Abstract
Many plant-pathogenic xanthomonads rely on Transcription Activator-Like (TAL) effectors to colonize their host. This particular family of type III effectors functions as specific plant transcription factors via a programmable DNA-binding domain. Upon binding to the promoters of plant disease susceptibility genes in a sequence-specific manner, the expression of these host genes is induced. However, plants have evolved specific strategies to counter the action of TAL effectors and confer resistance. One mechanism is to avoid the binding of TAL effectors by mutations of their DNA binding sites, resulting in resistance by loss-of-susceptibility. This article reviews our current knowledge of the susceptibility hubs targeted by Xanthomonas TAL effectors, possible evolutionary scenarios for plants to combat the pathogen with loss-of-function alleles, and how this knowledge can be used overall to develop new pathogen-informed breeding strategies and improve crop resistance.
Keywords: Xanthomonas, plant disease susceptibility S genes, hubs, TAL effectors, agricultural biotechnology, loss-of-function alleles
Introduction
Xanthomonas are a genus of plant pathogenic bacteria that cause devastating diseases on a wide range of hosts, leading to severe impact on yield quantity and quality of important crops such as rice, cassava, cotton, wheat, banana, mango, citrus, and cabbage (Schornack et al., 2013). In order to colonize their host, most of these pathogens rely on a type three secretion system (T3SS) specialized in the injection of virulence factors into the host cell. Also called type three effectors (T3Es), these proteins collectively promote the pathogen’s adaptation to specific host tissues, species, and genotypes. This adaptation includes suppression of plant immunity, nutrient acquisition, dispersal, or other virulence-related processes that benefit the pathogen. Upon translocation, T3Es localize to various subcellular compartments such as the plasma membrane or organelles.
The Transcription Activator-Like (TAL) effectors form a particular family of T3Es that act as bona fide plant transcription factors able to reprogram the host transcriptome following nuclear localization. They have a highly conserved structure that is modular and characterized by an N-terminal type three secretion signal, a C-terminal region bearing two to three nuclear localization signals and an acidic transcription activation domain. In between is a central region built of quasi-identical tandem direct repeats of 33–35 amino-acids forming a unique type of DNA-binding domain. Each repeat folds into a super-helical structure that wraps the DNA. Each repeat forms a two-helix bundle in which so-called repeat variable di-residues (RVDs) at positions 12 and 13 reside on the interhelical loop that projects into the DNA major groove. More precisely, RVDs mediate the binding of TAL effectors to the double-strand DNA in a sequence-specific manner with residue 12 stabilizing the overall interaction while residue 13 specifically contacts the DNA base (reviewed in Mak et al., 2013). Functional differences between TAL effectors are therefore mainly due to the nature of the string of RVDs that determine the sequence of the so-called TAL effector binding element (EBE) in host promoters. TAL effectors can be found in most Xanthomonas species and related proteins are also present in Ralstonia solanacearum (RipTALs; de Lange et al., 2013) and Burkholderia rhizoxinica (Bats; Juillerat et al., 2014; Lange et al., 2014).
Several Xanthomonas TAL effectors are known to play an essential role during infection, i.e., the deletion of the genes encoding them leads to reduced disease. Plant genes may contribute to disease susceptibility in different ways, notably by favoring pathogen attraction and attachment to the host, controlling the establishment of a favorable environment for host tissue penetration, colonization, or pathogen dispersal (reviewed by Lapin and Van den Ackerveken, 2013). In the case of plant pathogenic bacteria our knowledge on the diversity and function of disease determinants is still scarce. However, recent breakthroughs in the field of TAL effector biology are significantly improving our understanding of bacterial disease processes, thereby fostering innovative disease control strategies. In this review, we analyze host–pathogen interactions over evolutionary time using the popular video game Mortal Kombat1, in which combatants strike and counter to survive an epic battle, as a metaphor. We initially focus on the mechanisms underlying susceptibility caused by TAL effectors. Then we explore the strategies that plants have evolved to counter the action of TAL effectors, with emphasis on the strategy of “resistance through loss-of-susceptibility” achieved by avoiding the binding of TAL effectors to host DNA using mutations of their binding sites. Finally, we discuss how, in light of these findings, natural or engineered resistance can be bred to control TAL effector-mediated diseases.
Round 1: Bacteria Attack Using TAL Effectors to Induce Susceptibility Genes
Transcription Activator-Like effectors are found in many plant pathogenic Xanthomonas species where they significantly contribute to disease development but their exact function as minor or major virulence factors has been disentangled for only a few of them (Boch and Bonas, 2010). The number of TAL effectors genes contained in the genome of a Xanthomonas strain is highly variable between strains. For example, strains of Xanthomonas oryzae pv. oryzae (Xoo) contain from 9 up to more than 15 different TAL effectors. Yet it has been shown that only one or two of them play an essential role in pathogenicity and encode major virulence factors (Yang and White, 2004). For these strains the mutation of the corresponding genes leads to no development of the disease (Yang and White, 2004). The genes targeted by these TAL effectors are called (S) susceptibility genes because their induction is essential to the complete development of the disease. Unlike resistance (R) genes that are usually expressed only in the presence of the pathogen, most of the known S genes play roles in plant development, and are exploited by pathogens through their overexpression during the infection. The discovery of the TAL effector-DNA binding code combined with transcriptomic data has led to the identification of a S gene list for different Xanthomonas/host interactions (Table 1). Notably, many of these S genes encode either transporters (sugar or sulfate) or transcription factors, and their induction is hypothesized to facilitate bacterial colonization and symptom development.
Table 1.
Plant susceptibility or candidate S genes targeted by Xanthomonas Transcription Activator-Like (TAL) effectors.
| Target function | Target name | TAL name | Specie | Host | Symptoms | Effect | Reference | Loss S alleles | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Transcription factor | UPA20 | AvrBs3 | Xav | Pepper | Cell hypertrophy | Increased egress to the leaf surface | Kay et al. (2007) | ||
| CsLOB1 | PthA series | Xcc | Citrus | Cell expansion | Canker | Hu et al. (2014), Li et al. (2014) | |||
| PthB | Xca | Citrus | Cell expansion | Canker | Al-Saadi et al. (2007), Hu et al. (2014) | ||||
| PthC | Xca | Citrus | Cell expansion | Canker | |||||
| TFX1 | PthXo6 | Xoo | Rice | Water soaking | Increased growth and lesions | Sugio et al. (2007) | |||
| TFIIAγI | PthXo7 | Xoo | Rice | Water soaking | Increased growth and lesions | ||||
| Sugar Transporter | CsSWEET1 | PthA series | Xcc | Citrus | Unknown | Unknown | Hu et al. (2014) | ||
| OsSWEET11 | PthXo1 | Xoo | Rice | Water soaking | Increased growth and lesions | Chu et al. (2006), Yang et al. (2006) | xa13 | Chu et al. (2006), Yang et al. (2006) | |
| OsSWEET13 | PthXo2 | Xoo | Rice | Water soaking | Increased growth and lesions | Zhou et al. (2015) | xa25 | Liu et al. (2011), Zhou et al. (2015) | |
| OsSWEET14 | PthXo3 | Xoo | Rice | Water soaking | Increased growth and lesions | Yang and White (2004) | TALEN-based disruption | Li et al. (2012) | |
| AvrXa7 | Xoo | Rice | Water soaking | Increased growth and lesions | Antony et al. (2010) | ||||
| Tal5 | Xoo | Rice | Water soaking | Increased growth and lesions | Streubel et al. (2013) | ||||
| TalC | Xoo | Rice | Water soaking | Increased growth and lesions | Yu et al. (2011) | ||||
| MeSWEET10a | TAL20 | Xam | Cassava | Unknown | Cohn et al. (2014) | ||||
| Sulfate transporter | SULTR3;6 | Tal2g | Xoc | Rice | Water soaking | Increased lesions and egress to the leaf surface | Cernadas et al. (2014) | ||
| miRNA stability | OsHEN1 | PthXo8 | Xoo | Rice | Not reported | Moscou and Bogdanove (2009) | |||
| Tal1C | Xoc | Rice | None detected |
Xcv, Xanthomonas axonopodis pv. vesicatoria; Xcc, X. citri pv. citri; Xca, X. citri pv. aurantifolii; Xoo, X. oryzae pv. oryzae; Xoc, X. oryzae pv. oryzicola; Xam, X. axonopodis pv. manihotis.
