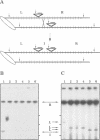
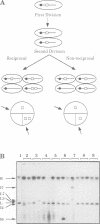
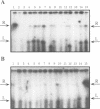
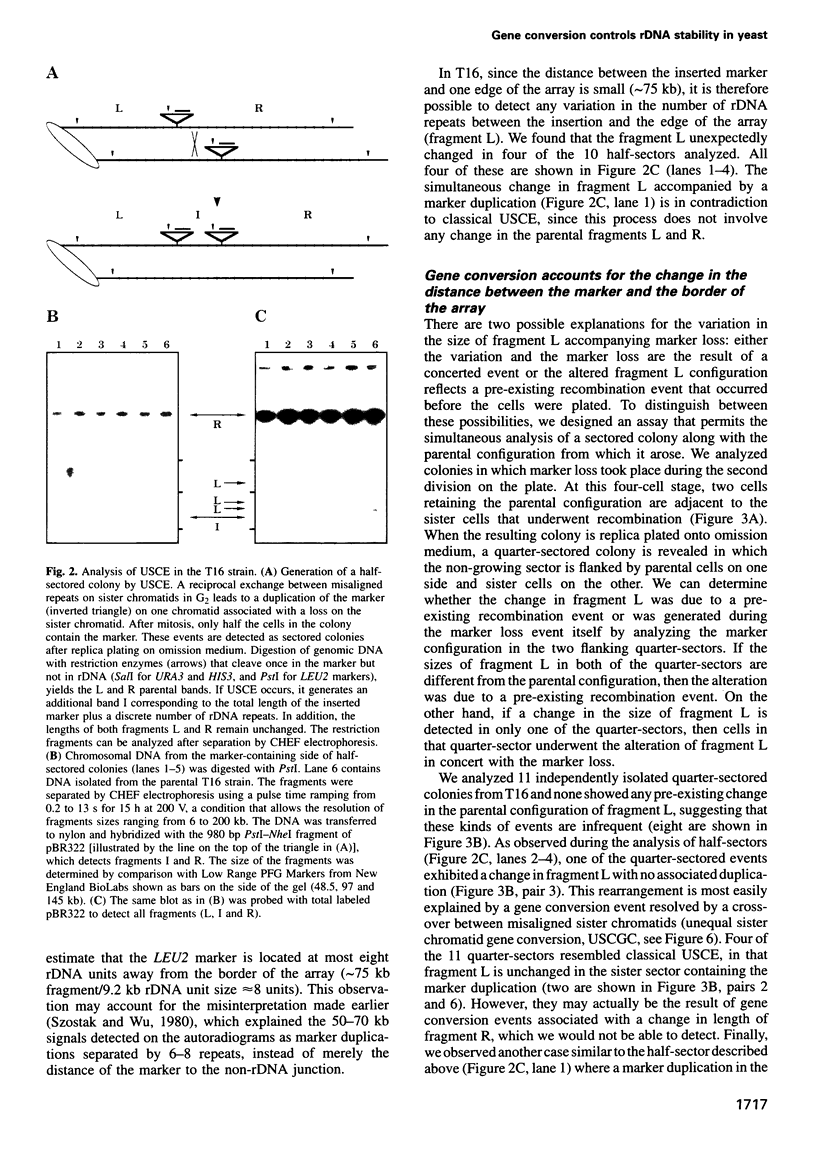
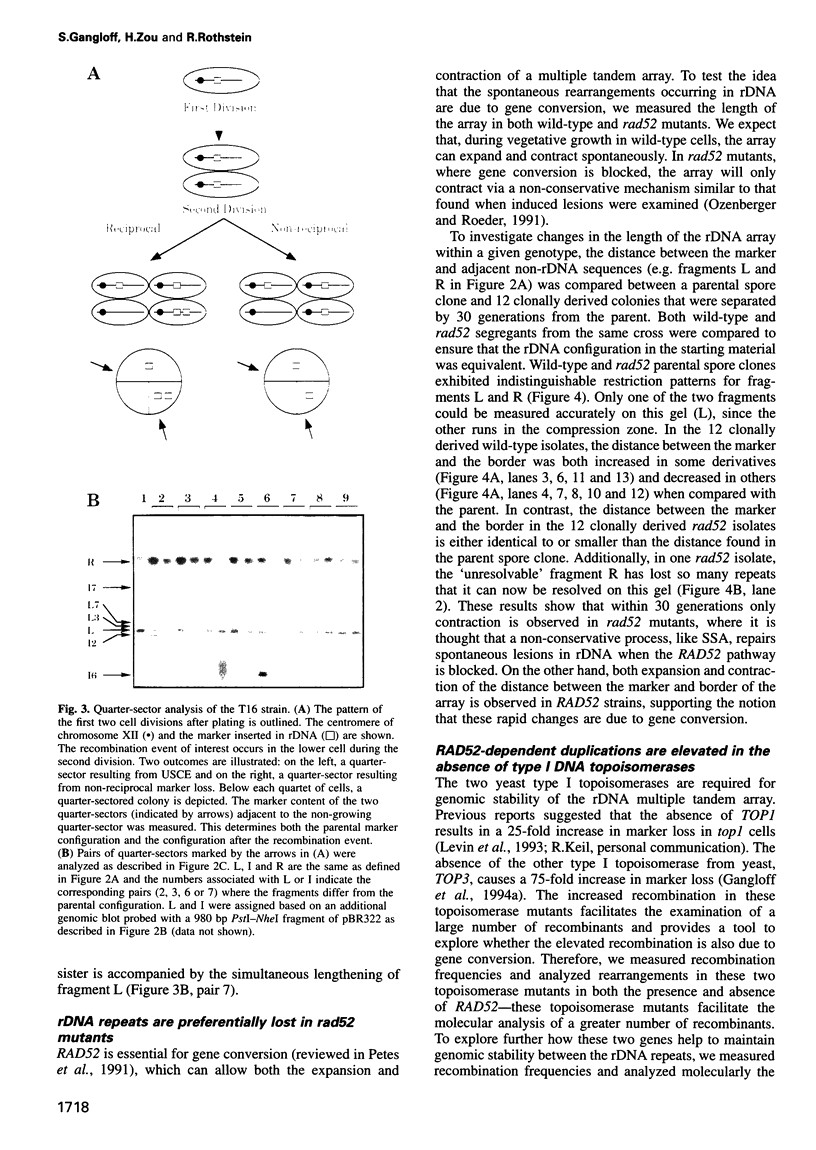
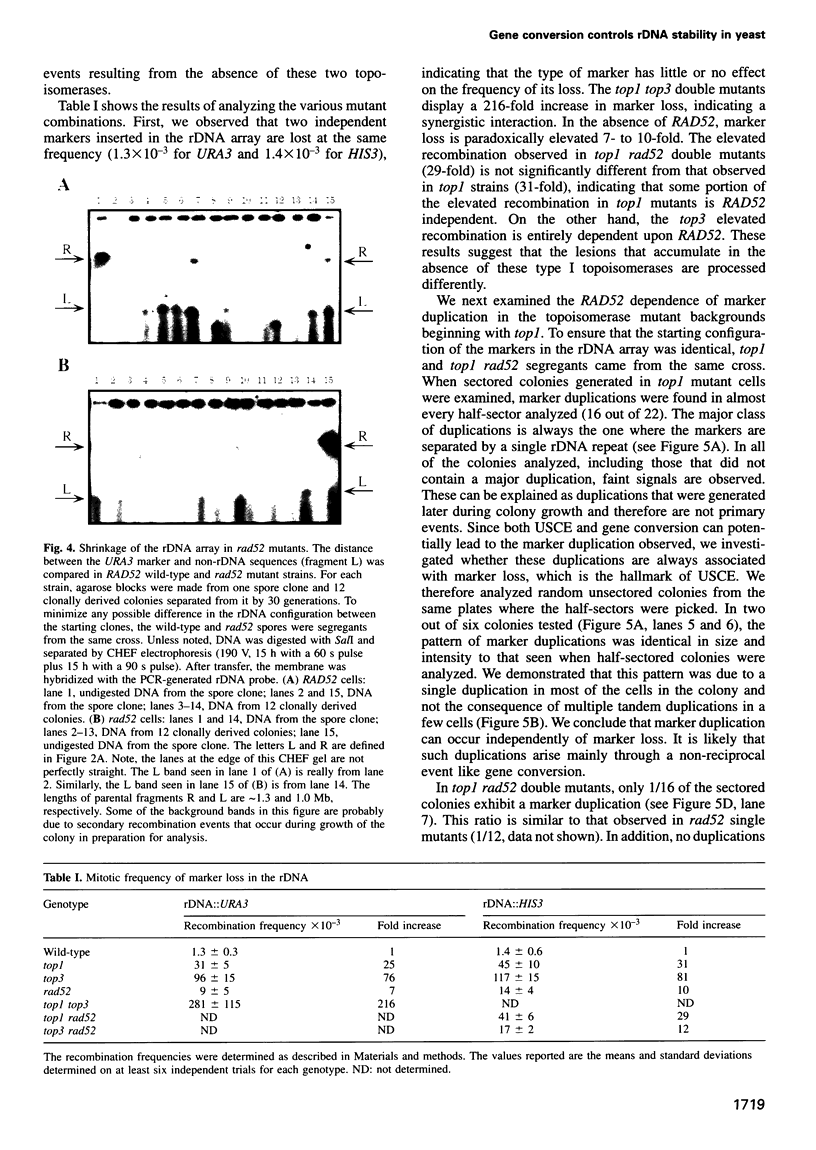
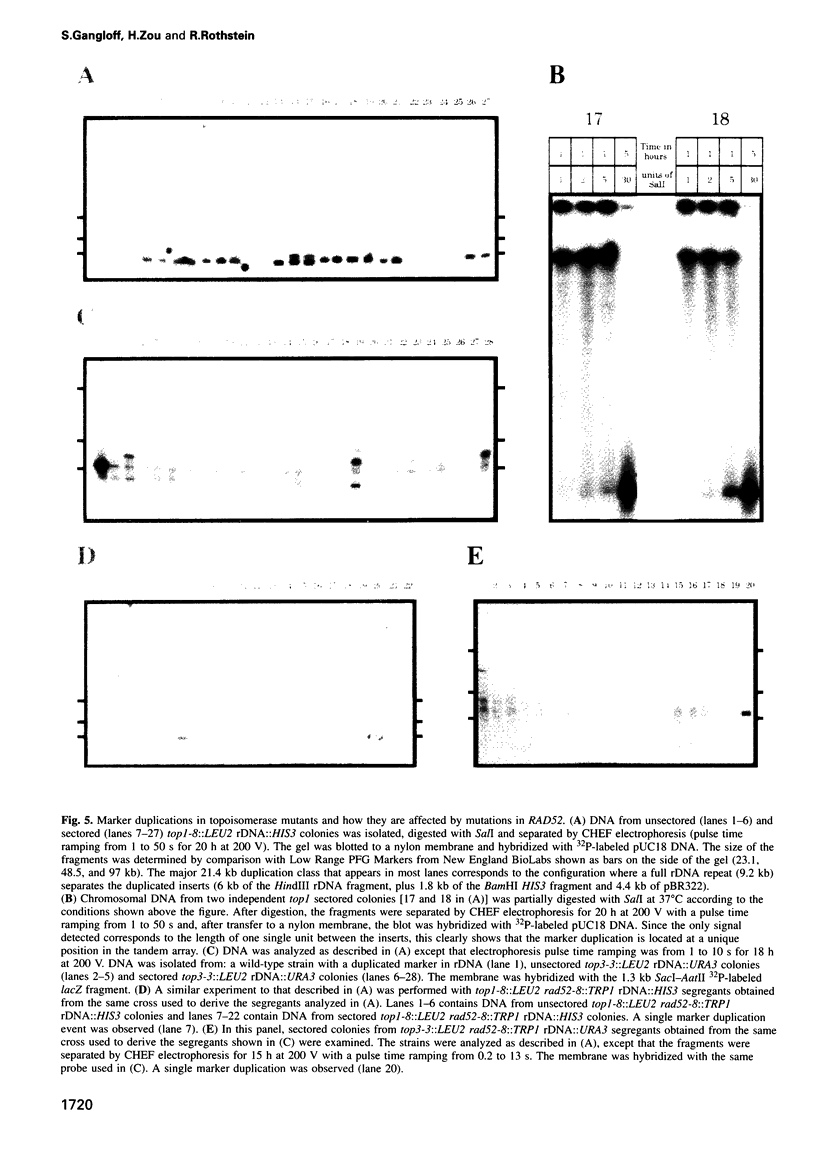
Abstract
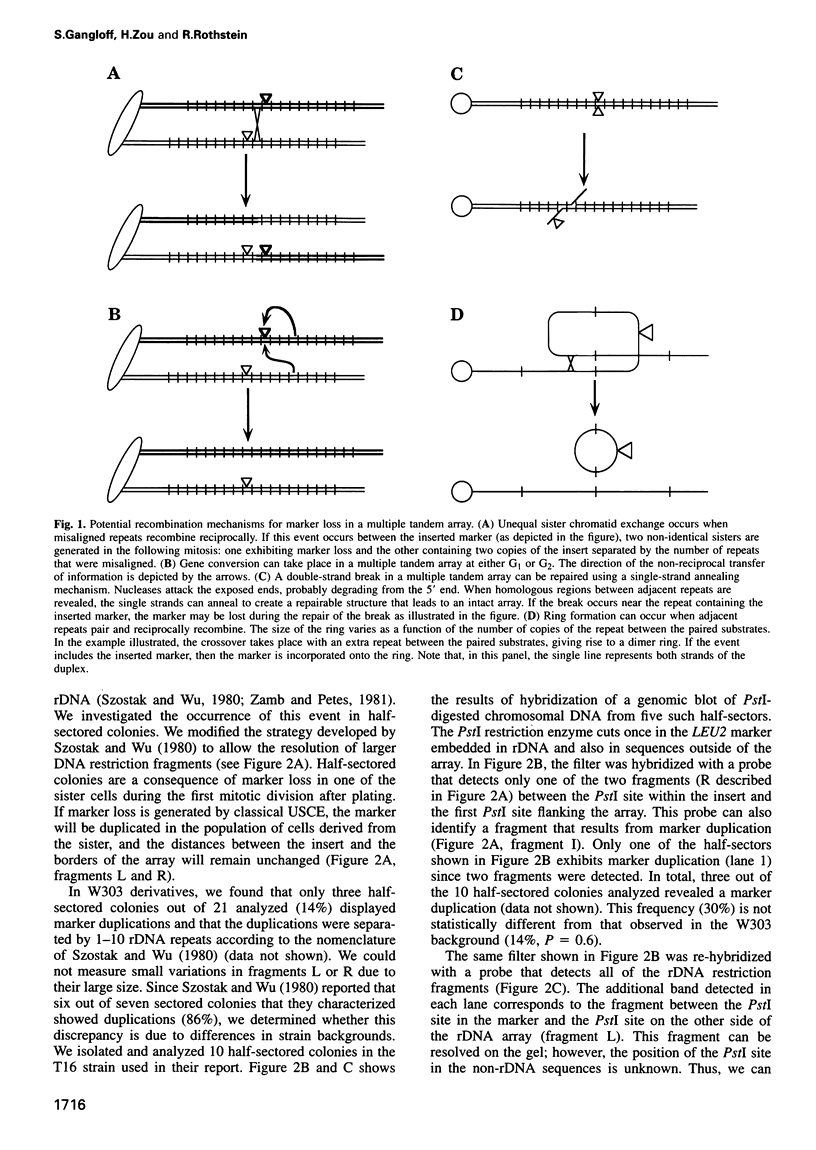
The genomic stability of the rDNA tandem array in yeast is tightly controlled to allow sequence homogenization and at the same time prevent deleterious rearrangements. In our study, we show that gene conversion, and not unequal sister chromatid exchange, is the predominant recombination mechanism regulating the expansion and contraction of the rDNA array. Furthermore, we found that RAD52, which is essential for gene conversion, is required for marker duplication stimulated in the absence of the two yeast type I topoisomerases. Our results have implications for the mechanisms regulating genomic stability of repetitive sequence families found in all eukaryotes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adzuma K., Ogawa T., Ogawa H. Primary structure of the RAD52 gene in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2735–2744. doi: 10.1128/mcb.4.12.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis A. M., Rothstein R. A defect in mismatch repair in Saccharomyces cerevisiae stimulates ectopic recombination between homeologous genes by an excision repair dependent process. Genetics. 1990 Nov;126(3):535–547. doi: 10.1093/genetics/126.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendixen C., Sunjevaric I., Bauchwitz R., Rothstein R. Identification of a mouse homologue of the Saccharomyces cerevisiae recombination and repair gene, RAD52. Genomics. 1994 Sep 1;23(1):300–303. doi: 10.1006/geno.1994.1503. [DOI] [PubMed] [Google Scholar]
