Abstract
The venom of each species of Conus contains different kinds of pharmacologically-active peptides which are mostly unique to that species. Collectively, the ~500 – 700 species of Conus produce a large number of these peptides, perhaps exceeding 140,000 different types in total. To date, however, only a small fraction of this diversity has been characterized via transcriptome sequencing. In addition, the sampling of this chemical diversity has not been uniform across the different lineages in the genus. In this study, we used high-throughput transcriptome sequencing approach to further investigate the diversity of Conus venom peptides. We chose a species, Conus tribblei, as a representative of a poorly studied clade of Conus. Using the Roche 454 and Illumina platforms, we discovered 136 unique and novel putative conopeptides belonging to 30 known gene superfamilies and 6 new conopeptide groups, the greatest diversity so far observed from a transcriptome. Most of the identified peptides exhibited divergence from the known conopeptides and some contained cysteine frameworks observed for the first time in cone snails. In addition, several enzymes involved in post-translational modification of conopeptides and also some proteins involved in efficient delivery of the conopeptides to prey were identified as well. Interestingly, a number of conopeptides highly similar to the conopeptides identified in a phylogenetically distant species, the generalist feeder Conus californicus, were observed. The high diversity of conopeptides and the presence of conopeptides similar to those in C. californicus suggest that C. tribblei may have a broad range of prey preferences.
Keywords: conopeptide, transcriptome, Conus tribblei, conotoxin, diversity
Introduction
Each Conus species has a unique and diverse complement of an estimated 50–200 peptides, known as conopeptides or conotoxins (Olivera 2006). Conopeptides, like most animal toxins (e.g. toxins of snakes and spiders), are encoded by multigene families (Nei and Rooney 2005) with highly conserved signal sequences and hypervariable mature toxins (Kordiŝ and Gubenŝek 2000). The toxins in a cone snail’s venom target cell surface-signaling components like ion channels, neurotransmitter transporters and receptors in the nervous system (Olivera 2006). These toxins have been utilized extensively in biomedical research because of their therapeutic potentials.
An assessment of the conopeptides identified in earlier studies revealed that peptides produced by closely related species are more likely to have similar structure and function (Olivera and Teichert 2007). To achieve a more efficient discovery of diverse conopeptides, phylogenetic approaches were used for the identification of distinct clades containing conopeptides with different functional and structural characteristics (Espiritu et al. 2001). Evidently, while some conopeptide gene superfamilies are widespread among Conus species, several gene superfamilies have been identified in a few clades only (Puillandre et al. 2012). Although cone snails have been intensively investigated for the past 30 years, only 100 species of Conus (Kaas et al. 2010) have been studied for the discovery of novel conopeptides. Most studies have specifically focused on fish-hunting cone snails, while several vermivorous clades have been overlooked (Kaas et al. 2010).
Recent studies on the transcriptome of the venom duct of several Conus species using high-throughput sequencing technologies have revealed new and novel cysteine patterns in several conopeptide gene superfamilies and more than 18 new conopeptide gene superfamilies have been identified (Hu et al. 2011; Hu et al. 2012; Lluisma et al. 2012; Terrat et al. 2012; Dutertre et al. 2013; Lavergne et al. 2013). In contrast, using traditional cDNA library construction, only few conopeptide gene superfamilies were identified in each cone snail.
Therefore, a comprehensive study for exploring the peptide repertoire of representative species of under-studied clades is important for identifying toxins that are clade-specific and may result in the identification of new conopeptides, novel cysteine frameworks or even new gene superfamilies. Discovery of new conopeptides and identification of novel structures may lead to potential sources of drug candidates with therapeutic applications and to a better understanding of the conopeptide diversity in general. High-throughput sequencing can achieve higher sequencing depth and greater coverage of transcriptome so that even rare transcripts with low expression levels can be identified, providing a more accurate resolution of the conopeptide diversity in cone snails. In fact, identification of diverse conopeptides is not limited to the species in the major clade of Conus; the survey of the Conus californicus venom duct showed significant conopeptide diversity in this phylogenetically distant species (Biggs et al. 2010; Elliger et al. 2011). Thus, identification of conopeptides in distinct clades of Conus will facilitate understanding the evolution and diversification mechanisms of the conopeptide genes.
Conus tribblei belongs to a poorly known clade of vermivorous species. Because this species is generally found in deep waters, its biology and ecology is poorly known and the biochemistry of its venom is still unexplored. This study presents the transcriptome analysis of the C. tribblei venom duct using both Roche 454 and Illumina sequencing technologies. In addition to the identification of new conopeptides and cysteine patterns, other components of the transcriptome including post-translational modification enzymes were also identified.
Materials and methods
mRNA extraction and transcriptome sequencing
Twenty specimens of C. tribblei (4–5 cm) were collected in Sogod, Cebu province in the Philippines. Each specimen was dissected; the venom duct was isolated, stored in RNAlater (Ambion, Austin, TX) and kept at −20 C. The mRNA was extracted from the venom ducts of specimens using Dynabeads® mRNA DIRECT™ kit (Invitrogen® Dynal AS Oslo, Norway). Venom ducts were homogenized with 0.5mm Zirconia/Silica beads (Biospec Products, Inc.) in a bead beater (Precellys, Berlin Technologies) until all the tissues were completely lysed. The rest of the mRNA isolation protocol was done according to the manufacturer’s recommendations. The extracted mRNA from all the specimens was pooled.
The pooled extracted mRNA was sequenced using both Roche 454 and Illumina technologies. Total of 8 μg mRNA was used for Illumina cDNA library construction; fragments with an average size of 260 bp (including 120 bp sequencing adaptors) were size-selected and paired-end sequenced using Illumina HiSeq 2000. Additionally, around 1.5 μg of the mRNA was fragmented using zinc chloride and double-stranded cDNA was synthesized using the Roche Primer ‘random’. Emulsion-based clonal amplification (emPCR) of the cDNA library and the subsequent single-end sequencing using GS Junior Roche was done according to the manufacturer’s recommendations.
Phylogenetic analysis
The whole body tissue of specimens of C. tribblei were stored in 95% ethanol and kept at room temperature. A small piece of the foot tissue was used for DNA extraction using DNeasy blood and tissue kit (Qiagen, USA) according to manufacturer’s recommendations and stored at −20 °C. A fragment of cytochrome oxidase c subunit 1 (COI) gene of mitochondrial DNA was amplified using universal primers: LCO1490 and HCO2198 (Folmer et al. 1994). The resulting PCR fragment was sequenced using the forward primer and the sequence was manually inspected and cleaned based on the sequencing chromatograms using BioEdit version 7.0.0 (Hall 1999). COI sequences of several species of Conus were downloaded from GenBank (Table 1S) and were aligned using MUSCLE (Edgar 2004). Parameters of the substitution model were estimated during the analysis (six substitution categories, a gamma-distributed rate variation across sites approximated in four discrete categories and a proportion of invariable sites) independently for each codon position of the COI gene. Bayesian analysis was performed in two parallel runs in MrBayes (Huelsenbeck & Ronquist 2001), each consisting of six Markov chains of 1,000,000 generations with a sampling frequency of one tree each ten thousand generations, and the chain temperature was set to 0.02. Convergence of each analysis was evaluated using Tracer1.4.1 (Drummond and Rambaut 2007) to check that all effective sample size (ESS) values exceeded 200. When log-likelihood scores were found to stabilize, the first 25% of trees were omitted as burn-in and a consensus tree was calculated.
