm). Similar findings have been observed in follicular lymphoma and colorectal cancer15,17.
In all 12 cancers, we identified at least one clonal somatic driver mutation or copy number event shared by all samples. In 4 cancers, we identified subclonal driver mutations, including recurrent TP53 missense mutations, MYC amplification, a canonical mutation in PIK3CA and a nonsense mutation in BRCA2. In these 4 examples, the subclonal driver mutation was absent from 5–7 of the 8 samples sequenced despite a collective coverage of around 1,000-fold. In 7 of 12 cases, some mutations were subclonal in the tumor as a whole but could be erroneously characterized as clonal if only a single biopsy were sequenced (Supplementary Fig. 3).
Subclonal growth in multifocal cancer
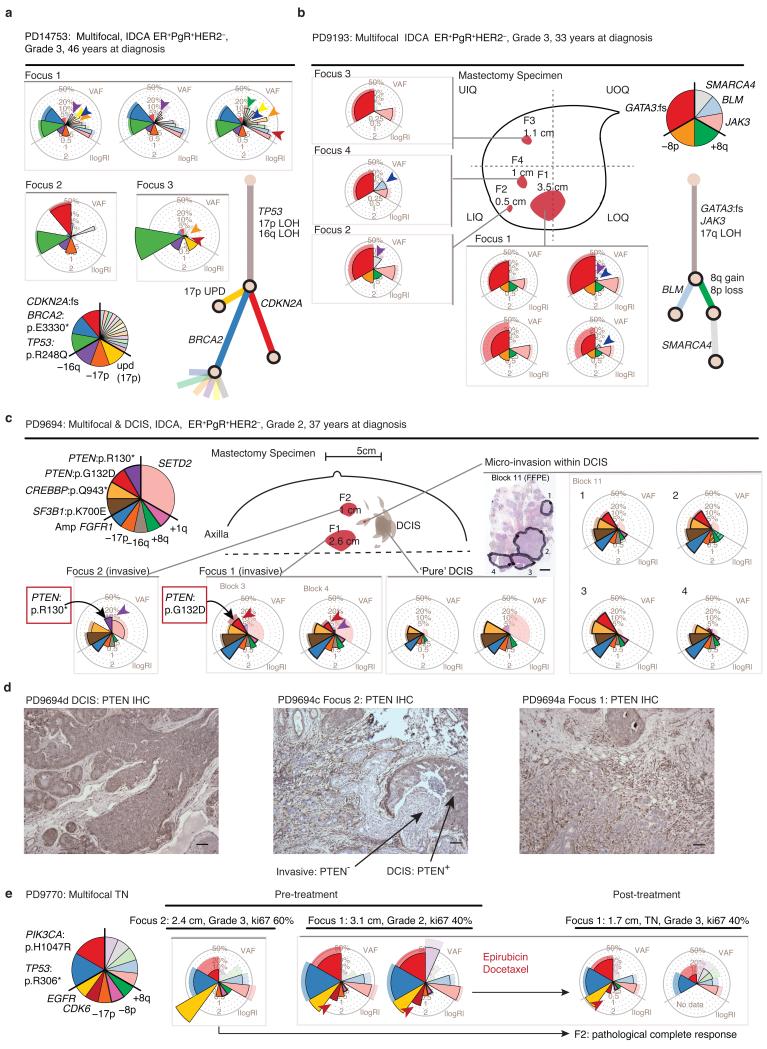
For 4 cancers, we sequenced samples from more than one focus (2–5) of a multifocal cancer. In each case, separate foci of disease were clonally related (Fig. 3). Within individual foci, we found that many private mutations had high variant allele fractions, indicating that during the growth of each focus, complete ‘clonal sweeps’ had occurred in which a clone completely replaced all other tumor cells in that focus. In 3 of 4 cases, mutations private to a disease focus included known driver events: BRCA2 and CDKN2A inactivation (Fig. 3a), PTEN point mutation (Fig. 3c-d) and CDK6 amplification (Fig. 3e).
Figure 3. Subclonal patterns in multifocal breast cancers.
(a–c,e) Targeted capture genomic analysis of subclonal structure in four subjects’ multifocal cancers. Coxcomb plots and mock phylogenetic trees are generated as described in Figure 2. Plots from multiple samples from the same tumor focus are grouped together within grey outlined boxes. Colored arrow-heads identify subclones that are shared by fewer than all invasive foci. (a) Case PD14753: Genotypes of 5 samples from 3 disease foci indicate deep branching of the tree, driver heterogeneity and subclone intermingling across foci. (b) Case PD9193: Genotypes of 7 samples from 4 disease foci demonstrate subclone intermingling. Orientation within mastectomy specimen: UIQ = Upper Inner Quadrant, UOQ = Upper Outer quadrant, LIQ = Lower Inner Quadrant, LOQ = Lower Outer Quadrant. (c) Case PD9694: Parallel evolution with 2 unique PTEN driver mutations in different foci. Schematic representation of pathological features in the mastectomy specimen. Dashed line represents the deep (chest wall) margin. Scale represents 3mm in a formalin fixed paraffin embedded tissue section. (d) Case PD9694: PTEN immunohistochemistry shows PTEN protein to be present in DCIS but lost in invasive disease foci 1 and 2. Scale = 100 microns. (e) Genotypes of 3 samples from 2 disease foci in PD9770, prior to chemotherapy and 2 samples from Focus 1 after neoadjuvant chemotherapy. Focus 2 exhibited a pathological complete response to 3 cycles of each chemotherapy agent.
The complex intermixing of minor subclones seen in some unifocal tumors also existed within and between multiple foci of disease (Fig. 3a–b, colored arrow-heads). By definition, lesions in multifocal breast cancer are separated by apparently normal breast tissue. Therefore, the fact that these distinct foci are clonally related shows that subclones in these developing tumors are capable of transiting considerable distances through normal breast tissue by the lymphatic, ductal or microcirculatory systems as has been demonstrated in metastatic prostate cancers35.
PD9694, a multifocal ER+HER2− cancer with two macroscopic foci and several microscopic foci of invasive disease occurring within a large region of scattered DCIS, embodies a remarkable example of subclonal dissemination (Fig. 3c–d). Two distinct PTEN driver mutations appeared in the different regions – these mutations had evolved in parallel during the tumor’s development (Fig. 3c) and were confined to disease with invasive potential (Fig. 3d). Critically, we detected one or the other of the PTEN mutations in discontiguous areas of microinvasive disease within predominant DCIS (Fig. 3c). The most plausible explanation for this is that the two PTEN-null subclones disseminated intraductally within the DCIS, setting up several new, discrete foci of invasion.
Subclonal driver mutations in multifocal cancers were not restricted to point mutations, with one sample showing a high-level CDK6 amplification in one focus absent from the other (Fig. 3e). The CDK6-amplified focus showed only a partial response to neoadjuvant chemotherapy, whereas the other focus underwent a complete pathological response.
