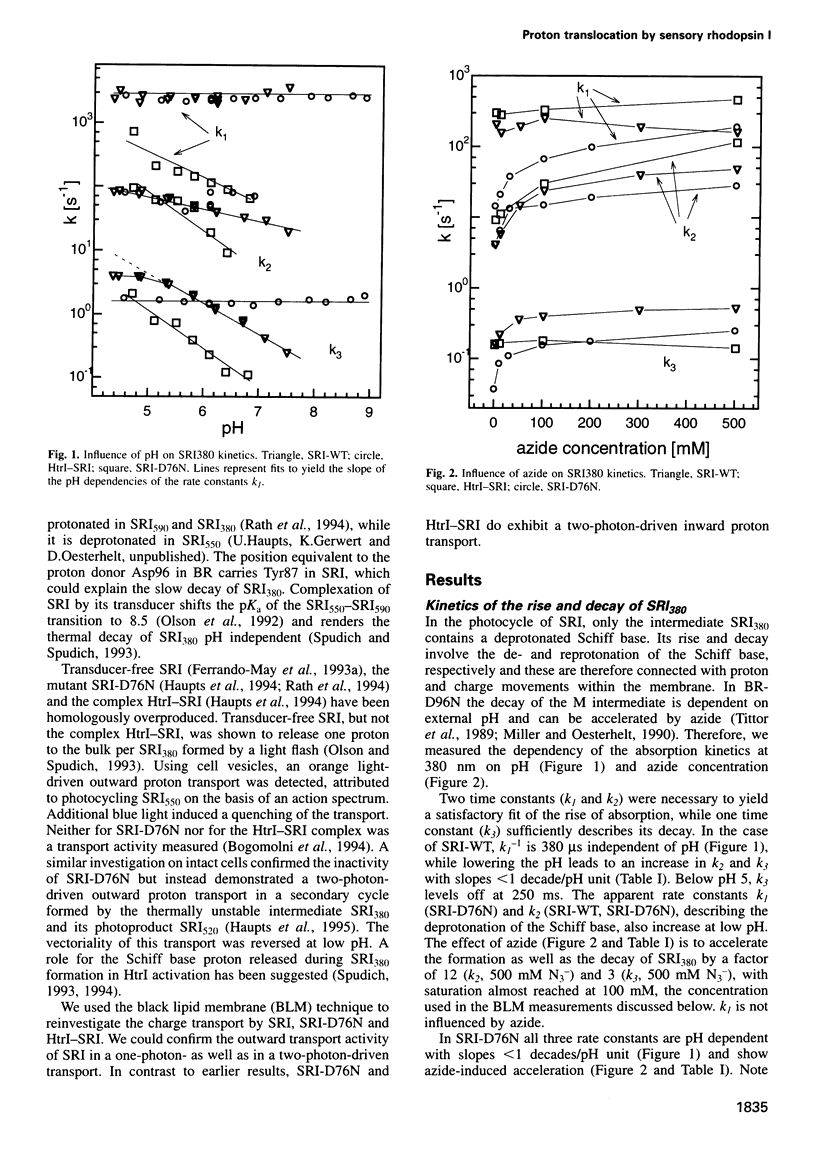
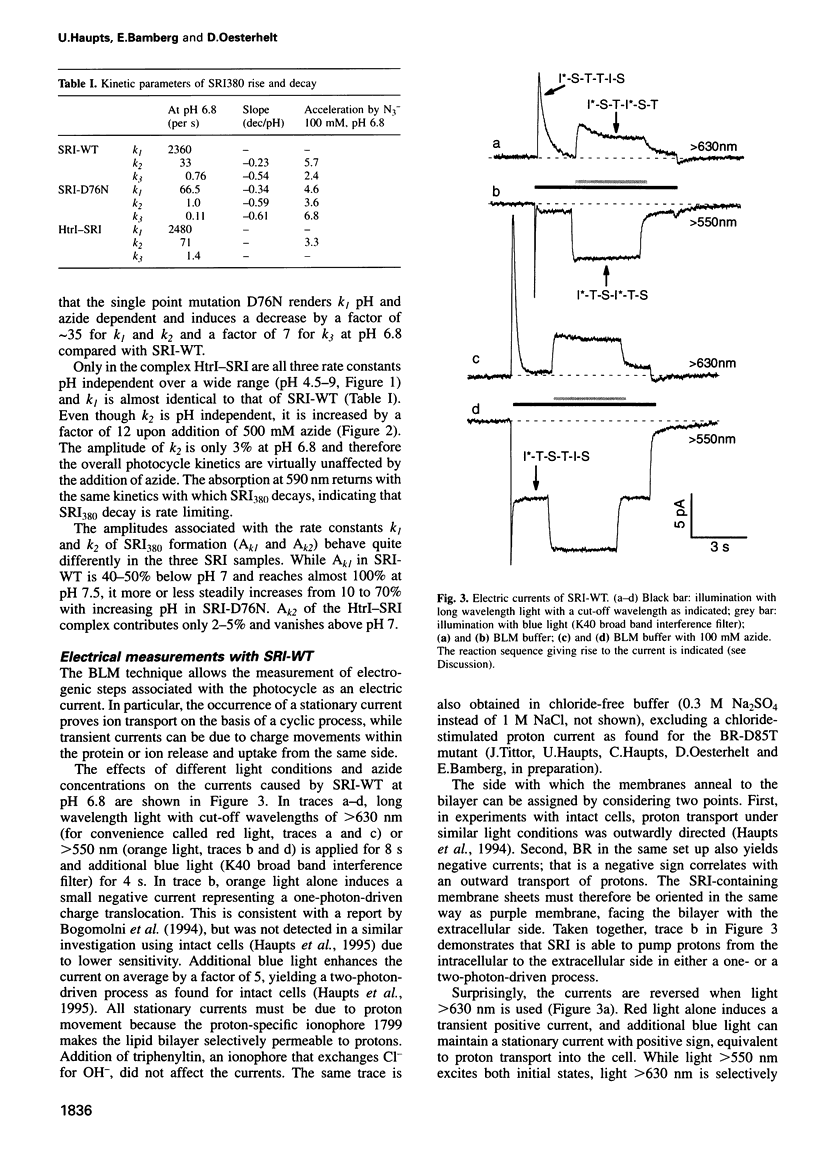
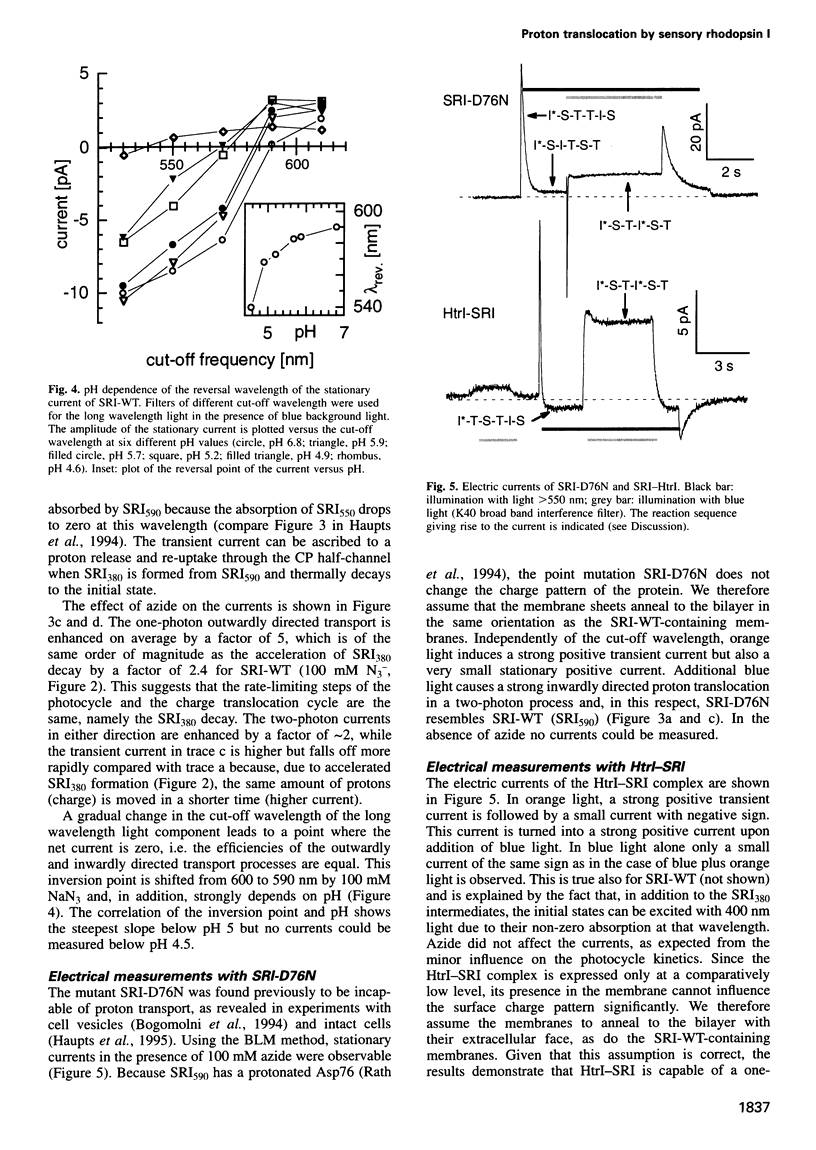
Abstract
The membrane-bound complex between sensory rhodopsin I (SRI) and its transducer HtrI forms the functional photoreceptor unit that allows transmission of light signals to the flagellar motor. Although being a photosensor, SRI, the mutant SRI-D76N and the HtrI-SRI complex can transport protons, as we demonstrate by using the sensitive and ion-specific black lipid membrane technique. SRI sustains an orange light-driven (one-photon-driven) outward proton transport which is enhanced by additional blue light (two-photon-driven). The vectoriality of the two-photon-driven transport could be reversed at neutral pH from the outward to the inward direction by switching the cut-off wavelength of the long wavelength light from 550 to 630 nm. The cut-off wavelength determining the reversal point decreases with decreasing pH. The currents could be enhanced by azide. A two-photon-driven inward proton transport by SRI-D76N (catalyzed by azide) and by the complex HtrI-SRI is demonstrated. The influence of pH and azide concentration on the rise and decay kinetics of the SRI380 intermediate is analyzed. The different modes of proton translocation of the SRI species are discussed on the basis of a general model of proton translocation of retinal proteins and in the context of signal transduction.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bamberg E., Tittor J., Oesterhelt D. Light-driven proton or chloride pumping by halorhodopsin. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):639–643. doi: 10.1073/pnas.90.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanck A., Oesterhelt D., Ferrando E., Schegk E. S., Lottspeich F. Primary structure of sensory rhodopsin I, a prokaryotic photoreceptor. EMBO J. 1989 Dec 20;8(13):3963–3971. doi: 10.1002/j.1460-2075.1989.tb08579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogomolni R. A., Stoeckenius W., Szundi I., Perozo E., Olson K. D., Spudich J. L. Removal of transducer HtrI allows electrogenic proton translocation by sensory rhodopsin I. Proc Natl Acad Sci U S A. 1994 Oct 11;91(21):10188–10192. doi: 10.1073/pnas.91.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braiman M. S., Mogi T., Marti T., Stern L. J., Khorana H. G., Rothschild K. J. Vibrational spectroscopy of bacteriorhodopsin mutants: light-driven proton transport involves protonation changes of aspartic acid residues 85, 96, and 212. Biochemistry. 1988 Nov 15;27(23):8516–8520. doi: 10.1021/bi00423a002. [DOI] [PubMed] [Google Scholar]