Transcriptome assembly
The quality of 100 bp-long paired-end reads generated by Illumina sequencing was evaluated using FastQC software v0.10.1 (www.bioinformatics.babraham.ac.uk/projects/fastqc/). FASTX-Toolkit version 0.0.6 (Pearson et al. 1997) was used for trimming low quality score bases (< 20 phred score) at both ends of the reads and subsequently removing the reads containing low quality score bases (< 20 phred score). Finally the reads with no pairs were separated and the remaining paired-end reads were assembled into transcripts using Trinity (Grabherr et al. 2011), Trans-ABySS 1.4.4 (Robertson et al. 2010), Oases 0.2.08 (Schulz et al. 2012) and SOAPdenovo-Trans 1.02 (Luo et al. 2012). To evaluate the quality of assemblies, the paired-end reads were aligned to each assembly using Bowtie2 2.1.0 (Langmead and Salzberg 2012). The default setting of end-to-end alignment with sensitive mode was used with no mismatches allowed (N=0) per seed (L=20). Around 89.84% of the Illumina reads aligned to the Trinity-assembled transcripts while only 60–75% of the reads aligned to the transcripts assembled by other assemblers (Trans-ABySS, Oases and SOAPdenovo-Trans). Of those reads that aligned back to the Trinity-assembled transcripts, 69.51% were aligned in the expected forward/reverse orientation and/or the expected distance (0–500 bp) between the mates of each paired-end read. In addition, the diversity of the identified conopeptide gene superfamilies in the Trinity assembly (30 conopeptide superfamilies) was higher than in other assemblies (16–20 conopeptides superfamilies) (data not shown). After evaluating the quality of each assembly and an assessment of the diversity of the identified conopeptides gene superfamilies in the assembled transcripts for each assembler, the Trinity-assembled transcripts were chosen for subsequent analysis.
The low quality bases at the 3′ end of the Roche 454 sequencing reads were trimmed using the ‘-trim’ option of Newbler 2.5p1 (Margulies et al. 2005) and the trimmed reads were assembled into contigs and further into isotigs using Newbler 2.5p1. The single-end reads of 454 sequencing were also assembled using Trinity. While one third of the identified conopeptides was similar to the conopeptides identified in the Newbler assembly of 454 reads, a large number of conopeptides in the Trinity assembly were truncated (data not shown). Therefore, for 454 sequencing reads, the isotigs assembled using Newbler were used for the subsequent analysis. The Newbler-assembled isotigs were translated into six reading frames and all the possible open reading frames (ORFs) were identified. The quality of this assembly was evaluated as explained above by aligning the single-end reads of Roche 454 to the assembled isotigs using Bowtie2.
Conopeptide identification and superfamily classification
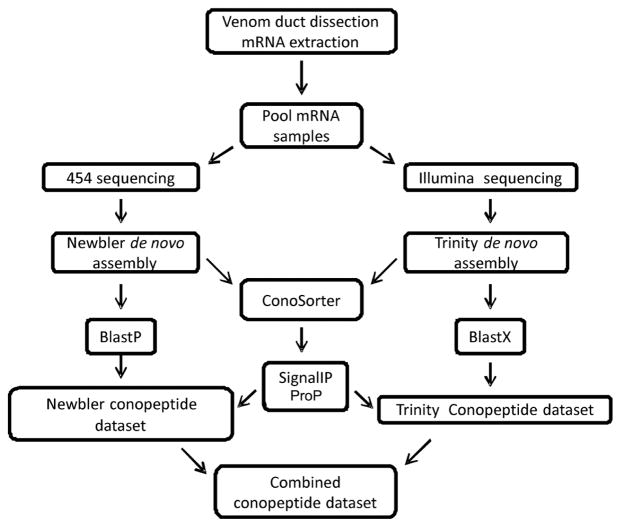
To identify conopeptide sequences, a BLASTX search of Stand-alone BLAST (Basic Local Alignment Search Tool) software version 2.2.29+ (Altschul et al. 1990) was conducted on the resulting transcripts assembled by Trinity against all the sequences in ConoServer update 2013-05-27 (Kaas et al. 2012) and UniProtKB-SwissProt release Feb 2014 (Apweiler et al. 2004) databases. Those transcripts that had a hit in BLASTX result were translated according to the reading frame identified by BLASTX and were manually inspected. The signal and mature regions of the conopeptides were predicted using ConoPrec (Kaas et al. 2012) and those transcripts with disrupted cysteine frameworks in the mature region were removed and the good quality putative conopeptide precursors were selected as the ‘Trinity conopeptide dataset’. In addition, a BLASTP search was performed on the identified ORFs in isotigs assembled by Newbler against the ConoServer and SwissProt databases, and those isotigs with hit in BLASTP result were extracted. The signal and mature regions of the selected isotigs were identified using ConoPrec. After manual inspection of the sequences, redundant isotigs and those with disrupted cysteine framework in the mature region or an abnormally long C terminal were removed. The remaining good quality putative conopeptide precursors were collected into the ‘Newbler conopeptide dataset’ (Fig. 1).
Fig. 1.
Sample preparation, sequencing and transcriptome analysis workflow. 1) mRNA extraction and sequencing using Illumina hi-seq 2000 and Roche 454 technologies, 2) de novo assembly using Trinity and Newbler, 3) conopeptide identification using BLAST, 4) putative conopeptide identification using ConoSorter, signal validation using SignalIP and propeptide cleavage site prediction using ProP, 5) pooling the conopeptide precursors in the ‘Combined conopeptide dataset’ (for details refer to Materials and Methods).
Additionally, to discover conopeptides with new signal sequences, the Newbler- and Trinity-assembled transcripts were analyzed by ConoSorter (Lavergne et al. 2013) and the results were filtered as described in Lavergne et al. (2013). In particular, those putative conopeptide sequences identified by ConoSorter having the length of least 50 amino acid residues and more than 60% hydrophobic amino acid residues in the signal region, and also showing matches for only the pro and mature regions were selected. The presence of canonical signal regions in the sequences was confirmed using SignalIP 4.1 (Petersen et al. 2011) and the propeptide cleavage sites of the putative conopeptides were detected using ProP 1.0 (Duckert et al. 2004). The selected Newbler- and Trinity-assembled transcripts with canonical signal regions and the propeptide cleavage sites were added to the Newbler and Trinity conopeptide datasets respectively (Fig. 1).
The conopeptide precursors in the Newbler and Trinity datasets were pooled; unique sequences were identified and collated in the ‘Combined conopeptide dataset’ (Fig. 1). Names were assigned to each sequence: a three letter word (Ctr) represented the species name, a randomly assigned number and finally a suffix: N (Newbler), T (Trinity) or TN (both Trinity and Newbler) indicating the original conopeptide dataset. The conopeptides present in both datasets with 100% sequence identity were given a suffix: TN. The conopeptides that were identified in both datasets having one or more amino acid difference or those that were present in only one dataset were given the suffix T (if present in Trinity) or N (if present in Newbler).
