Abstract
Advancements in high-throughput nucleotide sequencing techniques have brought with them state-of-the-art bioinformatics programs and software packages. Given the importance of molecular sequence data in contemporary life science research, these software suites are becoming an essential component of many labs and classrooms, and as such are frequently designed for non-computer specialists and marketed as one-stop bioinformatics toolkits. Although beautifully designed and powerful, user-friendly bioinformatics packages can be expensive and, as more arrive on the market each year, it can be difficult for researchers, teachers and students to choose the right software for their needs, especially if they do not have a bioinformatics background. This review highlights some of the currently available and most popular commercial bioinformatics packages, discussing their prices, usability, features and suitability for teaching. Although several commercial bioinformatics programs are arguably overpriced and overhyped, many are well designed, sophisticated and, in my opinion, worth the investment. If you are just beginning your foray into molecular sequence analysis or an experienced genomicist, I encourage you to explore proprietary software bundles. They have the potential to streamline your research, increase your productivity, energize your classroom and, if anything, add a bit of zest to the often dry detached world of bioinformatics.
Keywords: bioinformatics software, CLC bio, Geneious, genome assembly, nucleotide alignment, phylogenetics software
INTRODUCTION
Most mornings I wake up to a slew of spam email from biotech companies offering unbeatable bargains on next-generation sequencing (NGS). Yesterday, for example, Beckman Coulter kindly offered to ‘take the stress out of sequencing’ for only a few thousand dollars. Illumina recently provided me with ‘a glimpse into the future of genomics’, just by clicking on their buyer’s guide. And Macrogen, a South Korean sequencing conglomerate, dared me to race the HiSeq ‘Xpressway to the $1000 genome’. These irritating emails underscore an important point: massively parallel sequencing has arrived to the masses. NGS is now standard fare in almost all facets of life science research [1]. It is also big business and intimately tied to another burgeoning industry—bioinformatics [2].
Anyone who has ever had something sequenced, such as a genome, transcriptome, gene or PCR product, or used nucleotide or protein sequence data in their research has probably dabbled in bioinformatics. Not long after scientists started generating molecular sequence information, computer-savvy biologists and biology-savvy computer scientists began developing programs to analyse those data [3]. Given the breadth and depth of questions that can be addressed with primary biological sequence information, many of these programs have become immensely popular. For example, the journal article describing the basic local alignment search tool (BLAST), which allows a query nucleotide or amino acid sequence to be compared against a database of sequences, has been cited >50 000 times [4].
Today’s omics-obsessed scientific marketplace is overflowing with bioinformatics programs. Whatever your sequence analysis problem (assembling, aligning, annotating, folding, etc.), there is probably a program or online application to solve it—skim through the community-maintained list of bioinformatics software at SEQanswers.com to see what I mean: http://seqanswers.com/wiki/Software/list. The majority of these tools are open source, but they can be difficult to learn, install and run; some require an in-depth knowledge of computers [5]. There are, however, various commercial alternatives, which bring together multiple bioinformatics programs into user-friendly stand-alone packages. Although beautifully designed, these software suites can come with a hefty price tag, meaning that most researchers, teachers and students are lucky if they can afford just one. Like buying a car, choosing between different suites can be challenging, and there is surprisingly little information appraising the different programs. Here, I describe my own experiences with using commercial bioinformatics packages, focusing on their cost, functions and educational utility.
I have no affiliation, past or present, with any of the programs, software or companies described in this manuscript, but being a longtime genomics enthusiast, I use many of these applications daily, and I am a strong proponent of ease of use and accessibility in bioinformatics [5]. Although the focus of this article is commercial software, there are a number of free browser-based bioinformatics toolkits worth considering, e.g. [6, 7]. Two toolkits that I use regularly and recommend are MEGA [8] and Unipro UGENE [9]. See Vincent and Charette [10] for a succinct but compelling summary of the drawbacks of commercial tools and arguments for freedom in bioinformatics.