- Bezzubova O. Y., Schmidt H., Ostermann K., Heyer W. D., Buerstedde J. M. Identification of a chicken RAD52 homologue suggests conservation of the RAD52 recombination pathway throughout the evolution of higher eukaryotes. Nucleic Acids Res. 1993 Dec 25;21(25):5945–5949. doi: 10.1093/nar/21.25.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill S. J., DiNardo S., Voelkel-Meiman K., Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. 1987 Mar 26-Apr 1Nature. 326(6111):414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- Brill S. J., Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988 Jul 29;54(3):403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- Caskey C. T., Pizzuti A., Fu Y. H., Fenwick R. G., Jr, Nelson D. L. Triplet repeat mutations in human disease. Science. 1992 May 8;256(5058):784–789. doi: 10.1126/science.1589758. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Sniegowski P., Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994 Sep 15;371(6494):215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Christman M. F., Dietrich F. S., Fink G. R. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell. 1988 Nov 4;55(3):413–425. doi: 10.1016/0092-8674(88)90027-x. [DOI] [PubMed] [Google Scholar]
- Fu Y. H., Kuhl D. P., Pizzuti A., Pieretti M., Sutcliffe J. S., Richards S., Verkerk A. J., Holden J. J., Fenwick R. G., Jr, Warren S. T. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991 Dec 20;67(6):1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Fu Y. H., Pizzuti A., Fenwick R. G., Jr, King J., Rajnarayan S., Dunne P. W., Dubel J., Nasser G. A., Ashizawa T., de Jong P. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992 Mar 6;255(5049):1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- Gangloff S., Lieber M. R., Rothstein R. Transcription, topoisomerases and recombination. Experientia. 1994 Mar 15;50(3):261–269. doi: 10.1007/BF01924009. [DOI] [PubMed] [Google Scholar]
- Gangloff S., McDonald J. P., Bendixen C., Arthur L., Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994 Dec;14(12):8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley H. G., Brook J. D., Rundle S. A., Crow S., Reardon W., Buckler A. J., Harper P. S., Housman D. E., Shaw D. J. Expansion of an unstable DNA region and phenotypic variation in myotonic dystrophy. Nature. 1992 Feb 6;355(6360):545–546. doi: 10.1038/355545a0. [DOI] [PubMed] [Google Scholar]
- Huxley C., Green E. D., Dunham I. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 1990 Aug;6(8):236–236. doi: 10.1016/0168-9525(90)90190-h. [DOI] [PubMed] [Google Scholar]
- Jackson J. A., Fink G. R. Gene conversion between duplicated genetic elements in yeast. Nature. 1981 Jul 23;292(5821):306–311. doi: 10.1038/292306a0. [DOI] [PubMed] [Google Scholar]
- Jinks-Robertson S., Michelitch M., Ramcharan S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Jul;13(7):3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. P., Warner J. R. Unusual enhancer function in yeast rRNA transcription. Mol Cell Biol. 1989 Nov;9(11):4986–4993. doi: 10.1128/mcb.9.11.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R. A., Wang J. C. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J Mol Biol. 1989 Jul 20;208(2):257–267. doi: 10.1016/0022-2836(89)90387-2. [DOI] [PubMed] [Google Scholar]