For each identified conopeptide in the ‘Combined conopeptide dataset’, the two highest-scoring full-length conopeptide precursor hits in the BLAST results against the ConoServer and SwissProt databases were used as references to facilitate categorization of the putative conopeptides into gene superfamilies. Each putative conopeptide was assigned to a specific gene superfamily based on the similarity of its signal region to the highly conserved signal sequence of the known conopeptide gene superfamilies. The signal regions of the putative conopeptides in the ‘Combined conopeptide dataset’ were used as query sequences to search (using BLASTP) for similar signal regions of the reference conopeptides retrieved from databases and the percentage sequence identity (PID) was computed as the ratio of the number of identical amino acid residues to the length of alignment. Initially, the putative conopeptides having PID of 76 for the signal region (Kaas et al. 2010) were classified into gene superfamilies. However, the conservation of the signal region for some gene superfamilies is below this threshold. Therefore, a threshold value specific to each superfamily was computed (see Results section ‘Identification and classification of conopeptides’). Multiple sequence alignment was performed on the precursors of the putative conopeptides and the reference conopeptide sequences using ClustalW version 2.1 (Larkin et al. 2007) followed by manual refinement using BioEdit version 7.0.0.
Functional annotation of transcripts
The functional annotation of the assembly was performed using the pipeline version of BLAST2GO software (B2G4Pipe) (Götz et al. 2008). The local B2G MySQL database was installed and the result of the BLASTX search of transcripts assembled by Trinity against the UniProtKB-SwissProt database were loaded to the pipeline, and based on the best blast hit, GO terms were assigned to each transcript. The annotation results were further refined by GO-Slim function, and the plot of transcripts GO classifications was constructed using Web Gene Ontology Annotation Plot (WEGO) software (Ye et al., 2006).
Results
Assembly of transcriptome sequences
The Illumina sequencing produced 33,544,045 paired-end reads of 100 bp long from both ends of cDNA fragments with average insert size of 140 bp. After trimming low quality reads as described in Materials and Methods, total of 25,825,187 high quality paired-end reads with a minimum length of 77 bp and an average length of 92 bp were used for assembly. The reads were assembled into 163,513 transcripts with an average size of 513 bp (Table 1) using Trinity; the total assembled size was 83.97 Mbp (mega base pairs) with N50 of 614 bp. The shortest transcript assembled by Trinity was 201 bp and the longest one was 16,967 bp.
Table 1.
Statistics summary of the de novo transcriptome assemblies. Illumina and 454 sequence datasets were assembled using Trinity and Newbler respectively. N50 represents the minimum transcript length for which all transcripts longer than or equal to that threshold contain 50% of total length of all transcripts. Mbp: mega base pairs (106 base pairs)
| Assembly parameters | Trinity | Newbler |
|---|---|---|
| Total assembled length (Mbp) | 83.97 | 2.21 |
| Number of transcripts | 163,513 | 2,473 |
| N50 (bp) | 614 | 966 |
| Maximum transcript length (bp) | 16,967 | 9,602 |
| Minimum transcript length (bp) | 201 | 53 |
| Mean transcript length (bp) | 513 | 891 |
| Median transcript length (bp) | 326 | 603 |
The transcriptome sequencing by 454 Roche technology generated 121,139 single-end reads. After trimming the low quality bases at the 3′ end of the reads, length of the reads ranged from 40 to 1,196 bp with an average length of 434 bp. These single-end reads of 454 sequencing were assembled into contigs and subsequently into 2,473 isotigs using Newbler 2.5p1. The total assembled size was 2.21 Mbp with N50 of 966 bp and an average isotigs size of 891 bp. The length of isotigs assembled by Newbler ranged from 53 to 9,602 bp (Table 1). The 454 single-end reads were aligned to the transcripts assembled using Newbler under single-end mode. Nearly 63.47% of the 454 reads were mapped to the assembly.
The length distribution of the transcripts assembled from 454 and Illumina reads using Newbler and Trinity, respectively, is shown in Table 2. The majority (63%) of the Trinity-assembled transcripts were 201–400 bp long while 65% of the transcripts assembled using Newbler fell in the range of 401–800 bp.
Table 2.
The length distribution of transcripts assembled by Trinity (Illumina sequence dataset) and Newbler (454 sequence dataset). The percentage of transcripts in each length category is computed in relation to the total number of transcripts: 163,513 (Trinity) and 2,473 (Newbler).
| Length(bp) | No. of Transcripts | % of Transcripts | ||
|---|---|---|---|---|
|
| ||||
| Trinity | Newbler | Trinity | Newbler | |
| ≤200 | 0* | 32 | 0* | 1.29 |
| 201–400 | 102,920 | 73 | 62.94 | 2.95 |
| 401–800 | 39,171 | 1,607 | 23.96 | 64.98 |
| 801–1000 | 6,581 | 223 | 4.02 | 9.02 |
| 1001–5000 | 14,429 | 519 | 8.82 | 20.99 |
| ≥5001 | 412 | 19 | 0.25 | 0.77 |
The minimum length of transcripts in Trinity is 201bp
Identification and classification of conopeptides
The putative conopeptide sequences were identified by BLAST search against the ConoServer and Swiss-Prot databases. The BLASTX results of the Trinity-assembled transcripts showed that 331 transcripts had high sequence similarity to the conopeptides in databases. After removal of the duplicate and truncated transcripts, a total of 96 putative conopeptide precursors were identified as “Trinity conopeptide dataset”. The ORFs of isotigs assembled using Newbler were subjected to BLASTP search and 129 transcripts with high similarity to the conopeptides in the ConoServer and Swiss-Prot databases were detected. Duplicate transcripts and the sequences with disrupted cysteine frameworks in the mature region were removed and 72 putative conopeptides were identified as “Newbler conopeptide dataset”. The majority of the identified transcripts in both Trinity and Newbler conopeptide datasets were full-length or nearly full-length conopeptide precursors.
In addition, the transcripts assembled by Trinity and Newbler were analyzed using ConoSorter. The putative conopeptide sequences identified by ConoSorter were filtered by length, hydrophobicity of the signal region and the presence of match for the pro and mature regions as described in Materials and Methods. Total of 10 Trinity-assembled and 3 Newbler-assembled transcripts showing canonical signal regions and prepro cleavage sites were added to the Trinity and the Newbler conopeptide datasets respectively.
To identify the unique conopeptides in C. tribblei, the ‘Trinity conopeptide dataset’ and ‘Newbler conopeptide dataset’ were merged into the ‘Combined conopeptide dataset’ and the unique sequences were identified and named as explained in Materials and Methods. The ‘Combined conopeptide dataset’ contained 152 unique putative conopeptides of which 29 were present in both datasets (suffix: TN) while 77 were exclusive to the Trinity dataset (suffix: T) and 46 were only present in the Newbler dataset (suffix: N). All 152 putative conopeptides are unique sequences (at least one amino acid residue difference in the mature regions). However, 4 pairs of conopeptides were observed to have identical mature regions but differ (1–5 amino acid residues) either in the pro or the signal regions (Ctr_120_T and Ctr_125_N, Ctr_51_T and Ctr_69_N, Ctr_159_N and Ctr_161_T, and Ctr_16_T and Ctr_64_N in the supplementary figures).