A bioinformatics magic bullet
During my PhD I spent hours a day at the computer assembling and analysing organelle genomes. Friends and colleagues would poke their heads into my cubbyhole of an office and recoil at the sight of my bloodshot eyes and the genomic chaos playing out on the high-definition dual monitors that surrounded me. ‘Dave, how many analyses are you running?’ they would ask, gaping at the mosaic disarray of program windows scattered across the screens. Like any decent genomics junkie, I usually had half a dozen different bioinformatics applications running concurrently. I would be desperately editing and assembling Sanger sequences with Phred, Phrap and Consed [11, 12] while blasting the resulting contigs locally against custom databases and annotating the output on an ongoing GenBank entry. A chug of coffee and I would switch to gene and genome alignments with ClustalW [13] and MAUVE [14], which were pumped directly into PAML [15] to measure genetic diversity and substitution rates. A quick blink of the eyes and I was plotting sliding-window GC contents across entire organelle chromosomes, all while mainlining a medley of phylogenetic and tree-building programs, from MrBayes [16] to PhyML [17] to PAUP [18] to MacClade [19]. And because some of these applications worked only on a PC and others on a Mac, I had Windows and Apple operating systems running at the same time, with command-line terminals piled on top of graphical user interfaces (GUIs).
‘There’s got to be an easier, more efficient way of doing this’, I would say to myself, as I tossed another empty coffee cup into the trash. Sensing my angst, a colleague recommended that I invest in a commercial, cross-platform, GUI-based bioinformatics package, arguing that it would streamline and simplify my work. I was reluctant to take his advice. I felt that paying for such programs went against the spirit of academic research and that using GUI software would weaken my computational skills. However, after failing for the fourth time to correctly install and run an open-source genome assembly algorithm, I gave in and bought a user-friendly bioinformatics bundle, and have not regretted it.
Show me the money
In 2007, with the grant support of my former PhD supervisor, I purchased my first bioinformatics software package. At the time, there was a small but strong cohort of commercial options available, most of which offered a free 30-day trial—a practice that, with few exceptions, continues today, although in some cases the trial period has been reduced to ≤2 weeks. After testing an assortment of programs, I decided on Geneious (Biomatters Ltd., Auckland, New Zealand), which was first released in 2005 and is now among the more widely used cross-platform commercial bioinformatics packages (Table 1). I chose Geneious not because it was necessarily better than other software, but because the company offered, and continues to offer, student discounts. Seven years ago, I paid approximately $200 (all prices in US dollars) for a student license of Geneious, which allowed me to install the software on a single computer. As Geneious increases in popularity, so does its price tag. As of May 2014, a student license costs $395 (a standard academic one is $795), which still makes it among the least expensive all-in-one commercial suites on the market. In comparison, stand-alone academic licenses of the Lasergene Genomics Suite (DNASTAR, Madison, USA) and Sequencher (Gene Codes, Ann Arbor, USA) are around $6000 and $2500, respectively (Table 1).
Table 1.
Examples, features and comparisons of some commonly used commercial bioinformatics software suites
| Software | Company | Cost (USD)a | Free trial (days) | Platformb | NGS analysesc | Evolutionary analysesd | Database searchinge | Plug-ins | Workflows | Teaching suitability |
|---|---|---|---|---|---|---|---|---|---|---|
| Avadis NGS | Strand Scientific Intelligence | $4500 | 20 | M, W, L | ✓ | ✗ | ✗ | ✗ | ✓ | ✗ |
| CLC Genomics Workbench | ClC bio, Qiagen | $5500 | 30 | M, W, L | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| CodonCode Aligner | CodonCode | $720 | 30 | M, W | ✓ | ✓ | ✗ | ✗ | ✗ | ✓ |
| Genamics Expression | Genamics | $295 | 30 | W | ✗ | ✓ | ✓ | ✓ | ✗ | ✗ |
| Geneious | Biomatters | $795 | 14 | M, W, L | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Full Lasergene Suite | DNASTAR | $5950 | 30 | M, W | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| MacVector & Assembler | MacVector | $300 | 21 | M | ✓ | ✓ | ✓ | ✗ | ✗ | ✓ |
| NextGENe | Softgenetics | $4049 | 35 | W | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ |
| Sequencher | Gene Codes | $2500 | 30 | M, W | ✓ | ✓ | ✓ | ✓ | ✗ | ✓ |
| VectorNTI Advance | Life Technologies | $600 | 30 | W | ✗ | ✓ | ✓ | ✗ | ✓ | ✓ |
