Abstract
Prostate development, benign hyperplasia and cancer involve androgen and growth factor signaling as well as stromal–epithelial interactions. We review how DNA methylation influences these and related processes in other organ systems such as how proliferation is restricted to specific cell populations during defined temporal windows, how androgens elicit their actions and how cells establish, maintain and remodel DNA methylation in a time and cell specific fashion. We also discuss mechanisms by which hormones and endocrine disrupting chemicals reprogram DNA methylation in the prostate and elsewhere and examine evidence for a reawakening of developmental epigenetic pathways as drivers of prostate cancer and benign prostate hyperplasia.
Keywords: benign prostate hyperplasia, DNA methylation, epigenetics, lower urinary tract, prostate, prostate cancer
Overview of DNA methylation
Epigenetics is the study of chemical modifications to DNA and histone proteins that regulate gene expression without altering DNA sequence. A growing number of epigenetic marks have been identified including DNA methylation, histone tail modifications, non-coding RNAs and others. Here we focus on the epigenetic mark of DNA methylation which occurs by addition of a methyl group to the 5′ position of cytosine. DNA methylation most often occurs in the context of CpG dinucleotides, though non-CpG DNA methylation can occur [1,2]. DNA methylation is classically perceived as a stable and sometimes heritable mark. However, DNA methylation events can also be surprisingly dynamic and added or removed in a spatially and temporally defined context.
DNA methylation is catalyzed by DNA methyltransferases (Dnmts), which include maintenance (Dnmt1) and de novo methyltransferases (Dnmt3a, Dnmt3b). An additional member, Dnmt3L, lacks catalytic activity but participates in de novo methylation by interacting with Dnmt3a and Dnmt3b and other transcription factors [3–5] and uses the histone landscape to recruit de novo DNA methyltransferases [3]. DNA methylation can also be lost, either through passive mechanisms whereby the methylation pattern is not maintained upon subsequent cell divisions, or through active mechanisms involving base modification, substitution, excision or repair (reviewed in [6]).
DNA methylation typically regulates gene expression by repressing transcription but in some cases DNA methylation can be transcriptionally activating. For example, it has recently been demonstrated that non-neuronal-derived serotonin increases Shh mRNA abundance in mouse mammary gland by increasing DNA methylation at one site of the Shh locus and decreasing DNA methylation at another [7]. DNA methylation also acts in concert with methyl-CpG binding proteins (MBDs) and chromatin modifiers to change the chromatin landscape. These events can be influenced by hormones, environment, drugs and other chemicals, leading to derangement of DNA methylation marks which can perturb development or trigger inappropriate growth later in life, potentially contributing to a host of diseases, including cancer.
Here we focus on prostate and describe developmental processes that implicate DNA methylation as a critical gene expression regulatory mechanism and describe how aberration of DNA methylation events influences prostate disease processes. We highlight evidence in other systems which may help bridge the knowledge gap in understanding how prostate cells establish, maintain and remodel DNA methylation in a time and cell specific fashion during prostate development and the onset and progression of prostate disease.
Overview of prostate development
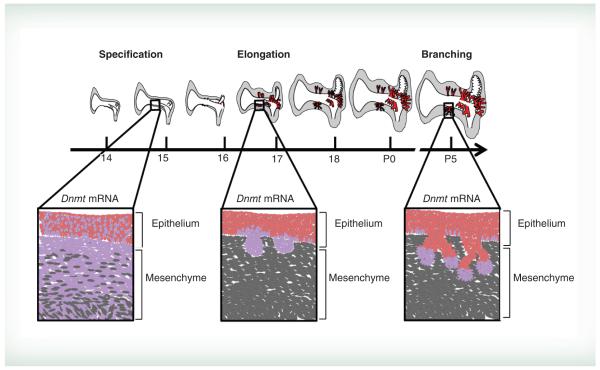
The prostate arises from a subcompartment of the lower urinary tract known as the urogenital sinus (UGS). Prostate formation is dependent upon androgen action as well as reciprocal stromal–epithelial interactions. Androgen signaling via androgen receptor (AR) in UGS mesenchyme instructs and initiates prostate ductal precursors, called prostate buds, to form from UGS epithelium. In mouse, testicular androgen synthesis occurs around 13 days post coitus (dpc) and epithelial prostate buds emerge from UGS epithelium about three days later, creating a lag between the onset of androgens and bud formation [8]. After prostate buds initiate they elongate into UGS mesenchyme and undergo branching morphogenesis, which continues postnatally (Figure 1) to give rise to the adult prostate ductal network.
Figure 1. Mouse prostate development and localization of Dnmt1 expression over time.
In mouse the prostate develops from the urogenital sinus (UGS). Prostate specification begins at 14 days post coitus after which time epithelial prostate buds (red) begin to emerge and elongate (16–18 dpc) into surrounding mesenchyme (grey). Prostate buds then undergo branching morphogenesis, which continues postnatally. Insets represent magnified images of prostate epithelial and mesenchymal morphology over the course of bud formation. Dnmt1 mRNA expression patterns are shown in purple displaying widespread expression in mesenchyme prior to bud formation before diminishing in mesenchyme and localizing to developing prostate buds and bud tips.
For color images please see online at: www.futuremedicine.com/doi/full/10.2217/EPI.15.8.
AR signaling in prostate mesenchyme is necessary for prostate epithelial morphogenesis, suggesting androgen-induced paracrine signaling factors guide prostate development. These factors have been termed andromedins. Several andromedins have been proposed but to date no single gene has been identified as the andromedin responsible for prostate development. Multiple gene families participate in prostate development, including Fgf, Tgfβ, Bmps, Shh and others (reviewed in [9]). KGF/FGF7 and FGF10 were the first identified candidate andromedins [9]. Wif1 was also identified as a candidate andromedin [10]. Wif1 expression is regulated by androgens and acts to promote androgen dependent bud formation, but is unable to stimulate prostate bud formation in the absence of androgens – a proposed characteristic of a true andromedin [10].
Therefore, additional mechanisms likely drive prostate morphogenesis. One such mechanism may involve DNA methylation. Recently, DNA methylation has been shown to play a critical role in regulating expression of key genes involved in prostate morphogenesis, including the Ar. Existing evidence for DNA methylation roles in prostate development is described below.
Role of DNA methylation in prostate development
In situ hybridization has been used to map mRNA expression patterns for DNA methylation modifying genes in developing mouse prostate at 14.5 dpc - P5 [11, 12]. Dnmts are expressed throughout prostate development, but change in abundance and spatial distribution over the course of prenatal and postnatal prostate morphogenesis [11]. Dnmts predominate in prostate mesenchyme prior to prostate bud outgrowth (14 dpc) and localize to prostate bud epithelium during elongation (17 dpc) and branching morphogenesis (P5) [11]. The spatially defined Dnmt expression patterns in developing prostate guided the search for gene targets within UGS epithelium and mesenchyme that are regulated by DNA methylation to drive morphogenesis. These studies have highlighted specific roles of DNA methylation in regulating Cdh1 and Ar gene expression during discrete windows of prostate development.