Variable extent of subclonal heterogeneity in breast cancer
Across the 50 cancers in cohorts 1 and 2, we assessed intratumoral heterogeneity in the targeted gene screen, taking into account fluctuations in normal cell contamination and sequence coverage. For 23 cancers, no significant difference in point mutations (Supplementary Table 7) existed across the different tumor subregions (Fig. 1a, Supplementary Table 6), although in 4 of these cases, there was heterogeneity in copy number changes. For three cancers, we detected profound heterogeneity, exemplified by private mutations in most of the samples (PD14753, PD9850, PD12334). Most cancers, however, had intermediate levels of intratumoral heterogeneity.
We created an index of heterogeneity based on discordance of mutation frequencies averaged across all possible pairs of samples from each cancer, after adjusting for normal cell contamination and differences in coverage (Online Methods). Our data indicated no correlation between the level of heterogeneity and histology, ER status, grade, intratumoral lymphocyte infiltration or Ki67 score of the tumor (Supplementary Fig. 5b–h). Heterogeneity in Ki67 scores across samples did not correlate with our index of genomic heterogeneity (Supplementary Fig. 5i). We detected a trend towards a greater degree of heterogeneity with increasing age at diagnosis (p = 0.05, F-test) and larger tumor size (p = 0.005, F-test) amongst triple negative cancers. Notably, response to neoadjuvant chemotherapy, typically anthracycline-based regimens with or without a taxane, did not correlate with the extent of intratumoral heterogeneity amongst pre-treatment samples in this cohort, albeit with limited sample size (p = 0.2, F-test) (Supplementary Fig. 5e).
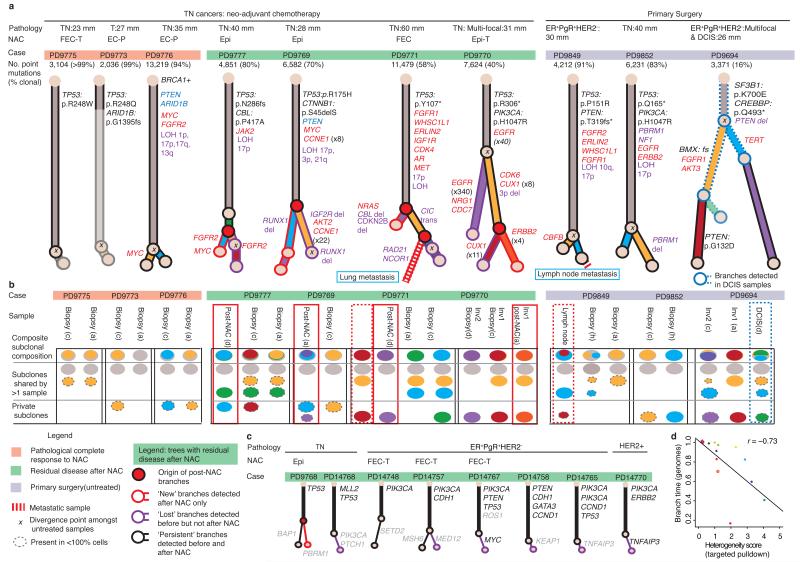
To test if genetic heterogeneity inferred from targeted capture data matches genome-wide distribution, we performed multiregion whole genome sequencing on 10 cancers (Fig. 4). For each, the thousands of somatic base substitutions allowed us to reconstruct phylogenetic trees and to determine the subclonal composition of each sampled region (Fig. 4, Supplementary Fig. 1). As for the targeted capture analysis, the extent of subclonal diversification varied markedly between tumors (Fig. 4a–b). We found good correlation between the branching time implied from whole genome data and the heterogeneity score determined from targeted capture analysis of samples from the same cancer (Fig. 4d).
Figure 4. The genome-wide spectrum of branching evolution.
(a) Phylogenetic trees generated by clustering genome-wide point mutation data from 10 multiregion sampled primary cancers. Relative branch lengths are determined from the proportion of mutations in each branch. An ‘x’ indicates the most recent common ancestor inferred from treatment-naive samples alone. (a and c) Cases where post-treatment samples are available (green highlighting bar above trees): Red node(s) indicate where subclones only detected after treatment (branches with red outlines) emerged within the tree. Branches only detected amongst pre-treatment samples are indicated by a purple outline, black branches indicate detection in both pre- and post-chemotherapy samples. Likely driver genes are colored according to mutation type: amplification (red text), homozygous deletion (blue text), point mutation (black text) and potentially relevant structural variants (purple text). Cancer type is specified: triple negative = TN, DCIS = ductal carcinoma in situ. Type of neo-adjuvant chemotherapy (NAC): Epi, Epirubicin; T, Docetaxel; P, Paclitaxel; (F)EC, (Fluorouracil), Epirubicin, Cyclophosphamide (b) The subclonal composition of individual samples where colors correspond to the tree branch in (a) and the area is proportional to the percentage of cells in that sample that contain the mutations in that branch. (c) Mock trees inferred from targeted capture data for samples with pre- and post-treatment samples. Six samples with no branching are not presented. Branches are colored as stated above for genome data. (d) Pearson’s correlation for heterogeneity estimates from whole genome and targeted capture data.
Resistant subclones may be unmasked by chemotherapy
For 18 cancers, we sequenced DNA from both diagnostic biopsies and residual, invasive disease after neoadjuvant chemotherapy. In 6 cases, mutations appeared to be subclonal (present in < 100% of tumor cells) in both pre- and post-chemotherapy samples indicating that some subclones persist despite treatment (Fig. 4a–c, black outlined branches: PD9768, PD9770, PD9771, PD9777, PD14748, PD14757). In 5 cancers, we identified a subclone in the post-chemotherapy residual tumor mass not evident in pre-chemotherapy samples (Fig. 4a–c, red outlined branches: PD9768, PD9769, PD9770, PD9771, PD9777). Within these treatment-resistant subclones, potential driver mutations included amplifications of CDK6 (PD9770), FGFR2 and MYC (PD9777) and a deletion within RUNX1 (PD9769).
Variants found only in post-chemotherapy samples could either represent mutations acquired during chemotherapy or mutations present in pre-existing subclones that were not sampled before therapy. For three cases, we had detailed phylogenies from samples before and after chemotherapy (PD9770, PD9777 and PD9771). In the latter 2, the branching point of the post-treatment subclone (Fig. 4a; red-filled circles) predated the branching point inferred from pre-treatment samples only (black cross). Post-treatment (purple outline) and pre-treatment subclone branches (purple outline) were of similar lengths, suggesting a similar molecular age. Furthermore, similarity in mutational signature profiles (Supplementary Fig. 6a) in the pre- and post-treatment branches suggests minimal contribution from chemotherapy-induced mutagenesis. Clones detected only in residual tumor mass after neoadjuvant chemotherapy are therefore likely to represent subclones in which most of the mutations are already present prior to treatment, a conclusion also reached by evolutionary simulations of breast cancers before and after chemotherapy36.