aApproximate price of a single-user academic license. Prices were taken directly from company websites (as of 1 June 2014) or were obtained by sales representatives sometime between January and June 2014. Many companies offer a range of pricing and licensing options, and frequently have promo deals. bRuns on the following platforms: Mac (M), Windows (W) and Linux (L). cCan store, organize and analyse (e.g. assemble or map to a reference sequence) next-generation sequencing data. In some cases, de novo assembly features are missing. dContains some tools for studying molecular evolution, such as those for performing multiple sequence alignments, phylogenetic analyses and/or repeat identification. eIs able to connect and interact with online sequence databases, such as GenBank. ✓ = yes, ✗ = no
I have since gone on to test, and in some instances purchase, a multitude of other commercial bioinformatics platforms, which have varied widely in price, usability and quality. In several cases, the costs of these software suites were not listed on the company websites or anywhere else online. To get pricing details, I had to request quotes from sales representatives. For example, after a successful 30-day trial of CLC Genomics Workbench (CLC Bio, Qiagen, Aarhus, Denmark), I filled out an online pricing request form and was contacted 2 days later by a sales agent who provided me with a formal quote (an estimated $5500 for a standard academic license). I went through similar processes to get pricing on Lasergene, Sequencher and various other bioinformatics programs. Since starting this article, CLC bio now posts some of their prices online (www.clcbio.com; accessible through the ‘buy online’ icon), but many companies still require potential customers to contact sales reps, making it difficult and time-consuming to compare prices of different software packages. On a number of occasions, after requesting quotes or free trial access, I was bombarded with emails and phone calls by sales agents asking whether I had come to any decisions about purchasing the software or whether I needed more information; one time a representative even called a laboratory where I used to work, asking for my current contact details—so if you request a quote, be prepared to be pestered.
What do you get for your money and for how long?
Purchasing a commercial sequence analysis suite is not as simple as a one-time payment followed by a lifetime of bioinformatics bliss. There can be hidden unexpected costs and clauses associated with running the software and continuing to use it in the future. Most commercial packages include 12 months of free maintenance, upgrades and support. Shortly after I bought my student license for Geneious, the firm released a new version of the software. Because this occurred within 1 year of my purchasing the program, I was able to upgrade to the newest version for free. Geneious and other bioinformatics manufacturers have recently switched to ‘version-based licensing’, meaning that users receive free updates for their version of the software (e.g. switching from v1.1 to v1.2), no matter when they are released, but access to newer versions (e.g. switching from v1 to v2) requires an upgrade, which typically costs anywhere from 25 to 75% of the software list price.
Last year, for approximately $6000, I purchased as part of a package deal a single academic license of CLC Genomics Workbench and a genome finishing plug-in (more on plug-ins later). Enrollment in the maintenance, upgrade and support program for the first 12 months, which was mandatory, was an additional $1500, making the initial cost of the software $7500. Renewal of the maintenance program was 25% of the purchase price per year, and, most importantly, was automatic, ‘unless terminated in writing by one of the involved parties (CLC bio or the customer) not later than 3 months before the beginning of the next calendar year’. In other words, 9 months after buying the software, I was sent an invoice for $1500, with 2% interest per month.
Although costly, subscribing to the maintenance agreement can be wise. Commercial bioinformatics programs (Table 1), such as Geneious, CLC Genomics Workbench and Lasergene, frequently undergo major changes, which can significantly improve the software. In the past, I have regretted not renewing certain software, and more than once I have bought programs anew at full price because I let the maintenance period expire.
Before investing in a bioinformatics package, there are other important details to consider. I suggest asking about the rules on moving the software to another computer, in case, for example, you buy a new laptop or your old one breaks down. I have found that most companies allow users to transfer their software license to a different computer. But doing so normally requires contacting user support for a new software activation key, and if you have let your maintenance agreement expire, then you might have to renew it before being able to migrate the software. Similarly, if you update your computer operating system—from Apple OS X 10.8 to 10.9, for instance—your bioinformatics package might have to be upgraded as well. Most bioinformatics companies offer their software for both Windows and Apple platforms, and some, including Geneious and CLC bio, have Linux versions too, so in most cases, it is possible to switch operating systems completely and continue running the program.