Each putative conopeptide precursor was assigned to a conopeptide gene superfamily based on the percentage of sequence identity to the highly conserved signal region of the conopeptide gene superfamilies present in ConoServer database as described in Materials and Methods. However, the conservation of the signal region for some gene superfamilies is below 76%. Therefore, for I1, I2, L, M, P and S superfamilies, the conopeptides of each superfamily were retrieved from ConoServer. The pairwise PID for the signal regions of the members of each superfamily was computed using MatGAT 2.02 (Campanella et al. 2003). Based on the average PID of each superfamily, the threshold value for assigning the conopeptides to I1-, I2-, L-, M-, P- and S-superfamilies were adjusted to 71.85%, 57.6%, 67.5%, 69.3%, 69.1% and 72.9% respectively. Additionally, for the con-ikot-ikot family, the signal regions of several complete precursors (Maricq et al. 2009) and the available precursor sequences in ConoServer were collected, the average PID of the signal regions were computed and the threshold value was set to 64.5% ± 20.2. The divergent superfamilies representing the conopeptides identified in C. californicus are mostly represented by only one sequence per superfamily or show low conservation in the signal regions. For example the conservation of the signal region was 64.22% ± 20.53 among members of the ‘divergent M---L-LTVA’ superfamily. Since the ‘divergent MSKLVILAVL’ and ‘divergent MKFPLLFISL’ superfamilies each contain one sequence only, the threshold value of ‘divergent M---L-LTVA’ superfamily (64.22% ± 20.53) was used for all these divergent superfamilies. The conopeptides were assigned to a superfamily if the percentage sequence identity for the signal region was above the threshold of that specific superfamily. Only ten conopeptide precursors in the ‘Combined conopeptide dataset’ contained partial signal region thus the whole precursor was used for the superfamily classification of these conopeptides. If the signal region of a conopeptide precursor showed percentage sequence identity below the threshold values defined for different superfamilies, the conopeptide was classified as a new conopeptide group. For the new conopeptide groups showing slight similarity either in the signal or the mature region to any known superfamily, the new groups were named after the most similar superfamily plus ‘-like’ suffix. Otherwise, the assigned name was ‘SF-‘ plus Arabic numbers such as ‘01’.
From the 152 conopeptides in the ‘Combined conopeptide dataset’, 127 conopeptides were classified into 30 known conopeptide gene superfamilies and families. The conopeptides in the ‘Trinity conopeptide dataset’ (30 gene superfamilies) were more diverse than those in the ‘Newbler conopeptide dataset’ (20 gene superfamilies) (Table 3). Conopeptides belonging to the majority of the gene superfamilies (D, F, H, I1, I2, L, M, N, O1, O2, O3, P and S) and several cysteine-rich families like conkunitzin, conodipine, conopressin/conophysin and con-ikot-ikot were present in both Newbler and Trinity conopeptide datasets. However, several gene superfamilies (B2, I3, J, K and Y) and a cysteine-poor family, conantokin (classified as B1 superfamily in ConoServer), were observed in the ‘Trinity conopeptide dataset’ only (Table 3). In addition, some conopeptides showing similarity to the newly identified R, W and Y2 superfamilies (Lavergne et al. 2013) were also discovered.
Table 3.
The abundance and cysteine framework of conopeptides in Trinity (T), Newbler (N) and Combined (T+N) conopeptide datasets. For each superfamily, the conopeptide sequences of Newbler and Trinity conopeptide datasets were pooled and unique conopeptides were represented in ‘Combined conopeptide dataset’. Superfamilies R, W and Y2 were identified in Lavergne et al. (2013).
| Superfamily | Cysteine framework | No. of conopeptides | Remarks | ||
|---|---|---|---|---|---|
| N | T | T+N | |||
| Conantokin (B1) | no-Cys | 0 | 2 | 2 | |
| B2 | no-Cys | 0 | 1 | 1 | |
| D | XX | 2 | 1 | 3 | |
| F | no-Cys | 1 | 1 | 1 | |
| H | XIV# | 3 | 1 | 4 |
|
| I1 | VI/VII, XI | 2 | 2 | 2 | |
| I2 | XI, XII, XIII# | 2 | 3 | 4 |
|
| I3 | XI | 0 | 1 | 1 | |
| J | XIV | 0 | 2 | 2 | |
| K | C-C-C-CC*† | 0 | 1 | 1 |
|
| L | XIV, XXIII# | 2 | 3 | 4 |
|
| M | C-C†, no-Cys, C-CC-C-C-C* XXV#, VIII#, IX, XVI, III |
7 | 13 | 18 |
|
| N | XV, C2-CC-CC-C4*1 | 4 | 2 | 5 |
|
| O1 | VI/VII, XI# | 2 | 6 | 6 |
|
| O2 | VI/VII, XV, XII# | 6 | 5 | 11 |
|
| O3 | VI/VII | 3 | 2 | 4 | |
| P | IX | 4 | 5 | 6 | |
| S | VIII, C4-CC-C4*2 | 2 | 2 | 3 |
|
| Y | XVII | 0 | 1 | 1 | |
| MSTLGMTLL- | XIX | 0 | 1 | 1 | |
| MSKLVILAVL | IX | 1 | 1 | 1 | |
| M---L-LTVA | IX, VI/VII | 0 | 2 | 2 | |
| MKFPLLFISL | VI/VII, IX# | 0 | 3 | 3 |
|
| Conkunitzin | C4, C8, C6-CC-C6 *3 | 4 | 4 | 6 |
|
| Con-ikot-ikot | CC-C5-CC-C54 C-CC-C6-CC-C3*5, CC-C10*6, CC-C11*7† CC-C11-CC-C5*8 CC-C10-CC-C5-CC-C5*9 CC-C5-CC-C5*10† |
17 | 5 | 20 |
|
| Conodipine | C10, C12 | 2 | 5 | 7 |
|
| Conopressin/Conophysin | C-C, C3-CC-C5-CC-C211 | 2 | 3 | 5 |
|
| R | no-Cys | 0 | 1 | 1 | |
| W | no-Cys | 1 | 1 | 1 | |
| Y2 | XII | 1 | 1 | 1 | |
Cysteine frameworks that are new in the conopeptides of Conus species but has been observed in other organisms,
The cysteine patterns that have already been identified in other conopeptide superfamilies but they’ve not been observed in this superfamily yet,
likely to form inter-chain disulfide bond. 1: C-C-CC-CC-C-C-C-C, 2: C-C-C-C-CC-C-C-C-C, 3: C-C-C-C-C-C-CC-C-C-C-C-C-C, 4: CC-C-C-C-C-C-CC-C-C-C-C-C, 5: C-CC-C-C-C-C-C-C-CC-C-C-C, 6: CC-C-C-C-C-C-C-C-C-C-C, 7: CC-C-C-C-C-C-C-C-C-C-C-C, 8: CC-C-C-C-C-C-C-C-C-C-C-C-CC-C-C-C-C-C, 9: CC-C-C-C-C-C-C-C-C-C-C-CC-C-C-C-C-C-CC-C-C-C-C-C, 10: C-CC-C-C-C-C-C-CC-C-C-C-C-C, 11: C-C-C-CC-C-C-C-C-C-CC-C-C, XX: C-CC-C-CC-C-C-C-C, XV: C-C-CC-C-C-C-C, XI: C-C-CC-CC-C-C, XII: C-C-C-C-CC-C-C, XIV: C-C-C-C, XXIII: C-C-C-CC-C, III: CC-C-C-CC, XXV: C-C-C-C-CC, VIII: C-C-C-C-C-C-C-C-C-C, IX: C-C-C-C-C-C, XVI: C-C-CC, VI/VII: C-C-CC-C-C, XVII: C-C-CC-C-CC-C, XIX: C-C-C-CCC-C-C-C-C, XIII: C-C-C-CC-C-C-C, C4: C-C-C-C, C8: C-C-C-C-C-C-C-C, C10: C-C-C-C-C-C-C-C-C-C, C12: C-C-C-C-C-C-C-C-C-C-C-C, no-Cys: no cysteine residues in the mature region.