Regulation of E-cadherin expression in developing prostate
Dnmt localization to elongating prostatic buds at 17 dpc, a time when prostatic buds are elongating and are actively invading the adjacent stroma, raises the hypothesis that DNA methylation may control cell adhesion and prostatic bud outgrowth. Targeted studies at this developmental stage reveal a role for DNA methylation in regulating Cdh1 mRNA expression [13]. Cdh1 mRNA and protein are abundant in prostatic epithelium prior to bud formation (14 dpc), then decrease in basal epithelium of developing prostatic buds (17 dpc) [13]. DNA methylation of Cdh1 increases during this time. Inhibition of DNA methylation in vitro increases CDH1 abundance in prostate buds and causes prostatic buds to be shorter, a phenotype reversed by disrupting homotypic interactions between CDH1 domains on adjacent epithelial cells [13]. Thus, DNA methylation contributes to prostate bud outgrowth at least in part by downregulating Cdh1, which presumably modifies cell adhesion and permits epithelial rearrangements needed for prostate ductal growth.
Regulation of androgen receptor expression in developing prostate
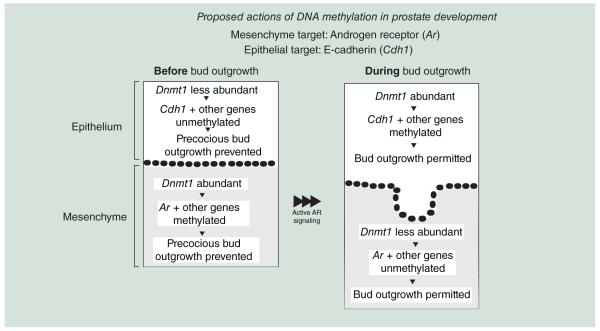
There is also evidence that DNA methylation is used to regulate androgen action at an early stage of prostate development. Prior to prostate bud outgrowth, during the period of prostate bud specification (14 dpc), Dnmts are present in developing prostate mesenchyme (Figure 1) [11]. Prostate mesenchyme is necessary for prostate development and is the site of androgen action [14,15]. This suggests that genes involved in AR signaling may be regulated by DNA methylation. It has recently been shown that Ar DNA methylation is lower in mesenchyme at 17 dpc compared with 14 dpc [16]. Further, inhibiting DNA methylation during prostate specification in vitro, when Dnmts predominate in UGS mesenchyme, uniquely enhances prostate bud formation [16]. Inhibiting DNA methylation decreases Ar DNA methylation as assessed by methylated DNA immunoprecipitation (MeDIP) and pyrosequencing of bisulfite converted DNA, increases AR expression, UGS androgen sensitivity and prostate bud formation rate [16]. A proposed model of two DNA methylation roles in guiding prostate morphogenesis is shown in Figure 2. DNA methylation of the Ar during early prostate development constrains prostate bud formation and prevents precocious growth. Later in development, when Dnmts predominate in epithelium, DNA methylation of Cdh1 facilitates prostate epithelial differentiation and outgrowth (Figure 2). There are still many unanswered questions in how androgens elicit their actions on prostate development and in how cells establish, maintain and remodel DNA methylation in a developmental time- and cell-specific fashion. For example, how is prostate DNA methylation established and removed during specific developmental stages? Are there other target genes regulated by DNA methylation during prostate organogenesis? Do androgens modulate DNA methylation? Can alterations in DNA methylation influence proliferative growth processes? In order to answer these questions in prostate, it is useful to examine known DNA methylation roles in other developmental processes. The following sections specifically focus on how DNA methylation is established, maintained and remodeled during embryogenesis. We also describe known roles of DNA methylation in development of nonprostate organs, how hormones influence DNA methylation and how endocrine disrupting chemicals and disease processes can alter DNA methylation and impact health.
Figure 2. Proposed mechanisms of action for DNA methylation during prostate development.
Early in prostate development, when DNA methyltransferase expression predominates in mesenchyme, DNA methylation of Ar acts to constrain prostate bud formation and prevents precocious growth. Later in development, when Dnmts predominates in epithelium, DNA methylation of Cdh1 facilitates prostate epithelial differentiation and outgrowth.
Ar: Androgen receptor; Cdhl: E-Cadherin.
Mechanisms of establishing & erasing DNA methylation marks in embryogenesis that may be conserved in prostate development
Understanding how the DNA methylome is rapidly and extensively remodeled during embryonic development may inform future studies on whether aspects of this process occur during prostate development. A rapid wave of paternal genome demethylation occurs shortly after fertilization in preimplantation embryos. DNA methylation patterns are then re-established largely through de novo methylation to permit embryonic specification in the blastocyst [17,18]. Rapid genomic reprogramming also occurs in primordial germ cells (PGCs) prior to and once reaching the embryonic gonad around 10–13 dpc in mouse [19–21]. Unlike the wave of DNA demethylation which occurs in male pronucleus during fertilization, PGC loss of DNA methylation occurs in both male and female PGCs mainly by inactivation of maintenance DNA methyltransferase activity [20,22] and downregulation of Dnmt3a, Dnmt3b and Dnmt3l [23]. Once reprogrammed, sexspecific DNA methylation marks are established in a time- and stage-specific fashion [6,20,22,24–25].
The identification of active mechanisms for removing DNA methylation marks filled a longstanding gap in understanding epigenetic programming. It was first thought that passive loss of DNA methylation or presence of a demethylase enzyme capable of removing methyl groups was the only mechanism to mediate demethylation. While some evidence points to Mbd2 as having independent demethylase activity [26] this is still controversial. In contrast, DNA demethylation has been shown to occur in a stepwise fashion, involving first modification of methylated cytosines and then replacement by DNA repair pathways. DNA hydroxymethylation is a cytosine modification catalyzed by ten eleven translocation gene family members, which leads to base excision repair and demethylation [27–29]. Deoxycytidine deamination occurs via the activation-induced cytidine deaminase (Aicda or Aid) and Apobec1, 2 and 3 family members [30,31]. Deoxycytidine deamination generates DNA mismatches which trigger DNA base excision repair, activation of mismatch repair pathways [32–34] or activation of DNA glycosylases which remove damaged bases or T:G and U:G mismatches [35]. Active mechanisms of DNA demethylation provide for potentially rapid and context-specific addition and removal of DNA methylation marks during specific morphogenetic events.
Like the mRNAs of DNMT enzymes responsible for establishing DNA methylation, the mRNAs for Tets, Aid, Apobecs, DNA glycosylases and other DNA repair enzymes, capable of removing DNA methylation, are expressed in male mouse UGS during prostate development [11]. Many of the mRNAs encoding these enzymes are abundant in UGS mesenchyme prior to prostate bud formation before diminishing therein and localizing instead to prostate bud epithelium [11]. A temporal change in localization and expression of the chromatin modifier, Ezh2 also occurs in developing prostate [36], suggesting that machinery for modifying histone structure, like DNA methylation machinery, is dynamically regulated during prostate development. These results raise the possibility that transient changes in DNA methylation and demethylation may contribute to morphogenetic changes associated with AR activation and onset of prostate development.
Roles of DNA methylation in organogenesis – conserved mechanisms & pathways important for prostate development
Developmental mechanisms, genes and pathways affected by DNA methylation in other developing organs inform future studies in developing prostate. How DNA methylation influences organogenesis is not as well understood as its developmental role in preimplantation embryos. This is largely because global DNA methylation is necessary for embryonic survival. Global inactivating mutations in mouse Dnmt1, Dnmt3b or Dnmt3a+Dnmt3b cause mid-gestational mortality (0.5–11.5 dpc) [37,38], the ultimate cause is still unknown. Early embryonic mortality and growth retardation of Dnmt mutant mice limits their utility in testing DNMT requirements in prostate development, which occurs late in gestation and relies on several prerequisites including successful cloacal septation, pathfinding of mesonephric and nephric ducts, cell differentiation and testicular testosterone synthesis [39].