Things get even more complicated when purchasing network (or ‘floating’) licenses of bioinformatics programs. Unlike a single computer license, which works only on one computer, a network/floating license allows multiple people to use a bioinformatics package simultaneously by logging on to a network computer (e.g. a powerful computer housed in the lab) and running the program from it. The number of people that can log on depends on the number of floating licenses that were purchased. Network/floating licenses are more expensive (typically twice the price) than their single-computer counterparts, but they can be more economical for big labs or classroom settings, where purchasing multiple single-user licenses makes less sense. Floating licenses can also be convenient for groups that have a high turnover—such as those with a lot of summer students and undergraduate volunteers—as they allow software key codes to be issued to individual lab members and then taken back once the member leaves. Sequencher (Table 1) offers a ‘hardkey’ option, whereby the user is sent a USB dongle after purchasing the software. Sequencher can then be loaded onto as many computers as the owner wants—all that is required to activate the software is plugging in the USB key. But, as I can attest, USB dongles are easy to misplace (and, if issued from Sequencher, expensive and inconvenient to replace).
Cloud computing has also arrived to bioinformatics [20]. Companies like DNAnexus, InterpretOmics, and others are selling bioinformatics as a service, whereby consumers buy online access to powerful computers and their associated software tools, analysis pipelines and data storage and sharing capabilities. The sequencing giant Illumina sells online access to their genomics cloud-computing infrastructure BaseSpace—10 terabytes of storage will run you $12 000 per year. Alternatively, the popular web-based platform Galaxy is a free, open-source, cloud-based bioinformatics tool. It is safe to assume that bioinformatics clouds will only grow larger and more popular over the next few years and are where the most innovative new software will be based.
But what does the software actually do?
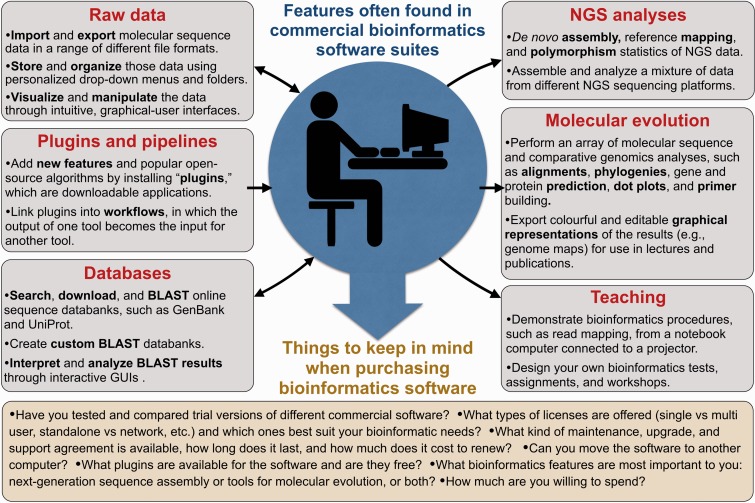
You have paid your money and decided on the best maintenance and licensing options for your needs, now what? Well, it is time to start examining molecular sequence data and making some big discoveries, of course. Commercial bioinformatics packages bring together, into a single browser-based platform, a diversity of nucleotide and protein analysis tools (Figure 1). These tools do everything from simple pairwise alignments to restriction site and gene predictions to whole genome and transcriptome assemblies. Given the prevalence of high-throughput sequencing in life science research, many of the tools are designed for analysing, visualizing and arranging NGS information.
Figure 1.
The tools and features commonly found in commercial bioinformatics software packages, and what to keep in mind when purchasing one.
One of the most sought after and marketed features of commercial bioinformatics software is their ability to perform fast, efficient and high-quality de novo assemblies of NGS data—taking millions, even billions, of single or paired-end sequencing reads and assembling them into contigs. Go to any of the big bioinformatics software websites and you will find statements like ‘Dominating the high-throughput sequencing data analysis challenge’, ‘Quick and accurate de novo assembly on a desktop computer’ and ‘Next-gen sequence assembly with a clear graphical interphase’. These kinds of claims are often associated with a white paper describing the software’s de novo assembler, including its algorithm, speed and accuracy, how well it performs on standard datasets, such as the human genome, and how it stacks up against other brand-name and open-source assemblers. White papers, however, do tend to present commercial software in an overly positive light and—unlike open-source programs—only a few of the widely used proprietary tools have undergone peer review.
Commercial browser-based assemblers once had a reputation for being slow, memory-expensive and inferior to the free open-source alternatives. Early on, I admittedly struggled to generate quality assemblies, even of small genomes, using commercial programs. In recent years, however, proprietary assembly algorithms have improved immensely and are now used by some of the top academic and industrial research laboratories in the world. With software like CLC Genomics Workbench v7, I have been able to assemble draft genome and transcriptome sequences of microalgae from my laptop computer, which has 16 GB of memory and an Intel Core i7 processor. Many teams are using proprietary tools to assemble complex eukaryotic nuclear genomes, including those of land plants. But these kinds of assemblies require large amounts of time, resources and computing power.