There are sequences in the ConoServer database that belong to new but still unnamed superfamilies (currently, in the ConoServer database, these superfamilies have been given temporary names based on the first ten amino acid residues in the signal regions and prefixed with the word ‘divergent’). Some of the identified conopeptides in C. tribblei were highly similar to these ‘divergent’ superfamilies. A sequence identified in both Newbler and Trinity conopeptide datasets was assigned to the ‘divergent MSKLVILAVL’ superfamily, whereas in the ‘Trinity conopeptide dataset’, six sequences were identified to belong to the ‘divergent MSTLGMTLL-’, ‘divergent M---L-LTVA’ and ‘divergent MKFPLLFISL’ superfamilies.
The cysteine frameworks of the identified conopeptides are shown in Table 3. Several conopeptides of the ‘divergent MKFPLLFISL’, H, I2-, K-, L-, M-, N-, O1-, O2- and S-superfamilies and also con-ikot-ikot and conkunitzin families showed cysteine patterns that are new in cone snails or exhibited known cysteine frameworks that have not been observed in these superfamilies yet.
Furthermore, the sequence identity in the signal regions of 9 putative conopeptides in the ‘Combined conopeptide dataset’ were below the threshold value specified for different superfamilies as described above. These novel putative conopeptides were classified into 6 groups (Table 4): N-like, G-like, A-like, Y2-like, V-like and SF01. In addition, 9 of the 152 sequences in the ‘Combined conopeptide dataset’, identified as putative conopeptide using ConoSorter, did not show sequence identity in the signal region to any known superfamily (Fig. S1). The predicted mature regions of three of these sequences contains no cysteine residues whereas other sequences have either two or odd number of cysteine residues. However, the presence of these peptides in the venom could not be confirmed due to absence of proteomics data for C. tribblei. Additionally, seven conopeptides containing only the pro and mature regions of A-, G- and O1-superfamilies were also identified in the ‘Combined conopeptide dataset’ (Fig. S2).
Table 4.
The abundance and cysteine framework of the new conopeptide groups. The notes are indicated in Table 3. I: CC-C-C, XXII: C-C-C-C-C-C-C-C
| Conopeptide Group | Cysteine framework | No. of conopeptides | ||
|---|---|---|---|---|
| N | T | T+N | ||
| N-like | XV | 1 | 1 | 1 |
| G-like | XX | 2 | 0 | 2 |
| A-like | I | 0 | 1 | 1 |
| Y2-like | XII | 0 | 2 | 2 |
| V-like | VI/VII, XXII | 1 | 2 | 2 |
| SF01 | C12 | 1 | 1 | 1 |
Conopeptide diversity
The conopeptide mixture in C. tribblei consisted of 136 new conopeptides that were classified into 30 known conopeptide gene superfamilies and 6 new conopeptide groups. Sequences of the M-, O-superfamilies and con-ikot-ikot family were among the most predominant conopeptide groups in C. tribblei. Despite the observed high diversity, C-, B3-, E-, T- and V-superfamilies were not observed in the C. tribblei transcriptome dataset. Some sequences of the mature regions of A-, G- and O1-superfamilies were also observed in C. tribblei (Fig. S2).
The M-superfamily is the most predominant superfamily both in terms of the number of conopeptides (18 peptides) and also the diversity of cysteine frameworks (6 patterns). Conopeptides of the cysteine-free conomarphin family (Fig. S3a) and the M-1 branch (Jacob and McDougal 2010) of M-conotoxins (Fig. 2) with only one amino acid between the fourth and fifth cysteine residues were identified. In addition, Ctr_87_T and Ctr_100_T sequences exhibited XXV (C-C-C-C-CC) (Aguilar et al. 2013) and VIII (C-C-C-C-C-C-C-C-C-C) (England et al. 1998) cysteine frameworks respectively (Fig. 2). Although framework VIII has previously been identified in conopeptides of S superfamily, this is the first time that this cysteine pattern was observed in an M superfamily peptide. Several conopeptides of M superfamily contained only two cysteine residues in their mature region (Ctr_46_T, Ctr_63_T and Ctr_110_T) suggesting the likely formation of inter-disulfide bridges. While cysteine pattern C-CC-C-C-C has been observed in other organisms, Ctr_48_N and few other conopeptides of M-superfamily (Fig. 2) are the first conopeptides showing this cysteine pattern in cone snails.
Fig. 2.

The putative conopeptide precursors of M superfamily. The conopeptides identified in C. tribblei are shown in black and the conopeptide nomenclature is described in Materials and Methods. The reference sequences are shown in green and cysteine residues are shown in bold italic red. The names of the reference sequences are derived from the ConoServer database unless noted otherwise. The signal regions are highlighted and the mature regions are underlined.
Although the average percentage sequence identity for the signal sequences in putative con-ikot-ikot sequences in C. tribblei was 59.6%, most con-ikot-ikot sequences had identical cysteine pattern (CC-C-C-C-C-C-CC-C-C-C-C-C) (Fig. 3). Sequence alignment revealed that these sequences separate into three clades (Fig. 3) and members of each clade had similarities both in the signal region and also in the identity and number of amino acids in the cysteine loops. All the conopeptides in clade I have five and two amino acids in their fifth (between the sixth and seventh cysteine residues) and sixth (between the 7th and 8th cysteine) loops, respectively. Furthermore, the peptides in clade I seem to bifurcate into two groups. While members of clade Ia have 18–19 amino acids between the 11th and 12th cysteine (ninth loop), the conopeptides in Ib have only 13 residues in the same position. On the other hand, the conopeptides in clade II and III have 7–8 amino acids in both the fifth and sixth cysteine loops. However, the conopeptides in clade II have 14 amino acids in the ninth loop while members of clade III have only 10 residues. It’s worth mentioning that except for the 5th, 6th and 9th loops, the number of amino acids in other cysteine loops is relatively comparable between all clades. Additionally, six new cysteine frameworks were also discovered in this peptide family (Fig. S3b).
Fig. 3.

The putative conopeptide precursors of con-ikot-ikot family. The notes are indicated in Fig 2. C.figulinus and C. planorbis sequences are from Mariq et al. (2009).
A number of conkunitzin sequences were also identified in C. tribblei dataset. The previously identified conkunitzin sequences in cone snails, despite having similarity to kunitz proteins, contained four cysteine residues exhibiting only two disulphide bonds (I–IV, II–III) (Bayrhuber et al. 2005). The conkunitzin sequences identified in C. tribblei are the first conkunitzin sequences identified in cone snails having two consecutive kunitz-like domains containing either four or six cysteine residues in each domain (Fig. S3c). Although these sequences show similarity to kunitz proteins, the disulphide connectivity of the cysteine residues may be different from the typical kunitz proteins (I–VI, II–IV, III–V) (Rawlings et al. 2004).
Several putative peptide sequences were observed to contain consecutive conopressin and conophysin domains. While most of the sequences showed high similarity in the conophysin domain to conophysin-R (Lirazan et al. 2002), the other sequence (Ctr_122_T) was more similar to the conophysin in Lymnaea stagnalis (Van Kesteren et al. 1995) (Fig. S4a). Like conophysin-R, the conophysin domains in Ctr_120_T, Ctr_121_T, Ctr_124_N and Ctr_125_N had four amino acids between the second and the third cysteine residues and also contained a long loop between Cys9 and Cys10. On the contrary, similar to other neurophysins, there were seven amino acids between the 2nd and 3rd cysteine residues in Ctr_122_T and the loop between the 9th and 10th cysteine residues was smaller as well. Moreover, a conopressin domain was exhibited preceding the conophysin domain (Fig. S4a). The presence of tandem domains of conopressin and conophysin in C. tribblei and conophysin-G (Hu et al. 2012) similar to the conopressin/neurophysin sequence in L. stagnalis may indicate that conopressin and conophysin are cleaved from one single precursor in Conus.