- Klar A. J., Strathern J. N., Hicks J. B. A position-effect control for gene transposition: state of expression of yeast mating-type genes affects their ability to switch. Cell. 1981 Aug;25(2):517–524. doi: 10.1016/0092-8674(81)90070-2. [DOI] [PubMed] [Google Scholar]
- Klein H. L. Different types of recombination events are controlled by the RAD1 and RAD52 genes of Saccharomyces cerevisiae. Genetics. 1988 Oct;120(2):367–377. doi: 10.1093/genetics/120.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein H. L. Lack of association between intrachromosomal gene conversion and reciprocal exchange. 1984 Aug 30-Sep 5Nature. 310(5980):748–753. doi: 10.1038/310748a0. [DOI] [PubMed] [Google Scholar]
- Klein H. L., Petes T. D. Intrachromosomal gene conversion in yeast. Nature. 1981 Jan 15;289(5794):144–148. doi: 10.1038/289144a0. [DOI] [PubMed] [Google Scholar]
- Kuhl D. P., Caskey C. T. Trinucleotide repeats and genome variation. Curr Opin Genet Dev. 1993 Jun;3(3):404–407. doi: 10.1016/0959-437x(93)90112-3. [DOI] [PubMed] [Google Scholar]
- Kulkens T., van der Sande C. A., Dekker A. F., van Heerikhuizen H., Planta R. J. A system to study transcription by yeast RNA polymerase I within the chromosomal context: functional analysis of the ribosomal DNA enhancer and the RBP1/REB1 binding sites. EMBO J. 1992 Dec;11(12):4665–4674. doi: 10.1002/j.1460-2075.1992.tb05568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada A. R., Wilson E. M., Lubahn D. B., Harding A. E., Fischbeck K. H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991 Jul 4;352(6330):77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- Larionov V. L., Grishin A. V., Smirnov M. N. 3 micron DNA - an extrachromosomal ribosomal DNA in the yeast Saccharomyces cerevisiae. Gene. 1980 Dec;12(1-2):41–49. doi: 10.1016/0378-1119(80)90014-1. [DOI] [PubMed] [Google Scholar]