Commercial assemblers, unlike certain open-source ones, are also great at handling data from different sequencing platforms, such as assembling a mixture of Illumina, 454, PacBio and Sanger reads (Table 1); in fact, for many researchers, this is a key selling point. In March 2014, for example, Northwestern University purchased an organization-wide license of Lasergene, providing all faculty, staff and students with access to the software [21]. Similarly, the J. Craig Venter Institute has been using ‘CLC bio’s enterprise platform since 2009 and currently uses it on more than 30 research grants, including their work as part of the Human Microbiome Project’ [22].
Read mapping, which is when sequencing reads are aligned to a reference, such as an entire chromosome or genome, is another core feature of commercial bioinformatics packages. Like with the de novo assemblers, bioinformatics companies regularly boast about their highly tuned, ultra-fast mapping algorithms for reference-guided alignments. CLC bio maintains that their ‘read mapper not only maps more than 1.3 billion Illumina reads (100 nt, paired-end) in less than 5 hours, but [that it] also achieves consistently high mapping accuracy even for complex read data, such [as those] originating from the PacBioRS system’ [23]. They go on to argue that the CLC ‘mapper consistently outperforms the market in all major disciplines’, including the open-source peer-reviewed mapping algorithms Bowtie 2 and BWA [23]. Geneious makes similar claims about their proprietary mapper: ‘Six read mapping algorithms were evaluated on Illumina HiSeq and Ion Torrent sequence data from an Escherichia coli—BWA (0.6.2-r126), Bowtie 1 (0.12.8), Bowtie 2 (2.0.0-beta7), SMALT (0.6.4), SOAP2 (2.20) and Geneious (6.0.3). The results demonstrate that the Geneious Read Mapper produces superior results to the other mapping algorithms on these data sets’ [24]. The claims can be overstated, but in my experience commercial read mappers are as good as or outperform many of the open-source alternatives.
The ultimate test for any assembler or read mapper is whether it is cited in peer-reviewed journals. There is no question that open-source programs are cited more than proprietary ones. The paper presenting the mapper Bowtie 2, for instance, has received 700 citations in just 2 years [25]. But citations for commercial software suites, especially their assembly and mapping algorithms, are on the rise and catching up to their open-source counterparts. A keyword search of ‘CLC Genomics’ in Google Scholar returns >2000 hits. Visit the Geneious blog (http://blog.geneious.com) and you will find a section called ‘Citation Sunday’, highlighting peer-reviewed research that used Geneious. Click the ‘publications’ link on the DNASTAR homepage (www.dnastar.com) and you will see a long list of papers and the following bold statement: ‘Every year for the last 28 years, more researchers have cited DNASTAR's software in scientific journals than any other sequence analysis software’ (italics their own). Skimming through these publications, it is obvious that most papers citing proprietary programs reference a range of open-source ones as well, and that contemporary genomics research often involves a hodgepodge of commercial and free bioinformatics software. Lizzy Sollars, a PhD student at CLC bio, put it best when describing her work on the Ash Tree Genome Project: ‘Using CLC bio's de novo assembler, along with the open-source scaffolding tool SSPACE, we produced our best de novo assembly so far’ [26]. Visit the Broad Institute Software Archive (www.broadinstitute.org/scientific-community/software) for a list of widely used open-source tools for analysing large genome-related datasets.
More than just browser-based assemblers and mappers
Commercial sequence analysis suites, in addition to assembling and mapping NGS data, are designed to carry out the day-to-day bioinformatics tasks involved in molecular, evolutionary and genome biology (Figure 1). Although it might sound trivial, one of the more useful features of commercial packages is visualizing, organizing and storing molecular sequence information. The intuitive graphical interfaces of commercial software allow users to easily build folder hierarchies and drop-down lists of sequence data, move or export these data to different folders and change file formats for use in other applications. In most cases, the software can connect to online resources, such as the National Centre for Biotechnology Information (NCBI) and UniProt, providing quick direct access to vast amounts of nucleotide and protein sequence information, which can then be downloaded, interpreted and analysed through interactive sequence viewers. Many commercial programs also give users the ability to BLAST [4] their data directly against NCBI and UniProt databases, or custom databases, and view and analyse the results through GUIs. My research on organelle DNA has benefited greatly from these types of search tools—in minutes, using commercial software, I can download all of the completely sequenced mitochondrial and chloroplast genomes from GenBank, extract their annotations, sort and search them based on a range of features and transfer them to subfolders for downstream analyses.