The growing number of genetically modified mice that express cell type specific cre drivers facilitates an organ- and cell-type specific approach toward understanding the role of DNA methylation in development. Using currently available mouse strains, several studies have revealed context-dependent roles for DNA methylation. A role for Dnmt1 and DNA methylation has been established in progenitor cell populations of developing pancreas, hematopoetic system and neurons, where conditional Dnmt1 deletion impairs cell survival and differentiation, progenitor cell self renewal and lineage appropriate gene expression [40–42]. DNA methylation is not only important for progenitor cell survival and differentiation, but also morphogenesis. Selective Dnmt1 deletion in developing excitatory neurons and astroglia of cortex and hippocampus causes cortical degeneration and persistent learning and memory deficits [43]. Interestingly, a fraction of hypomethylated neurons survive postnatally but exhibit increased dendritic branching and impaired excitability [43]. Developmental mechanisms responsible for these phenotypes include changes to genes responsible for cell layer-specification, cell death and ion transport [43]. Dnmt1 deletion in retinal pigment epithelium or neural retina disrupts retinal pigment epithelia morphology and polarization, photoreceptor development and neural retinal differentiation [44]. These events are accompanied by disrupted actin cytoskeleton morphology, decreased retinal adhesion and DNA methylation changes in Hedgehog, Notch and Wnt pathway genes [44].
DNA methylation also mediates key events in mouse hair follicle development and regeneration. Epidermal Dnmt1 deletion causes uneven epidermal and hair fiber thickness, altered follicle size and an aging-related decline in hair fiber quantity [45]. Focal cell proliferation defects partially account for uneven epidermal thickness [45]. Molecular mechanisms responsible for changes in hair follicle size are unknown largely in part because few molecules are known to influence hair size. Proposed targets include Bmp and Wnt pathway members which can influence hair diameter and thickness [46,47]. Together these studies indicate that DNA methylation is not only important for cell survival, but can also influence structural morphogenesis and function.
Developing prostate relies upon several developmental mechanisms regulated by DNA methylation in retina, skin and hair follicle. Cytoskeleton rearrangement, cell adhesion and epithelial polarity are all essential for prostate epithelial development and differentiation [48]. Signaling pathways including Hedgehog, Notch, Bmp and Wnt have all been linked to prostate epithelial budding, branching and differentiation [49–51]. Whether DNA methylation regulates these pathways in prostate, as it does in retina and hair follicles, is an area of future study.
Interactions between DNA methylation & hormones
DNA methylation regulates estrogen receptor and AR expression in adult prostate and other tissues [52,53]. Estrogen receptor and AR signaling both influence prostate development (reviewed in [54]), but it is not yet known if and how DNA methylation acts in concert with these hormones during prostate morphogenesis.
There is growing evidence that hormone action can alter Dnmt expression, activity and DNA methylation levels. For example, changes in estrogen and progesterone levels during the female menstrual cycle alter endometrial Dnmt expression [55–57] influencing DNA methylation patterns and mRNA abundance of endometrial genes involved in transcription, apoptosis, proliferation, extracellular matrix and blood vessel morphogenesis [58]. Neonatal testosterone exposure can masculinize DNA methylation at sexually dimorphic CpG sites in female mouse brain [59].
Evidence links hormone-induced DNA methylation patterns to specific anatomical and physiological endpoints. Estrogen and androgens are responsible for mouse gender dependent differences in DNA methylation and expression of cardiomyocyte gene, Csx2 in cardiac ventricle, leading to sexually dimorphic cardiac structure and function [60]. Further, DNA methylation, ventricle structure and function are reversed upon gonadectomy [60]. Together these results reveal that hormones can influence expression of DNA methylation regulators, DNA methylation patterns, expression of target genes and physiological responses elicited by target genes. Further investigation into these interactions will likely shed light on both developmental and disease processes driven by steroid hormones. These results will also inform future studies to determine whether hormones influence DNA methylation in developing prostate. DNA methylation could be a means by which androgens or andromedins (genes responsible for mediating the actions of androgens) alter gene expression to drive prostate development.
DNA methylation & the fetal basis of adult disease in prostate
The fact that the epigenomic landscape is established during embryonic development also makes it a period of susceptibility to perturbations by environmental and endocrine disrupting chemicals. Chemically induced epigenomic reprogramming during embryonic development can lead to changes in gene expression and altered histology and physiology later in life. This is one aspect of a fetal basis of adult disease (reviewed in [61]). Perhaps the best examples come from the field of toxicology, where for decades, researchers have been studying how in utero exposure to environmental and endocrine disrupting chemicals influence health in adulthood. Two estrogenic chemicals receiving considerable research focus are diethylstilbesterol (DES) and bisphenol A (BPA). Some mouse and rat strains exposed in utero to DES and BPA exhibit altered developmental morphogenesis, reprogrammed DNA methylation patterns and altered adult histology/physiology of the prostate.
The developing prostate is extremely sensitive to endocrine disrupting chemicals. CD-1 mice exposed in utero to estradiol, DES or BPA exhibit changes in the number of prostate buds formed and fetal UGS morphology [62]. Mice exposed during fetal and neonatal development to high doses of DES (200 μg/kg) generally develop fewer prostatic buds, while mice exposed to low doses of BPA (10 μg/kg) and DES (0.1 μg/kg) form more prostatic buds and exhibit increased UGS AR expression [62–64]. In Sprague–Dawley rats, in utero BPA exposure increases incidence of carcinogen-induced hyperplastic and precancerous lesions in adult prostate [65]. Molecular mechanisms driving abnormal prostate development and altered histology in adulthood are still under investigation. Evidence exists to support the hypothesis that BPA-induced changes in DNA methylation alter gene expression to disrupt homeostatic regulation of prostatic growth later in life. Male rats exposed in utero to BPA exhibit increases in prostatic Dnmt3a and 3b expression at day 10 and 90 which diminishes by day 200 [66]. This in utero BPA exposure induces hypo- or hyper-methylation in a gene specific context in prostate. Some of these marks persist throughout life, while others are not observed until after sexual maturity or stimulation with pro-oncogenic factors in adulthood [66]. These studies provide evidence that early life exposure to endocrine disruptors, and potentially endogenous hormones, do not necessarily elicit immediately detectable changes in DNA methylation or gene expression. In some cases, these responses are not detectable until later in life-puberty or in response to a ‘second hit’ stimulus such as changes in hormone levels. Together the results of these studies have important implications in assessing potential mechanisms of hormone action and health risks associated with endocrine disrupting chemical exposures in that timing of assessment is potentially as important as doses tested and endpoints measured.
Collectively, in prostate there are links between endocrine disruptor exposure and altered development, changes in DNA methylation and abnormal organ function and histology associated with disease later in life. These observations can help to inform future studies to understand how DES and BPA act to impair development of prostate and other organs. One potential mechanism is that in utero exposure to endocrine disrupting chemicals could influence prostate morphogenesis and homeostasis throughout life by altering DNA methylation during the embryonic period when DNA methylation guides prostate ductal development. This mechanism may extend to other environmental chemicals. For example, embryonic exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) also alters the number of prostate buds formed and the anatomical morphology of the UGS in fetal mice [8]. TCDD has been shown to increase DNA methyltransferase enzyme activity and alter DNA methylation in preimplantation mouse blastocysts [67]. Whether TCDD induces changes in DNA methylation to alter prostate bud formation is yet to be determined.