Metastases can derive from subclones detectable in the primary tumor
For two cases, we studied whole genomes from primary tumor biopsies and a metastatic deposit. In the first case (PD9771), the lung metastasis and prechemotherapy biopsies all arose from a subclone that contained over 800 base substitutions (yellow branch, Fig. 4a–b) and 43 structural variants. Notably, the residual disease sample, which also represents a chemotherapy-resistant population of cells, arose from a separate subclone (purple branch). In the second cancer (PD9849), we found that an axillary lymph node metastasis arose from a defined subclonal lineage detected within the primary tumor (Fig. 4a–b, Supplementary Fig. 1, PD9849: cluster 3), and not from the trunk of the phylogenetic tree.
This finding has clinical relevance – if metastatic disease arose from a very early branch of the phylogenetic tree, before all subclonal diversification within the primary tumor, treating actionable mutations that were subclonal in the primary tumor would not help prevent disease relapse. Although needing confirmation in larger studies, our results corroborate FISH-based studies of aneuploidy in metastatic breast cancer, which have also suggested that metastases arise from subclones of the primary cancer37.
Subclonal driver mutations and parallel evolution
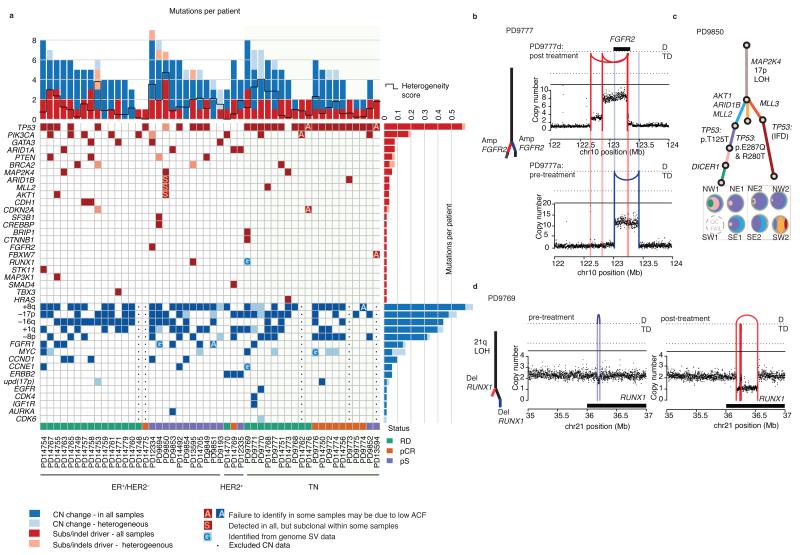
Across the cohort, the majority of driver point mutations and copy number changes were present in all lesions sequenced, suggesting they occurred before emergence of the cancer’s most recent common ancestor (Fig. 5a). Although numbers of subclonal driver mutations are too low to reach definitive conclusions about individual genes, and phylogenetic reconstruction gives only relative, not absolute, timing of driver mutations, it is clear that many of the common breast cancer genes can be mutated either early or late in disease. Driver mutations in TP53, PIK3CA, PTEN, BRCA2, CDKN2A were subclonal in some tumors in our study and fully clonal in others. Likewise, amplifications of MYC, CDK6 and FGFR1 sometimes occurred late in evolution. In 13 of 50 cancers, subclonal mutations affected genes that are potential targets of systemic therapies in clinical use or in development (Supplementary Fig. 6b).
Figure 5. Subclonal driver mutations and parallel evolution.
(a) Heatmap of somatic driver mutations and copy number changes identified from genomic sequencing of 50 tumors. Single base substitutions and small insertions and deletions are reported by red squares, intense red when detected in all associated samples from the tumor (omnipresent), pink when present in less than all samples, or clearly subclonal. Omnipresent and heterogeneous copy number changes are reported by dark-blue and light-blue squares respectively. (b–d) Three examples of parallel evolution, see fourth example in Figure 3c–d and 4a (PD9694). (c) One possible phylogenetic tree and sample subclonal compositions inferred from targeted capture data (as described in Fig. 2 and 4c legends) with TP53 mutations arising on 3 branches. See also Supplementary Figure 3 (PD9850). (b) Multiple independent episomal amplification events in FGFR2 and (d) two independent deletions in RUNX1 detected in 2 samples from the same cancer. In copy number graphs (b and d) the black dots reflect the number of copies of genomic DNA from that specific locus, with a level greater than 2 reflecting a net gain and a value less than 2 reflecting a loss. Reconstructed rearrangement breakpoints are represented by colored lines according to whether they are detectable in pre (purple) or post (red) chemotherapy samples only. The type of event is indicated by the position of the arc joining the breakpoints: D, deletion; TD, tandem duplication.
We found four cancers with parallel evolution of driver mutations, including the case with convergent PTEN mutations discussed above (Fig. 4a–b, Supplementary Fig. 6c: PD9694, PD9777, PD9769, PD9850). In a triple negative cancer (PD9777), we found three small, amplified episomal circles containing FGFR2, each present subclonally within the cancer and at variable proportions across the different samples, at least two of which must have arisen independently (Fig. 5b). In an ER+/HER2− cancer (PD9850), three separate subclonal lineages each carried different TP53 driver mutations (including a recurrent, silent mutation affecting TP53 splicing, Fig. 5c, Supplementary Fig. 6d). In a triple negative cancer (PD9769), we found distinct focal genomic rearrangements specifically deleting coding exons of RUNX1 in two subclonal branches (Fig. 5d, Supplementary Fig. 6e). Three of the four examples of parallel evolution represent the second hit in a tumor suppressor gene, with the first hit located on the trunk of the phylogenetic tree (Supplementary Fig. 6c–e).