- Butt H. J., Fendler K., Bamberg E., Tittor J., Oesterhelt D. Aspartic acids 96 and 85 play a central role in the function of bacteriorhodopsin as a proton pump. EMBO J. 1989 Jun;8(6):1657–1663. doi: 10.1002/j.1460-2075.1989.tb03556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druckmann S., Ottolenghi M., Rousso I., Friedman N., Sheves M. Time-resolved titrations of the Schiff base and of the Asp85 residue in artificial bacteriorhodopsins. Biochemistry. 1995 Sep 19;34(37):12066–12074. doi: 10.1021/bi00037a050. [DOI] [PubMed] [Google Scholar]
- Ehrlich B. E., Schen C. R., Spudich J. L. Bacterial rhodopsins monitored with fluorescent dyes in vesicles and in vivo. J Membr Biol. 1984;82(1):89–94. doi: 10.1007/BF01870735. [DOI] [PubMed] [Google Scholar]
- Eisfeld W., Pusch C., Diller R., Lohrmann R., Stockburger M. Resonance Raman and optical transient studies on the light-induced proton pump of bacteriorhodopsin reveal parallel photocycles. Biochemistry. 1993 Jul 20;32(28):7196–7215. doi: 10.1021/bi00079a017. [DOI] [PubMed] [Google Scholar]
- Ferrando-May E., Brustmann B., Oesterhelt D. A C-terminal truncation results in high-level expression of the functional photoreceptor sensory rhodopsin I in the archaeon Halobacterium salinarium. Mol Microbiol. 1993 Sep;9(5):943–953. doi: 10.1111/j.1365-2958.1993.tb01224.x. [DOI] [PubMed] [Google Scholar]
- Ferrando-May E., Krah M., Marwan W., Oesterhelt D. The methyl-accepting transducer protein HtrI is functionally associated with the photoreceptor sensory rhodopsin I in the archaeon Halobacterium salinarium. EMBO J. 1993 Aug;12(8):2999–3005. doi: 10.1002/j.1460-2075.1993.tb05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwert K., Souvignier G., Hess B. Simultaneous monitoring of light-induced changes in protein side-group protonation, chromophore isomerization, and backbone motion of bacteriorhodopsin by time-resolved Fourier-transform infrared spectroscopy. Proc Natl Acad