Prostate cancer & benign prostate hyperplasia (BPH): a reawakening of developmental signals
A full discussion of epigenomics as it relates to prostate cancer and BPH is beyond the scope of this review, interested readers are directed toward several reviews on the topic [68–70]. There are numerous genes regulated by DNA methylation in prostate cancer [52,68–71]. In the following section we specifically focus on genes known to be regulated by DNA methylation during prostate development (Ar and Cdh1) which may be inappropriately reactivated in prostate cancer and BPH.
Prostate cancer
It has been proposed that a reawakening of developmental signals drives inappropriate growth in prostate cancer and BPH [72]. Since DNA methylation is critical for controlling expression of key prostate development genes (Cdh1 and Ar), it is possible that aberrant DNA methylation patterns of these genes and others reawakens prostate growth mechanisms to drive onset and progression of BPH and prostate cancer. This notion is supported by multiple studies. Recently, a chromatin remodeling gene (Hmga2) was identified as being selectively activated during prostate development [73]. When Hmga2 was over-expressed in adult prostate stromal cells, combined with prostate epithelium and grown as xenografts, the grafts formed high-grade prostate intraepithelial neoplasia [73]. Prostate intraepithelial neoplasia precursor lesions precede cancer formation in predisposed mice and in humans [74]. This study provides evidence that inappropriate expression of epigenetic modifiers in prostate stroma may be sufficient to alter epithelial proliferative activity. DNMT protein abundance and activity are inappropriately elevated in human prostate cancer tissue and cell lines versus nonmalignant controls [75,76]. DNMT protein expression also increases in prostate intraepithelial neoplasia and well-differentiated tumors in transgenic adenocarcinoma of mouse prostate mice that are genetically predisposed to prostate cancer [77].
Androgen receptor
DNA methylation of Ar as assessed by methylated DNA immunoprecipitation and pyrosequencing of bisulfite converted DNA occurs during prostate development to control onset and rate of androgen dependent prostate bud formation, confining initiation of prostate development to a specific embryonic development window [16]. There is evidence that this developmental role of Ar DNA methylation could also be hijacked to drive prostate cancer. DNA methylation silences AR gene expression in primary human prostate cancer tissues [78,79] as well as advanced (androgen-insensitive) prostate cancer cell lines [52] as assessed by methylation specific PCR and bisulfite sequencing. Whether these changes are also observed with other highly quantitative approaches such as pyrosequencing of bisulfite converted DNA in human prostate cancer tissues is not clear. Chemical inhibition of Ar DNA methylation can restore AR expression and androgen responsiveness [80]. Administration of DNA methylation inhibitor 5′-aza-2′deoxycitidine to transgenic adenocarcinoma of mouse prostate mice impairs progression to androgen independent prostate cancer and when combined with castration increases survival [81]. These findings raise new hypotheses about the role of DNA methylation in progression of prostate cancer to an aggressive form that loses its growth dependence on androgens and can be lethal.
E-cadherin
DNA methylation of cell adhesion molecule, Cdh1, allows for prostate epithelial outgrowth during prostate development [13]. Epigenetic regulation of Cdh1 also influences epithelial cell migration and behavior in prostate cancer. CDH1 is inappropriately silenced by DNA methylation in prostate cancer [82]. Silencing of CDH1 is associated with tumor invasion and metastasis and is predictive of poor patient outcome [83]. Treatment with DNA methylation inhibitor restores Cdh1 expression in prostate cancer cell lines and decreases invasion potential [84]. Together these results suggest that DNA methylation is not only an important aspect of development but inappropriate DNA methylation activity is associated with prostate disease and progression.
Benign prostate hyperplasia
Benign growth of the prostate associates with lower urinary tract symptoms (LUTS) which include irritative symptoms like increased frequency, urgency, pain and nocturia as well as voiding or obstructive symptoms like weak stream, hesitancy, dribbling, incomplete emptying and overflow incontinence. LUTS decrease quality of life and represent a significant healthcare burden to the aging population [85]. Approximately 70% of men over the age of 70 experience BPH and/or LUTS [86]. Prostate volume positively associates with risk of developing bothersome LUTS, however men with small prostates can experience LUTS and men with large prostates can be relatively asymptomatic, making diagnosis and treatment complex [87]. While these diseases have a complex etiology, few studies have focused on the epigenome as a contributing factor.
It was first hypothesized by the pathologist John McNeal that the proliferative growth pattern in BPH nodules resembled developing prostate ducts undergoing branching morphogenesis [88]. He hypothesized that inappropriate reawakening of prostate development signaling pathways could contribute to BPH pathogenesis [89]. Evidence now exists to support the hypothesis that epigenetic changes co-occur with histological prostate hyperplasia. Global 5-methylcytosine levels are reduced [90] and localized DNA hypermethylation is observed in tumor suppressor genes [91] in benign hyperplasic prostate compared with histologically normal prostate tissue.
Inflammation is one of the major factors commonly observed in prostates from patients with BPH/LUTS. Men with chronic prostatic inflammation are 6.8 times more likely to have histological BPH than individuals without [92]. Men with higher prostate inflammation also report more bothersome LUTS [93]. The cause of inflammation is not well understood and since not all incidences are accompanied by histological or culturable presence of bacteria [94], multiple inflammatory mediators are likely involved. Evidence now exists that epigenetic abnormalities in the immune system may contribute to BPH associated with LUTS (symptomatic BPH). The innate antiviral immune response is activated in prostate tissue from patients with BPH and LUTS versus those with histologic BPH but few LUTS (asymptomatic) [95]. The cytosine deaminase, APOBEC3G, is upregulated in prostates from patients undergoing surgery for symptomatic BPH compared with patients with asymptomatic BPH or histologically normal prostate tissue [95]. Concurrently, demethylation and increased expression of LINE-1 retrotransposable elements occurs in BPH tissues from patients with symptomatic BPH compared with histologically normal tissues [95]. LINE-1 demethylation activates the immune response in autoimmune disorders [96]. Together these results raise the hypothesis that Apobec mediated demethylation of LINE-1 may occur in prostate to drive inflammation contributing to BPH and symptomatic LUTS.
Changes in the hormonal mileau have also been hypothesized as a driving factor in onset and progression of BPH and LUTS. As men age, testosterone levels decline while estrogen levels remain the same or even rise [97]. Severe bladder outlet obstruction and increased prostate wet weight is observed in rodent models mimicking this hormonal change [98]. Interplay between the epigenome and hormone action is also evident in BPH. Testosterone is converted to the more potent dihydrotestosterone by SRD5A2 within prostate, driving proliferative growth in development, BPH and prostate cancer. 5 alpha reductase inhibitors like finasteride are commonly used in men to alleviate symptoms of BPH with varying success. Currently, it is not understood why some men respond to these therapies better than others. Improved understanding of SRD5A2 regulation may shed light on factors responsible for patient drug responsiveness. SRD5A2 has CpG regions and DNA methylation has been shown to regulate its expression in prostate [99]. Interestingly, men exhibit varying levels of SRD5A2 expression in prostate [99]. Why some adult men express low levels of prostatic SRD5A2 is not known [99]. The possibility that men with highly methylated SRD5A2 may be more resistant to 5 alpha reductase inhibitor therapy has been raised [100]. What regulates SRD5A2 DNA methylation and why it is only seen in some men experiencing BPH is unknown but a promising field of study for designing personalized therapies.
