Abstract
Transcription activator-like effector nucleases (TALENs) emerged as powerful tools for locus-specific genome engineering. Due to the ease of TALEN assembly, the key to streamlining TALEN-induced mutagenesis lies in identifying efficient TALEN pairs and optimizing TALEN mRNA injection concentrations to minimize the effort to screen for mutant offspring. Here we present a simple methodology to quantitatively assess bi-allelic TALEN cutting, as well as approaches that permit accurate measures of somatic and germline mutation rates in Drosophila melanogaster. We report that percent lethality from pilot injection of candidate TALEN mRNAs into Lig4 null embryos can be used to effectively gauge bi-allelic TALEN cutting efficiency and occurs in a dose-dependent manner. This timely Lig4-dependent embryonic survival assay also applies to CRISPR/Cas9-mediated targeting. Moreover, the somatic mutation rate of individual G0 flies can be rapidly quantitated using SURVEYOR nuclease and capillary electrophoresis, and germline transmission rate determined by scoring progeny of G0 outcrosses. Together, these optimized methods provide an effective step-wise guide for routine TALEN-mediated gene editing in the fly.
Keywords: engineered endonuclease, genome engineering, mutagenesis, screening, TALEN
Abbreviations: TALENs, Transcription activator-like effector nucleases; TALEs, TAL effectors; ZFNs, Zinc Finger Nucleases; CRISPR, Clustered Regularly Interspersed Short Palindromic Repeats; Cas9, CRISPR-associated; RVDs, repeat-variable diresidues; DSBs, double-stranded breaks; NHEJ, non-homologous end joining; HR, homologous recombination; RFLP, restriction fragment length polymorphism; HRMA, high resolution melt analysis.
Introduction
Targeting specific genomic loci using customizable endonucleases expanded the horizon of genome engineering. Three types of targetable-endonuclease systems, Zinc Finger Nucleases (ZFNs), 1,2 Transcription Activator-Like Effector Nucleases (TALENs), 3-5 and Clustered Regularly Interspersed Short Palindromic Repeats (CRISPR)/CRISPR-associated (Cas9) 6-12 were applied to modify genomic loci of interest in Drosophila melanogaster. Design and construction of TALENs is considerably easier than ZFNs, 5,13 and TALENs have the additional advantage of high targeting specificity 14,15 without significant off-target DNA cleavages. Although the application of TALENs made mutagenesis much easier, establishing a mutant strain still requires a considerable effort. While current TALEN assembly methods facilitate rapid production of TALENs, the key to streamlining TALEN-induced mutagenesis lies in identifying efficient TALEN pairs and optimizing TALEN mRNA injection concentrations to minimize the effort required to screen for mutant offspring. Once an efficient TALEN pair is identified, the subsequent mutagenesis screening workload is greatly reduced, because the number of offspring needed is minimal.
The modular composition of the DNA-binding units of TAL effectors (TALEs) was applied in genome engineering since the DNA-binding “code” of TALEs was deciphered. 16,17 An engineered TALEN monomer comprises a DNA-binding domain and endonuclease domain ( Fig. 1A ). The DNA-binding domain we adopted for TALEN design contains modular nucleotide recognition units repeated 14.5 times for optimal DNA recognition and cleavage efficiencies. 18 Each modular unit consists of 34 amino acid residues, the twelfth and thirteenth of which are highly variable (repeat-variable diresidues; RVDs) and are responsible for the one-to-one binding between a modular DNA binding unit and a nucleotide in the DNA molecule. 16,17,19 The type IIS endonuclease, FokI, is fused to the C-terminal end of the DNA-binding domain of TALEs, 20-22 as was first accomplished in ZFNs. 23 Two TALEN monomers are required to make a DNA cut, since FokI cleaves a DNA molecule as an obligate dimer. 24,25 This dimeric requirement increases the specificity of TALEN pairs. In successful applications published to date, TALENs have generated loss-of-function mutations (frame-shift or large deletion mutations) 4,21,26 or introduced desirable sequences such as recombinase integration sequence, 27 reporters, 28 and sequence corrections for gene therapy. 29 However, even with these successful applications, the rates of somatic and germline mutations induced by any given TALEN pair varied widely. 3,18,30,31 The varying rates of mutagenesis may come from intrinsic properties of the TALEN proteins, epigenetic environments of genomic loci, extraneous experimental conditions, or other unknown causes. Currently, detecting an efficient TALEN pair usually depends on sophisticated molecular assays such as restriction fragment length polymorphism (RFLP) and high resolution melt analysis (HRMA). Thus, a more convenient method to identify highly efficient TALEN pairs will facilitate the utility of TALEN applications. Here we present a genetic test for assessing TALEN efficiency, which relies on a DNA Ligase IV (Lig4) knockout fly line.
Figure 1.
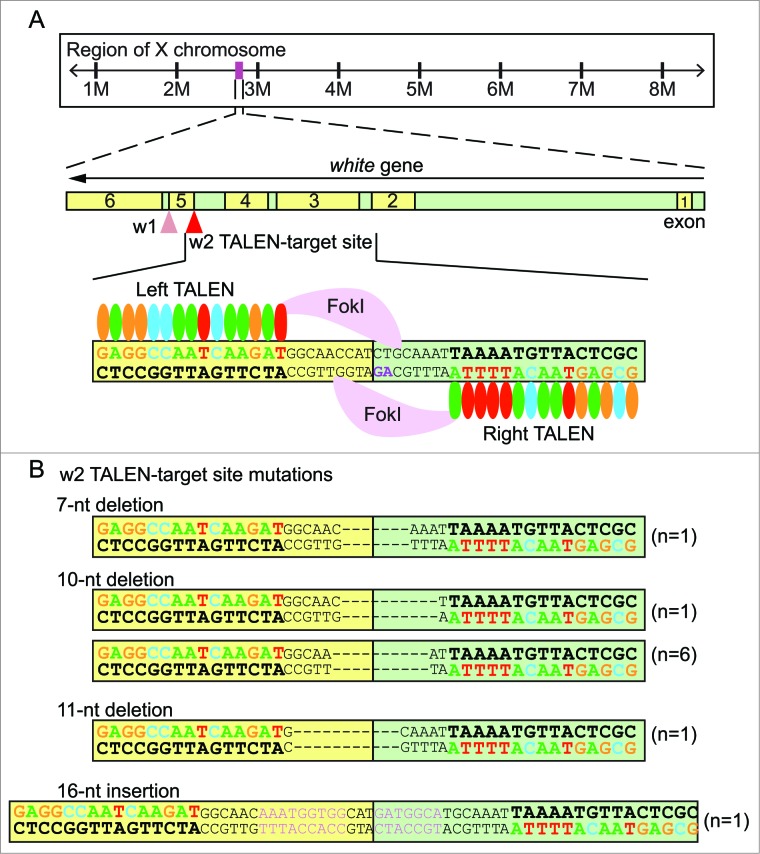
White gene TALEN design and mutagenesis. (A) The white gene locus on X chromosome and design of white TALEN pair 1 (w1 TALEN) and pair 2 (w2 TALEN). w1 TALEN targets the junction between exon 5 (yellow) and intron 5 (green). w2 TALEN targets the junction between intron 4 and exon 5. w2 TALEN arms (left and right) are represented as simplified RVD repeats: each colored oval representing a particular RVD recognizing a given nucleotide, the DNA recognition sequences are highlighted in matching color, and the conserved splicing acceptor dinucleotide 5′-AG-3′ sequence is highlighted in magenta. (B) Nature of mutations introduced by w2 TALEN. Among the 10 independent single G1 flies, 9 showed short deletion mutations of varying lengths (7-, 10-, and 11-nucleotide deletions). One G1 fly showed a short insertion of 16 nucleotides (pink).