Sci U S A. 1990 Dec 15;87(24):9774–9778. doi: 10.1073/pnas.87.24.9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupts U., Eisfeld W., Stockburger M., Oesterhelt D. Sensory rhodopsin I photocycle intermediate SRI380 contains 13-cis retinal bound via an unprotonated Schiff base. FEBS Lett. 1994 Dec 12;356(1):25–29. doi: 10.1016/0014-5793(94)01226-1. [DOI] [PubMed] [Google Scholar]
- Haupts U., Haupts C., Oesterhelt D. The photoreceptor sensory rhodopsin I as a two-photon-driven proton pump. Proc Natl Acad Sci U S A. 1995 Apr 25;92(9):3834–3838. doi: 10.1073/pnas.92.9.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelka W. A., Henderson R., Heymann J. A., Oesterhelt D. Projection structure of halorhodopsin from Halobacterium halobium at 6 A resolution obtained by electron cryo-microscopy. J Mol Biol. 1993 Dec 5;234(3):837–846. doi: 10.1006/jmbi.1993.1629. [DOI] [PubMed] [Google Scholar]
- Hegemann P., Oesterbelt D., Steiner M. The photocycle of the chloride pump halorhodopsin. I: Azide-catalyzed deprotonation of the chromophore is a side reaction of photocycle intermediates inactivating the pump. EMBO J. 1985 Sep;4(9):2347–2350. doi: 10.1002/j.1460-2075.1985.tb03937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., Downing K. H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990 Jun 20;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Heytler P. G. Uncouplers of oxidative phosphorylation. Methods Enzymol. 1979;55:462–442. doi: 10.1016/0076-6879(79)55060-5. [DOI] [PubMed] [Google Scholar]
- Krah M., Marwan W., Verméglio A., Oesterhelt D. Phototaxis of Halobacterium salinarium requires a signalling complex of sensory rhodopsin I and its methyl-accepting transducer HtrI. EMBO J. 1994 May 1;13(9):2150–2155. doi: 10.1002/j.1460-2075.1994.tb06491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyi J. K. Mechanism of base-catalyzed Schiff base deprotonation in halorhodopsin. Biochemistry. 1986 Oct 21;25(21):6706–6711. doi: 10.1021/bi00369a057. [DOI] [PubMed] [Google Scholar]