The applications within commercial bioinformatics suites that I tend to use most often are for evolutionary analyses and comparative genomics. Most packages come with software for aligning nucleotide and amino acid sequences (and entire chromosomes) as well as tools for inferring evolutionary relationships among sequences and constructing phylogenetic trees and distance matrices. Other useful tools include protein structure prediction, nucleotide repeat and motif finders and primer prediction software. An advantage to performing these kinds of analyses within commercial software is that the results—be they genome maps, alignments, nucleotide sequence dot plots or phylogenetic trees—are depicted in colourful and editable graphics, which can be exported and used for figures in lectures and publications. I regularly build genome maps with Geneious and then export them to a graphics-editing program for further polishing. All of the genome maps in Smith et al. [27, 28], for example, were constructed with Geneious. The interactive graphical visualization tools of commercial suites are excellent for exploring large genomic data sets (often depicted in stacked views) and allow for quick navigation to regions or contigs of interest. Many of these features parallel those of popular freely available NGS viewers, like the Interactive Genomics Viewer [29] and Tablet [30].
If you purchase a bioinformatics package and discover that a particular function is missing, do not panic because there is probably a ‘plug-in’ that can do the job. Plug-ins are downloadable applications that provide additional features to software packages—similar to apps for smartphones and tablets. For bioinformatics software, plug-ins add an array of new sequence analysis tools (ones that complement existing tools or that add novel functions), greatly improving the package. Companies are constantly designing new plug-ins for their software, which means that the repertoire of tools within bioinformatics packages is continually expanding. Plug-ins work in two ways: they allow users to add more features to the software, but they also allow developers to design their own apps for the software. Bioinformatics plug-ins can bring some of the most commonly used open-source software to proprietary programs, giving users the benefits of a user-friendly GUI and the power of peer-reviewed algorithms. A cursory scan through the plug-in list for Geneious reveals programs for phylogenetics (e.g. GARLI [31], MrBayes [16] and RAxML [32]), NGS assembly and mapping (e.g. Velvet [33], TopHat [34] and Bowtie [25]), sequence alignment (e.g. ClustalW [13], MAUVE [14] and Muscle [35]) and other molecular analysis procedures (e.g. Glimmer Gene Prediction [36], Phobos Tandem Repeat Finder (e.g. [37]) and DualBrothers Recombination Detection [38]). More plug-ins means more functions and sometimes more money. CLC bio provides a wide range of plug-ins for their Genomics Workbench package (www.clcbio.com/clc-plugin), many of which are free, but some can cost hundreds even thousands of dollars—the Shannon Human Splicing Pipeline plug-in is around $4000.
Once you have found the tools and plug-ins to suit your needs, you can start linking them together into ‘workflows’ and pipelines. As CLC bio puts it: ‘A workflow consists of a series of tools where the output of one tool is connected as the input to another tool. This way you can set up a workflow to go through (for example) read mapping, using the mapped reads as input for variant detection, and perform filtering of the variant track’. Workflows can save researchers huge amounts of time and are becoming more widespread among commercial bioinformatics packages. If you do not want to fork out the big bucks, check out The Galaxy Project (http://galaxyproject.org)—a free, web-based and user-friendly bioinformatics workflow management system, which provides access to a large number of data integration and analysis programs.
Bringing bioinformatics into the classroom
Students today are reared on a digital diet of smartphones, tablets and ultra-sleek retina-display laptops filled with intuitive software apps, which integrate seamlessly across platforms and devices. Thus, when these students are introduced to bioinformatics and molecular evolution, one would expect them to engage more easily and enthusiastically with easy-to-use GUI software than with barebones command-line-driven tools.