Ongoing structural variation in subclonal diversification
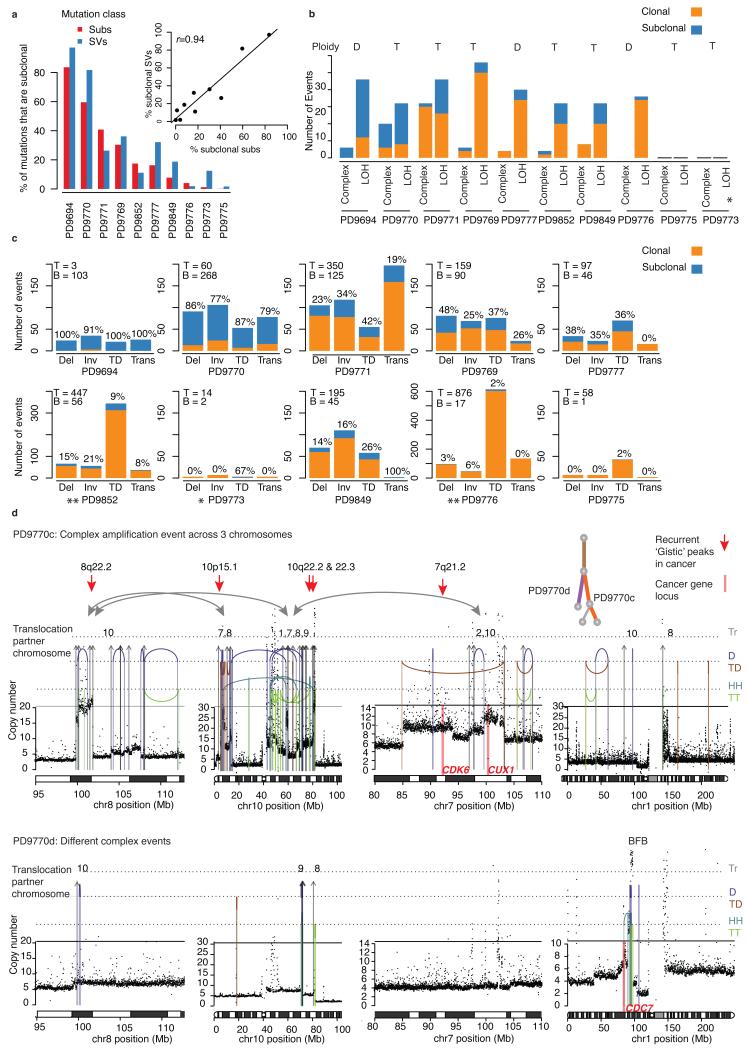
We assessed the relative activity of mutational processes over time for the 10 multi-region whole genomes (Fig. 6b–d). The proportion of structural variants that are subclonal broadly matched the proportion of subclonal substitutions (r = 0.94), although in some cancers, such as PD9777, late structural variants are the predominant driver of subclonal diversification (Fig. 6b–e).
Figure 6. Structural variants shape cancer evolution.
(a) Comparison of the proportion of substitutions (subs) and structural variants (SVs) that are subclonal in each cancer. Inset graph shows scatterplot and Pearson’s correlation coefficient (r). (b) Clonal and subclonal complex rearrangements (as described in the Supplementary Note section) and arm level loss of heterozygosity (LOH) events. The average genome-wide ploidy is indicated: T = tetraploid (4 copies), D = diploid (2 copies). (c) Breakdown of clonal and subclonal structural variants by category (inversion = Inv, deletion = Del, inter-chromosomal translocation = Trans, tandem duplication = TD). For each cancer the total number of mutations assigned to the trunk (T) or branches (B) is indicated in the top left corner, while the proportion of each mutation type that is subclonal (i.e. within the branches) is added as a percentage above each bar. (d) Case PD9770: Examples of two subclonal, complex structural rearrangements arising on separate branches of the phylogenetic tree. In PD9770c structural rearrangements link multiple regions of amplification across 3 chromosomes. Amplifications include multiple genomic regions that have been previously identified as recurrently amplified in cancers and are represented by red arrows while the locations of known oncogenes are marked by pink bars. In PD9770d these events are not seen but a breakage fusion-bridge event amplifies segments including the CDC7 gene. Rearrangement types include: Interchromosomal translocations (Tr), tail-to-tail inversions = TT (green), head-to-head inversions = HH (orange), tandem duplication = TD (orange), deletions = D (purple)
Similar to point mutational signatures (Supplementary Note, Supplementary Fig. 6b), rearrangement processes active early in tumor evolution tended to continue later in disease (Fig. 6c–d). For some cancers, tandem duplications dominated the structural variant landscape, sometimes numbering hundreds, and these continued to accumulate late in disease (Fig. 6d). Complex chromosomal events are a frequent feature being present in 7 of 10 cancers, and include 4 breakage-fusion-bridge cycles, a chromothripsis event followed by amplification and complex, amplification-associated rearrangements (Fig. 6c). In 5 tumors, complex events occurred both early and late in tumorigenesis and in some cases resulted in subclonal amplification of oncogenes (Fig. 6e, Supplementary Table 8), suggesting that catastrophic events can remodel the genome late in evolution and provide the phenotypic diversity upon which selection may operate.
Discussion
Most breast cancers are diagnosed at an early stage and are considered curable. Once established, distant metastatic disease is incurable, meaning that prevention of metastasis represents our best opportunity to improve breast cancer cure rates. We find that metastases can derive from subclones in the primary cancer, emphasizing the importance of understanding the patterns, extent and nature of subclonal diversification in primary tumors.
We find variable degrees of genomic heterogeneity across breast cancers, notwithstanding the fact that targeted gene sequencing may underestimate subclones. This contrasts with the profound heterogeneity seen almost universally in clear cell renal cell carcinoma (RCC), where subclonal diversification occurs early after VHL mutation18,19. In non-small cell lung cancer, subclonal heterogeneity is less marked38, and minimal in early (stage IA-IIIA) tumors31. Kidney cancers are often large (> 10cm) when diagnosed, whereas breast and lung cancers are typically smaller. Indeed, we find a correlation between tumor size and degree of heterogeneity in triple negative breast cancer. The direction of causality is unclear – it may be that tumors with profound heterogeneity grow to larger sizes, or it may be that beyond a certain size, complete clonal sweeps, where an especially fit clone expands to replace all other subclones in the tumor, become unlikely. In colon cancer, there is evidence that the latter can explain observed patterns of subclonal heterogeneity17.
Transcriptome and histological studies have shown that breast cancer comprises many subtypes39-42, with distinct biological, prognostic and therapeutic implications. We find that subclonal heterogeneity can be present in all major immunohistological subgroups of breast cancer, but our ‘all-comers’ study design prevents us from drawing definitive conclusions about any particular subtype. Heterogeneity may explain cases of borderline ER and HER2 positivity43,8, where survival benefits from anti-endocrine therapies extend to cancers with nuclear ER staining in as few as 1% of tumor cells8,44. FISH-based studies have found that heterogeneity of copy number changes predicts response to neoadjuvant chemotherapy36, something we did not observe. Resolving this discrepancy will require studies focusing on specific molecular subtypes of breast cancer with larger sample sizes, potentially in the setting of clinical trials of neoadjuvant therapies.
Understanding subclonality is fundamental to improving cancer care but will require prospective integration of genomics studies into clinical trials45. Important issues such as which subclones give rise to metastasis and the potential clinical benefits of treating subclonal actionable mutations can be addressed, provided sample size, sample acquisition and sample analysis are carefully planned. Drug development is increasingly ‘rational’, based around improved understanding of each tumor’s individual biology: drug testing should follow this lead, incorporating the biology of cancer evolution into trial design and evaluation.