The SWEET family of sugar transporters represents the best characterized group of S genes induced by TAL effectors (Chen, 2014). In rice, these include OsSWEET11 (a.k.a Xa13/xa13) activated by PthXo1 from strain Xoo PXO99A, OsSWEET13 (a.k.a Xa25/xa25) activated by PthXo2 from Xoo strains JXO1A and MAFF311018, and OsSWEET14 activated by AvrXa7, PthXo3, TalC, and Tal5 from Xoo strains PXO86, PXO61, BAI3, and MAI1, respectively (Yang and White, 2004; Chu et al., 2006; Antony et al., 2010; Yu et al., 2011; Streubel et al., 2013; Zhou et al., 2015). They are all members of clade III of the SWEET gene family. It was shown that in rice, all five members of clade III can act as a major S gene (Li et al., 2013; Streubel et al., 2013). Interestingly, related genes are also induced by Xanthomonas TAL effectors in other hosts. For instance MeSWEET10a and CsSWEET1 are, respectively, induced by TAL20 from X. axonopodis pv. manihotis (Xam) in cassava and the PthA series from X. citri ssp. citri in citrus (Cohn et al., 2014; Hu et al., 2014). Xam-mediated induction of MeSWEET10a which also belongs to clade III is essential for full development of watersoaking as infiltration of strain Xam668ΔTAL20 leads to reduced symptoms (Cohn et al., 2014). Intriguingly the induction of MeSWEET10a is necessary for symptom expansion but not for bacterial growth. In contrast, CsSWEET1 belongs to clade I and no function for citrus canker has been demonstrated to date (Hu et al., 2014). Similarly, AvrBs3 from X. axonopodis pv. vesicatoria induces UPA16 from pepper which also belongs to the SWEET family but there is no evidence for this gene to play a role for bacterial spot of pepper and tomato (Kay et al., 2009).
At least two members of the SWEET family in rice were shown to play a crucial role in plant development. Homozygous lines containing a T-DNA insertion in OsSWEET14 display delayed growth and seeds smaller than the wild type plant (Antony et al., 2010), and plants silenced for OsSWEET11 have reduced fertility (Yang et al., 2006). They encode sugar transporters that allow the efflux of carbohydrates from the cell to the intercellular space. Thus, their induction could make carbohydrates easily available for bacteria (Chen et al., 2012), though to date it has not been clearly demonstrated that these carbohydrates help bacterial growth directly. SWEET genes may play a role in multiple plant pathogen interactions (Lemoine et al., 2013). VvSWEET4 in grapevine is up regulated during the interaction with the gray mold fungus Botrytis cinerea and a knockout mutant in Arabidopsis for the orthologous gene AtSWEET4 becomes less susceptible to this pathogen (Chong et al., 2014). Also in Arabidopsis, differential expression of SWEET genes is observed after infection by Pseudomonas syringae pv. tomato, Golovinomyces cichoracearum, and Plamodiophora brassicae (Siemens et al., 2006; Chen et al., 2010) suggesting that numerous pathogens target the sugar transport mechanisms of the host upon infection.
While SWEET genes are essential for the vascular pathogen Xoo to cause Bacterial Blight in rice, they are curiously not targeted by the closely related non-vascular pathovar X. oryzae pv. oryzicola (Xoc) that is responsible for bacterial leaf streak of rice. It is then tempting to speculate that induction of S genes by TAL effectors may explain tissue specificity to some extent. However, transformation of Xoc with OsSWEET-inducing TAL effectors from Xoo does not allow vascular colonization by Xoc (Verdier et al., 2012). Nevertheless, the role of TAL effectors in defining host-ranges and colonization mechanisms is still a field worth exploring. Recently, the systematic mutagenesis of each TAL effector gene of Xoc strain BLS256 completed with the functional analysis of their potential targets unmasked a new S gene specifically required for leaf streak. OsSULTR3;6 encodes a sulfate transporter targeted by Tal2g and its induction is necessary for lesion expansion and bacterial exudation but not for bacterial growth (Cernadas et al., 2014). How this sulfate transporter contributes to disease has yet to be elucidated. One hypothesis is that its induction modifies the redox potential in a favorable manner for symptom establishment and bacterial expansion. Some orthologous genes have been identified to play a similar role in other pathosystems and in establishment of symbiosis (Cernadas et al., 2014).
Another group of functionally related genes targeted by TAL effectors are transcription factors. The first identified was UPA20 in pepper. This transcription factor induced by AvrBs3 from X. axonopodis pv. vesicatoria belongs to the bHLH family and trans-activates the secondary target UPA7 (Kay et al., 2007). The expression of UPA7 leads to enlargement of mesophyll cells of pepper leaves that could promote propagation of bacteria (Kay et al., 2007). Another transcription factor acting as a major susceptibility gene is CsLOB1, required for pustule formation during citrus bacterial canker. This gene is targeted by five different TAL effectors, including PthA4, PthAw, and PthA∗ from X. citri ssp. citri and PthB and PthC from X. citri ssp. aurantifolii (Hu et al., 2014). Interestingly an Arabidopsis protein belonging to the LOB family is described as a susceptibility gene for the fungal pathogen Fusarium oxysporum (Thatcher et al., 2009). The specific functions in development of CsLOB1 are not known but it was shown that proteins with LOB domain are responsive to numerous phytohormones, play a role in anthocyanin and nitrogen metabolism and are involved in the regulation of lateral organ development (Majer and Hochholdinger, 2011; Gendron et al., 2012). OsTFXI a bZIP transcription factor and OsTFIIAγ1 a general transcription factor, are targeted, respectively, by PthXo6 and PthXo7 from Xoo strain PXO99A (Sugio et al., 2007). OsTFXI is particularly interesting as it is induced by all Xoo strains assessed to date (White and Yang, 2009). In contrast to the wild type, a transgenic plant overexpressing OsTFXI is fully susceptible to a pthXo6 mutant confirming that it is an S gene (Sugio et al., 2007). How OsTFXI promotes disease is not yet understood.
In summary, for Xoo species, the SWEET transporters are a major target in rice and among them OsSWEET14 is targeted by four different TALs from genetically and geographically distant Xoo strains. The SWEET genes are targeted by at least three other Xanthomonas species in three other hosts. There is undoubtedly an evolutionary convergence for the induction of this family inter species and intra pathovar. This convergence occurs also for the transcription factors. Five different TAL effectors with different RVD content are able to induce CsLOB1, which seems to be the single susceptibility gene induced by both X. citri pathovars for the establishment of citrus canker (Hu et al., 2014). Thus, convergence seems to be a good indicator for the discovery of S genes (Figure 1). A similar behavior of convergence toward a selected group of targets in the host has also been found for non-TAL effectors from different pathogen groups in Arabidopsis (Mukhtar et al., 2011). In addition, computational predictions suggest that targeting of functional “hubs” is indeed a common feature for TAL effectors (Perez-Quintero et al., 2013).