Five of the identified conodipine sequences in the C. tribblei transcriptome were highly similar to the alpha and beta chains of conodipine-M in Conus magus (McIntosh et al. 1995), and they had 21 amino acid residues long signal region at the N terminal (Fig. S4b). The tandem presence of alpha and beta chains of conodipine in one transcript and the observed signal sequence are the first to be reported. Another sequence of this family (Ctr_119_T) showed more similarity to the Crassostrea gigas conodipine (Zhang et al. 2012) in terms of the number of cysteine residues (10) and also the identity and number of amino acids between cysteine residues (Fig. S4b).
Furthermore, three cysteine patterns (C-C-CC-CC-C-C-C-C, C-C-C-CC and C-C-C-C-CC-C-C-C-C) that have already been identified in other organisms were observed for the first time in cone snails: the N-, K- and S-superfamily conopeptides, respectively (Fig. S5). The odd number of cysteine residues in cysteine pattern C-C-C-CC may suggest formation of inter-chain disulfide bond. The conopeptides of I2-, L-, H-, O1-, O2- (Fig. S6), D-, I1-, I3- (Fig. S7), O3-, J, Y-, F-, B2- (Fig. S8), P-, R-, W- and Y2 superfamilies and conantokin family (B1 superfamily) (Fig. S9) were also discovered in the C. tribblei transcriptome. The occurrence of these transcripts (and hence their genes) was inferred based on the presence of at least 5 reads of Roche 454 or 5 reads of Illumina (Table 2S).
New conopeptide groups
In addition to 127 conopeptides classified into 30 known superfamilies, a total of 9 conopeptides showing divergent signal regions were assigned to 6 new conopeptide groups (Fig. 4 and Table 4); whether these groups should be recognized as distinct superfamilies or subgroups within a superfamily is not clear considering that the similarity of the sequences in the signal region with known ones is below but close to the threshold values used to ascertain superfamily membership. Putative conopeptide Ctr_13_TN showed 58.8% identity to N superfamily but it had different pro region. Ctr_13_TN also has XV (C-C-CC-C-C-C-C) cysteine framework (Peng et al. 2008) that is observed in members of the N superfamily. In addition, several conopeptides were classified as G-like due to the sequence similarity of 65% to the signal regions of G superfamily. However, G-like conopeptides had different pro regions from G-superfamily and also exhibited different cysteine pattern. Interestingly, the signal region of Ctr_19_T showed 76.2% identity to PuSG1.1, a conopeptide identified in the salivary gland of Conus pulicarius (biggs et al. 2008) whereas its mature region showed 75% sequence identity to vc1.3, a member of α4/7 subfamily of A-superfamily identified in the venom duct of Conus victoriae (Safavi-Hemami et al. 2011). Although the signal region of PuSG1.1 is different from A-superfamily, due to having the typical α4/7 subfamily cysteine pattern XIV and also the same spacing between cysteine residues (C-CX4CX7C), it was identified as α-like conopeptide; therefore Ctr_19_T was classified as A-like group. Two conopeptides showing 41–44% sequence identity to the signal region of V-superfamily were classified as V-like group. These conopeptides contained cysteine patterns VI/VII (C-C-CC-C-C) (Olivera et al. 1984) and XXII (C-C-C-C-C-C-C-C) (Elliger et al. 2011). Also, two sequences showing similarity at the mature region to the conopeptide recently classified as Y2 superfamily (Lavergne et al. 2013) were also identified. While these conopeptides have different signal regions, they all show similarity to molluscan insulin-related protein identified in L.stagnalis (Smit et al. 1993). Additionally, a conopeptide sequence, Ctr_52_TN, showing similarity to an unidentified conopeptide from Conus caracteristicus was discovered (SF01).
Fig. 4.
The putative conopeptide precursors of the new conopeptide groups. The notes are indicated in Fig. 2. Uniprot ID P80090 is molluscan insulin-related peptide 3 from Lymnaea stagnalis; Chain B (30–66 residues) and chain A (99–122 residues) of P80090 are underlined. Uniprot ID U3L0H2 from Conus flavidus. Mr_156 with Genbank accession number AB850850 from Conus marmoreus.
Divergent superfamilies
Seven putative sequences present in the C. tribblei dataset were found to be similar to conopeptide sequences that so far have only been reported for early divergent species, C. californicus and Conus distans (Fig. 5). These conopeptides are currently classified as ‘divergent’ superfamilies in ConoServer and are mostly represented by only one or two sequences per superfamily. In addition, one sequence (Ctr_32_T) displayed cysteine pattern IX (C-C-C-C-C-C) that has not been previously observed in other conopeptides of this superfamily (Fig. 5).
Fig. 5.

The putative conopeptide precursors of ‘divergent MSTLGMTLL-’, ‘divergent MSKLVILAVL’, ‘divergent M---L-LTVA’ and ‘divergent MKFPLLFISL’ superfamilies. The notes are indicated in Fig. 2.
Post-translational modification enzymes
Transcripts of genes that encode enzymes that are potentially involved in the post-translational modification of conopeptides were also found in the C. tribblei transcriptome dataset (Table 5). Sequences of the protein disulfide-isomerase (PDI) family which mediates formation of disulfide bonds showing high similarity to PDI (PDIA) isolated from cone snails (Wang et al. 2007; Safavi-hemami et al. 2012) were identified in C. tribblei. In addition, other PDI proteins with varying domain architecture showing high similarity to protein disulfide-isomerase A3 (ERP57), A4 (ERP72), A5 (PDIR) and A6 (P5) families were also discovered. Other chaperones and co-chaperones in protein folding such as calnexin, calreticulin, hsp40 (DNAJ), sacsin, hsp20, hsp70, hsp60, hsp90, hsp105, Grp78 (binding immunoglobulin protein (BiP)) and T-complex protein 1 were identified as well. In addition, two families of peptidylprolyl cis-trans isomerases (PPIases), cyclophilin and FKBP (FK506 binding protein), that enhance the oxidative folding of conopeptides in the presence of PDI and BiP (Safavi-hemami et al. 2012) were identified.
Table 5.