Results
Construction of TALENs targeting the white gene
Two independent pairs of TALENs targeting the junctions between exon 5 and intron 5 (w1), and intron 4 and exon 5 (w2) in the white gene were constructed (white TALENs) (shown in Fig. 1A). Disrupting the exon-intron junction is an effective strategy to abolish gene function because mutations in the splicing donor or acceptor site interfere with splicing events of the mRNA (mRNA). If the splicing events are not affected, insertion- or deletion-mutations (indels) in the coding region result in frame-shift mutations, rendering the targeted protein non-functional. The targeted sequences fall within the first α-helix of the 6 α-helices bundle in the transmembrane domain of the white protein.32 Disrupting the white protein in its transmembrane domain is likely to affect its membrane organization, molecular assembly and function. By generating mutations in the white gene, we intended to take advantage of the white null phenotype, i.e. white eyes. Indeed, the white TALENs generated eyes with white patches (mutagenesis in the somatic photoreceptor cells) in G0 flies (Fig. 2A). Importantly, these mutations were readily transmittable to subsequent generations, and the nature of predicted mutations have been confirmed by sequencing the mutant G1 flies (Fig. 1B, see following result sections for details). We first present targeting data using w2 TALEN pairs throughout these experiments since it showed a higher efficiency than w1 TALEN pairs (shown in a later section).
Figure 2.
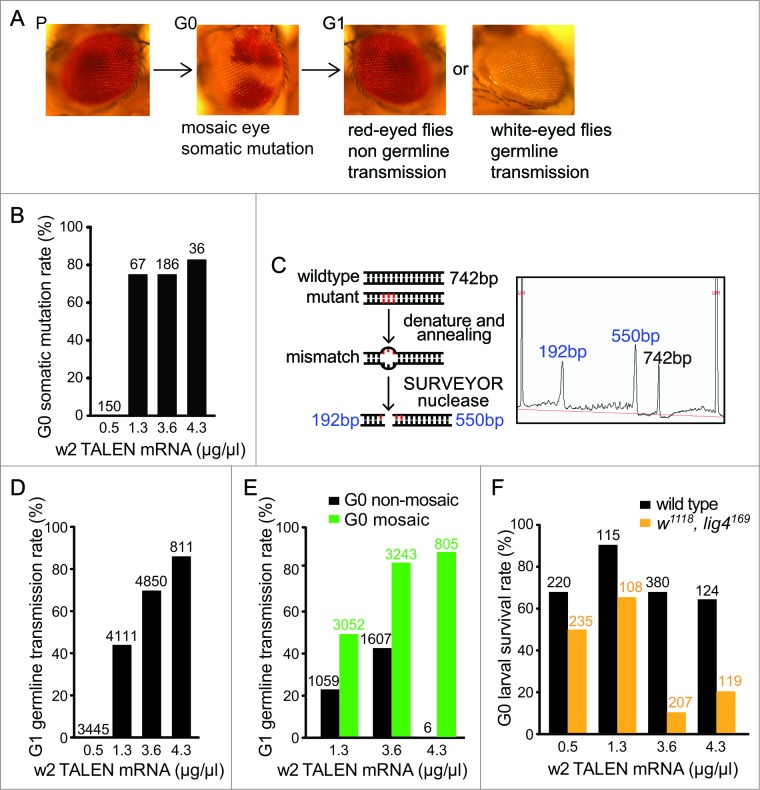
w2 TALEN-introduced somatic mutations and germline transmission. (A) The scheme of eye-color change introduced by w2 TALEN. From parental red-eyed flies (P), embryos injected w2 TALEN developed to G0 adults with mosaic eyes containing white patches. In G1 generation, white-eyed flies carried germline-transmitted mutations. (B) G0 somatic mutation rates at various w2 TALEN concentrations quantified by mosaic-eye phenotype. Numbers of G0 flies counted are listed on top of each bar. (C) The rational of quantifying global somatic mutations in individual G0 flies using SURVEYOR nuclease assay. Re-annealed amplicons around the predicted cut site undergone SURVEYOR nuclease treatment. The insert shows a representative result of digested product separation using Fragment Analyzer. Peaks at lower molecular weights (green) represent shorter DNA fragments generated by nuclease cut at a specific mutated site. See Figure S1 for more analysis and quantifications of gene modification rate. (D) G1 germline transmission rates at various w2 TALEN concentrations. The rate was represented by the percentage of white-eyed flies among total number of G1 progeny at each condition. Numbers of G1 flies counted are listed on top of each bar. (E) Comparisons of G1 germline transmission rates between progenies of non-mosaic eye (black bars) and mosaic eye G0 flies (green bars) at various w2 TALEN concentrations. Numbers of G1 flies counted are listed on top of each bar. (F) G0 larval survival rates at various w2 TALEN concentrations. This rate was represented by the percentage of injected embryos developed into first-instar larvae within 48 hours post injection from either wild-type (black bars) or lig4169 embryos (orange bars). Numbers of G0 embryos counted are listed on top of each bar.
High rates of somatic mutations in the G0 generation
We first tested the w2 TALEN mRNAs at the concentration of 4.3 μg/μl/TALEN-arm. At this concentration, we observed a high rate of somatic mutations in the injected G0 flies (83%; 30 red-white-mosaic-eyed flies out of a total 36 adult flies eclosed) ( Fig. 2B and Table S1). The white gene locates on the X chromosome, so white patches in the eyes of female G0 flies were reflective of bi-allelic cleavage of the white gene locus. Since the injected TALEN mRNAs were introduced at the posterior tips of syncytial embryos, mosaicism observed in the developed eyes showed that TALEN mRNAs diffused to the anterior of embryos, where photoreceptor cells originate. After the success in observing a high frequency of mutagenesis in injected G0 flies, we began titrating the concentration of injected TALEN mRNAs. The survival and fertility rates of the embryos injected at the concentration of 4.3 μg/μl/TALEN-arm were 64% (79 larvae surviving out of a total 124 embryos injected) and 39% (14 flies were able to produce offspring out of a total 36 adult flies), respectively ( Fig. 2F and Table S2). We set out to identify the optimal concentration of TALEN mRNAs that would lead to a high rate of G0 somatic mutations with higher fertility rates, since diminished fertility might mean toxicity that could involve non-specific DNA cleavages.
We tested 3 other concentrations of w2 TALEN mRNAs (0.5, 1.3, and 3.6 μg/μl/TALEN-arm; Fig. 2B). At 0.5 μg/μl/TALEN-arm, we did not observe any G0 adult flies carrying somatic mutations (n = 150). As the concentrations of TALEN mRNAs increased, more G0 flies showed somatic mutations in their eyes. At 1.3 and 3.6 μg/μl/TALEN-arm concentrations, we observed similar rates of the flies carrying mosaic eyes (75%; 50/67 and 140/186 flies, respectively, Fig. 2B). However, the size of white patches in the eyes were generally larger at 3.6 μg/μl/TALEN-arm than at 1.3 μg/μl/TALEN-arm (data not shown); in most flies injected at 3.6 μg/μl/TALEN-arm, the majority of photoreceptor cells are white rather than red. When we quantified the survival and fertility rates at these concentrations, the survival rate of the embryos injected at 1.3 μg/μl/TALEN-arm was 90% (104/115) and fertility rate was 87% (58/67) (Table S2). At 3.6 μg/μl/TALEN-arm, the survival rate was 69% (262/380) and fertility rate was 33% (61/186). While this set of data clearly demonstrated that w2 TALENs were able to induce somatic mutations in injected flies, we also found that at 1.3 μg/μl/TALEN-arm concentration w2 TALEN could generate a high rate of somatic mutations (75%) in G0 flies with relatively low toxicity evidenced by high survival and fertility rates.