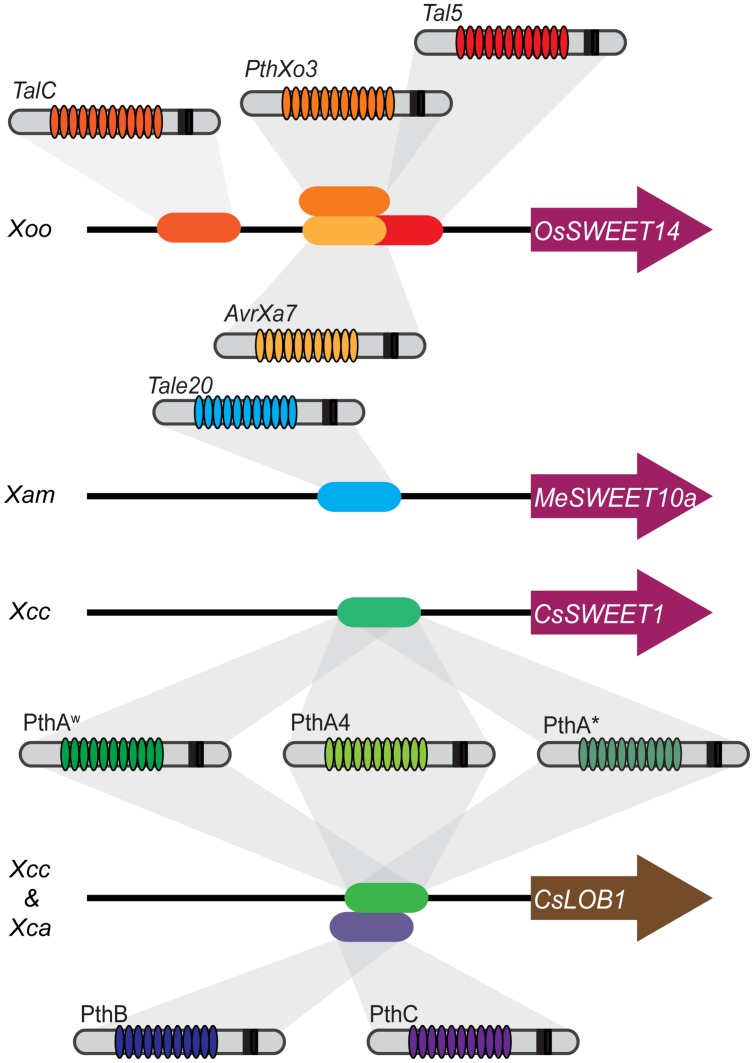
FIGURE 1.
Functional convergence in bacterial Transcription Activator-Like (TAL) effectors and plant targets. Several susceptibility targets of Xanthomonas oryzae pv. oryzae belong to Clade III of the SWEET family. Four TAL effectors from diverse Xoo strains belonging to different lineages and from distinct geographical origins target OsSWEET14. TalC and Tal5 from the two Xoo African strains BAI3 (Burkina Faso) and MAI1 (Mali) bind to two distinct effector binding elements (EBEs). In addition, the OsSWEET14 promoter is targeted by two Xoo Philippine strains, PXO86 and PXO61 through the TAL effectors AvrXa7 and PthXo3. The corresponding EBEs partially overlap with the Tal5 EBE. The repeat variable di-residue (RVD) arrays and the number of repeats (not represented in the figure) for these three TAL effectors are significantly different, highlighting evolutionary convergence. Not shown in the figure is the case of the resistance gene Xa7 that recognizes AvrXa7 but not PthXo3 that is slightly divergent from AvrXa7. A similar case of convergence is observed for the induction of CsLOB1 which is targeted by PthB and PthC from X. citri ssp. aurantifolii and the PthA series from X. citri ssp. citri. PthA4, PthAw, and PthA∗ (but not PthB and PthC) induce CsSWEET1 belonging to the Clade I of the SWEET family. However, a role in disease development has not been shown. A third species, X. axonopodis pv. manihotis, targets through TAL20 a member of the SWEET family, MeSWEET10a, that belongs to Clade III and acts as a susceptibility gene.
Taking this into account, new potential S genes arise, including for example OsHEN1 in rice. OsHEN1 encodes a methyl transferase and is the only common target of Xoo and Xoc via, respectively, PthXo8 and Tal1c (Moscou and Bogdanove, 2009). Although a tal1c mutant showed no reduction in virulence (Cernadas et al., 2014), and data for PthXo8 are yet to be reported, the convergence of different pathovars onto this target may suggest an important role in the respective diseases. OsHEN1 plays important roles in different physiological processes through the stabilization of small RNAs, introducing the exciting question of how induction of this gene aids in bacterial colonization. On the other hand, there are some TAL effector for which a role in pathogenicity has been shown but the target remains unknown or unconfirmed (Yang et al., 1994; Castiblanco et al., 2012; Munoz-Bodnar et al., 2014).
Round 2: Plant Dodges Bacteria’s TAL Effectors Through Mutation in Promoters of S Genes
The evolutionary convergence of TAL effector activity on a restricted set of key host genes of physiological importance (Figure 1) implies that some allelic variants (those that can avoid binding on the effector) could confer broad-spectrum resistance to Xanthomonas. To date, EBE variants that abolish TAL effector binding have been found in the promoter of two major susceptibility genes: OsSWEET11 and OsSWEET13, resulting in the recessive resistance alleles known, respectively, as xa13 and xa25 (Chu et al., 2006; Yang et al., 2006; Liu et al., 2011; Zhou et al., 2015).
The promoter of the susceptible allele of OsSWEET11:Xa13 but not of xa13 is induced directly by the TAL effector PthXo1 from PXO99A (Yang et al., 2006; Romer et al., 2010). Varieties containing xa13 are resistant to PXO99A and the recessive resistance observed is not characterized by the typical hypersensitive response phenotype as it results only on the loss of the induction of a gene essential to disease development (Yuan et al., 2009, 2011). There is a collection of rice varieties naturally presenting an xa13 allele. All these alleles differ from the Xa13 genotype by insertion, deletion, or substitution in the PthXo1 EBE (Romer et al., 2010). Interestingly a single substitution in the second nucleotide of the EBE is sufficient to avoid induction of OsSWEET11 by PthXo1. In the case of OsSWEET13, the susceptible allele Xa25 but not the race specific recessive resistance gene xa25 is induced by the Xoo strain PXO339 (Liu et al., 2011). Transformation of Xa25 in a resistant variety with a xa25 genotype led to susceptibility to PXO339 (Liu et al., 2011). Recently it was shown that PthXo2 directly induces Xa25. Xa25 differs from xa25 by a one nucleotide deletion in the EBE of PthXo2. Promoter mutation generated by CRISPR/Cas9 technology confirmed that OsSWEET13 induction is essential for strains that depend on PthXo2 to lead to disease susceptibility (Zhou et al., 2015).
For OsSWEET14, no naturally occurring resistance alleles have been reported yet, but they have been genetically engineered. Li et al. (2012) generated loss of susceptibility alleles altered in the box targeted by AvrXa7 and PthXo3 using the TALEN strategy, an approach based on TAL effector binding domains coupled with nucleases (reviewed in Bogdanove and Voytas, 2011; Carroll, 2014). They showed that a deletion of 4 bp is sufficient for loss-of-susceptibility to AvrXa7 and PthXo3. As discussed below Round 4), this genome editing strategy could be extensively used to develop resistant varieties to Xanthomonas in various systems.
It is expected that more naturally occurring loss-of-susceptibility alleles can be found by promoter screening of S genes in different host germplasm collections (see Round 4). An example of a resource for this type of screening is the recently released SNP data from rice obtained from the sequencing of the genomes of 3000 rice varieties, available in the SNPseek database (Alexandrov et al., 2015). Surprisingly when using this data to look for SNPs in a set of known EBEs in rice (not all from S genes) very few SNPs are found within the EBEs (Table 2). It is worth noting, that no indels are included in the SNPseek data and that all the SNPs were obtained from mapping NGS reads onto only one reference genome (Alexandrov et al., 2015). So it is possible that more SNPs in these regions will be found using different tools and datasets.
Table 2.