Commercial bioinformatics suites, given their browser-based point-and-click interface, lend themselves to teaching and learning. From a lecturer’s perspective, the high-end graphics, visual aids and tutorials built into proprietary software are great for communicating bioinformatics topics, themes and procedures, from sequence alignments to contig assemblies to blasting proteins against GenBank. I regularly incorporate bioinformatics software suites into my undergraduate lectures and conference presentations. With my notebook computer connected to a projector, I can use a program like Geneious to effectively communicate to a large audience the procedures and output of various bioinformatics analyses. For example, using a bioinformatics package, it takes me ∼10 min to import a set of Illumina sequencing reads, download a reference genome from GenBank, map the reads to the reference and then zoom in to the resulting alignment, showing the class where the reads mapped onto the genome, the polymorphic sites, paired-end distances and an assortment of other statistics. With the same software, I can design, distribute and evaluate bioinformatics assignments to be completed inside or outside of the classroom. These assignments typically involve a range of sequence analysis tools where the results of one tool are used as input for another. I almost always receive positive feedback from students when using user-friendly bioinformatics—some students have even said that it has inspired them to pursue a career in bioinformatics.
Obviously, the biggest barrier to bringing commercial software into the classroom is the high financial cost of the programs. It is unreasonable to ask students to pay hundreds of dollars for proprietary software, and most undergraduate departments are unable or unwilling to invest thousands of dollars into bioinformatics teaching resources—although with institutes like Northwestern buying campus-wide access to proprietary programs, this might be changing.
One strategy for using commercial bioinformatics in a course is to get all of the students to apply for a free trial version of the software. Their access to the software will be limited to ≤30 days, but this should be long enough for them to complete a few assignments or workshops. Alternatively, some commercial bioinformatics packages can be downloaded and used for free on a ‘basic’ or ‘test’ mode, which means that certain operations are turned off (e.g. assemblies cannot be exported or saved). However, even with limited functions, the software can still provide enough processes for teaching and developing assignments [39]. Again, there is nothing preventing instructors from investing in a personal copy of the software and using it for lectures.
Give it try and give us your feedback
Going forward, innovations in molecular sequencing techniques will result in ever more sophisticated bioinformatics programs, and it is crucial that these programs are accessible to a broad range of users. We might soon be at a point where walk-in medical clinics have genome sequencing and bioinformatics desks, where patients can play an active role in interpreting their gene sequences and contributing to genetic treatments, and where high-school students assemble and analyse genomes for homework. The increasingly integral role of bioinformatics in research, medicine and society also means that it will become an increasingly larger, more lucrative industry and one where users will have to pay for the best products.
My own experiences with proprietary bioinformatics software have been positive. The tools I have purchased have made my laboratory group and me more productive, and I certainly enjoy using stand-alone GUI-based programs more than command-line driven ones. This productivity and ease of use, however, has come at a cost, both intellectually and financially. Although I use sequence analysis tools almost every day, my bioinformatics skills, in certain respects, have plateaued. Moreover, the licensing and upgrading costs of using commercial software represent a significant proportion of my laboratory’s operating budget. Another downside to commercial bioinformatics is that the user can lose touch with what the programs/algorithms are actually doing (they can be a ‘black box’), whereas it is simple to look ‘under the hood’ of open-source tools, which makes them easy to modify and develop. But as bioinformatics software and algorithms become increasingly complex, it might be unrealistic to expect students to have a strong grasp of the math, theory and computer science that underpin those processes.
If you are considering commercial programs, I recommend taking advantage of the free trials that most of the bioinformatics companies offer. You may find that these programs streamline your research and invigorate your classroom, or that they are a waste of time and resources and you are better off using open-source and/or freeware alternatives. Wherever you stand on the topic, I urge you to share your opinions and experiences with others—and best of luck with all of your bioinformatics endeavours.
Key Points.
Innovations in molecular sequencing techniques, and the popular use of these technologies, have given rise to a range of user-friendly commercial bioinformatics software suites.
Often marketed as one-stop bioinformatics toolkits, these software packages can be expensive, and it can be difficult for consumers to choose between the different programs.
This review explores some of the currently available proprietary bioinformatics packages, comparing their prices, usability, functions and suitability for teaching.
Some commercial bioinformatics programs are arguably overpriced and overhyped, but many are well designed, sophisticated and, in my opinion, worth the investment.
I encourage readers to explore commercial bioinformatics packages; they have the potential to streamline your research, increase your productivity and energize your classroom.
Acknowledgment
The author thanks four anonymous reviewers whose feedback greatly improved the manuscript.
Biography
David Roy Smith is an assistant professor of biology at the University of Western Ontario, where he studies genome evolution of eukaryotic microbes. He can be found online at www.arrogantgenome.com and @arrogantgenome.