Online Methods
Sample acquisition
In this exploratory study, we analyzed a total of 303 multi-region tumor samples from 50 subjects’ breast cancers and a matched normal sample, derived from blood (n = 49) or adjacent normal breast tissue (n = 1). Cohort 1 consists of 98 samples from 12 cancers (average of 8.2 samples per cancer, range = 8–10). Cohort 2 comprises 205 samples from 38 cancers (average of 5.4 per cancer, range = 2–21) (Supplementary table 1). All subjects are female and in cohort 1 and 2 the average age at diagnosis is 67 years (range = 44-90 years) and 49 years (range = 29-67 years) respectively (Supplementary table 1). Sample collection and management complies with local institutional review board approvals and details are provided in the Supplementary Note. Samples in cohort 1 represent those from 12 patients undergoing primary surgery, who provided informed consent to participate in a prospective study. They encompass 12 geographically pre-determined tissue samples (15-20 mg) obtained using a 14G Tru-cut needle from fresh surgical specimens following the map in Figure 1b. In cohort 2 we studied de-identified residual tissue samples collected during routine clinical care. Samples in cohort 1 are derived from fresh needle biopsy specimens (n= 98) while those in cohort 2 are from a combination of diagnostic tumor biopsies (n = 95) and surgical specimen tissue blocks (n = 110) fixed in FFPE (n = 104) or fresh frozen at acquisition (n = 101) (Supplementary Table 1). Experienced local pathologists performed histopathological review of all primary tumors including IHC for ER and PgR Allred scores, and HER2 status with FISH confirmation for HER2 IHC scores of 2+ or 3+ (Supplementary Table 1). Pathologists assessed intra-tumoral and stromal lymphocytes according to the criteria previously described46 and Ki67 staining as described in the Supplementary Note and presented in Supplementary Table 1). Sample size was chosen to ensure that genes mutated in > 10% of tumors were sampled on average 5 times in the cohort.
DNA extraction
We performed DNA extraction from serial thick sections cut from tumor tissue samples. Pathologist guided macro-dissection ensured tumor cell enrichment in cases where the invasive tumor content was estimated to be less than 50% of cells. For two patients with multi-focal disease (PD9193, PD9694) we used histopathologically guided needle dissection of FFPE samples. We isolated tumor DNA from fresh or fresh frozen tissues using the DNeasy® Blood and Tissue Kit and from FFPE tissues using QIAamp® DNA FFPE Tissue Kit or Argylla Technologies® DNA nanoPurify kit. We used QIAGEN’s® QIAamp® DNA Blood Maxi Kit, QIAamp® DNA Mini Kit or DNeasy® Blood and Tissue Kit to isolate DNA from whole blood. In all cases we followed protocols according to the manufacturer’s recommendations.
Genomic sequencing
We created targeted capture pull-down (average insert size 150bp) and genome-wide, shot-gun (insert size 300-600bp) libraries from native DNA using previously described workflows1,14,47 (details in Supplementary Note) and generated paired-end sequence data (75bp and 100bp respectively) using Illumina HiSeq® machines. The sequence data, aligned to the human reference genome (NCBI build37) using BWA48 is deposited in the European Genome-Phenome Archive (https://www.ebi.ac.uk/ega/ at the EBI) with accession numbers EGAD00001000965 and EGAD00001000898. Within the custom targeted capture experiment we sequenced 290 tumor samples from 50 subjects’ cancers to a mean target coverage of 160x with 63% of exonic regions achieving >= 100-fold coverage (Supplementary Table 2). We sequenced to whole genome level 29 tumor and 13 matched normal samples with average sequence coverage of 40 and 31 fold respectively (Supplementary Table 2). We used both sequencing approaches for 16 tumor samples.
We used 2 in-house cancer gene panels (CGP, versions v1 and v2) designed to pull down a selection of genes (454 and 360 genes respectively) that are known, or suspected to play a role in cancer (Supplementary Table 3). The panel targets genes from the Cancer Gene Census (COSMIC)24, genes recurrently amplified or over-expressed in cancer22,23 and candidate cancer genes such as kinases from the MAP Kinase signaling pathway. All genes in CGP v2 are also in CGP v1 - only genes present in both CGP v1 and v2 are presented in these analyses. We performed custom RNA bait design following manufacturers’ guidelines (SureSelect®, Agilent®, UK) to create designs of approximately 2Mbp in size. The data from 63 and 240 tumor samples is derived form CGP v1 and v2 respectively Supplementary Table 1.
Somatic mutation calling
Comprehensive lists of all somatic substitutions, small insertions and deletions (indels) structural variants including variant allele frequencies from both whole genome and targeted capture analysis are available for download at ftp://ftp.sanger.ac.uk/pub/cancer/YatesEtAl/. All high confidence mutation calls within the scope of the cancer gene panel are presented in Supplementary Table 7. Coding substitution and indel calls and structural variants with potential oncogenic effects, identified in whole genome data, are summarized in Supplementary Table 8. Mutation calling algorithms used in the analysis are freely available at https://github.com/cancerit/ and are described in the Supplementary Note.
Validation approaches
When exploring heterogeneity determining when a mutation is absent is at least as important as determining when the mutation is present. To address this we performed validation using custom pull-down and sequencing of mutations identified in any sample (Illumina HiSeq® or MiSeq®) in all related samples from the same cancer. We enriched the validation experiment with mutations that appeared to be heterogeneous (from the branches of phylogenetic trees) or that defied the consensus tree. Across the 39 whole genome tumour samples we selected in excess of 2,000 somatic substitution locations for validation and created a 473kbp custom capture probe design using Agilent® Technologies freely available online software ‘Sure Select Design Wizard’ using high-stringency repeat masking, a tiling density of 2X and balanced boosting. We created DNA capture (paired-end, average insert size 150bp) libraries using native DNA where resources permitted, or if necessary using whole-genome amplification (WGA). We sequenced multiplexed libraries to an average depth of 265X using the Illumina MiSeq® platform. When a variant is called present in the tumor sample and absent in the matched normal sample in both discovery and validation experiments for one or more related sample we report it as validated somatic (see Supplementary Note for details of validation calls). Using these criteria 99% (2,217 out of 2,235) of substitutions validated somatic (Supplementary Table 4). The remaining calls are not detectable in any relevant sample’s validation data (false positive, n=10) or detected in the matched normal at validation (germline, n=7). We confirmed absence of 1,301 out of 1,683 (77%) of mutations. Overall concordance between the 2 experiments (true positives and true negatives/ all validation calls) was 90% (5,003 out of 5,527). The overall level of concordance for the targeted capture experiment is higher, with consistency between 189 out of 191 validation and discovery calls (99% concordance), likely reflecting the higher coverage in this experiment (Supplementary Table 4, Supplementary Note).