Age, one of the best known risk factors for BPH/LUTS, is also a predictor of DNA methylation status [101–103]. Epigenetic drift, or age associated changes in the DNA methylation landscape, occurs in prostate as it does in other organs [101]. Typically, a global loss of DNA methylation is associated with aging [104]. However, gene specific changes in DNA methylation (including hypo- and hypermethylation) as well as prostate tissue region specific changes in DNA methylation have also been observed [101]. Whether changes in DNA methylation contribute to BPH/LUTS onset or progression and whether interventions to enhance DNA methylation will be therapeutically beneficial has not been examined.
Conclusion
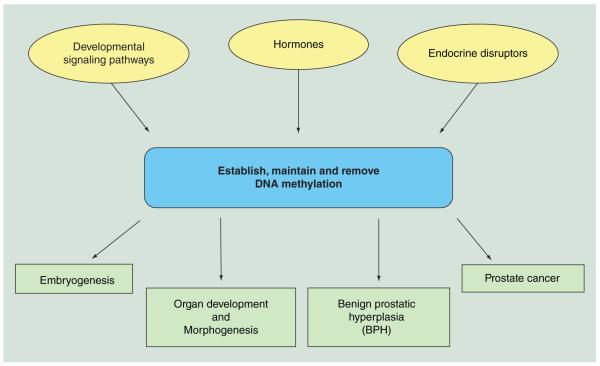
Figure 3 encompasses a summary of events in prostate and other organs which are capable of establishing, maintaining or removing DNA methylation marks, which in turn have downstream actions on embryogenesis, organogenesis and disease in prostate and other organs.
Figure 3. Dynamic DNA methylation events in development and disease.
Summary of events in prostate and other organs which are capable of establishing, maintaining or removing DNA methylation marks, which in turn have downstream actions on embryogenesis, organogenesis and disease.
Future perspective
Given the complexity in studying roles of DNA methylation, new tools and models will likely emerge. This will refine the intricate study of DNA methylation in carrying out actions of hormones, remodeling tissue structure and function in response to aging or endocrine disrupting chemicals. While significant progress has been made, it is still often difficult to manipulate the epigenome in a cell and DNA locus selective fashion. Global inhibitors of DNA methylation are available but have off target and widespread effects. Specific DNMT inhibitors are also becoming more widely used but they too cannot be targeted to a specific organ, specific genetic locus or specific time. Deletion of Dnmts is currently possible and continued expansion of available cre drivers will facilitate investigating the role of Dnmts in specific cell populations at critical periods of time and in specific cell populations. Development of new techniques to study DNA methylation in a gene, cell or developmental stage specific context will exponentially expand the field of epigenetics.
Executive summary.
Role of DNA methylation in prostate development
Androgens regulate prostate development but how they elicit their actions is not completely understood.
DNA methylation acts during specific developmental stages to guide prostate morphogenesis.
DNA methylation of e-cadherin contributes to prostate epithelial outgrowth.
DNA methylation of androgen receptor guides prostate bud patterning and rate of formation.
Understanding how DNA methylation is remodeled in other developmental context may shed light on how DNA methylation acts in prostate.
Mechanisms of establishing & erasing DNA methylation marks in embryogenesis that may be conserved in prostate development
DNA methylation is rapidly and extensively remodeled during embryonic development.
DNA demethylation occurs through base modifications, base excision repair, mismatch repair and DNA damage repair pathways.
Genes involved in these processes are expressed in developing prostate.
Roles of DNA methylation in organogenesis – conserved mechanisms & pathways important for prostate development
Transient changes in DNA methylation can regulate developmental pathways guiding organ morphogenesis in a stage and cell type specific fashion.
Mouse models employing conditional deletion of Dnmts have increased our ability to probe the roles of DNA methylation during organogenesis.
Conditional deletion of DNMT1 has revealed developmental pathways which rely on appropriate DNA methylation, including cell polarity, adhesion and signaling pathway members like Notch, Bmp and Wnt.
These developmental mechanisms all contribute to prostate development, whether DNA methylation regulates them in developing prostate is an area of future study.
Interactions between DNA methylation & hormones
DNA methylation acts to regulate hormone receptor expression in prostate and elsewhere.
Hormones are capable of influencing DNA methylation machinery and altering gene specific DNA methylation leading to changes in tissue structure and function.
Whether hormones influence DNA methylation in developing prostate is unknown.
DNA methylation & the fetal basis of adult disease in prostate
Endocrine disrupting chemicals such as diethylstibesterol and bishenol A are capable of altering prostate development.
Developmental exposure to these chemicals in prostate and other organs is capable of modifying DNA methylation, which has been linked to altered histology/physiology later in life.
This raises the hypothesis that these chemicals may act by altering DNA methylation in developing prostate.
Prostate cancer & benign prostate hyperplasia: a reawakening of developmental signals
Reawakening of developmental signaling pathways has been proposed as a mediator of prostate disease onset and progression.
Inappropriate activation of DNA methylation pathway genes as well as changes in DNA methylation are observed in prostate cancer and benign prostate hyperplasia.
Future perspective
New techniques and animal models capable of manipulating DNA methylation at specific cell types and specific stages will enhance our understanding of the complex and intricate regulation of DNA methylation in development and disease.
Acknowledgments
This work was supported by grants from the National Institutes of Health DK099328, DK096074 and ES001332.
No writing assistance was utilized in the production of this manuscript.
Footnotes
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as:
• of interest;
•• of considerable interest
- 1.Arand J, Spieler D, Karius T, et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 2012;8(6):e1002750. doi: 10.1371/journal.pgen.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barres R, Osler ME, Yan J, et al. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10(3):189–198. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Ooi SK, Qiu C, Bernstein E, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448(7154):714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pacaud R, Sery Q, Oliver L, Vallette FM, Tost J, Cartron PF. DNMT3L interacts with transcription factors to target DNMT3L/DNMT3B to specific DNA sequences: role of the DNMT3L/DNMT3B/p65-NFkappaB complex in the (de-) methylation of TRAF1. Biochimie. 2014;104:36–49. doi: 10.1016/j.biochi.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 2004;279(26):27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 6.Feng S, Cokus SJ, Zhang X, et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl Acad. Sci. USA. 2010;107(19):8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laporta J, Keil KP, Weaver SR, et al. Serotonin regulates calcium homeostasis in lactation by epigenetic activation of Hedgehog signaling. Mol. Endocrinol. 2014;28(11):1866–1874. doi: 10.1210/me.2014-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin TM, Rasmussen NT, Moore RW, Albrecht RM, Peterson RE. Region-specific inhibition of prostatic epithelial bud formation in the urogenital sinus of C57BL/6 mice exposed in utero to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 2003;76(1):171–181. doi: 10.1093/toxsci/kfg218. [DOI] [PubMed] [Google Scholar]
- 9.Thomson AA. Mesenchymal mechanisms in prostate organogenesis. Differentiation. 2008;76(6):587–598. doi: 10.1111/j.1432-0436.2008.00296.x. [DOI] [PubMed] [Google Scholar]
- 10.Keil KP, Mehta V, Branam AM, et al. Wnt inhibitory factor 1 (Wif1) is regulated by androgens and enhances androgen-dependent prostate development. Endocrinology. 2012;153(12):6091–6103. doi: 10.1210/en.2012-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keil KP, Altmann HM, Mehta V, Abler LL, Elton EA, Vezina CM. Catalog of mRNA expression patterns for DNA methylating and demethylating genes in developing mouse lower urinary tract. Gene Expr. Patterns. 2013;13(8):413–424. doi: 10.1016/j.gep.2013.07.008. •• The mRNAs for genes encoding enzymes responsible for establishing, maintaining and remodeling DNA methylation are expressed in developing prostate and localize to different tissue compartments over time.