FUNDING
This work was supported by a Discovery Grant to DRS from the Natural Sciences and Engineering Research Council (NSERC) of Canada.
References
- 1.Metzker ML. Sequencing technologies — the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 2.Kumar S, Dudley J. Bioinformatics software for biologists in the genomics era. Bioinformatics. 2007;23:1713–17. doi: 10.1093/bioinformatics/btm239. [DOI] [PubMed] [Google Scholar]
- 3.Moody G. Digital Code of Life: How Bioinformatics is Revolutionizing Science, Medicine, and Business. Hoboken: Willey and Sons, Inc.; 2004. [Google Scholar]
- 4.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Smith DR. The battle for user-friendly bioinformatics. Front Genet. 2013;4:187. doi: 10.3389/fgene.2013.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carver T, Harris SR, Berriman M, et al. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–9. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pabinger S, Dander A, Fischer M, et al. A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform. 2013;15:256–78. doi: 10.1093/bib/bbs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamura K, Stecher G, Peterson D, et al. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okonechnikov K, Golosova O, Fursov M. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–7. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 10.Vincent AT, Charette SJ. Freedom in bioinformatics. Front Genet. 2014;5:259. doi: 10.3389/fgene.2014.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewing B, Green P. Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–94. [PubMed] [Google Scholar]
- 12.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 13.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 14.Darling AC, Mau B, Blattner FR, et al. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–91. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 16.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 17.Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 18.Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (*and other methods) Version 4.0b10a. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- 19.Maddison WP, Maddison DR. MacClade Version 3. Sunderland, MA: Sinauer Associates; 1992. [Google Scholar]
- 20.Stein LD. The case for cloud computing in genome informatics. Genome Biol. 2010;11:207. doi: 10.1186/gb-2010-11-5-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. DNASTAR press release, 31 March 2014: Northwestern University adopts DNASTAR Lasergene software. http://www.dnastar.com/t-NorthwesternPress.aspx (1 June 2014, date last accessed)
- 22. CLC bio press release, 8 Jan 2013: J. Craig Venter Institute extends CLC bio site license through 2017. http://www.clcbio.com/news/jcvi-extends-site-license/(1 June 2014, date last accessed)
- 23. CLC bio White Paper, Read Mapping. 2012. http://www.clcbio.com/files/whitepapers/whitepaper-on-CLC-read-mapper.pdf (1 June 2014, date last accessed)
- 24.Kearse M, Sturrock S, Meintjes P. The Geneious 6.0.3 read mapper. http://assets.geneious.com/documentation/geneious/GeneiousReadMapper.pdf (1 June 2014, date last accessed) [Google Scholar]
- 25.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. CLC bio press release, 26 Sep 2013: CLC bio and UK scientists assemble ash tree genome. http://www.clcbio.com/news/clc-bio-and-uk-scientists-assemble-ash-tree-genome/(1 June 2014, date last accessed)
- 27.Smith DR, Kayal E, Yanagihara AA, et al. First complete mitochondrial genome sequence from a box jellyfish reveals a highly fragmented linear architecture and insights into telomere evolution. Genome Biol Evol. 2012;4:52–8. doi: 10.1093/gbe/evr127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith DR, Hua J, Archibald, et al. Palindromic genes in the linear mitochondrial genome of the nonphotosynthetic green alga Polytomella magna. Genome Biol Evol. 2013;5:1661–7. doi: 10.1093/gbe/evt122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2012;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milne I, Bayer M, Cardle L, et al. Tablet—next generation sequence assembly visualization. Bioinformatics. 2010;26:401–2. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD diss., The University of Texas at Austin, 2006. [Google Scholar]
- 32.Stamatakis A. RAxML Version 8: a tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics. 2014;30:1312–13. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delcher AL, Bratke KA, Powers EC, et al. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–9. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer C, Leese F, Tollrian R. Genome-wide analysis of tandem repeats in Daphnia pulex-a comparative approach. BMC Genomics. 2010;11:277. doi: 10.1186/1471-2164-11-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minin VN, Dorman KS, Fang F, et al. Dual multiple change-point model leads to more accurate recombination detection. Bioinformatics. 2005;21:3034–42. doi: 10.1093/bioinformatics/bti459. [DOI] [PubMed] [Google Scholar]
- 39.Kearse M, Moir R, Wilson A, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]