After establishing w2 TALEN generated somatic mutations that resulted in a mosaic eye phenotype in the injected G0 flies, we sought to determine the precise somatic mutation rates in individual flies. Beyond the white patches that we could observe from the mosaic eyes, there could be lesions on the white locus in other somatic cells that were not reflected by the morphological readout. We reasoned that unrestricted TALEN activity throughout an embryo is likely, given the observed mutant patches from the anterior derived photoreceptor cells. We quantified the global somatic mutation rates in G0 adult flies using the SURVEYOR™ nuclease assay (Fig. 2C).33,34 In this assay, the genomic DNA around the TALEN-target site was amplified with flanking primers, and subsequently underwent denaturation and re-annealing. SURVEYOR nuclease digestion was performed with the re-annealed DNA; when the double-stranded DNA contained a mismatch (arising from hybridization between a wild type strand and a complementary strand bearing mutation), the nuclease generated site-specific cleavage at the mismatched site. The cleaved, two smaller fragments of DNAs were separated and subsequently quantitated using a fragment analyzer together with the single full length DNA fragment (Fig. 2C). Among the individual G0 flies randomly selected for the assay were those injected at 3.6 and 4.3 μg/μl/TALEN-arm concentrations. The rate of site-specific modification was as high as 40%, and ranged between 25% and 40% (Fig. S1). This meant that up to 80% of all somatic cells in the injected G0 flies could have at least one mutated chromosome, which was consistent with our observation of flies with mosaic eyes.
High rates of germline transmission in the G1 generation
Being able to transmit mutations to the next generation is one of the most critical requirements for most genome engineering applications. After confirming a high activity of the w2 TALENs in generating somatic mutations, we asked whether the mutagenesis was also efficient in germline cells and heritable. To evaluate the efficiency of germline transmission of TALEN-induced mutations, we crossed the TALEN-injected OR flies with w1118 flies (FlyBase ID: FBal0018186, carrying a deletion on 5′ part of white gene) and quantified the number of white-eyed flies among the total G1 flies (Fig. 2D). Male G1 flies from TALEN-injected male G0 flies were not scored because males contribute no X chromosome to their male offspring.
At 0.5 μg/μl/TALEN-arm concentration, the number of mutant flies in the G1 generation was negligible (0.05%; two white-eyed flies/3445 flies). However, the number of mutant flies with white eyes sharply increased to 43% (1758/4111) at 1.3 μg/μl/TALEN-arm. At 3.6 μg/μl/TALEN-arm, the mutant progeny was 69% (3337/4850) and, at 4.3 μg/μl/TALEN-arm, it reached 86% (699/811) (Fig. 2D). The mutation rate obtained through germline transmission was TALEN-dosage dependent. Once we found that there was an association between the concentration of TALEN mRNAs and the percentage of G1 mutant progeny, we asked whether there could be a direct association between the rates of somatic and germline mutations. Indeed, when we broke down the population of G0 flies into groups of detectable and undetectable white-eyed patches, we found the association that G0 flies with mosaic eyes gave rise to a significantly higher percentage of G1 mutant progeny, compared to those from G0s without visible somatic mutations (Fig. 2E and Table S3). For example, at 1.3 μg/μl/TALEN-arm, G0 flies with mosaic eyes produced 50% of mutant G1 progeny (1514/3052) whereas G0 flies without mosaic eyes produced only 23% of mutant G1 progeny (244/1059). At 3.6 and 4.3 μg/μl/TALEN-arm, 82% (2651/3243) and 87% (699/805) of the G1 progeny from G0 flies with mosaic eyes were mutants whereas 43% (686/1607) and 0% (0/6) of the G1 progeny from G0 flies without mosaic eyes were mutants, respectively.
In agreement with our observation that germline mutation could be passed from G0 flies without visually detectable mosaicism, though at a lower rate, we have confirmed that the vast majority of the TALEN-injected G0 flies carried mutations in their germline. Around 86% of the fertile G0 flies injected with 1.3 μg/μl/TALEN-arm (51/59) and 4.3 μg/μl/TALEN-arm (12/14) gave rise to at least one mutant progeny (Fig. S2). Therefore, our findings demonstrate a highly potent germline transmittable mutagenesis approach, though obtaining a mutant fly strain is more easily accomplished from the G0 flies with detectable mosaicism.
Once the association between somatic and germline mutations was uncovered, we sought to identify the nature of mutations transmitted in G1 flies. We sequenced 10 independent mutant G1 flies (Fig. 1B). Three out of 10 mutations were in-frame deletions removing only 3 nucleotides of exon 5 (n = 2) or inserting 9 nucleotides (n = 1). The other 7 were frame-shift mutations, deleting 4 (n = 6) or 8 (n = 1) nucleotides of exon 5. All ten mutants showed a disruption in the 3′ splice site (AG) at the end of intron 4 (highlighted purple in Fig. 1A). Thus, in the cases of the in-frame mutations, non-functional proteins might be produced from the mis-splicing events at the exon 5-intron 4 junction.
Rapid assessment of TALEN efficiency using the Lig4 null fly line
Though we were able to observe TALEN cutting at the white gene by mosaic eye phenotypes in G0 flies, many mutations in other target genes produce no morphologically observable phenotypes. If we could quickly and conveniently assess the efficiency of a TALEN pair without depending on phenotypic markers or molecular assays, the application of TALENs to generate mutant fly strains could be even easier. It is thought that TALEN-induced double-stranded breaks (DSBs) are repaired mainly by two independent mechanisms: non-homologous end joining (NHEJ) and homologous recombination (HR).35,36 When homologous sequences are not available, NHEJ is the preferred pathway to repair DSBs. DNA Ligase IV (Lig4) in a complex with the XRCC4 (X-ray repair complementing defective repair in Chinese hamster cells 4) protein takes part in the last step of the NHEJ process37 by sealing the nicks in the phosphodiester bond between 2 adjacent nucleotides. Lig4 null flies are viable whereas mammals that lack Lig4 are embryonic lethal due to cell death in the central nervous system.38-40 Although there could be Lig4-independent pathways that contribute to the NHEJ in Drosophila,40 Lig4 plays a critical role in various developmental stages to repair DSBs.39,41 Moreover, Lig4 null mutations were reported to suppress NHEJ31,41 and increase HR.43,44 In the case of bi-allelic cuts generated by TALENs, homologous donors are absent to repair DSBs through HR. Thus, we hypothesized that, in such a scenario, Lig4 null flies would be less capable of fixing the DSBs induced by TALENs due to the simultaneous blockage of NHEJ repair. A more severe embryonic lethality was expected specifically under circumstances wherein TALENs induced bi-allelic lesions.
Affirming our hypothesis, a higher percentage of lig4169 G0 flies died during embryonic development after w2 TALEN mRNA injection, specifically at higher concentrations (3.6 and 4.3 μg/μl/TALEN-arm) (Fig. 2F and Table S2). The survival rates in the lig4169 background were 10% (20/207) and 20% (25/119) at 3.6 and 4.3 μg/μl/TALEN-arm compared with a 47% baseline larval survival rate as observed from water-only injected lig416 flies (42/90). Given the approximately 40% G0 global somatic mutagenesis rate in the wild-type background under these conditions (Fig. S1), we expected that in a significant portion of somatic cells (predicted around 16% given 2 copies of same chromosome per cell in females) TALENs would induce cuts simultaneously on both chromosomes. However, at 1.3 μg/μl/TALEN-arm, we did not find a significant increase lethality in lig4169 background, with the larval survival rate at 65% (70/108). We speculated that, at a lower concentration, the chance of a TALEN-induced bi-allelic cut was much smaller and thus lesions could be mostly repaired with homology-based repair independent of Lig4. It is important to note that this significant concentration-dependent reduction in survival rate was only observed in the lig4169 background, since the lethality rates in w2 TALEN-injected wild-type flies in all concentrations tested were not significantly different from water injected controls (Fig. 2F and Table S2). Our findings demonstrate that the reduction in survival rate of Lig4 null flies was a rapid and reliable indication of efficient TALEN-induced bi-allelic cuts. To validate whether one could assess the efficiency of TALENs rapidly by performing a small-scale injection at high concentrations in lig4169 flies, we tested 2 other pairs of TALENs targeting 2 different positions in the Drosophila CG42678 gene using this methodology. The larval survival rates of corresponding TALEN-injected lig4169 flies were 1% (2/220) and 7% (15/225) at 6 μg/μl/TALEN-arm and 2% (2/118) at 8 μg/μl/TALEN-arm. Subsequent molecular analysis using SURVEYOR nuclease confirmed that these 2 independent pairs of TALENs indeed introduced site-specific indel mutations at high frequency (Lee et al., unpublished). Therefore, lethality scoring of TALEN mRNA injected Lig4 mutant embryos likely reflects the TALEN cutting efficiency.