SNPs found in 3000 rice genomes at the EBEs of known TAL effector targets in rice.
| TAL | Xo Strain | Reference | Gene | Effector binding element (EBE) sequence | SNPs EBE | SNPs promoter | SNPs gene |
|---|---|---|---|---|---|---|---|
| PthXo3 | PXO61 | Yang and White (2004) | LOC_Os11g31190 (SWEET14) | TATATAAACCCCCTCCAACCAGGTGCTAAG | 2 | 20 | 28 |
| AvrXa7 | PXO86 | Antony et al. (2010) | LOC_Os11g31190 (SWEET14) | TATATAAACCCCCTCCAACCAGGTGCT | 2 | 20 | 28 |
| Tal3b | BLS256 | Cernadas et al. (2014) | LOC_Os07g36430 | TATAAGAAGCCCTCACTCG | 1 | 22 | 3 |
| Tal9b | BLS256 | LOC_Os01g51040 | TAGAAACTGCTCTTCCT | 1 | 2 | 22 | |
| TalC | BAI3 | Yu et al. (2011) | LOC_Os11g31190 (SWEET14) | CATGCATGTCAGCAGCTGGTCAT | 0 | 20 | 28 |
| Tal5 | MAI1 | Streubel et al. (2013) | LOC_Os11g31190 (SWEET14) | TAAGCTCATCAAGCCTTCA | 0 | 20 | 28 |
| PthXo6 | PXO99A | Sugio et al. (2007) | LOC_Os09g29820 (OsTFX1) | TATAAAAGGCCCTCACCAACCCAT | 0 | 5 | 3 |
| PthXo7 | PXO99A | LOC_Os01g73890 (OsTFIIAγ1) | TATAATCCCCAAATCCCCTCCTC | 0 | 8 | 8 | |
| PthXo1 | PXO99A | Yang et al. (2006) | LOC_Os08g42350 (SWEET11) | TGCATCTCCCCCTACTGTACACCAC | 0 | 8 | 17 |
| Tal9A | PXO99A | Moscou and Bogdanove (2009) | LOC_Os07g06970 (HEN1) | TCCCTTCCCTAAACCCCACTT | 0 | 4 | 20 |
| Tal1c | BLS256 | Moscou and Bogdanove (2009) | LOC_Os07g06970 (HEN1) | TCCCCCTCGCTTCCCTT | 0 | 4 | 20 |
| Tal2c | BLS256 | Cernadas et al. (2014) | LOC_Os03g03034 | TCCGGCCCCTCTCCCCCCGCCACCTGAC | 0 | 5 | 25 |
| Tal2d | BLS256 | LOC_Os04g49194 | TATTCCCTCGCGTGATC | 0 | 18 | 78 | |
| Tal3b | BLS256 | LOC_Os02g34970 | TATAAGTAGCACTCGCTCA | 0 | 4 | 9 | |
| Tal3b | BLS256 | LOC_Os05g27590 | TATAAATAGCCCTCACTCT | 0 | 3 | 0 | |
| Tal4a | BLS256 | LOC_Os03g37840 | TATATATCTCGGTCAGGCCAGGCCCCT | 0 | 12 | 28 | |
| Tal3c | BLS256 | LOC_Os03g07540 | TACACGTTCCCTCCACC | 0 | 3 | 9 | |
| Tal3c | BLS256 | LOC_Os02g47660 | TACACATTAGCTACCAT | 0 | 2 | 11 | |
| Tal6 | BLS256 | LOC_Os09g29100 | TACAAAGAGAACGCATCCCCC | 0 | 11 | 24 | |
| Tal6 | BLS256 | LOC_Os12g42970 | TAAAAGCGAAACCCTCCCTCC | 0 | 16 | 8 | |
| Tal5a | BLS256 | LOC_Os02g15290 | TAAATCCCCCCTGCACAGGAAAGCT | 0 | 31 | 2 | |
| Tal2g | BLS256 | LOC_Os01g52130 | TGGCCCGTAGCCTCTCCT | 0 | 0 | 89 | |
| Tal2g | BLS256 | LOC_Os06g46500 | TGGCAAGTGACCTCAGCT | 0 | 9 | 14 | |
| Tal4c | BLS256 | LOC_Os06g37080 | TATAAAACCTGGACAAGCCTCTCT | 0 | 7 | 46 | |
| Tal4b | BLS256 | LOC_Os09g32100 | TGTATAGTATCCCCT | 0 | 14 | 14 |
SNPs found elsewhere in the promoter regions (1 kb upstream TSS) and the genes (including intrond) are also indicated. Position of the SNPs in the EBE sequences is indicated by bold and underlined nucleotides. SNP data were obtained from the Rice SNPseek database (Alexandrov et al., 2015).
Nonetheless, this raises the question: how common are loss-of-susceptibility alleles of S genes?
Gaining resistance through loss-of-susceptibility alleles requires the plant to mutate a region of the promoter to avoid binding of the TAL effector without altering physiologically important regulatory elements in the promoter. This might be particularly hard to achieve given that many TAL effectors seem to bind to crucial elements of the promoter. As a matter of fact, most of the known functional binding sites for TAL effectors are located in a region overlapping with that of core promoter elements of the gene (between 300 bp upstream and 200 bp downstream of the transcription start site). Indeed, TAL effectors seem to bind predominantly to functional motifs, such as the TATA box and the TC box (Grau et al., 2013; Pereira et al., 2014).
Regulatory elements in plant promoters are usually very short; median observed length of 8 bp and notoriously conserved (Reineke et al., 2011). Point mutations (transversions and transitions) in these elements may prevent binding of endogenous plant TFs and probably will not be selected unless they offer a great improvement in fitness (i.e, resistance to pathogenic bacteria). However, point mutations in an EBE could fail to prevent binding since the way TAL effectors bind to DNA allows for enough flexibility to tolerate “mismatches”. For instance some RVDs are able to indiscriminately accommodate different nucleotides, particularly those RVDs with weaker interactions with DNA (Streubel et al., 2012). Also, RVDs in the C-terminal region of TAL effectors seem to be particularly tolerant of mismatches (Meckler et al., 2013; Perez-Quintero et al., 2013). So a theoretical scenario for point mutations in an EBE to be effectively selected would be if they occur in a position normally bound by a strong-effect RVD in the N-terminal region. This seems to be the case of the xa13 alleles found in the rice varieties Tepa1, BJ1, Chinsurah 11484, Chinsurah 11760, and Chinsurah 50930 (Romer et al., 2010) where a single nucleotide substitution in the position matching the first RVD of PthXo1 is enough to prevent binding of the TAL effector. It would be interesting to test whether the SNPs found in the EBEs in Table 2 can prevent the binding of the corresponding TAL effector.
An alternative route to produce loss-of-susceptibility alleles is through deletion and insertion. Since cis-elements are often redundant, modular, and can exert their function regardless of their order in the promoter (Vedel and Scotti, 2011), indels and recombination events could in theory occur in some areas in the promoter between cis-elements or in areas including redundant elements without affecting gene regulation. These events could be more effective in defeating binding of TAL effectors since, with some exceptions (discussed under Round 3, below), TAL effectors do not tolerate gaps in their binding site as shown in numerous assays (Boch et al., 2009; Streubel et al., 2012; Richter et al., 2014). Indeed, many of the resistant xa13 alleles found in various rice varieties contain insertions or deletions in the promoter that disrupts the EBE found in susceptible varieties (Chu et al., 2006; Yang et al., 2006; Romer et al., 2010) and the resistant allele of xa25 is caused by a one-nucleotide deletion in the promoter in resistant varieties (Zhou et al., 2015). It is possible that indels in the promoters of S genes are a more common way of generating resistance, which would explain why such low variability is found in the SNPseek data (Table 2) that does not consider indels.
It is worth noting that there are other ways for the plant to defend itself against TAL effectors than through loss-of-susceptibility alleles, notably by using the TAL effector machinery against bacteria and “tricking” them into inducing resistance genes: the mechanism of the so-called executor genes is reviewed elsewhere (Boch et al., 2014). Additionally, plants can also directly recognize TAL effectors using “traditional” resistance genes, as is the case of Bs4 from tomato, encoding a NBS-LRR protein that directly recognizes the TAL effectors AvrBs4, Hax3, and Hax4 and subsequently activates defenses (Schornack et al., 2004; Kay et al., 2005).