- Le Coutre J., Tittor J., Oesterhelt D., Gerwert K. Experimental evidence for hydrogen-bonded network proton transfer in bacteriorhodopsin shown by Fourier-transform infrared spectroscopy using azide as catalyst. Proc Natl Acad Sci U S A. 1995 May 23;92(11):4962–4966. doi: 10.1073/pnas.92.11.4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi T., Stern L. J., Marti T., Chao B. H., Khorana H. G. Aspartic acid substitutions affect proton translocation by bacteriorhodopsin. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4148–4152. doi: 10.1073/pnas.85.12.4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D., Tittor J., Bamberg E. A unifying concept for ion translocation by retinal proteins. J Bioenerg Biomembr. 1992 Apr;24(2):181–191. doi: 10.1007/BF00762676. [DOI] [PubMed] [Google Scholar]
- Olson K. D., Deval P., Spudich J. L. Absorption and photochemistry of sensory rhodopsin--I: pH effects. Photochem Photobiol. 1992 Dec;56(6):1181–1187. doi: 10.1111/j.1751-1097.1992.tb09743.x. [DOI] [PubMed] [Google Scholar]
- Olson K. D., Spudich J. L. Removal of the transducer protein from sensory rhodopsin I exposes sites of proton release and uptake during the receptor photocycle. Biophys J. 1993 Dec;65(6):2578–2585. doi: 10.1016/S0006-3495(93)81295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath P., Olson K. D., Spudich J. L., Rothschild K. J. The Schiff base counterion of bacteriorhodopsin is protonated in sensory rhodopsin I: spectroscopic and functional characterization of the mutated proteins D76N and D76A. Biochemistry. 1994 May 10;33(18):5600–5606. doi: 10.1021/bi00184a032. [DOI] [PubMed] [Google Scholar]
- Rudolph J., Oesterhelt D. Chemotaxis and phototaxis require a CheA histidine kinase in the archaeon Halobacterium salinarium. EMBO J. 1995 Feb 15;14(4):667–673. doi: 10.1002/j.1460-2075.1995.tb07045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobert B., Lanyi J. K. Halorhodopsin is a light-driven chloride pump. J Biol Chem. 1982 Sep 10;257(17):10306–10313. [PubMed] [Google Scholar]
- Seidel R., Scharf B., Gautel M., Kleine K., Oesterhelt D., Engelhard M. The primary structure of sensory rhodopsin II: a member of an additional retinal protein subgroup is coexpressed with its transducer, the halobacterial transducer of rhodopsin II. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):3036–3040. doi: 10.1073/pnas.92.7.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. O., Lugtenburg J., Mathies R. A. Determination of retinal chromophore structure in bacteriorhodopsin with resonance Raman spectroscopy. J Membr Biol. 1985;85(2):95–109. doi: 10.1007/BF01871263. [DOI] [PubMed] [Google Scholar]