- 12.Gudmap www.gudmap.org.
- 13.Keil KP, Abler LL, Mehta V, et al. DNA methylation of E-cadherin is a priming mechanism for prostate development. Dev. Biol. 2014;387(2):142–153. doi: 10.1016/j.ydbio.2014.01.020. •• DNA methylation of the e-cadherin promoter is methylated during prostate bud elongation, which is associated with decreased e-cadherin mRNA and protein expression and prostate bud elongation.
- 14.Cunha GR, Donjacour AA, Cooke PS, et al. The endocrinology and developmental biology of the prostate. Endocr. Rev. 1987;8(3):338–362. doi: 10.1210/edrv-8-3-338. [DOI] [PubMed] [Google Scholar]
- 15.Lasnitzki I, Mizuno T. Role of the mesenchyme in the induction of the rat prostate gland by androgens in organ culture. J. Endocrinol. 1979;82(1):171–178. doi: 10.1677/joe.0.0820171. [DOI] [PubMed] [Google Scholar]
- 16.Keil KP, Abler LL, Laporta J, et al. Androgen receptor DNA methylation regulates the timing and androgen sensitivity of mouse prostate ductal development. Dev. Biol. 2014;396(2):237–245. doi: 10.1016/j.ydbio.2014.10.006. •• DNA methylation of the androgen receptor prior to prostate bud outgrowth plays a role in regulating the number, rate and onset of prostate bud formation.
- 17.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99(3):371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 18.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 2002;241(1):172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 19.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat. Rev. Mol. Cell. Biol. 2009;10(8):526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 2008;9(2):129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 21.Surani MA. Germ cells: the eternal link between generations. C. R. Biol. 2007;330(6–7):474–478. doi: 10.1016/j.crvi.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139(1):15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- 23.Kurimoto K, Yamaji M, Seki Y, Saitou M. Specification of the germ cell lineage in mice: a process orchestrated by the PR-domain proteins, Blimp1 and Prdm14. Cell Cycle. 2008;7(22):3514–3518. doi: 10.4161/cc.7.22.6979. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi H, Sakurai T, Miura F, et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res. 2013;23(4):616–627. doi: 10.1101/gr.148023.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henckel A, Nakabayashi K, Sanz LA, Feil R, Hata K, Arnaud P. Histone methylation is mechanistically linked to DNA methylation at imprinting control regions in mammals. Hum. Mol. Genet. 2009;18(18):3375–3383. doi: 10.1093/hmg/ddp277. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397(6720):579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 27.Ito S, D’alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139(11):1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 2004;279(50):52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 31.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463(7284):1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franchini DM, Schmitz KM, Petersen-Mahrt SK. 5-methylcytosine DNA demethylation: more than losing a methyl group. Annu. Rev. Genet. 2012;46:419–441. doi: 10.1146/annurev-genet-110711-155451. [DOI] [PubMed] [Google Scholar]
- 33.Modrich P. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 2006;281(41):30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popp C, Dean W, Feng S, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463(7284):1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cortellino S, Xu J, Sannai M, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146(1):67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duan Z, Zou JX, Yang P, et al. Developmental and androgenic regulation of chromatin regulators EZH2 and ANCCA/ATAD2 in the prostate via MLL histone methylase complex. Prostate. 2013;73(5):455–466. doi: 10.1002/pros.22587. [DOI] [PubMed] [Google Scholar]
- 37.Lei H, Oh SP, Okano M, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122(10):3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 38.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 39.Timms BG. Prostate development: a historical perspective. Differentiation. 2008;76(6):565–577. doi: 10.1111/j.1432-0436.2008.00278.x. [DOI] [PubMed] [Google Scholar]
- 40.Fan G, Martinowich K, Chin MH, et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132(15):3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- 41.Georgia S, Kanji M, Bhushan A. DNMT1 represses p53 to maintain progenitor cell survival during pancreatic organogenesis. Genes Dev. 2013;27(4):372–377. doi: 10.1101/gad.207001.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5(4):442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hutnick LK, Golshani P, Namihira M, et al. DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum. Mol. Genet. 2009;18(15):2875–2888. doi: 10.1093/hmg/ddp222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nasonkin IO, Merbs SL, Lazo K, et al. Conditional knockdown of DNA methyltransferase 1 reveals a key role of retinal pigment epithelium integrity in photoreceptor outer segment morphogenesis. Development. 2013;140(6):1330–1341. doi: 10.1242/dev.086603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Jiang TX, Hughes MW, et al. Progressive alopecia reveals decreasing stem cell activation probability during aging of mice with epidermal deletion of DNA methyltransferase 1. J. Invest. Dermatol. 2012;132(12):2681–2690. doi: 10.1038/jid.2012.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enshell-Seijffers D, Lindon C, Kashiwagi M, Morgan BA. Beta-catenin activity in the dermal papilla regulates morphogenesis and regeneration of hair. Dev. Cell. 2010;18(4):633–642. doi: 10.1016/j.devcel.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharov AA, Mardaryev AN, Sharova TY, et al. Bone morphogenetic protein antagonist noggin promotes skin tumorigenesis via stimulation of the Wnt and Shh signaling pathways. Am. J. Pathol. 2009;175(3):1303–1314. doi: 10.2353/ajpath.2009.090163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chambers KF, Pearson JF, Aziz N, O’toole P, Garrod D, Lang SH. Stroma regulates increased epithelial lateral cell adhesion in 3D culture: a role for actin/cadherin dynamics. PloS ONE. 2011;6(4):e18796. doi: 10.1371/journal.pone.0018796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vezina CM, Allgeier SH, Fritz WA, et al. Retinoic acid induces prostatic bud formation. Dev. Dyn. 2008;237(5):1321–1333. doi: 10.1002/dvdy.21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Omori A, Miyagawa S, Ogino Y, et al. Essential roles of epithelial bone morphogenetic protein signaling during prostatic development. Endocrinology. 2014;155(7):2534–2544. doi: 10.1210/en.2013-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mehta V, Schmitz CT, Keil KP, et al. Beta-catenin (CTNNB1) induces Bmp expression in urogenital sinus epithelium and participates in prostatic bud initiation and patterning. Dev. Biol. 2013;376(2):125–135. doi: 10.1016/j.ydbio.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jarrard DF, Kinoshita H, Shi Y, et al. Methylation of the androgen receptor promoter CpG island is associated with loss of androgen receptor expression in prostate cancer cells. Cancer Res. 1998;58(23):5310–5314. [PubMed] [Google Scholar]