Round 3: Bacteria Strike Back With New Weapons
On the bacterial side, resistance through loss-of-susceptibility could be overcome in a couple of ways. On one hand, it is possible that bacteria could overcome the effect of mismatches between TAL effectors and their cognate EBEs. For example, Richter et al. (2014) showed that some TAL effectors contain aberrant-length repeats that can “loop-out” when the TAL effector binds to the EBE thus allowing the recognition of target DNA sequences with a -1 nucleotide frame-shift, which allows for more flexibility in binding.
PthXo3 contains an aberrant repeat that apparently helps it to bind to the promoter of OsSWEET14 by disengaging from the interaction to shift the interaction register downstream. AvrXa7 also has an aberrant repeat that might function the same way at OsSWEET14, though in this case EBEs are predicted both with and without the register shift (Richter et al., 2014). Furthermore, by using artificial TAL effectors it was shown that indeed a TAL effector containing an aberrant repeat could target both the “susceptible” and “resistant” version of the promoter of the susceptibility gene Xa25 (OsSWEET13; Richter et al., 2014).
Another way for bacteria to counteract loss-of-susceptibility is by acquiring and harboring functionally redundant TAL effectors. Different TAL effectors could target dissimilar EBEs or variants of the promoter susceptibility gene. Alternatively several TAL effectors can activate redundant susceptibility genes. One example of the latter can be found in the Xoo strain MAFF311018 that contains AvrXa7 that can induce OsSWEET14, as well as PthXo2 that induce the susceptible allele of OsSWEET13 (Perez-Quintero et al., 2013; Zhou et al., 2015). TAL effectors could also diverge to avoid recognition by resistance genes as exemplified by the case of AvrXa7 and PthXo3, where, despite both effectors binding to overlapping EBEs in the OsSWEET14 promoter (Antony et al., 2010), PthXo3 presumably avoids Xa7-triggered resistance (White and Yang, 2009).
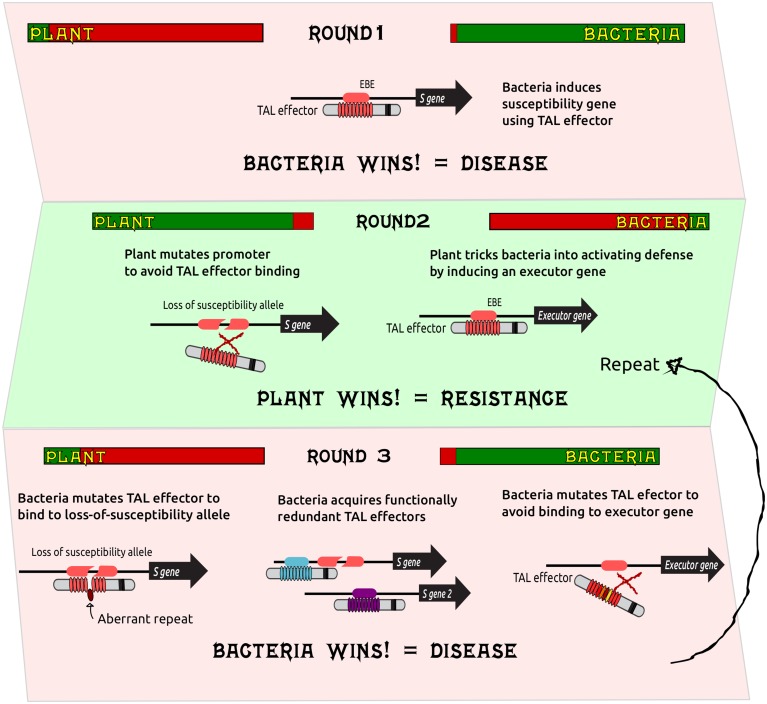
We have proposed a simplified model for the pathogen-host combat (Figure 2) where, in response to disease caused by the induction of S genes by TAL effectors (Round 1), plants evolve loss-of-susceptibility alleles predominantly through the selection of mutations that impede binding of the TAL effectors to their cognate EBEs but that ideally do not disrupt plant transcription factor binding sites. Alternatively, plants can trick bacteria into activating defenses by acquiring EBEs for bacterial TAL effectors in the promoter regions of resistance genes, the so-called executor genes (Round 2). It is also possible that executor genes arise from susceptibility genes that become resistance genes through selection. In response, bacteria can mutate TAL effectors to recognize the loss-of-susceptibility alleles (for example, by acquiring aberrant repeats) as well as mutate TAL effectors to avoid the induction of executor genes, or they can incorporate an arsenal of functionally redundant TAL effectors giving bacteria multiple ways to induce S genes, or to induce functionally equivalent S gene paralogs.
FIGURE 2.
A simplified model for combat involving bacterial TAL effectors and plant susceptibility genes. To cause disease, bacteria use TAL effectors that bind to the promoter regions of susceptibility (S) genes. The plant might defend itself against these effectors through mutations in the promoter that prevent binding of the TAL effector(s): so called loss-of-susceptibility alleles. Alternatively, plants can trick bacteria into inducing defenses via an executor gene. Bacteria can in turn counter by mutating TAL effectors to recognize the loss-of-susceptibility alleles using for example aberrant repeats that can accommodate single base deletions in the EBE or by mutating TAL effectors to avoid binding to the promoters of executor genes. Bacteria can also acquire new TAL effectors to redundantly induce S genes, the plant can evolve new loss-of-susceptibility alleles and executor genes, and the process becomes cyclical. Bars for plant and bacterium represent “health bars” typically used in video games to indicate the extent to which a combatant is winning or losing. Bold black arrows represent genes.
Of course, for each new S gene induced by bacteria, new selection pressure arises for the plant to avoid binding, and the rounds can repeat themselves indefinitely. Maybe the large repertoires of TAL effectors found in some strains are constituted by fallen players in the combat beaten by the plants defenses, and/or by tag team players, waiting to get into the ring and overkill (i.e., strike a fatality).
Round 4: Researchers Enter the Ring
Genetic engineering strategies to improve disease resistance are based on the knowledge of the mechanism employed by plants to recognize pathogen proteins. Although resistance to virus and to some extent Xanthomonas is mediated by recessive genes, efforts mainly focused on the identification of dominant R genes and the transformation of susceptible varieties using these genes. This type of resistance is often strain-specific and can be easily overcome by the pathogen through mutations in unique and specific avirulence genes (Dangl et al., 2013). In contrast, the knowledge of TAL effector mechanisms offers new exciting opportunities for researchers aiming to develop resistant varieties. A research strategy that exploits TAL effector knowledge to generate broad-spectrum and durable resistance (Figure 3) might include:
FIGURE 3.