Variant annotation
To identify likely driver events we first identified from the literature the genes that are most likely to contribute to breast cancer oncogenesis. For each individual mutation that fell in one of these 45 high confidence breast cancer genes we assigned a likely oncogenic status as follows: Presumed oncogenic – Mutations that meet any of the following criteria:
Canonical oncogenic mutations in recurrent hotspots
Recurrent mutations in a known oncogene: >= 2 confirmed non-synonymous or in frame deletion, somatic mutations have previously been confirmed at this locus in COSMIC
Likely damaging events in a known tumor suppressor: Truncating, frameshift, essential splice variant or within a mutation hotspot (>= 2 somatic mutations) or Synonymous mutation in a known recurrent splice site hotspot49
Possible oncogenic – Previously unreported variant in a high confidence breast cancer gene that occurs within 3 amino acids of >= 2 confirmed somatic mutations or truncating events in ‘medium confidence’ tumor suppressors (defined as known tumour suppressor role in cancers other than breast cancer). All other non-synonymous mutations are assigned a status of unknown relevance. Using these criteria the 260 mutations identified across the dataset are annotated as follows: 87 oncogenic, 8 possible oncogenic, 124 of unknown oncogenicity and 41 nononcogenic (synonymous) (Supplementary Table 3).
Copy number analysis
Likely driver copy number changes are reported for individual samples within the targeted gene capture experiment in Supplementary Table 7, and for whole genome samples in Supplementary Table 8. Segmental copy number information was derived for each of the 29 tumor samples for which we had whole genome NGS data using the ASCAT algorithm (allele-specific copy number analysis) of tumors as previously described32. The algorithm simultaneously determines and utilizes aberrant cell fraction and ploidy estimates to determine allele specific copy number from NGS data. A segment is considered amplified if it is present at more than twice the estimated average ploidy across the whole genome. Homozygous deletions are identified as segments where total copy number equals zero (subclonal homozygous deletions if copy number is less than 1). Visual inspection of copy number transitions and reconstructed, associated rearrangement breakpoints are used to validate driver copy number events as described in the Supplementary Note.
Within the targeted capture experiment we evaluated copy number using libraries from the ASCAT algorithm and used LogR and BAF values to identify five of the most frequent arm level copy number changes in breast cancer – 16p and 17p loss and 1q, 8q, 16p gain32 and amplification of 12 genes frequently identified as amplified in breast cancers (FGFR1, MYC, CCND1, CCND3, CCNE1, CDK4, CDK6, IGF1R, ZNF217, AURKA, EGFR and ERBB2)33,34. Details are provided in the Supplementary Note alongside the targeted capture copy number validation approach that employed multiplex ligation dependent probe amplification (Supplementary Table 4).
Statistical and informatics approaches
We performed statistical analysis and produced graphics using R version 3.0.1: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/). The stars() function generated coxcomb plots. Other packages used include RColorBrewer, xlxs , lme4, mgcv, as well as packages from bioconductor50. All hypothesis tests performed in the manuscript are 2-sided where appropriate.
Measuring heterogeneity in targeted capture data
(i) Estimating variant allele frequencies and confidence intervals
In a sequencing experiment with finite coverage it is likely to completely miss mutations present at low variant allele frequency (VAF). Our point estimate of the VAF is
where x is the observed number of reads reporting the variant and n is the coverage. This is the maximum likelihood estimated under a simple binomial sampling model,
It is, however, also possible to observe x reads if the true VAF is greater than the ML estimate. To decide what is the maximal allele frequency compatible with our data, we defined a one-sided 95% confidence interval CI to be that VAF beyond which the probability to observe x or less reads is smaller than 5%,
where F denotes the cumulative density function of the binomial distribution.
(ii) Testing for presence or absence of mutations
For the purposes of determining if an individual mutation is present or absent, as displayed in the driver mutation heatmap (Figure 5) we determined the presence of each mutation in a dichotomous fashion in each sample. A mutation is considered to be: (i) Present if found at positive variant allele frequency (VAF > 0); (ii) Indeterminate in cases with no detectable VAF = 0, but 95% confidence intervals spanning CI > 5% allele frequency, as the absence of such mutations could not be ruled out with sufficient certainty; (iii) Absent if undetectable (VAF = 0) and the 95% confidence interval CI < 5%. Only in such cases are mutations reported as heterogeneous.
(iii) Measuring Heterogeneity
In addition to sampling fluctuations, the observed VAF is confounded by the tumor cell fraction T. Any comparison of VAF between samples should therefore normalize for T. A low tumor cell fraction T will rescale all observed variant allele frequencies by a factor of T. For the computation of the heterogeneity index, we hence use the average VAF in sample j as an estimate of Tj and rescale all VAF values by 1/Tj. To quantify and compare heterogeneity between cancers we calculated a continuous index of heterogeneity across all data from the targeted gene screen. This index measures the average discordance of mutation frequencies between any two pairs of samples, after adjusting for the extent of tumor cell content. The distance in tumor cell fraction adjusted VAF of gene i between sample j and k is computed as
Note that the above distance uses the distance to the CI if that appears closer to the observed VAF.
The heterogeneity index is then defined as the average distance between all genes and samples
where g is the number of genes and b the number of samples.
A heterogeneity value of 0 indicates perfect concordance of all samples and a value of 1 would correspond to a situation, in which one sample has one additional fully clonal mutation. The heterogeneity index shows a strong inverse correlation with the branch time derived from whole-genome sequencing data in the sense that late-branching tumors display higher levels of geographic heterogeneity (rho=−0.73; Supplementary Fig. 3j).
(iv) Testing for heterogeneity
We used generalized linear models (glm’s) with an overdispersed binomial family to test whether the observed differences in variant allele frequencies between genes and samples in a given cancer can be explained by sampling fluctuations and differences in tumor cellularity alone. In a binomial glm the expected count of mutation i in sample j is given by
where f is the inverse logit function. Here αi sets the average frequency of each gene i and βj the common factor by which the gene frequencies change in sample j due to changes in tumor cellularity across samples. The parameter γij reflects the deviation of gene i in sample j from the trend imposed by the gene-specific allele frequency and cellularity in sample j. Note that there can be maximally (g-1) × (b − 1) γij’s because of the g+b shared factors αi and βj; the total number of observations is g × b.