- 53.Kerdivel G, Flouriot G, Pakdel F. Modulation of estrogen receptor alpha activity and expression during breast cancer progression. Vitam. Horm. 2013;93:135–160. doi: 10.1016/B978-0-12-416673-8.00004-6. [DOI] [PubMed] [Google Scholar]
- 54.Mcpherson SJ, Ellem SJ, Risbridger GP. Estrogen-regulated development and differentiation of the prostate. Differentiation. 2008;76(6):660–670. doi: 10.1111/j.1432-0436.2008.00291.x. [DOI] [PubMed] [Google Scholar]
- 55.Yamagata Y, Asada H, Tamura I, et al. DNA methyltransferase expression in the human endometrium: down-regulation by progesterone and estrogen. Hum. Reprod. 2009;24(5):1126–1132. doi: 10.1093/humrep/dep015. [DOI] [PubMed] [Google Scholar]
- 56.Vincent ZL, Farquhar CM, Mitchell MD, Ponnampalam AP. Expression and regulation of DNA methyltransferases in human endometrium. Fertil. Steril. 2011;95(4):1522–1525. e1521. doi: 10.1016/j.fertnstert.2010.09.030. [DOI] [PubMed] [Google Scholar]
- 57.Van Kaam KJ, Delvoux B, Romano A, D’hooghe T, Dunselman GA, Groothuis PG. Deoxyribonucleic acid methyltransferases and methyl-CpG-binding domain proteins in human endometrium and endometriosis. Fertil. Steril. 2011;95(4):1421–1427. doi: 10.1016/j.fertnstert.2011.01.031. [DOI] [PubMed] [Google Scholar]
- 58.Houshdaran S, Zelenko Z, Irwin JC, Giudice LC. Human endometrial DNA methylome is cycle-dependent and is associated with gene expression regulation. Mol. Endocrinol. 2014;28(7):1118–1135. doi: 10.1210/me.2013-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghahramani NM, Ngun TC, Chen PY, et al. The effects of perinatal testosterone exposure on the DNA methylome of the mouse brain are late-emerging. Biol. Sex Differ. 2014;5:8. doi: 10.1186/2042-6410-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sebag IA, Gillis MA, Calderone A, et al. Sex hormone control of left ventricular structure/function: mechanistic insights using echocardiography, expression, and DNA methylation analyses in adult mice. Am. J. Physiol. Heart Circ. Physiol. 2011;301(4):H1706–H1715. doi: 10.1152/ajpheart.00088.2011. [DOI] [PubMed] [Google Scholar]
- 61.Barker DJ. Maternal and fetal origins of coronary heart disease. J. R. Coll. Physicians Lond. 1994;28(6):544–551. [PMC free article] [PubMed] [Google Scholar]
- 62.Timms BG, Howdeshell KL, Barton L, Bradley S, Richter CA, Vom Saal FS. Estrogenic chemicals in plastic and oral contraceptives disrupt development of the fetal mouse prostate and urethra. Proc. Natl Acad. Sci. USA. 2005;102(19):7014–7019. doi: 10.1073/pnas.0502544102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vom Saal FS, Timms BG, Montano MM, et al. Prostate enlargement in mice due to fetal exposure to low doses of estradiol or diethylstilbestrol and opposite effects at high doses. Proc. Natl Acad. Sci. USA. 1997;94(5):2056–2061. doi: 10.1073/pnas.94.5.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richter CA, Taylor JA, Ruhlen RL, Welshons WV, Vom Saal FS. Estradiol and bisphenol A stimulate androgen receptor and estrogen receptor gene expression in fetal mouse prostate mesenchyme cells. Environ. Health Perspect. 2007;115(6):902–908. doi: 10.1289/ehp.9804. • In utero exposure to bisphenol A (BPA) is capable of increasing androgen receptor (AR) expression in developing mouse prostate.
- 65.Ho SM, Tang WY, Belmonte De Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66(11):5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang WY, Morey LM, Cheung YY, Birch L, Prins GS, Ho SM. Neonatal exposure to estradiol/bisphenol A alters promoter methylation and expression of Nsbp1 and Hpcal1 genes and transcriptional programs of Dnmt3a/b and Mbd2/4 in the rat prostate gland throughout life. Endocrinology. 2012;153(1):42–55. doi: 10.1210/en.2011-1308. • Evidence exists to support the hypothesis that BPA-induced changes in DNA methylation alter gene expression to disrupt homeostatic regulation of prostate growth later in life.
- 67.Wu Q, Ohsako S, Ishimura R, Suzuki JS, Tohyama C. Exposure of mouse preimplantation embryos to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) alters the methylation status of imprinted genes H19 and Igf2. Biol. Reprod. 2004;70(6):1790–1797. doi: 10.1095/biolreprod.103.025387. • Environmental chemical TCDD, which is capable of altering prostate development is also able to alter DNA methylation of genes in utero.
- 68.Dobosy JR, Roberts JLW, Fu VX, Jarrard DF. The expanding role of epigenetics in the development, diagnosis and treatment of prostate cancer and benign prostatic hyperplasia. J. Urol. 2007;177(3):822–831. doi: 10.1016/j.juro.2006.10.063. [DOI] [PubMed] [Google Scholar]
- 69.Blute ML, Jr, Damaschke NA, Jarrard DF. The epigenetics of prostate cancer diagnosis and prognosis: update on clinical applications. Curr. Opin. Urol. 2015;25(1):83–88. doi: 10.1097/MOU.0000000000000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ngollo M, Dagdemir A, Karsli-Ceppioglu S, et al. Epigenetic modifications in prostate cacner. Epigenomics. 2014;6(4):415–426. doi: 10.2217/epi.14.34. [DOI] [PubMed] [Google Scholar]
- 71.Yang M, Park JY. DNA methylation in promoter region as biomarkers in prostate cancer. Methods Mol. Biol. 2012;863:67–109. doi: 10.1007/978-1-61779-612-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bierhoff E, Walljasper U, Hofmann D, Vogel J, Wernert N, Pfeifer U. Morphological analogies of fetal prostate stroma and stromal nodules in BPH. Prostate. 1997;31(4):234–240. doi: 10.1002/(sici)1097-0045(19970601)31:4<234::aid-pros4>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 73.Zong Y, Huang J, Sankarasharma D, et al. Stromal epigenetic dysregulation is sufficient to initiate mouse prostate cancer via paracrine Wnt signaling. Proc. Natl Acad. Sci. USA. 2012;109(50):E3395–E3404. doi: 10.1073/pnas.1217982109. • Inappropriate expression of epigenetic modifiers in adult prostate stroma may be sufficient to alter epithelial proliferative activity.