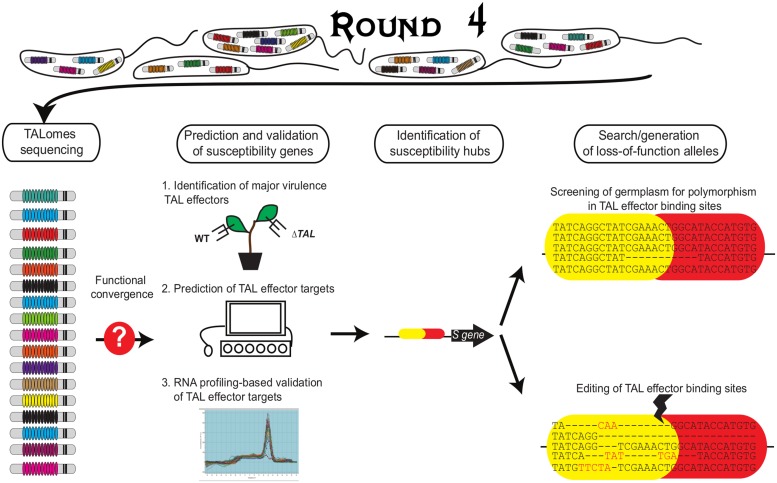
Exploiting TAL effector knowledge to identify S genes and loss-of-function alleles. Based on diversity studies conducted on a particular Xanthomonas species, it is possible to direct the research for virulence targets from the most prevalent TAL effectors. Field surveys allow the collection of strains at a regional or national scale. All or representative ones are selected to characterize the TALomes, i.e., their complete repertory of TAL effectors. The corresponding S genes can be identified employing three complementary strategies. First, the expected major virulence role of a prevalent TAL effector has to be assessed in planta (by loss of function mutational analysis or heterologous expression for example). Second, different bioinformatics algorithms (TALVEZ, TAL Effector Nucleotide Targeter, Talgetter) based on the TAL-DNA binding specificity code are employed to predict TAL effector targets in the host genome (assuming that at least a reference genome is available). Finally, RNA profiling strategies are employed to identify TAL effector-dependent differentially expressed genes. Any such genes with a predicted EBE for the TAL effector are strong candidates. For these, functional characterization using designer TAL effectors is the next step (Boch et al., 2014). Then, bioinformatics and functional analyses can be used to identify S hubs. The identification of natural variants in the EBEs from germplasm databases or Ecotilling or the generation of EBE mutations by genome editing in specific EBEs will lead to loss-of-function alleles as new plant disease resistance sources.
The Identification of Major TAL Effectors in Pathogen Populations of Interest
The characterization of a complete or near-complete TAL effector repertoire for a large group of strains of a particular Xanthomonas species, collected in geographically diverse regions and through different time periods, allows for the establishment of the more prevalent TAL effectors for that population (Figure 3). The sequencing of TAL effectors is a very complicated task given their repetitive nature. However, through the employment of new, relatively low cost, long-read sequencing technologies such as single molecule, real-time sequencing (Pacific Biosciences), it is possible to accurately assemble complete TAL effector sequences from whole genome data (A. Bogdanove, personal communication). A more difficult and time-consuming strategy is to clone and sequence TAL effectors individually. A non-tested way could be to generate TAL effector-specific libraries and sequence them. The knowledge gained on “TALome” diversity will allow estimation of the durability of a loss-of-susceptibility allele or an executor gene, which can be anticipated depending on the conservation of a particular TAL effector or a group of functionally equivalent TAL effectors in a given population. Also, a prevalent TAL effector can be taken as candidate virulence factor and prioritized for functional characterization. These TAL effectors can then be tested experimentally to confirm whether or not they have a role in virulence. This knowledge about TALome diversity will be key to design the best breeding resistance strategy. The next step would be:
The Prediction and Validation of Susceptibility Hubs in the Host
Any host gene representing a benefit to bacteria is a potential plant susceptibility gene and can be considered as a candidate for transformation strategies. However, considering that the aim is to provide broad and durable resistance, those S genes that constitute convergence hubs for various TAL effectors (like the SWEET family) are ideal. These could be identified by combining EBE prediction using the RVD sequences for the major virulence TAL effectors identified and expression data (RNA-seq or microarray). Functional characterization of the S gene would also be ideal.
Once a key S gene targeted by one or more major TAL effectors is identified, a logical step is:
The Screening for Natural Loss-of-Susceptibility Alleles in the Germplasm
For this, researchers can take advantage of the new sequencing technologies and the public genome sequence databases. In addition to the Rice SNPSeek data (Alexandrov et al., 2015), and other genomic databases for rice (Yonemaru et al., 2014; Zhao et al., 2015), similarly large datasets exist for other hosts worth exploring. In Arabidopsis, the 1001 project as of September 2014 includes the sequencing of over 1000 lines, representing a valuable resource to find resistance sources for X. campestris (Weigel and Mott, 2009). In cassava, an important resource consisting of the partial genome for 600 accessions is available and can be exploited to identify polymorphism in EBEs for targets of TAL effectors from X. axonopodis pv. manihotis2. In tomato the sequence of 84 accessions has been recently reported (Tomato Genome Sequencing Consortium et al., 2014). For important crops with no such databases, alternatives like ecotilling or amplicon sequencing can be considered for identifying natural loss-S alleles (Wang et al., 2012). These alleles could then be introgressed into susceptible lines. An alternative to breeding is:
The Application of Genome Editing Strategies to Achieve Resistance
Once an S gene targeted by a particular TAL effector is identified, genome editing using tools such as TALENs and CRISPR-Cas9 can be employed to modify the EBE to prevent TAL effector binding. As mentioned before, this poses the risk of altering the endogenous regulation of the gene, so ideally several variants should be created and screened for any deleterious effects. The introduction of loss-of-function alleles to commercial, agronomically important varieties using these genome edition strategies can be expected to be subject to less restrictive regulation and greater public acceptance than GMOs (Araki and Ishii, 2015).
Alternatively, an artificial executor gene can be created by engineering the promoter of a classical R gene to have several EBEs corresponding to different TAL effectors. This approach has already been applied in rice and was shown to successfully confer resistance to the desired strains (Hummel et al., 2012; Zeng et al., 2015). A predicted, but not tested, alternative involves silencing of the S gene upon pathogen infection. For example an artificial microRNA can be designed directed specifically against the S host gene. However, to avoid the above-mentioned problem of affecting endogenous function, the expression of this microRNA should be under the control of a pathogen-inducible promoter. In this scenario, the EBE for a particular, prevalent TAL effector could be employed. The identification of naturally occurring and selected mutations presents a major advantage over de novo edited EBE knockouts since it avoids the risk of affecting endogenous cis-regulatory elements and thus plant gene functions. Yet, one should also raise the possibility that viability of such alleles might be genotype-dependent, they need therefore to be tested when moved to any elite recipient variety. In summary, if bacteria have evolved TAL effectors to take advantage of the presence of host genes to induce their expression for their benefit, plant breeders can counter with strategies directed at these TAL effectors to combat these plant pathogens.
Conclusion
The discovery of the TAL effector-DNA binding code is greatly increasing our ability to identify new S proteins and better understand how diseases caused by Xanthomonas species occur in several major crops. Because of the highly conserved structure of TAL effectors and their mode of action to promote disease, standardized experimental pipelines can be developed in various Xanthomonas-host interactions to search for major S genes, provided that TAL effectors are at play. There is no doubt that in the near future, notably with the help of new sequencing technologies, extensive lists of new S genes will be generated to guide the search for pathogen-adapted loss-of-function alleles useful for marker-assisted breeding of loss-of-susceptibility. Less straightforward will be the challenge of finding such recessive resistance genes that cannot be easily broken, which will require better understanding of TAL effector diversity and evolution in pathogen populations.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to J. Lang for edits and comments and to L. Lamy for his help in retrieving SNP data. MH and AP-Q are supported by doctoral fellowships awarded by the MESR and the Erasmus Mundus Action 2 PRECIOSA program of the European Community, respectively. This project was supported by a grant from Agence Nationale de la Recherche (ANR-14-CE19-0002) and from Fondation Agropolis (#1403-073).
Footnotes
http://www.mortalkombat.com/, Warner Bros. Interactive Entertainment.