An overall test for heterogeneity in a given cancer can then be conducted by testing whether all γij are zero, the alternative being that there is variation in any gene. This can be achieved by means of a likelihood ratio test (LRT) with (g-1) × (b − 1) degrees of freedom. We used the following R commands to derive a P-value for each sample: # x is a vector of variant allele counts for all lesions and biopsies; the length of x is g*b
# n is a vector of the corresponding coverage
# genes is a factor() determining which gene x and n refer to
# biopsies is a factor() determining the biopsy
y <- cbind(x, n-x)
fit1 <- glm(y ~ genes + biopsies −1 + genes:biopsies, family=quasibinomial)
Ppatient <- anova(fit1, test=“LRT”, dispersion=1.5)[4,5]
P-values for each patient are subsequently corrected for multiple testing using the Benjamini-Hochberg procedure. P- and Q-values for each patient are reported in Supplementary Table 6. Similary we tested for variation in a particular gene i using an interaction term for this gene only:
fit.gene <- glm(y ~ genes + biopsies −1 + (genes==gene):biopsies, family=quasibinomial)
glm.p.value <- anova(fit.gene, test=“LRT”, dispersion=1.5)[4,5]
Additionally, we used glm’s with random effects, implemented in mgcv::gam(), to compute estimates of the variation of allele frequencies across samples. Gene-wise P-values from glm and gam models are also listed in Supplementary Table 6 (glm.p.value, gam.stddev, gam.p.value).
(v) Testing of clinical associations
Possible associations between clinical or pathological factors and genetic heterogeneity as a response are fitted using R’s lm() function. F-tests for overall association are then computed using the anova() command.
Basic principles of phylogenetic tree construction
For ten patients with multi-sample whole genome sequencing data, to model the subclonal structure we employed a number of bioinformatic and deductive reasoning approaches. The intellectual framework for our methods has been previously described1 and this approach has been extended and reinterpreted by many others since, using the original data51,52. All the conclusions we derive follow from three basic principles that also underlie the ‘mock’ trees derived from targeted capture data: (i) Cancer cells divide by asexual reproduction; (ii) The exact same mutation does not occur more than once during the evolution of the cancer (note that this so-called ‘infinitely many sites’ assumption is potentially not true for hot-spot mutations in, say, PIK3CA but will be true for virtually all passenger mutations given the size of the genome and the relative paucity of somatic mutations); (iii) Sequencing reads from massively parallel sequencing data are a random sample from the alleles present in the DNA.
The approach used in this paper followed 3 main steps: (i) Identification of large-scale subclonal copy number changes using the Battenberg algorithm as previously described1. Code is publically available at https://github.com/cancerit/cgpBattenberg; (ii) Clustering of subclonal somatic substitutions in whole genome data using a Bayesian Dirichlet process in multiple dimensions across related samples as previously described47; (iii) Hierachical clustering across multiple samples by applying the ‘pigeon hole principle’ (PHP). Next, we performed validation of mutations in individual branches by targeted pulldown and validation of tree structures by independent clustering of indels and targeted pulldown substitutions following steps i–iii (Supplementary Fig. 1).
Each step is described in further detail below and for individual patients all potential solutions and reasoning are represented in Supplementary Figure 1, while individual cases are discussed in the Supplementary Note. Branch length, cluster sizes and poster confidence intervals are provided in Table 5.
(i) Whole genome data: mutation copy number and cancer cell fraction
For each mutation we calculated the mutation copy number as previously described27, using the mutant allele burden and the aberrant cell fraction and the locus specific copy number in the tumor and matched normal from ASCAT 32. The mutation copy number reflects the percentage of tumor cells within a sample carrying that mutation, and permits the cross-comparison of the mutation in related samples despite differences in tumor purity and/ or copy number profiles as previously demonstrated47.
Mutations present on multiple copies of a chromosomal segment will have a mutation copy number greater than 1. To group mutations according to the percentage of cells containing it, the number of chromosomes carrying the mutation must be determined. For all mutations within amplified regions with a major allele copy number of C, the observed fraction of mutated reads is compared to the expected fraction of mutated reads resulting from a mutation present on 1,2,3,…C copies, assuming a binomial distribution. The fraction of cancer cells reporting the mutation, or ‘cancer cell fraction’, is then determined as the mutation copy number divided by the value of C with the maximum likelihood. Mutations are determined as clonal if reported by ~100% or tumor cells and subclonal if present in significantly less than 100% of cells.
For the purpose of comparing multiple related samples we excluded mutations from clustering analysis when they occur in a region of different copy number between samples and where the absence or altered copy number may explain the loss or different allele burden in the related samples. This approach is essential to reduce overestimation of inter-sample heterogeneity. Large-scale losses, including those at the arm or whole chromosome level are frequent during evolution (Fig. 6b). Allelic loss can therefore be accompanied by loss of large numbers of point mutations and indels, which could be misinterpreted as gained events (i.e. ongoing evolution) in related samples. We placed the few individual driver mutations that occurred in regions of differential copy number state on the reconstructed tree post hoc. This then allowed them to be included in the temporal ordering inference.
(ii) Mutational clustering
For individual samples we inferred the number of subclones and the fraction of cells within each subclone using a previously described Bayesian Dirichlet process (DP) to cluster mutations according to their cancer cell fraction1,47. We extended this process into multiple dimensions for the 10 patients with multiple related samples where the number of mutant reads obtained from multiple related samples are modeled as independent binomial distributions. Clusters are identified as local peaks in the posterior mutation density obtained from the DP. For each cluster, a region representing a ‘basin of attraction’ is defined by a set of planes running through the point of minimum density between each pair of cluster positions. Mutations are assigned to the cluster in whose basin of attraction they are most likely to fall, using posterior probabilities from the DP. The R code required to sample clustering of mutations from a Dirichlet process and to make density plots of the clustering for each pair of samples is released as a supplementary material alongside this paper.
(iii) Hierarchical ordering of mutation clusters using the ‘Pigeon Hole Principle’
To determine the most likely phylogenetic tree we applied the PHP to determine the order in which mutational clusters arose in time and in relation to each other1. This principle operates upon the premise that if the fraction of cells reporting 2 different mutations adds up to >100%, then at least one tumor cell must contain both mutations. By the same principle one can determine if clusters of mutations are collinear i.e. on the same branch of the phylogenetic tree, and often the temporal order in which they arose. For all clonally related samples the same underlying phylogenetic tree must exist. This exerts a greater stringency to the inferred ordering of subclonal clusters – firstly the PHP must be fulfilled within all individual related samples and the ordering of events cannot be contradictory across related samples.