List of the identified post-translational modification enzymes in the C. tribblei transcriptome
| Post-translational modification enzymes | Function |
|---|---|
protein disulfide isomerase (PDI)
|
disulfide bond formation |
| thioredoxin-related transmembrane protein 1 and 2 families, thioredoxin domain containing protein 5 and 12 families, 15 kDa selenoprotein and sulfhydryl oxidase 1 | protein folding |
| hsp40 (DNAJ), Sacsin, hsp20, hsp70, hsp60, hsp90, hsp105, Grp78 (BiP), Calnexin, Calreticulin and T-complex protein 1 | chaperone and co-chaperone in protein folding and disulfide bond formation |
peptidylprolyl cis-trans isomerases (PPIases)
|
cis-trans isomerization of proline |
| Vitamin K dependent gamma-carboxylase Vitamin K epoxide reductase |
conversion of glutamic acid to γ-carboxyglutamic acid |
| glutaminyl-peptide cyclotransferase | cyclization of N-terminal glutamine to pyroglutamatein |
peptidyl-glycine alpha-amidating monooxygenase:
|
alpha-amidation |
| cysteine-rich secretory protein (CRISP) | cleavage of the mature conopeptides from their propeptide precursors |
| conoporin | haemolytic activities |
| conohyal | degradation of extracellular matrix |
| metalloprotease, C-type lectin, serine protease and serine protease inhibitor | antihemostatic role |
Sequences of vitamin K dependent gamma-carboxylase and glutaminyl-peptide cyclotransferase were identified in the C. tribblei dataset. A sequence containing both domains of peptidyl-glycine alpha-amidating monooxygenase: peptidylglycine alpha-hydroxylating monooxygenamse (PHM) and peptidyl-alpha-hydroxyglycine alpha-amidating lyase (PAL) was also identified. A sequence with high similarity to the cysteine-rich secretory protein (CRISP) family of a pathogenesis-related protein superfamily and sequences with high similarity to conoporin and conohyal were discovered in the C. tribblei transcriptome demonstrating the involvement of these proteins in envenomation.
Functional classification of transcripts
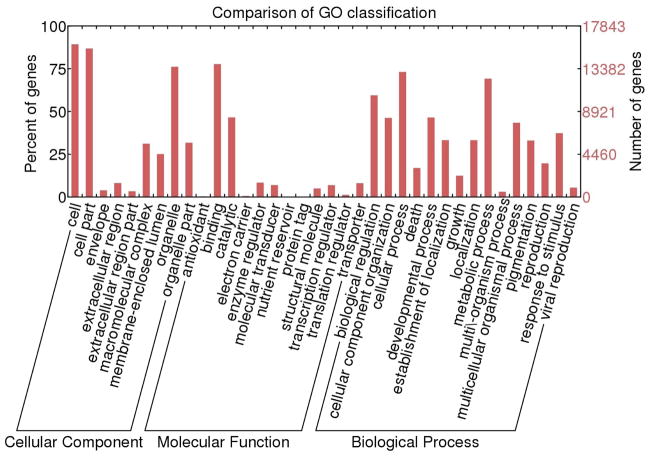
The BLASTX results showed that 21,069 transcripts had significant similarities to proteins in UniprotKB Swiss-Prot database. Using Blast2Go software, gene ontology terms were assigned to 17,843 transcripts (84.7%). In the molecular function category, binding and catalytic activity were the most represented terms (Fig. 6) while in the biological process category cellular process, metabolic process and biological regulation had the highest number of assigned terms. In cellular component category, cell and organelle were the most abundant terms. The most highly expressed transcripts are presented in Table 3S.
Fig. 6.
GO annotations of the venom duct transcripts of C. tribblei
Discussion
Next-generation transcriptome sequencing has been employed previously to investigate the diversity of conopeptides in several Conus species (Hu et al. 2011; Hu et al. 2012; Lluisma et al. 2012; Terrat et al. 2012; Dutertre et al. 2013; Lavergne et al. 2013). The number of conopeptide sequences discovered ranged from 30- 263; these peptides were reported to belong to 10–26 gene superfamilies (Table 6a). The diversity of conopeptide gene superfamilies in C. tribblei, as revealed by our analysis of the venom duct transcriptome using a combination of Illumina and Roche 454 sequencing technologies, is by far the highest discovered from a Conus species to date. A total of 136 putative conopeptides which could be classified into 30 known gene superfamilies were identified. In addition, conopeptides showing divergent signal regions were also found.
Table 6.
Comparison of the transcriptome studies of Conus species venom duct through next generation sequencing a The total number of identified conopeptides and gene superfamilies for each species b The relative abundance of conopeptide superfamilies for each species. Diets are specified as P: piscivorous, M: molluscivorous and V: vermivorous.
| Conus Species | C.bul | C.con | C.geo | C.pul | C.mar§ | C.tri |
|---|---|---|---|---|---|---|
| (a) | ||||||
| Diet | P | P | P | V | M | V |
| No. of superfamilies | 10 | 11 | 16 | 14 | 26 | 30 (+6†) |
| No. of conopeptides | 30* | 61 | 63 | 82 | 263 | 127 (+9‡) |
|
| ||||||
| (b) | ||||||
| A | 10 | 14 | 12 | 2 | ||
|
| ||||||
| Conantokin (B1) | 1 | 2 | 3 | 2 | ||
|
| ||||||
| B2 | 1 | 1 | ||||
|
| ||||||
| Contulakin (C) | 4 | 1 | 1 | |||
|
| ||||||
| D | 3 | |||||
|
| ||||||
| E | 1 | |||||
|
| ||||||
| F | 1 | 1 | ||||
|
| ||||||
| H | 7 | 4 | ||||
|
| ||||||
| I & | 1 | 10 | 30 | 7 | ||
|
| ||||||
| J | 1 | 4 | 2 | |||
|
| ||||||
| K | 1 | |||||
|
| ||||||
| L | 1 | 5 | 4 | |||
|
| ||||||
| M | 3 | 8 | 2 | 4 | 75 | 18 |
|
| ||||||
| N | 3 | 5 | ||||
|
| ||||||
| O # | 15 | 15 | 20 | 47 | 82 | 21 |
|
| ||||||
| P | 1 | 3 | 6 | |||
|
| ||||||
| R | 1 | 1 | ||||
|
| ||||||
| S | 3 | 5 | 2 | 3 | ||
|
| ||||||
| T | 5 | 6 | 5 | 26 | ||
|
| ||||||
| V | 1 | |||||
|
| ||||||
| W | 2 | 1 | ||||
|
| ||||||
| Y | 1 | |||||
|
| ||||||
| Y2 | 2 | 1 | ||||
|
| ||||||
| MSTLGMTLL- | 1 | 1 | ||||
|
| ||||||
| MKFPLLFISL | 3 | |||||
|
| ||||||
| M---L-LTVA | 2 | |||||
|
| ||||||
| MSKLVILAVL | 1 | |||||
|
| ||||||
| Con-ikot-ikot | 7 | 20 | ||||
|
| ||||||
| Conkunitzin | 1 | 7 | 1 | 2 | 6 | |
|
| ||||||
| Conodipine | 2 | 7 | ||||
|
| ||||||
| Conopressin/conophysin | 1 | 1 | 5 | |||
Number of putative full-length and partial conopeptides presented in this study,
O superfamily includes O1, O2 and O3 superfamilies and contryphans,
: I superfamily includes I1, I2 and I3 superfamilies.
16 of the 26 superfamilies identified in this species are shown in (b).
number of the new conopeptide groups,
number of conopeptides in the new conopeptide groups. C.bul: Conus bullatus (Hu et al. 2011), C.con: Conus consors (Terrat et al. 2012), C.geo: Conus geographus (Hu et al. 2012), C.pul: Conus pulicarius (Lluisma et al. 2012), C.mar: Conus marmoreus (Dutertre et al. 2013; Lavergne et al. 2013) and C.tri: Conus tribblei (this study).
The relative abundances of the different conopeptide gene superfamilies are known to vary in the venom duct of Conus species. Similar to Conus bullatus, Conus marmoreus and C. pulicarius, the M- and O-superfamilies are among the most abundant superfamilies in C. tribblei. In contrast to Conus consors, Conus geographus and C. bullatus, where A-superfamily sequences are the most abundant, this superfamily is under represented in the C. tribblei transcriptome. What is remarkable about C. tribblei is the unexpectedly great abundance of con-ikot-ikot family (20 sequences) (Table 6b). In total, the major proportion of the identified conopeptides in C. tribblei belonged to the M- and O-superfamilies and con-ikot-ikot family.