References
- Alexandrov N., Tai S., Wang W., Mansueto L., Palis K., Fuentes R. R., et al. (2015). SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 43 D1023–D1027. 10.1093/nar/gku1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Saadi A., Reddy J. D., Duan Y. P., Brunings A. M., Yuan Q., Gabriel D. W. (2007). All five host-range variants of Xanthomonas citri carry one pthA homolog with 17.5 repeats that determines pathogenicity on citrus, but none determine host-range variation. Mol. Plant Microbe Int. 20 934–943. 10.1094/MPMI-20-8-0934 [DOI] [PubMed] [Google Scholar]
- Antony G., Zhou J., Huang S., Li T., Liu B., White F., et al. (2010). Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell 22 3864–3876. 10.1105/tpc.110.078964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki M., Ishii T. (2015). Towards social acceptance of plant breeding by genome editing. Trends Plant Sci. 20 145–149. 10.1016/j.tplants.2015.01.010 [DOI] [PubMed] [Google Scholar]
- Boch J., Bonas U. (2010). Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 48 419–436. 10.1146/annurev-phyto-080508-081936 [DOI] [PubMed] [Google Scholar]
- Boch J., Bonas U., Lahaye T. (2014). TAL effectors–pathogen strategies and plant resistance engineering. New Phytol. 204 823–832. 10.1111/nph.13015 [DOI] [PubMed] [Google Scholar]
- Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., et al. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326 1509–1512. 10.1126/science.1178811 [DOI] [PubMed] [Google Scholar]
- Bogdanove A. J., Voytas D. F. (2011). TAL effectors: customizable proteins for DNA targeting. Science 333 1843–1846. 10.1126/science.1204094 [DOI] [PubMed] [Google Scholar]
- Carroll D. (2014). Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83 409–439. 10.1146/annurev-biochem-060713-035418 [DOI] [PubMed] [Google Scholar]
- Castiblanco L. F., Gil J., Rojas A., Osorio D., Gutiérrez S., Muñoz-Bodnar A., et al. (2012). TALE1 from Xanthomonas axonopodis pv. manihotis acts as a transcriptional activator in plant cells and is important for pathogenicity in cassava plants. Mol. Plant Pathol. 14 84–95. 10.1111/j.1364-3703.2012.00830.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernadas R. A., Doyle E. L., Nino-Liu D. O., Wilkins K. E., Bancroft T., Wang L., et al. (2014). Code-assisted discovery of TAL effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PLoS Pathog. 10:e1003972 10.1371/journal.ppat.1003972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Q. (2014). SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 201 1150–1155. 10.1111/nph.12445 [DOI] [PubMed] [Google Scholar]
- Chen L. Q., Hou B. H., Lalonde S., Takanaga H., Hartung M. L., Qu X. Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468 527–532. 10.1038/nature09606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Q., Qu X. Q., Hou B. H., Sosso D., Osorio S., Fernie A. R., et al. (2012). Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335 207–211. 10.1126/science.1213351 [DOI] [PubMed] [Google Scholar]
- Chong J., Piron M. C., Meyer S., Merdinoglu D., Bertsch C., Mestre P. (2014). The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J. Exp. Bot. 65 6589–6601. 10.1093/jxb/eru375 [DOI] [PubMed] [Google Scholar]
- Chu Z., Yuan M., Yao J., Ge X., Yuan B., Xu C., et al. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20 1250–1255. 10.1101/gad.1416306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M., Bart R. S., Shybut M., Dahlbeck D., Gomez M., Morbitzer R., et al. (2014). Xanthomonas axonopodis virulence is promoted by a transcription activator-like effector-mediated induction of a SWEET sugar transporter in cassava. Mol. Plant Microbe Interact. 27 1186–1198. 10.1094/MPMI-06-14-0161-R [DOI] [PubMed] [Google Scholar]
- Dangl J. L., Horvath D. M., Staskawicz B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341 746–751. 10.1126/science.1236011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange O., Schreiber T., Schandry N., Radeck J., Braun K. H., Koszinowski J., et al. (2013). Breaking the DNA-binding code of Ralstonia solanacearum TAL effectors provides new possibilities to generate plant resistance genes against bacterial wilt disease. New Phytol. 199 773–786. 10.1111/nph.12324 [DOI] [PubMed] [Google Scholar]
- Gendron J. M., Liu J.-S., Fan M., Bai M.-Y., Wenkel S., Springer P. S., et al. (2012). Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109 21152–21157. 10.1073/pnas.1210799110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau J., Wolf A., Reschke M., Bonas U., Posch S., Boch J. (2013). Computational predictions provide insights into the biology of TAL effector target sites. PLoS Comput Biol 9:e1002962 10.1371/journal.pcbi.1002962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Zhang J., Jia H., Sosso D., Li T., Frommer W. B., et al. (2014). Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. U.S.A. 111 E521–E529. 10.1073/pnas.1313271111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel A. W., Doyle E. L., Bogdanove A. J. (2012). Addition of transcription activator-like effector binding sites to a pathogen strain-specific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak. New Phytol. 195 883–893. 10.1111/j.1469-8137.2012.04216.x [DOI] [PubMed] [Google Scholar]
- Juillerat A., Bertonati C., Dubois G., Guyot V., Thomas S., Valton J., et al. (2014). BurrH: a new modular DNA binding protein for genome engineering. Sci Rep. 4 3831 10.1038/srep03831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay S., Boch J., Bonas U. (2005). Characterization of AvrBs3-like effectors from a Brassicaceae pathogen reveals virulence and avirulence activities and a protein with a novel repeat architecture. Mol. Plant Microbe Interact. 18 838–848. 10.1094/MPMI-18-0838 [DOI] [PubMed] [Google Scholar]
- Kay S., Hahn S., Marois E., Hause G., Bonas U. (2007). A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318 648–651. 10.1126/science.1144956 [DOI] [PubMed] [Google Scholar]
- Kay S., Hahn S., Marois E., Wieduwild R., Bonas U. (2009). Detailed analysis of the DNA recognition motifs of the Xanthomonas type III effectors AvrBs3 and AvrBs3Deltarep16. Plant J. 59 859–871. 10.1111/j.1365-313X.2009.03922.x [DOI] [PubMed] [Google Scholar]
- Lange O. D., Wolf C., Dietze J., Elsaesser J., Morbitzer R., Lahaye T. (2014). Programmable DNA-binding proteins from Burkholderia provide a fresh perspective on the TALE-like repeat domain. Nucleic Acids Res. 42 7436–7449. 10.1093/nar/gku329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapin D., Van den Ackerveken G. (2013). Susceptibility to plant disease: more than a failure of host immunity. Trends Plant Sci. 18 546–554. 10.1016/j.tplants.2013.05.005 [DOI] [PubMed] [Google Scholar]
- Lemoine R., La Camera S., Atanassova R., Dedaldechamp F., Allario T., Pourtau N., et al. (2013). Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 4:272 10.3389/fpls.2013.00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Huang S., Zhou J., Yang B. (2013). Designer TAL effectors induce disease susceptibility and resistance to Xanthomonas oryzae pv. oryzae in rice. Mol. Plant 6 781–789. 10.1093/mp/sst034 [DOI] [PubMed] [Google Scholar]
- Li T., Liu B., Spalding M. H., Weeks D. P., Yang B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30 390–392. 10.1038/nbt.2199 [DOI] [PubMed] [Google Scholar]
- Li Z., Zou L., Ye G., Xiong L., Ji Z., Zakria M., et al. (2014). A potential disease susceptibility gene CsLOB of citrus is targeted by a major virulence effector PthA of Xanthomonas citri subsp. citri. Mol. Plant 7 912–915. 10.1093/mp/sst176 [DOI] [PubMed] [Google Scholar]
- Liu Q., Yuan M., Zhou Y., Li X., Xiao J., Wang S. (2011). A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 34 1958–1969. 10.1111/j.1365-3040.2011.02391.x [DOI] [PubMed] [Google Scholar]
- Majer C., Hochholdinger F. (2011). Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 16 47–52. 10.1016/j.tplants.2010.09.009 [DOI] [PubMed] [Google Scholar]