It is possible that significant off-targeted lesions might be generated when TALEN is introduced at a high dosage. While nonspecific lesions could be potentially toxic to cells, we postulate that the Lig4 depletion dependent embryonic lethality is not likely due to the potential off-targeted lesions, since similar levels of off-targeted lesions were introduced to Lig4+ embryos without a significant increase of lethality. To further validate the cause of Lig4 embryonic lethality, we examined the dosage-dependent survival rate after injecting a far less potent TALEN pair against the white gene (w1). When injected into wild-type embryos at 3 concentrations (1, 2.2, and 4 μg/μL/TALEN-arm), we did not observe any developed G0 flies with mosaic eyes (n = 104) as in the case for w2 TALEN pair. Among the progenies of fertile G0 flies, only 0.77% of G1 lost eye color (35 out of 4532 total progeny). Consistent with the apparent low targeting efficiency reflected by eye color morphological marker, we confirmed that w1 is indeed a poor pair of TALEN targeting the white gene since no obvious indel-dependent surveyor nuclease cleavage product was detected from 8 randomly selected G0 individuals (Fig. S3) at the highest TALEN injection dosage. Consistent with our hypothesis that the Lig4-dependent embryo lethality is associated with efficient bi-allelic targeting, we did not observe significant lower embryonic survival rate of lig4169 embryos compared with those of wild-type embryos receiving the same dosage of w1 TALEN mRNA. Thus the lig4169 embryo lethality test is indeed a true and rapid indicator of TALEN targeting efficiency.
Cleavage-dependent lig4169 embryo lethality assay can be applied to the CRISPR/Cas9 system
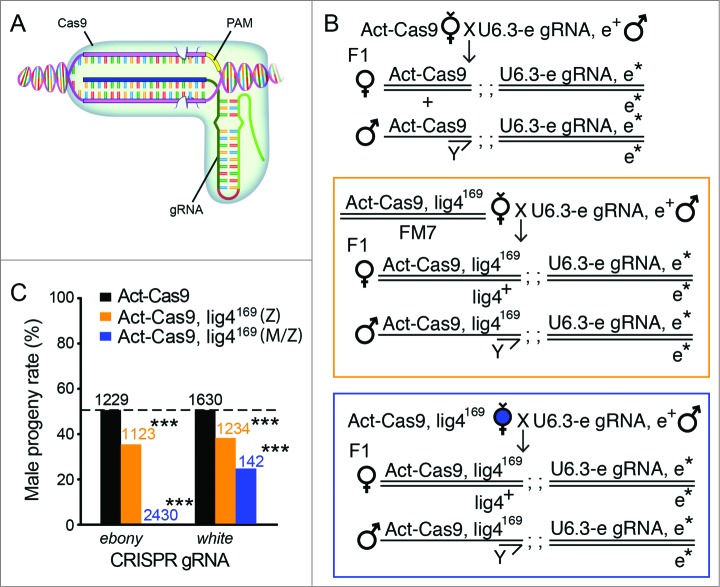
Our analysis has indicated that the efficiency of gene targeting for a TALEN pair can be rapidly assayed by injecting TALENs into lig4169 embryos. To generalize the usefulness of this approach, we examined whether similar cleavage-dependent lethality readouts can be applied to other engineered nuclease systems, such as the CRISPR/Cas9 system. CRISPR/Cas9 system employs a short guide RNA (gRNA)-mediated target recognition and subsequent gRNA-loaded Cas9 endonuclease mediated DNA cleavage (Fig. 3A).45 Recently, we and others have developed simplified CRISPR/Cas9 platforms in Drosophila without directly injecting the targeting designer nuclease into the embryos, by using endogenously expressed source of Cas9 protein and gRNAs through transgenesis.10,12,46 Through genetic crosses, Cas9 and gRNA expressing transgenes can be brought together to achieve gene targeting. For example, crossing strain ubiquitously expressing Cas9 protein (Act-Cas9) with the one containing a gRNA-expressing construct against the ebony gene (U6.3-ebony gRNA) produced dark coloration of the adult cuticle in almost all progenies (Fig. 3B, top cross scheme, all F1 had dark cuticle),46 suggesting highly efficient bi-allelic Cas9 targeting at the ebony locus. We also crossed the Act-Cas9 strain with a strain expressing 2 gRNA-expressing constructs against the white gene (U6.2-white gRNAx2).12 In F1 generation, all male progenies contained mosaic eyes with white patches of varying sizes, this indicated the single copy of functional white gene on the single male X chromosome was frequently disrupted; much fewer female progenies contained mosaic eyes, likely due to lower efficient bi-allelic disruption of the white gene (one copy at the white locus and one copy in the Act-Cas9 transgene).
Figure 3.
Cleavage-dependent lig4169 embryo lethality with the CRISPR/Cas9 system. (A) A cartoon diagram of the CRISPR/Cas9 system, which recognizes specific DNA using a RNA/DNA/protein complex (modified from our recent review article61). The 5′ end of gRNA sequence is used for target recognition on genomic DNA around a tri-nucleotide protospacer adjacent motif (PAM). Two tooth-shape structures represent Cas9 endonuclease active sites responsible for DNA cleavage on either stand of the double strand DNA. (B) Fly genetic crossing schemes for ubiquitous gene targeting using endogenously expressed Cas9 (Act-Cas9) and gRNAs (U6.3-ebony gRNA as an example here). F1 progenies bearing both transgenes activate the CRISPR/Cas9 system ubiquitously. Bi-allelic cuts on the target gene disrupt both copies of the locus (ebony, donated with asterisk). On the middle and bottom panel, lig4169 was recombined to Act-Cas9 chromosome to assay the contribution of Lig4 in animal survival in the gene-targeted progenies. While gene targeting is predicted to be equivalently efficient in progenies of either sex, the male progeny alone in both cases are zygotic hemizygous for lig4169 mutation. Sharing the same progeny genotypes, the middle (orange box) and bottom panel (blue box) differ by their maternal genotypes: heterozygous of lig4169 for the middle panel, and homozygous of lig4169 for the bottom panel. The homozygous mothers generate both maternal and zygotic null male progenies for Lig4. (C) The percentage of male progenies surviving ubiquitous CRISPR-mediated gene targeting 2 independent loci (ebony or white gRNAs), using Act-Cas9 (blank bars); Act-Cas9, lig4169 zygotic mutant only (Z, orange bars) or Act-Cas9, lig4169 maternal and zygotic mutant (M/Z, blue bars). The statistic deviation from the theoretical 50% percent perfect ratio was tested by student t-test (***P < 0.0001).