We attempted to reconstruct phylogenetic trees from the whole genome discovery substitution data using all clusters that are estimated to contain at least 150 substitutions or >=2% of all clustered substitutions. To reflect the lower overall numbers in validation and indel data this threshold requirement is set at 5%. In tree construction the percentage of all mutations in a cluster determined the relative branch length. Within an individual sample the cancer cell fraction fraction of a given cluster ‘X’, is the fraction of tumor cells reporting the mutations in cluster X. Credible intervals for the cancer cell fraction are typically small reflecting high numbers of mutations in most clusters. We allowed 5% variation in either direction to the assigned cluster sizes when determining ordering
In 7 out of 10 cases we derived a single, unambiguous phylogenetic tree solution from the whole genome discovery data (Supplementary Fig. 1). In two cases we identified one or more alternative tree using the discovery data (PD9773, PD9694) while in another case (PD9777), a soloution could only be deconvoluted using revised VAFs from high depth validation data. The validation clustering data identified additional tree branches in 2 cases (PD9775, PD9849). In cases where uncertainty as to the position or size of a branch the relevant branch(es) are ‘faded out’ in Fig. 4a. Our approaches are described in detail in the Supplementary Notes section where we focus on patient PD9694 and cases where solutions are less clear-cut.
Mutational signature analysis
Mutational signatures are detected in two independent ways: (i) de novo extraction based on somatic substitutions and their immediate sequence context and (ii) refitting of previously identified consensus signatures of mutational processes. We used a previously developed theoretical model and its corresponding computational framework to perform de novo extraction53. Details of mutational signature analysis can be found in the Supplementary Note.
Supplementary Material
Acknowledgements
This work is supported by the Wellcome Trust. P.J. Campbell is a Wellcome Trust Senior Clinical Fellow (103858/Z/14/Z). L.R. Yates, Y. Li and L.B. Alexandrov are funded by Wellcome trust PhD fellowships. S. Nik-Zainal is funded by a Wellcome Trust Intermediate Clinical Research Fellowship (WT100183MA). P. Van Loo is a postdoctoral researcher of the Research Foundation Flanders (FWO). Work within the project is supported by the Belgian Cancer Plan-Ministry of Health, the Breast Cancer Research Foundation, the Brussels Region, the Norwegian Cancer Society, the Norwegian Health Region West and the Bergen Research Foundation. Some samples in this publication will be included in the Breast Cancer Genome Analyses for the International Cancer Genome Consortium (ICGC) Working Group led by the Wellcome Trust Sanger Institute. BASIS is a part of the ICGC working group and is funded by the European Community’s Seventh Framework Programme (FP7/2010-2014) under the grant agreement number 242006. This working group also encompasses a triple negative breast cancer project funded by the Wellcome Trust (grant reference 077012/Z/05/Z) and a HER2+ breast cancer project funded by Institut National du Cancer (INCa). We thank B. Leirvaag, D. Ekse, N. K. Duong and C. Eriksen for technical assistance. Research performed at Los Alamos National Laboratory was carried out under the auspices of the National Nuclear Security Administration of the US Department of Energy.
Footnotes
Accession Codes
The sequence data, aligned to the human reference genome (NCBI build37) using BWA is deposited in the European Genome-Phenome Archive (https://www.ebi.ac.uk/ega/ at the EBI) with accession numbers EGAD00001000965 and EGAD00001000898.
Competing Financial Interests
Peter J Campbell and Michael R Stratton are founders, stock-holders and consultants for 14M Genomics Ltd, a genomics diagnostic company.
References
- 1.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah SP, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meric-Bernstam F, et al. Concordance of genomic alterations between primary and recurrent breast cancer. Molecular cancer therapeutics. 2014;13:1382–1389. doi: 10.1158/1535-7163.MCT-13-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell reports. 2013;4:1116–1130. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammond ME, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seol H, et al. Intratumoral heterogeneity of HER2 gene amplification in breast cancer: its clinicopathological significance. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2012;25:938–948. doi: 10.1038/modpathol.2012.36. [DOI] [PubMed] [Google Scholar]
- 10.Simon R, Roychowdhury S. Implementing personalized cancer genomics in clinical trials. Nature reviews. Drug discovery. 2013;12:358–369. doi: 10.1038/nrd3979. [DOI] [PubMed] [Google Scholar]
- 11.Sleijfer S, Bogaerts J, Siu LL. Designing transformative clinical trials in the cancer genome era. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:1834–1841. doi: 10.1200/JCO.2012.45.3639. [DOI] [PubMed] [Google Scholar]
- 12.Moskaluk CA, Hruban RH, Kern SE. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer research. 1997;57:2140–2143. [PubMed] [Google Scholar]
- 13.Powell SM, et al. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 14.Papaemmanuil E, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green MR, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121:1604–1611. doi: 10.1182/blood-2012-09-457283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yachida S, Iacobuzio-Donahue CA. Evolution and dynamics of pancreatic cancer progression. Oncogene. 2013;32:5253–5260. doi: 10.1038/onc.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sottoriva A, et al. A Big Bang model of human colorectal tumor growth. Nature genetics. 2015;47:209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerlinger M, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nature genetics. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper CS, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nature genetics. 2015 doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nature reviews. Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 23.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Futreal PA, et al. A census of human cancer genes. Nature reviews. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez-Perez A, et al. IntOGen-mutations identifies cancer drivers across tumor types. Nature methods. 2013;10:1081–1082. doi: 10.1038/nmeth.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens PJ, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellis MJ, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banerji S, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nature genetics. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balko JM, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer discovery. 2014;4:232–245. doi: 10.1158/2159-8290.CD-13-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gundem G, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Almendro V, et al. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell reports. 2014;6:514–527. doi: 10.1016/j.celrep.2013.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Almendro V, et al. Genetic and phenotypic diversity in breast tumor metastases. Cancer research. 2014;74:1338–1348. doi: 10.1158/0008-5472.CAN-13-2357-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Bruin EC, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ali H, et al. Genome-driven integrated classification of breast cancer validated in over 7,500 samples. Genome biology. 2014;15:431. doi: 10.1186/s13059-014-0431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen TO, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 41.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 43.Rakha EA, Ellis IO. Breast cancer: updated guideline recommendations for HER2 testing. Nature reviews. Clinical oncology. 2014;11:8–9. doi: 10.1038/nrclinonc.2013.230. [DOI] [PubMed] [Google Scholar]
- 44.Early Breast Cancer Trialists’ Collaborative, G. et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–784. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan Y, et al. Assessing the clinical utility of cancer genomic and proteomic data across tumor types. Nature biotechnology. 2014;32:644–652. doi: 10.1038/nbt.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Online methods references
- 46.Denkert C, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:105–113. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 47.Bolli N, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nature communications. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Supek F, Minana B, Valcarcel J, Gabaldon T, Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell. 2014;156:1324–1335. doi: 10.1016/j.cell.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 50.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischer A, Vazquez-Garcia I, Illingworth CJ, Mustonen V. High-definition reconstruction of clonal composition in cancer. Cell reports. 2014;7:1740–1752. doi: 10.1016/j.celrep.2014.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oesper L, Mahmoody A, Raphael BJ. THetA: inferring intra-tumor heterogeneity from high-throughput DNA sequencing data. Genome biology. 2013;14:R80. doi: 10.1186/gb-2013-14-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell reports. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.