- Mak A. N., Bradley P., Bogdanove A. J., Stoddard B. L. (2013). TAL effectors: function, structure, engineering and applications. Curr. Opin. Struct. Biol. 23 93–99. 10.1016/j.sbi.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckler J. F., Bhakta M. S., Kim M. S., Ovadia R., Habrian C. H., Zykovich A., et al. (2013). Quantitative analysis of TALE-DNA interactions suggests polarity effects. Nucleic Acids Res. 41 4118–4128. 10.1093/nar/gkt085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou M. J., Bogdanove A. J. (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326 1501–1501. 10.1126/science.1178817 [DOI] [PubMed] [Google Scholar]
- Mukhtar M. S., Carvunis A. R., Dreze M., Epple P., Steinbrenner J., Moore J., et al. (2011). Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333 596–601. 10.1126/science.1203659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Bodnar A., Perez-Quintero A. L., Gomez-Cano F., Gil J., Michelmore R., Bernal A., et al. (2014). RNAseq analysis of cassava reveals similar plant responses upon infection with pathogenic and non-pathogenic strains of Xanthomonas axonopodis pv. manihotis. Plant Cell Rep. 33 1901–1912. 10.1007/s00299-014-1667-7 [DOI] [PubMed] [Google Scholar]
- Pereira A. L., Carazzolle M. F., Abe V. Y., De Oliveira M. L., Domingues M. N., Silva J. C., et al. (2014). Identification of putative TAL effector targets of the citrus canker pathogens shows functional convergence underlying disease development and defense response. BMC Genomics 15:157 10.1186/1471-2164-15-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Quintero A. L., Rodriguez R. L., Dereeper A., Lopez C., Koebnik R., Szurek B., et al. (2013). An improved method for TAL effectors DNA-binding sites prediction reveals functional convergence in TAL repertoires of Xanthomonas oryzae strains. PLoS ONE 8:e68464 10.1371/journal.pone.0068464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke A. R., Bornberg-Bauer E., Gu J. (2011). Evolutionary divergence and limits of conserved non-coding sequence detection in plant genomes. Nucleic Acids Res. 39 6029–6043. 10.1093/nar/gkr179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A., Streubel J., Blucher C., Szurek B., Reschke M., Grau J., et al. (2014). A TAL effector repeat architecture for frameshift binding. Nat. Commun. 5 3447 10.1038/ncomms4447 [DOI] [PubMed] [Google Scholar]
- Romer P., Recht S., Strauss T., Elsaesser J., Schornack S., Boch J., et al. (2010). Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytol. 187 1048–1057. 10.1111/j.1469-8137.2010.03217.x [DOI] [PubMed] [Google Scholar]
- Schornack S., Ballvora A., Gurlebeck D., Peart J., Baulcombe D., Ganal M., et al. (2004). The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 37 46–60. 10.1046/j.1365-313X.2003.01937.x [DOI] [PubMed] [Google Scholar]
- Schornack S., Moscou M. J., Ward E. R., Horvath D. M. (2013). Engineering plant disease resistance based on TAL effectors. Annu. Rev. Phytopathol. 51 383–406. 10.1146/annurev-phyto-082712-102255 [DOI] [PubMed] [Google Scholar]
- Siemens J., Keller I., Sarx J., Kunz S., Schuller A., Nagel W., et al. (2006). Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development. Mol. Plant Microbe Interact. 19 480–494. 10.1094/MPMI-19-0480 [DOI] [PubMed] [Google Scholar]
- Streubel J., Blucher C., Landgraf A., Boch J. (2012). TAL effector RVD specificities and efficiencies. Nat. Biotechnol. 30 593–595. 10.1038/nbt.2304 [DOI] [PubMed] [Google Scholar]
- Streubel J., Pesce C., Hutin M., Koebnik R., Boch J., Szurek B. (2013). Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 200 808–819. 10.1111/nph.12411 [DOI] [PubMed] [Google Scholar]
- Sugio A., Yang B., Zhu T., White F. F. (2007). Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAgamma1 and OsTFX1 during bacterial blight of rice. Proc. Natl. Acad. Sci. U.S.A. 104 10720–10725. 10.1073/pnas.0701742104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher L. F., Manners J. M., Kazan K. (2009). Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J. 58 927–939. 10.1111/j.1365-313X.2009.03831.x [DOI] [PubMed] [Google Scholar]
- Tomato Genome Sequencing Consortium. Aflitos S., Schijlen E., De Jong H., De Ridder D., Smit S.et al. (2014). Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. 80 136–148. 10.1111/tpj.12616 [DOI] [PubMed] [Google Scholar]
- Vedel V., Scotti I. (2011). Promoting the promoter. Plant Sci. 180 182–189. 10.1016/j.plantsci.2010.09.009 [DOI] [PubMed] [Google Scholar]
- Verdier V., Triplett L. R., Hummel A. W., Corral R., Cernadas R. A., Schmidt C. L., et al. (2012). Transcription activator-like (TAL) effectors targeting OsSWEET genes enhance virulence on diverse rice (Oryza sativa) varieties when expressed individually in a TAL effector-deficient strain of Xanthomonas oryzae. New Phytol. 196 1197–1207. 10.1111/j.1469-8137.2012.04367.x [DOI] [PubMed] [Google Scholar]
- Wang T. L., Uauy C., Robson F., Till B. (2012). TILLING in extremis. Plant Biotechnol. J. 10 761–772. 10.1111/j.1467-7652.2012.00708.x [DOI] [PubMed] [Google Scholar]
- Weigel D., Mott R. (2009). The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 10 107 10.1186/gb-2009-10-5-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White F. F., Yang B. (2009). Host and pathogen factors controlling the rice-Xanthomonas oryzae interaction. Plant Physiol. 150 1677–1686. 10.1104/pp.109.139360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Sugio A., White F. F. (2006). Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. U.S.A. 103 10503–10508. 10.1073/pnas.0604088103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., White F. F. (2004). Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Mol. Plant Microbe Interact. 17 1192–1200. 10.1094/MPMI.2004.17.11.1192 [DOI] [PubMed] [Google Scholar]
- Yang Y., De Feyter R., Gabriel D. W. (1994). Host-specific symptoms and increased release of Xanthomonas citri and X. campestris pv. malvacearum from leaves are determined by the 102-bp tandem repeats of pthA and avrb6, respectively. Mol. Plant Microbe Interact. 7 345–355. 10.1094/MPMI-7-0345 [DOI] [Google Scholar]
- Yonemaru J., Ebana K., Yano M. (2014). HapRice, an SNP haplotype database and a web tool for rice. Plant Cell Physiol. 55:e9 10.1093/pcp/pct188 [DOI] [PubMed] [Google Scholar]
- Yu Y. H., Streubel J., Balzergue S., Champion A., Boch J., Koebnik R., et al. (2011). Colonization of rice leaf blades by an African Strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice nodulin-3 Os11N3 gene. Mol. Plant Microbe Interact. 24 1102–1113. 10.1094/MPMI-11-10-0254 [DOI] [PubMed] [Google Scholar]
- Yuan M., Chu Z., Li X., Xu C., Wang S. (2009). Pathogen-induced expressional loss of function is the key factor in race-specific bacterial resistance conferred by a recessive R gene xa13 in rice. Plant Cell Physiol. 50 947–955. 10.1093/pcp/pcp046 [DOI] [PubMed] [Google Scholar]
- Yuan T., Li X., Xiao J., Wang S. (2011). Characterization of Xanthomonas oryzae-responsive cis-acting element in the promoter of rice race-specific susceptibility gene Xa13. Mol. Plant 4 300–309. 10.1093/mp/ssq076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Tian D., Gu K., Zhou Z., Yang X., Luo Y., et al. (2015). Genetic engineering of the Xa10 promoter for broad-spectrum and durable resistance to Xanthomonas oryzae pv. oryzae. Plant Biotechnol. J. 10.1111/pbi.12342 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Zhao H., Yao W., Ouyang Y., Yang W., Wang G., Lian X., et al. (2015). RiceVarMap: a comprehensive database of rice genomic variations. Nucleic Acids Res. 43 D1018–D1022. 10.1093/nar/gku894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Peng Z., Long J., Sosso D., Liu B., Eom J. S., et al. (2015). Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82 632–643. 10.1111/tpj.12838 [DOI] [PubMed] [Google Scholar]