Both gRNAs for ebony and white were potent, we next used these gRNAs to test whether Lig4 is critical for fly survival with CRISPR-mediated bi-allelic double stranded breaks. Since both Lig4 locus and Act-Cas9 transgene resided on the X-chromosome, we recombined Act-Cas9 and lig4169 onto the same X-chromosome (Act-Cas9, lig4169) for simplifying further genetic analysis. Male progenies from female Act-Cas9, lig4169 flies would be zygotic hemizygous for lig4169 mutation, whereas female progenies would be heterozygous for lig4169 (Fig. 3B, middle and bottom cross scheme). The functional Lig4 gene in female progenies was expected to repair CRISPR-mediated double stranded breaks efficiently even without homologous sequences, then generate survival advantage compared to the male siblings. As expected, using Act-Cas9 to drive gRNAs targeting ebony and white genes, the male: female progeny ratios were not significantly deviant from 1:1 theoretical rate (male progeny rates: 50.7% N = 1229 for ebony gRNA; 50.9% N = 1630 for white gRNA, Fig. 3C black bars). However, under zygotic Lig4 null background (Act-Cas9, Lig4169), the male progeny rates were significantly lower (35.2% N = 1123 for ebony gRNA, and 37.5% N = 1234 for white gRNA, Fig. 3C orange bars). Proportional inference analysis concluded that the occurrence of targeted male progeny under Lig4 null background was statistically lower than 50% theoretical ratio (with P values of 1.0E-25 and 2.5E-17, respectively), whereas no sex preference was observed without Lig4 mutation, or the maternal Act-Cas9, lig4169/FM7 strain itself (with P values of 0.34, 0.30 and 0.41 respectively).
The significant residual survival of zygotic lig4 mutant under efficient CRISPR targeting inspired us to further establish a rigorous relationship between embryonic survival under efficient bi-allelic designer nuclease targeting and the integrity of the Lig4 dependent canonic NHEJ DNA repair machinery. In the previous experiment, even though no functional Lig4 protein can be made in all male progeny bearing the recombinant Act-Cas9, lig4169 chromosome, we speculated residual maternally contributed Lig4 proteins deposited to the zygotic mutant embryos might be responsible for their survival. Thus we repeated the cross using homozygous Act-Cas9, lig4169 females. While lig4169 homozygous flies are viable and fully fertile, they don't contribute Lig4 proteins directly to their progenies. Similar to our observation on lig4 embryonic lethality injected with high dosage of W2 TALENs, we recover fewer lig4 mutant male progenies after CRISPR targeting using white gRNAs (24.6%, N = 142, Fig. 3C, right blue bar), compared to when maternal Lig4 pool is not depleted (37.5%, N = 1234; Fig. 3C, right orange bar). Strikingly, with the more potent ebony gRNA, not a single surviving male progeny can be recovered with repeated crosses from homozygous Act-Cas9, lig4169 females (0%, N = 2430; Fig. 3C, left blue bar). While this result highlights the significant contribution of maternal deposited Lig4 protein, we found the zygotic transcription is also largely capable to rescue the embryonic lethality, since we obtain a roughly normal number of female progenies from such ebony targeting crosses. In summary, we concluded Lig4 null fly embryos are sensitive to efficient gene targeting using designer nucleases (either TALEN or CRISPR), especially when the targeting efficiency is potent enough that bi-allelic cuts are prevalent. The lethality rates of Lig4 mutant embryos correlate well with targeting efficiency (comparing W1 and W2 TALENs for direct embryonic injection; white and ebony gRNAs for genetic manipulations), especially with the complete absence of residual maternal Lig4 protein. Thus we recommend this method to be a quick, economic and reliable assay to estimate the efficiency of gene targeting in vivo via designer nucleases.
Discussion
Different methods to evaluate TALEN efficiency
There are several methods to evaluate the efficiency of site-specific mutagenesis induced by customizable nucleases in G0 animals: restriction fragment length polymorphism (RFLP),18,27 high-resolution melt analysis (HRMA),5,7,47 T7 endonuclease assay,48 single strand annealing (SSA) assay,4,49 LacZ assay,50 and SURVEYOR nuclease assay.51-53 A common step in these molecular assays is a PCR amplification of the locus of interest, followed by hybridization, enzymatic digestion, reporter assays, or gel electrophoresis. We applied the SURVEYOR nuclease assay followed by a fragment analysis because the assay provides 2 advantages over other methods. The SURVEYOR nuclease assay is a modified RFLP, and reveals both the presence of mutations and the position of mutations unlike the HRMA methods that only informs the presence of sequence variants and requires highly specific amplification of the target DNA. Moreover, it does not depend on specific restriction sites, unlike the RFLP technique, because it cleaves DNAs at mismatch or bulging sites, making TALEN-design less confining.
Although these molecular assays are indispensable in quantifying the rates of somatic mutations in G0 animals, a convenient methodology to rapidly assess the efficiency of designer nucleases is lacking. We addressed this deficit by using percent lethality of Lig4 null flies as a readout for mutagenic efficiency of TALEN pairs. Taking advantage of the high lethality of Lig4 null flies upon TALEN-induced bi-allelic cuts, we counted the number of surviving larvae 2 days after TALEN microinjection and demonstrated that the decreased survival rate of injected embryos is highly dependent on TALEN mRNA dosage (Fig. 2F).
However, if targeted genes are essential for embryonic development, TALENs may cause a significant reduction in the survival rate of both wild-type flies and Lig4 null flies. Therefore, for these genes, percent lethality upon TALEN injection in wild-type flies will be sufficient to indicate TALEN efficiency.
Factors that may affect TALEN efficiency
There are several factors that can affect the efficiency of a TALEN pair including TALEN mRNA concentrations, chromatin accessibility of a locus, polymorphisms of the DNA-binding sites of TALENs, and extraneous experimental conditions. It seems that the efficiency of a TALEN pair does not only depend on DNA-binding module binding strength (repeat-variable diresidues; RVDs) of TALENs18,19,21,53; but a combination of factors determine mutagenic efficiency. Several studies that applied TALENs in Drosophila melanogaster used a range of TALEN mRNA concentrations from 0.2 μg/μl to 0.4 μg/μl5 and approximately 0.5 μg/μl.3,4 In contrast to these applications, we used a much higher concentration of TALENs ranging from 0.5 to approximately 4 μg/μl/TALEN-arm. We did not observe any phenotypes in G0 flies with a TALEN mRNA concentration of 0.5 μg/μl/TALEN-arm (Fig. 2B) whereas Beumer et al. reported the phenotypic detection in approximately 45% of G0 flies injected with a TALEN mRNA (yT1-63).5 The difference may arise from the different target loci or chromatin accessibility of a particular locus.
In addition, the difference in TALEN mRNA concentrations is dependent on the animal model used. In crickets (Gryllus bimaculatus), 1 μg/μl/TALEN-arm of TALEN mRNAs was able to reliably generate mutations without notable toxic effects.53 In zebrafish (Danio rerio), the concentrations of TALEN mRNAs tolerated were approximately 20 ng/μl /TALEN-arm delivering 40-80 pg of mRNAs,18,27 which is much lower than the lowest TALEN mRNA concentration (200 ng/μl) applied in Drosophila and that in Gryllus (1 μg/μl). In medaka (Oryzias latipes), TALEN mRNA concentrations from 10 ng/μl to 300 ng/μl per TALEN arm were used.55 In murine embryos, TALEN mRNAs with concentrations of 20 and 50 ng/μl successfully generated mutations.56
The possibility of off-target effects caused by higher concentrations of TALEN mRNAs has raised some concern in the scientific community. The survival rates of TALEN-injected embryos did not show a significant difference with increasing TALEN concentration in the wild-type background (Fig. 2F). However, the fertility of the TALEN-injected G0 flies decreased at higher concentrations than 1.3 μg/μl/TALEN-arm (Table S2). At 3.6 and 4.3 μg/μl/TALEN-arm, the fertility of G0 flies were 33% and 39%, respectively, compared to 88% at 1.3 μg/μl/TALEN-arm. Thus, there might be undefined effects at higher concentrations of TALEN mRNAs such as non-specific DNA cleavages or more generalized toxic effects caused by high concentrations of TALEN mRNAs. To achieve both high somatic mutation and germline transmission rates and reduce non-specific or toxic effects, we propose a carefully titrated injection dosage with a starting concentration at 1∼1.5 μg/μl/TALEN-arm.