- Lehrman M. A., Goldstein J. L., Russell D. W., Brown M. S. Duplication of seven exons in LDL receptor gene caused by Alu-Alu recombination in a subject with familial hypercholesterolemia. Cell. 1987 Mar 13;48(5):827–835. doi: 10.1016/0092-8674(87)90079-1. [DOI] [PubMed] [Google Scholar]
- Levin N. A., Bjornsti M. A., Fink G. R. A novel mutation in DNA topoisomerase I of yeast causes DNA damage and RAD9-dependent cell cycle arrest. Genetics. 1993 Apr;133(4):799–814. doi: 10.1093/genetics/133.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskay R. M., Stachelek J. L. Information transfer between duplicated chromosomal sequences in mammalian cells involves contiguous regions of DNA. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1802–1806. doi: 10.1073/pnas.83.6.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone R. E., Esposito R. E. The RAD52 gene is required for homothallic interconversion of mating types and spontaneous mitotic recombination in yeast. Proc Natl Acad Sci U S A. 1980 Jan;77(1):503–507. doi: 10.1073/pnas.77.1.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack W. T., Thompson C. B. Chicken IgL variable region gene conversions display pseudogene donor preference and 5' to 3' polarity. Genes Dev. 1990 Apr;4(4):548–558. doi: 10.1101/gad.4.4.548. [DOI] [PubMed] [Google Scholar]
- Muris D. F., Bezzubova O., Buerstedde J. M., Vreeken K., Balajee A. S., Osgood C. J., Troelstra C., Hoeijmakers J. H., Ostermann K., Schmidt H. Cloning of human and mouse genes homologous to RAD52, a yeast gene involved in DNA repair and recombination. Mutat Res. 1994 Nov;315(3):295–305. doi: 10.1016/0921-8777(94)90040-x. [DOI] [PubMed] [Google Scholar]