- Spudich E. N., Spudich J. L. The photochemical reactions of sensory rhodopsin I are altered by its transducer. J Biol Chem. 1993 Aug 5;268(22):16095–16097. [PubMed] [Google Scholar]
- Spudich J. L., Bogomolni R. A. Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature. 1984 Dec 6;312(5994):509–513. doi: 10.1038/312509a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L. Color sensing in the Archaea: a eukaryotic-like receptor coupled to a prokaryotic transducer. J Bacteriol. 1993 Dec;175(24):7755–7761. doi: 10.1128/jb.175.24.7755-7761.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L. Protein-protein interaction converts a proton pump into a sensory receptor. Cell. 1994 Dec 2;79(5):747–750. doi: 10.1016/0092-8674(94)90064-7. [DOI] [PubMed] [Google Scholar]
- Takei H., Gat Y., Sheves M., Lewis A. Low temperature FTIR study of the Schiff base reprotonation during the M-to-bR backphotoreaction: Asp 85 reprotonates two distinct types of Schiff base species at different temperatures. Biophys J. 1992 Dec;63(6):1643–1653. doi: 10.1016/S0006-3495(92)81757-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittor J., Soell C., Oesterhelt D., Butt H. J., Bamberg E. A defective proton pump, point-mutated bacteriorhodopsin Asp96----Asn is fully reactivated by azide. EMBO J. 1989 Nov;8(11):3477–3482. doi: 10.1002/j.1460-2075.1989.tb08512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl R., Meyer B., Desel H. A polychromatic flash photolysis apparatus (PFPA). J Biochem Biophys Methods. 1984 Nov;10(1-2):35–48. doi: 10.1016/0165-022x(84)90048-4. [DOI] [PubMed] [Google Scholar]
- Váró G., Lanyi J. K. Pathways of the rise and decay of the M photointermediate(s) of bacteriorhodopsin. Biochemistry. 1990 Mar 6;29(9):2241–2250. doi: 10.1021/bi00461a006. [DOI] [PubMed] [Google Scholar]
- Yan B., Spudich J. L. Evidence that the repellent receptor form of sensory rhodopsin I is an attractant signaling state. Photochem Photobiol. 1991 Dec;54(6):1023–1026. doi: 10.1111/j.1751-1097.1991.tb02125.x. [DOI] [PubMed] [Google Scholar]
- Yao V. J., Spudich J. L. Primary structure of an archaebacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]