Dependence of designer nuclease targeted embryo survival on Lig4 dependent canonic NHEJ DNA repair pathway
While DNA Ligase IV (Lig4) /XRCC4 complex catalyze the final step of the NHEJ process37 by sealing the nicks in the phosphodiester bond between 2 adjacent nucleotides at DNA DSBs, Lig4 null flies are viable and fully fertile, albeit with elevated sensitivity to radiation and other DNA damaging agents.39,57,58 Now we have shown Lig4 dependent canonic NHEJ repair is indispensible for fly embryonic survival with highly efficient gene targeting. When bi-allelic cuts are prominent, the genome loses the backup template to repair DNA DSB via Homologous recombination, and thus depends on the NHEJ machinery to rejoin the broken chromosome. The embryonic lethality could be directly due to the failure for rejoining broken DNA without both canonic NHEJ and HR repair, or via stress signals triggered by prolonged broken DNA ends. Since even in the case of maternal and zygotic Lig4 mutant, when targeting with high dose W2 TALENs or ubiquitously activated white-targeting CRISPR system, we can obtain a small number of progeny bearing mosaic eyes. Once white gene is targeted in this case, the cell cannot fix the broken X chromosome either with canonic NHEJ pathway (due to the lack of Lig4 protein), or with homologous recombination in males (due to the lack of a homologous X chromosome). The survival of these cells (demonstrated as white patches on the photoreceptors) argues against that DSB can only be exclusively fixed by one of these 2 major DNA repair pathways. In contrast, our results favor the contribution from a poorly characterized Lig4-independent non-canonic NHEJ pathway to rejoin the broken DNA eventually, to ensure cell survival.59 This back-up DNA repair mechanism would be less efficient, leaving the DSB unfixed over prolonged periods compared to cells retaining HR and canonic NHEJ mechanism. We argue the widespread stress response from unfixed DNA damage from many cells in the targeted embryo leads to the lethality we observe.60 Comparing the white and ebony targeting CRISPR experiments in maternal and zygotic Lig4 null embryos, we demonstrate that more efficient targeting leads to significant less chance of the embryo to fix the breaks and continue its development. Thus Lig4 null embryo lethality is truly a very sensitive assay to estimate the efficiency of gene targeting. It is also interesting to note that fly embryos might have a specific sensitive period for DSB triggered stress, which could overlap with the perdurance of maternal Lig4 proteins. The less severe phenotype of zygotic null mutant alone could also be explained by the error-prone canonic NHEJ activity contributed by maternal Lig4 proteins might have changed the targeting locus in the majority of cells to prevent further efficient targeting.
Conclusions
We report that Lig4 null flies can be used as a strain to estimate the efficiency of a TALEN pair at higher concentrations. The same general rational also apply to other designer nuclease mediated genome engineering approaches such as CRISPR/Cas9 system. A rapid in vivo estimation of TALEN efficiency streamlines the process of TALEN-induced mutagenesis and establishment of mutant strains in Drosophila melanogaster. We also showed that TALEN-induced mutant progeny can be rapidly identified without toxic effects by doubling the concentration of TALEN mRNAs from those applied in other studies (approximately 0.5 μg/μl/TALEN-arm) to approximately 1.0 to 1.5 μg/μl/TALEN-arm. Drosophila is a premiere model organism that offers a number of genetic tools, fast generation time, and the advantage of statistical power due to numerous progeny. Our study optimized the application of TALENs in Drosophila, facilitating rapid mutagenesis and establishment of mutant strains, thereby substantiating TALEN technology as a valuable addition to the fly community.
Materials and Methods
Fly stock management and microinjection
All the fly stocks were maintained at room temperature (23-25°C). The fly strains used were Oregon R (FlyBase ID: FBst0000005), w1118 (FlyBase ID: FBal0018186), and w1118, lig4169 (FlyBase ID: FBal0176089). lig4169 is a null allele and was generated by imprecise excision of the EP385 P-element.40 Act-Cas9 and U6.3-ebony gRNA lines were generous gifts from Dr. Fillip Port at the University of Cambridge.46 U6.2-white gRNAx2 was obtained from the Japan National Institute of Genetics.4 The characteristics of the fly strains can be found at http://flybase.org. The microinjection of TALEN mRNAs was performed by Rainbow Transgenic Flies, Inc. (Camarillo, CA USA) following standard injection procedures.
Construction of TALENs
TALEN DNA binding domains were designed using a free TALEN designing tool, Mojo Hand at http://talendesign.com/mojohand_main.php.61 TALENs were constructed following the Golden Gate TALEN Assembly protocol.54 All TALENs comprised a 15-mer DNA binding domain and FokI nuclease domain. The first 10 DNA binding modules containing the repeat-variable diresidues (RVDs) were cloned into the pFUS_A vector and 4 RVDs were cloned into the pFUS_B4 vector. The assembled pFUS_A and pFUS_B4 vectors along with the last RVD (pLR) were cloned into the pT3TS-GoldyTALEN mRNA expression vector. After linearization of the expression vectors with SacI endonuclease (New England Biolabs Inc., Catalog number R0156S), the constructed GoldyTALENs were in vitro transcribed to TALEN mRNAs using the mMessage mMachine® T3 kit (Life Technologies, Catalog number AM1348). The four concentrations tested were 0.5, 1.3, 3.6, and 4.3 μg/μl/TALEN-arm. To obtain higher concentrations of TALENs (e.g. 3.6 and 4.3 μg/μl/TALEN arm), we reconstituted eluted TALEN mRNAs from multiple in vitro transcription reactions in deionized water. Designs of the 2 pairs of TALENs targeting the white gene are below:
white TALEN Pair 1 (w1 TALEN)
left arm (15 bp): g t a t t c t a a a c a t g a
NN NG NI NG NG HD NG NI NI NI HD NI NG NN NI
spacer (14 bp): cttacATTTATCGT
right arm (15 bp): C A A A A C G T C T T T G C C
HD NI NI NI NI HD NN NG HD NG NG NG NN HD HD
white TALEN Pair 2 (w2 TALEN)
left arm (15 bp): G A G G C C A A T C A A G A T
NN NI NN NN HD HD NI NI NG HD NI NI NN NI NG
spacer (17 bp): GGCAACCATctgcaaat
right arm (15 bp): g c g a g t a a c a t t t t a
NN HD NN NI NN NG NI NI HD NI NG NG NG NG NI
Single fly genomic DNA polymerase chain reaction
The genomic DNA (gDNA) from flies was isolated using a buffer solution following a standard molecular biology protocol: Tris-HCl (10 mM, pH 8.2), EDTA (1 mM), NaCl (3 M), and proteinase K (200 μg/ml). An adult or larval fly was homogenized with a pipet tip in the buffer, and the homogenate was incubated at 25-37°C for 30-60 min followed by a 95°C incubation for 2 min to inactivate proteinase K. From the single fly gDNA preparation, the w2-TALEN target site was amplified by polymerase chain reaction (PCR) with the primers below:
Forward: 5′ CTAAATCGAATCGATTCATT 3′
Reverse: 5′ AACATCTCAACTCCTATCCA 3′
The size of the amplicon was 742 nt, and the 2 digested DNA fragments produced by SURVEYOR nuclease cleavage after mutagenesis were approximately 192 nt and 550 nt.
SURVEYOR nuclease assay
The PCR product was subject to re-annealing reactions according to the manufacturer's guideline (Transgenomic, Inc., catalog number 706020). The hybridization product was subject to SURVEYOR nuclease digestion at 42°C for 1 h followed by an addition of Stop Solution.
Fragment analysis
The SURVEYOR nuclease digestion products were subject to fragment analysis using High Sensitivity NGS fragment Diluent Marker Solution (DNF-394-0003) following the manufacturer's protocol (Fragment AnalyzerTM Automated CE System, Advanced Analytical Technologies, Inc.).
Analysis of somatic mutation rates
Somatic mutation rates were quantified by analyzing the outcomes from fragment analysis readouts. The formula used was from 34:
In addition to fragment analysis outcomes, somatic mutation rates were also obtained by counting the number of G0 adult flies with mosaic eye phenotypes out of the total number of eclosed G0 adult.