- Muscarella D. E., Vogt V. M. A mobile group I intron from Physarum polycephalum can insert itself and induce point mutations in the nuclear ribosomal DNA of saccharomyces cerevisiae. Mol Cell Biol. 1993 Feb;13(2):1023–1033. doi: 10.1128/mcb.13.2.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K., Hall B. D., Astell C., Smith M. Physical analysis of mating-type loci in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):961–981. doi: 10.1101/sqb.1981.045.01.113. [DOI] [PubMed] [Google Scholar]
- Ostermann K., Lorentz A., Schmidt H. The fission yeast rad22 gene, having a function in mating-type switching and repair of DNA damages, encodes a protein homolog to Rad52 of Saccharomyces cerevisiae. Nucleic Acids Res. 1993 Dec 25;21(25):5940–5944. doi: 10.1093/nar/21.25.5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozenberger B. A., Roeder G. S. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol Cell Biol. 1991 Mar;11(3):1222–1231. doi: 10.1128/mcb.11.3.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D. Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell. 1980 Mar;19(3):765–774. doi: 10.1016/s0092-8674(80)80052-3. [DOI] [PubMed] [Google Scholar]
- Petes T. D. Yeast ribosomal DNA genes are located on chromosome XII. Proc Natl Acad Sci U S A. 1979 Jan;76(1):410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Machin N., Stahl F. W. A DNA double chain break stimulates triparental recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6225–6229. doi: 10.1073/pnas.86.16.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M. A. Genetic control of radiation sensitivity in Saccharomyces cerevisiae. Genetics. 1969 Jul;62(3):519–531. doi: 10.1093/genetics/62.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. M., Axel R. Gene amplification and gene correction in somatic cells. Cell. 1982 May;29(1):109–119. doi: 10.1016/0092-8674(82)90095-2. [DOI] [PubMed] [Google Scholar]
- Seifert H. S., Chen E. Y., So M., Heffron F. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Feb;83(3):735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980 Apr 3;284(5755):426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989 Feb 24;56(4):619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Wallis J. W., Chrebet G., Brodsky G., Rolfe M., Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989 Jul 28;58(2):409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases: why so many? J Biol Chem. 1991 Apr 15;266(11):6659–6662. [PubMed] [Google Scholar]
- Warburton P. E., Waye J. S., Willard H. F. Nonrandom localization of recombination events in human alpha satellite repeat unit variants: implications for higher-order structural characteristics within centromeric heterochromatin. Mol Cell Biol. 1993 Oct;13(10):6520–6529. doi: 10.1128/mcb.13.10.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993 May;134(1):63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J. W., Maloney D. H., Fogel S. Unequal crossing-over and gene conversion at the amplified CUP1 locus of yeast. Mol Gen Genet. 1990 Jul;222(2-3):304–310. doi: 10.1007/BF00633833. [DOI] [PubMed] [Google Scholar]
- Willis K. K., Klein H. L. Intrachromosomal recombination in Saccharomyces cerevisiae: reciprocal exchange in an inverted repeat and associated gene conversion. Genetics. 1987 Dec;117(4):633–643. doi: 10.1093/genetics/117.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]