- 74.Lee TK, Miller JS, Epstein JI. Rare histological patterns of prostatic ductal adenocarcinoma. Pathology. 2010;42(4):319–324. doi: 10.3109/00313021003767314. [DOI] [PubMed] [Google Scholar]
- 75.Gravina GL, Ranieri G, Muzi P, et al. Increased levels of DNA methyltransferases are associated with the tumorigenic capacity of prostate cancer cells. Oncol. Rep. 2013;29(3):1189–1195. doi: 10.3892/or.2012.2192. [DOI] [PubMed] [Google Scholar]
- 76.Patra SK, Patra A, Zhao H, Dahiya R. DNA methyltransferase and demethylase in human prostate cancer. Mol. Carcinog. 2002;33(3):163–171. doi: 10.1002/mc.10033. [DOI] [PubMed] [Google Scholar]
- 77.Morey Kinney SR, Smiraglia DJ, James SR, Moser MT, Foster BA, Karpf AR. Stage-specific alterations of DNA methyltransferase expression, DNA hypermethylation, and DNA hypomethylation during prostate cancer progression in the transgenic adenocarcinoma of mouse prostate model. Mol. Cancer Res. 2008;6(8):1365–1374. doi: 10.1158/1541-7786.MCR-08-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamanaka M, Watanabe M, Yamada Y, et al. Altered methylation of multiple genes in carcinogenesis of the prostate. Int. J. C. 2003;106(3):382–387. doi: 10.1002/ijc.11227. [DOI] [PubMed] [Google Scholar]
- 79.Kinoshita H, Shi Y, Sandefur C, et al. Methylation of the androgen receptor minimal promoter silences transcription in human prostate cancer. Cancer Res. 2000;60(13):3623–3630. [PubMed] [Google Scholar]
- 80.Izbicka E, Macdonald JR, Davidson K, Lawrence RA, Gomez L, Von Hoff DD. 5,6 Dihydro-5′-azacytidine (DHAC) restores androgen responsiveness in androgen-insensitive prostate cancer cells. Anticancer Res. 1999;19(2A):1285–1291. [PubMed] [Google Scholar]
- 81.Zorn CS, Wojno KJ, Mccabe MT, Kuefer R, Gschwend JE, Day ML. 5-aza-2′-deoxycytidine delays androgen-independent disease and improves survival in the transgenic adenocarcinoma of the mouse prostate mouse model of prostate cancer. Clin. Cancer Res. 2007;13(7):2136–2143. doi: 10.1158/1078-0432.CCR-06-2381. [DOI] [PubMed] [Google Scholar]
- 82.Graff JR, Herman JG, Lapidus RG, et al. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55(22):5195–5199. [PubMed] [Google Scholar]
- 83.Li LC, Zhao H, Nakajima K, et al. Methylation of the E-cadherin gene promoter correlates with progression of prostate cancer. J. Urol. 2001;166(2):705–709. [PubMed] [Google Scholar]
- 84.Kwon O, Jeong SJ, Kim SO, et al. Modulation of E-cadherin expression by K-Ras; involvement of DNA methyltransferase-3b. Carcinogenesis. 2010;31(7):1194–1201. doi: 10.1093/carcin/bgq071. [DOI] [PubMed] [Google Scholar]
- 85.Saigal CS, Joyce G. Economic costs of benign prostatic hyperplasia in the private sector. J. Urol. 2005;173(4):1309–1313. doi: 10.1097/01.ju.0000152318.79184.6f. [DOI] [PubMed] [Google Scholar]
- 86.Berry SJ, Coffey DS, Walsh PC, Ewing LL. The development of human benign prostatic hyperplasia with age. J. Urol. 1984;132(3):474–479. doi: 10.1016/s0022-5347(17)49698-4. [DOI] [PubMed] [Google Scholar]
- 87.Turkbey B, Huang R, Vourganti S, et al. Age-related changes in prostate zonal volumes as measured by high-resolution magnetic resonance imaging (MRI): a cross-sectional study in over 500 patients. BJU Int. 2012;110(11):1642–1647. doi: 10.1111/j.1464-410X.2012.11469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mcneal JE. Origin and evolution of benign prostatic enlargement. Invest. Urol. 1978;15(4):340–345. [PubMed] [Google Scholar]
- 89.Cunha GR, Ricke WA. A historical perspective on the role of stroma in the pathogenesis of benign prostatic hyperplasia. Differentiation. 2011;82(4–5):168–172. doi: 10.1016/j.diff.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bedford MT, Van Helden PD. Hypomethylation of DNA in pathological conditions of the human prostate. Cancer Res. 1987;47(20):5274–5276. [PubMed] [Google Scholar]
- 91.Henrique R, Jeronimo C, Hoque MO, et al. Frequent 14–3–3 sigma promoter methylation in benign and malignant prostate lesions. DNA Cell Biol. 2005;24(4):264–269. doi: 10.1089/dna.2005.24.264. [DOI] [PubMed] [Google Scholar]
- 92.Zlotta AR, Egawa S, Pushkar D, et al. Prevalence of inflammation and benign prostatic hyperplasia on autopsy in asian and caucasian men. Eur. Urol. 2014;66(4):619–622. doi: 10.1016/j.eururo.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 93.Nickel JC, Roehrborn CG, O’leary MP, Bostwick DG, Somerville MC, Rittmaster RS. The relationship between prostate inflammation and lower urinary tract symptoms: examination of baseline data from the REDUCE trial. Eur. Urol. 2008;54(6):1379–1384. doi: 10.1016/j.eururo.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nickel JC, Downey J, Young I, Boag S. Asymptomatic inflammation and/or infection in benign prostatic hyperplasia. BJU Int. 1999;84(9):976–981. doi: 10.1046/j.1464-410x.1999.00352.x. [DOI] [PubMed] [Google Scholar]
- 95.Madigan AA, Sobek KM, Cummings JL, Green WR, Bacich DJ, O’keefe DS. Activation of innate anti-viral immune response genes in symptomatic benign prostatic hyperplasia. Genes Immun. 2012;13(7):566–572. doi: 10.1038/gene.2012.40. • The cytosine deaminase, APOBEC3G, is upregulated in prostates from patients undergoing surgery for symptomatic benign prostatic hyperplasia (BPH) compared with patients with asymptomatic BPH or histologically normal prostate tissue.
- 96.Mavragani CP, Crow MK. Activation of the type I interferon pathway in primary Sjogren’s syndrome. J. Autoimmun. 2010;35(3):225–231. doi: 10.1016/j.jaut.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 97.Harman SM, Metter EJ, Tobin JD, Pearson J, Blackman MR. Longitudinal effects of aging on serum total and free testosterone levels in healthy men. Baltimore Longitudinal Study of Aging. J. Clin. Endocrinol. Metab. 2001;86(2):724–731. doi: 10.1210/jcem.86.2.7219. [DOI] [PubMed] [Google Scholar]
- 98.Nicholson TM, Ricke EA, Marker PC, et al. Testosterone and 17beta-estradiol induce glandular prostatic growth, bladder outlet obstruction, and voiding dysfunction in male mice. Endocrinology. 2012;153(11):5556–5565. doi: 10.1210/en.2012-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Niu Y, Ge R, Hu L, et al. Reduced levels of 5-alpha reductase 2 in adult prostate tissue and implications for BPH therapy. Prostate. 2011;71(12):1317–1324. doi: 10.1002/pros.21348. • SRD5A2 is regulated by DNA methylation, men exhibit varying levels of SRD5A2 expression in adulthood, raising a potential mechanism by which some men may be more resistant to 5 alpha reductase inhibitor therapy.
- 100.Bechis SK, Otsetov AG, Ge R, Olumi AF. Personalized medicine for the management of benign prostatic hyperplasia. J. Urol. 2014;192(1):16–23. doi: 10.1016/j.juro.2014.01.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Damaschke NA, Yang B, Bhusari S, Svaren JP, Jarrard DF. Epigenetic susceptibility factors for prostate cancer with aging. Prostate. 2013;73:1721–1730. doi: 10.1002/pros.22716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Teschendorff AE, West J, Beck S. Age-associated epigenetic drift: implications, and a case of epigenetic thrift? Hum. Mol. Genet. 2013;22:R7–R15. doi: 10.1093/hmg/ddt375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.West J, Widschwendter M, Teschendorff AE. Distinctive topology of age-associated epigenetic drift in the human interactome. Proc. Natl Acad. Sci. USA. 2013;110:14138–14143. doi: 10.1073/pnas.1307242110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gonzalo S. Epigenetic alterations in aging. J. Appl. Physiol. 2010;109:586–597. doi: 10.1152/japplphysiol.00238.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]