Sequencing
Genomic DNA samples prepared from a single fly were used for sequencing to characterize the mutations induced by the w2 TALEN. We crossed the TALEN-injected OR flies with w1118 flies (FlyBase ID: FBal0018186, carrying a deletion on 5′ part of white gene), and quantified the number of white-eyed flies among the total G1 flies. Male G1 flies from TALEN-injected male G0 flies were not scored because male G0 flies do not contribute X chromosomes to male progeny. The sequencing primer was the same as the forward primer (5′ CTAAATCGAATCGATTCATT 3′) used for PCR. All the sequencing services were provided by the Mayo Clinic Advanced Genomics Technology Center (Rochester, MN USA).
Acknowledgments
We thank Rainbow Transgenic Flies, Inc. for excellent embryonic injection services and Qianqian Guo, Angela Gregor for fly stock management. We thank Dr. Stephen C. Ekker and Dr. Jeong Heon Lee for experimental advice, Melissa McNulty and the Mayo Clinic Center for Cell Signaling in Gastroenterology Genetics and Model System Core and Mayo Clinic Center for Individualized Medicine Epigenetics Development Laboratory for reagents and technical support, and Dr. Ma Chun Hang, Patrick R. Blackburn, and Jarryd M. Campbell for providing advice on TALEN designs. We thank Susan R. Miller for assistance with manuscript preparation.
Author Contributions
H. B. L. and Y. P. conceived and designed the experiments, H. B. L., Z. L. S., and Y. P. performed the experiments, H. B. L., Z. L. S., and Y. P. analyzed the data, H. B. L., Z. L. S., Y. P. and Y. G. wrote the paper.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The work was funded by a Mayo Clinic Summer Undergraduate Research Fellowship to Zachary L. Sebo, a Mayo Graduate School Fellowship to Han B. Lee, a Mayo Clinic New Investigator Startup Fund, a Richard F. Emslander Career Development Award, a Mayo Clinic Center for Individualized Medicine Epigenomics Program fund, and Mayo Clinic Center for Cell Signaling in Gastroenterology Pilot and Feasibility Award (NIDDK P30DK084567) to Dr. Yi Guo.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- 1.Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics 2006; 172:2391-403; PMID:16452139; http://dx.doi.org/ 10.1534/genetics.105.052829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beumer KJ, Trautman JK, Mukherjee K, Carroll D. Donor DNA Utilization during Gene Targeting with Zinc-finger Nucleases. G3 (Bethesda) 2013; 3:657-64; PMID:23550125; http://dx.doi.org/ 10.1534/g3.112.005439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, Li C, Yu Z, Huang P, Wu H, Wei C, Zhu N, Shen Y, Chen Y, Zhang B, et al. Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J Genet Genomics 2012; 39:209-15; PMID:22624882; http://dx.doi.org/ 10.1016/j.jgg.2012.04.003 [DOI] [PubMed] [Google Scholar]
- 4.Kondo T, Sakuma T, Wada H, Akimoto-Kato A, Yamamoto T, Hayashi S. TALEN-induced gene knock out in Drosophila. Dev Growth Differ 2014; 56:86-91; PMID:24172335; http://dx.doi.org/ 10.1111/dgd.12097 [DOI] [PubMed] [Google Scholar]
- 5.Beumer KJ, Trautman JK, Christian M, Dahlem TJ, Lake CM, Hawley RS, Grunwald DJ, Voytas DF, Carroll D. Comparing zinc finger nucleases and transcription activator-like effector nucleases for gene targeting in Drosophila. G3 (Bethesda) 2013; 3:1717-25; PMID:23979928; http://dx.doi.org/full_text [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baena-Lopez LA, Alexandre C, Mitchell A, Pasakarnis L, Vincent JP. Accelerated homologous recombination and subsequent genome modification in Drosophila. Development 2013; 140:4818-25; PMID:24154526; http://dx.doi.org/ 10.1242/dev.100933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep 2013; 4:220-8; PMID:23827738; http://dx.doi.org/ 10.1016/j.celrep.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O'Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 2013; 194:1029-35; PMID:23709638; http://dx.doi.org/ 10.1534/genetics.113.152710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Z, Ren M, Wang Z, Zhang B, Rong YS, Jiao R, Gao G. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics 2013; 195:289-91; PMID:23833182; http://dx.doi.org/ 10.1534/genetics.113.153825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebo ZL, Lee HB, Peng Y, Guo Y. A simplified and efficient germline-specific CRISPR/Cas9 system for Drosophila genomic engineering. Fly (Austin) 2013; 8:52-7; PMID:24141137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O'Connor-Giles KM. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in drosophila. Genetics 2014; 196:961-71; PMID:24478335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo S, Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013 Nov; 195(3):715-21. doi: 10.1534/genetics.113.156737 Epub 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell JM, Hartjes KA, Nelson TJ, Xu X, Ekker SC. New and TALENted genome engineering toolbox. Circ Res 2013; 113:571-87; PMID:23948583; http://dx.doi.org/ 10.1161/CIRCRESAHA.113.301765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blackburn PR, Campbell JM, Clark KJ, Ekker SC. The CRISPR system–keeping zebrafish gene targeting fresh. Zebrafish 2013; 10:116-8; PMID:23536990; http://dx.doi.org/ 10.1089/zeb.2013.9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaj T, Gersbach CA, Barbas CF, 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 2013; 31:397-405; PMID:23664777; http://dx.doi.org/ 10.1016/j.tibtech.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009; 326:1509-12; PMID:19933107; http://dx.doi.org/ 10.1126/science.1178811 [DOI] [PubMed] [Google Scholar]
- 17.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science 2009; 326:1501; PMID:19933106; http://dx.doi.org/ 10.1126/science.1178817 [DOI] [PubMed] [Google Scholar]
- 18.Ma AC, Lee HB, Clark KJ, Ekker SC. High efficiency In Vivo genome engineering with a simplified 15-RVD GoldyTALEN design. PLoS One 2013; 8:e65259; PMID:23734242; http://dx.doi.org/ 10.1371/journal.pone.0065259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Streubel J, Blucher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat Biotechnol 2012; 30:593-5; PMID:22781676; http://dx.doi.org/ 10.1038/nbt.2304 [DOI] [PubMed] [Google Scholar]
- 20.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010; 186:757-61; PMID:20660643; http://dx.doi.org/ 10.1534/genetics.110.120717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 2011; 29:143-8; PMID:21179091; http://dx.doi.org/ 10.1038/nbt.1755 [DOI] [PubMed] [Google Scholar]
- 22.Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res 2011; 39:359-72; PMID:20699274; http://dx.doi.org/ 10.1093/nar/gkq704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A 1996; 93:1156-60; PMID:8577732; http://dx.doi.org/ 10.1073/pnas.93.3.1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res 2000; 28:3361-9; PMID:10954606; http://dx.doi.org/ 10.1093/nar/28.17.3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol 2001; 21:289-97; PMID:11113203; http://dx.doi.org/ 10.1128/MCB.21.1.289-297.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol 2011; 29:699-700; PMID:21822242; http://dx.doi.org/ 10.1038/nbt.1939 [DOI] [PubMed] [Google Scholar]
- 27.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, Tan W, Penheiter SG, Ma AC, Leung AY, et al. In vivo genome editing using a high-efficiency TALEN system. Nature 2012; 491:114-8; PMID:23000899; http://dx.doi.org/ 10.1038/nature11537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zu Y, Tong X, Wang Z, Liu D, Pan R, Li Z, Hu Y, Luo Z, Huang P, Wu Q, et al. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat Methods 2013; 10:329-31; PMID:23435258; http://dx.doi.org/ 10.1038/nmeth.2374 [DOI] [PubMed] [Google Scholar]
- 29.Choi SM, Kim Y, Shim JS, Park JT, Wang RH, Leach SD, Liu JO, Deng C, Ye Z, Jang YY. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology 2013; 57:2458-68; PMID:23325555; http://dx.doi.org/ 10.1002/hep.26237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol 2011; 29:697-8; PMID:21822241; http://dx.doi.org/ 10.1038/nbt.1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, Joung JK, Sander JD, Peterson RT, Yeh JR. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res 2012; 40:8001-10; PMID:22684503; http://dx.doi.org/ 10.1093/nar/gks518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ewart GD, Cannell D, Cox GB, Howells AJ. Mutational analysis of the traffic ATPase (ABC) transporters involved in uptake of eye pigment precursors in Drosophila melanogaster. Implications for structure-function relationships. J Biol Chem 1994; 269:10370-7; PMID:8144619 [PubMed] [Google Scholar]
- 33.Qiu P, Shandilya H, D'Alessio JM, O'Connor K, Durocher J, Gerard GF. Mutation detection using Surveyor nuclease. Biotechniques 2004; 36:702-7; PMID:15088388 [DOI] [PubMed] [Google Scholar]
- 34.Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol 2010; 649:247-56; PMID:20680839; http://dx.doi.org/ 10.1007/978-1-60761-753-2_15 [DOI] [PubMed] [Google Scholar]
- 35.Goodarzi AA, Jeggo PA. The repair and signaling responses to DNA double-strand breaks. Adv Genet 2013; 82:1-45; PMID:23721719; http://dx.doi.org/ 10.1016/B978-0-12-407676-1.00001-9 [DOI] [PubMed] [Google Scholar]
- 36.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 2012; 47:497-510; PMID:22920291; http://dx.doi.org/ 10.1016/j.molcel.2012.07.029 [DOI] [PubMed] [Google Scholar]
- 37.Pastink A, Eeken JC, Lohman PH. Genomic integrity and the repair of double-strand DNA breaks. Mutat Res 2001; 480-481: 37-50; PMID:11506797 [DOI] [PubMed] [Google Scholar]
- 38.Karran P. DNA double strand break repair in mammalian cells. Curr Opin Genet Dev 2000; 10:144-50; PMID:10753787; http://dx.doi.org/ 10.1016/S0959-437X(00)00069-1 [DOI] [PubMed] [Google Scholar]
- 39.Gorski MM, Eeken JC, de Jong AW, Klink I, Loos M, Romeijn RJ, van Veen BL, Mullenders LH, Ferro W, Pastink A. The Drosophila melanogaster DNA Ligase IV gene plays a crucial role in the repair of radiation-induced DNA double-strand breaks and acts synergistically with Rad54. Genetics 2003; 165:1929-41; PMID:14704177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McVey M, Radut D, Sekelsky JJ. End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent. Genetics 2004; 168:2067-76; PMID:15611176; http://dx.doi.org/ 10.1534/genetics.104.033902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei DS, Rong YS. A genetic screen for DNA double-strand break repair mutations in Drosophila. Genetics 2007; 177:63-77; PMID:17660539; http://dx.doi.org/ 10.1534/genetics.107.077693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson-Schlitz DM, Flores C, Engels WR. Multiple-pathway analysis of double-strand break repair mutations in Drosophila. PLoS Genet 2007; 3:e50; PMID:17432935; http://dx.doi.org/ 10.1371/journal.pgen.0030050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beumer KJ, Trautman JK, Bozas A, Liu JL, Rutter J, Gall JG, Carroll D. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc Natl Acad Sci U S A 2008; 105:19821-6; PMID:19064913; http://dx.doi.org/ 10.1073/pnas.0810475105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bozas A, Beumer KJ, Trautman JK, Carroll D. Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila. Genetics 2009; 182:641-51; PMID:19380480; http://dx.doi.org/ 10.1534/genetics.109.101329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014; 157:1262-78; PMID:24906146; http://dx.doi.org/ 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci U S A 2014; 111:E2967-76; PMID:25002478; http://dx.doi.org/ 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet 2012; 8:e1002861; PMID:22916025; http://dx.doi.org/ 10.1371/journal.pgen.1002861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 2012; 30:460-5; PMID:22484455; http://dx.doi.org/ 10.1038/nbt.2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res 2009; 19:1279-88; PMID:19470664; http://dx.doi.org/ 10.1101/gr.089417.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hisano Y, Ota S, Arakawa K, Muraki M, Kono N, Oshita K, Sakuma T, Tomita M, Yamamoto T, Okada Y, et al. Quantitative assay for TALEN activity at endogenous genomic loci. Biol Open 2013; 2:363-7; PMID:23616919; http://dx.doi.org/ 10.1242/bio.20133871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 2007; 25:778-85; PMID:17603475; http://dx.doi.org/ 10.1038/nbt1319 [DOI] [PubMed] [Google Scholar]
- 52.Carmon A, Guertin MJ, Grushko O, Marshall B, MacIntyre R. A molecular analysis of mutations at the complex dumpy locus in Drosophila melanogaster. PLoS One 2010; 5:e12319; PMID:20811586; http://dx.doi.org/ 10.1371/journal.pone.0012319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe T, Ochiai H, Sakuma T, Horch HW, Hamaguchi N, Nakamura T, Bando T, Ohuchi H, Yamamoto T, Noji S, et al. Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nat Commun 2012; 3:1017; PMID:22910363; http://dx.doi.org/ 10.1038/ncomms2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 2011; 39:e82; PMID:21493687; http://dx.doi.org/ 10.1093/nar/gkr218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ansai S, Sakuma T, Yamamoto T, Ariga H, Uemura N, Takahashi R, Kinoshita M. Efficient targeted mutagenesis in medaka using custom-designed transcription activator-like effector nucleases. Genetics 2013; 193:739-49; PMID:23288935; http://dx.doi.org/ 10.1534/genetics.112.147645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim JS, Lee HW. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol 2013; 31:23-4; PMID:23302927; http://dx.doi.org/ 10.1038/nbt.2477 [DOI] [PubMed] [Google Scholar]
- 57.Romeijn RJ, Gorski MM, van Schie MA, Noordermeer JN, Mullenders LH, Ferro W, Pastink A. Lig4 and rad54 are required for repair of DNA double-strand breaks induced by P-element excision in Drosophila. Genetics 2005; 169:795-806; PMID:15545651; http://dx.doi.org/ 10.1534/genetics.104.033464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.White TB, Lambowitz AM. The retrohoming of linear group II intron RNAs in Drosophila melanogaster occurs by both DNA ligase 4-dependent and -independent mechanisms. PLoS Genet 2012; 8:e1002534; PMID:22359518; http://dx.doi.org/ 10.1371/journal.pgen.1002534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frit P, Barboule N, Yuan Y, Gomez D, Calsou P. Alternative end-joining pathway(s): bricolage at DNA breaks. DNA Repair (Amst) 2014; 17:81-97; PMID:24613763; http://dx.doi.org/ 10.1016/j.dnarep.2014.02.007 [DOI] [PubMed] [Google Scholar]
- 60.Alvarez-Quilon A, Serrano-Benitez A, Lieberman JA, Quintero C, Sanchez-Gutierrez D, Escudero LM, Cortes-Ledesma F. ATM specifically mediates repair of double-strand breaks with blocked DNA ends. Nat Commun 2014; 5:3347; PMID:24572510; http://dx.doi.org/ 10.1038/ncomms4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neff KL, Argue DP, Ma AC, Lee HB, Clark KJ, Ekker SC. Mojo Hand, a TALEN design tool for genome editing applications. BMC Bioinformatics 2013; 14:1; PMID:23323762; http://dx.doi.org/ 10.1186/1471-2105-14-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peng Y, Clark KJ, Campbell JM, Panetta MR, Guo Y, Ekker SC. Making designer mutants in model organisms. Development 2014; 141:4042-54; PMID:25336735; http://dx.doi.org/ 10.1242/dev.102186 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.