Diversity of conopeptides in a species is associated with its prey preferences (Duda and Palumbi 2004; Duda et al. 2009). It is believed that the generalist feeding behaviour of C. californicus has promoted the expression of extremely diverse conopeptides in this species that could not be classified according to the typical established gene superfamilies (Elliger et al. 2011), while the specialized diet of Conus leopardus has significantly decreased the diversity of the expressed conopeptides (Remigio and Duda 2008). Therefore, the presence of diverse conopeptides in the venom duct of C. tribblei may reflect its wide range of prey preferences.
Although several specimens were used in the mRNA extraction for this transcriptome study, the observed diversity in terms of gene superfamilies in the C. tribblei venom duct transcriptome is not likely due to intra-specific variation. A study by Dutertre et al. (2010) has shown that peptide masses in the venom profile vary among individuals of a species and have suggested the presence of intra-specific variability in the venom of cone snails. However, recently using parallel next generation sequencing and high sensitivity mass spectrometry, it was shown that thousands of peptide fragments may be derived from only a hundred conopeptide precursors through variable post-translational modification processing (Dutertre et al. 2013). While the contribution of intra-specific variability to the high diversity observed in this study remains to be determined in future studies, a comparison of the venom duct transcriptome of three individuals of C. tribblei has shown the presence of 27–28 gene superfamilies in each individual (manuscript in preparation).
The employed sequencing technologies may have also contributed to discovery of the outstanding conopeptide diversity in the C. tribblei venom duct. The 454 sequencing data contributed almost half of the conopeptides in the identified conopeptide repertoire of C. tribblei but 10 of the 30 known gene superfamilies were not present in this dataset and were retrieved from Illumina sequences only (Table 3). Evidently, the high-throughput paired-end sequences of Illumina have facilitated the identification of conopeptides that probably were transcribed less than the predominant conopeptides. On the other hand, discovery of the majority of con-ikot-ikot sequences from the 454 dataset shows that the long reads of 454 sequencing were more suitable for identification of these sequences that are longer that the typical conopeptides. Advantages of combinatorial high-throughput sequencing platforms have proven to be remarkable and the transcriptome analysis of the C. tribblei venom duct is another example.
Interestingly, a number of transcripts containing two consecutive conopeptide domains were identified in the C. tribblei transcriptome. Several sequences having tandem domains of conopressin and conophysin were observed (Fig. S4a). Although the presence of tandem domains of conopressin and neurophysin has been documented previously in L. stagnalis (Van Kesteren et al. 1995), the identified transcripts in C. tribblei are the first to be reported in any Conus species. In addition, several transcripts containing both alpha and beta chain sequences of Conodipine-M were identified in the C. tribblei dataset (Fig. S4b). Conodipine-M is the only phospholipase A2 isolated from cone snails that inhibits the binding of isradipine to the L-type calcium channel (McIntosh et al. 1995).
Another surprising and unexpected characteristic of the C. tribblei transcriptome is the presence of four ‘divergent’ superfamilies that are assigned to the conopeptides identified in C. californicus which is phylogenetically distant from other Conus species (Fig. 7). The conopeptide belonging to ‘divergent M---L-LTVA’ gene superfamily in the C. tribblei dataset displayed slightly different signal sequence despite having the typical cysteine pattern of this superfamily. Similarly, other conopeptides of this superfamily isolated from C. californicus have shown divergence in the signal sequence. The slight incongruence between conopeptides of the ‘divergent’ superfamilies from C. californicus and the ‘divergent’ conopeptides of C. tribblei is quite expected since C. californicus is a phylogenetically distant species. Conservatively, classical conopeptide gene superfamily names are not assigned to these conopeptides mainly because these divergent peptides have only been observed in C. californicus. However, two more similar conopeptides were identified in C. distans (Chen et al. 2008) and C. pulicarius (Lluisma et al. 2012), and the identification of additional sequences in C. tribblei may indicate the need for a more consistent classification of these peptides.
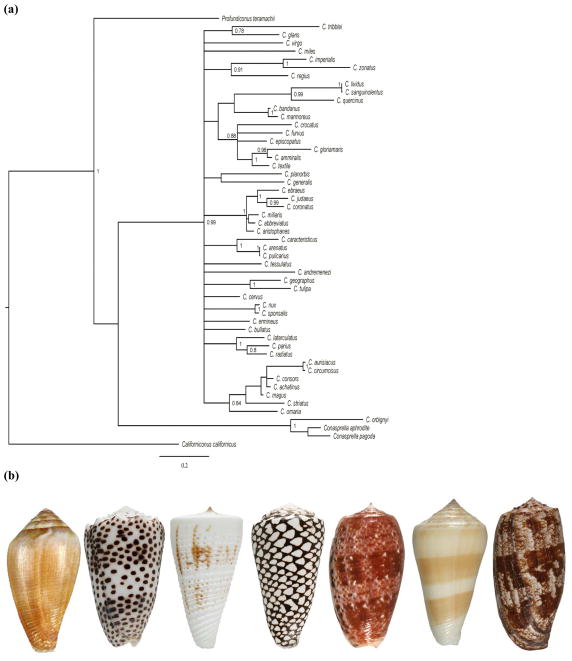
Fig. 7.
The phylogenetic relationship of Conus species (a) Shells of the cone snails in table 6: (left to right): Conus californicus, Conus pulicarius, Conus tribblei, Conus marmoreus, Conus bullatus, Conus consors and Conus geographus. The phylogenetic tree is inferred from the partial cytochrome oxidase I sequences using Bayesian analysis. Posterior probabilities are indicated for each node. Conasprella, Profundiconus and Californiconus have recently been classified as separate genera in the family Conidae (Olivera et al. 2014). Conus californicus is now Californiconus californicus.
It is proposed that some similar conopeptide genes, likely the result of allelic duplications before speciation, may still be present in the genome of Conus species although not expressed at high level (Duda and Palumbi 2004). Conticello et al. (2001) hypothesized that these genes will have enhanced expression after shifting to a new prey type or development of resistance in prey. The presence of conopeptides in the C. tribblei transcriptome that are similar to the conopeptides expressed in the early divergent species C. californicus may also suggest the persistence of some conopeptide genes in these two species long after the separation of C. californicus lineage from the rest of species in this genus.
While the conopeptides of C. californicus represent an ancient lineage, sequences that are apparently unique to C. tribblei represent peptides that evolved recently. These include not only those with novel mature regions but also those with divergent signal region including those assigned to new conopeptide groups (Fig. 4, Table 4).
Although, the biological functions and molecular targets of the conopeptides in C. tribblei still remains to be demonstrated in future studies, diversity of the identified conopeptides in this species suggests the wide range of molecular targets and biological functions for these peptides. Conopeptides of complex superfamilies despite having identical cysteine patterns are known to target different receptors, for example the different conopeptides of O-superfamily target Ca2+ channels (ω-conotoxins), K+ channels (κ-conotoxins) and Na+ channels (μO- and δ-conotoxins) while the M-superfamily conotoxins show affinity to Na+ channels (μ-conotoxins), K+ channels (κM-conotoxins) and nicotinic acetylcholine receptors (nAChRs) (ψ-conotoxins) (Olivera 2006). Although a speculation of putative targets of the M- and O-superfamily conopeptides identified in C. tribblei cannot be made, presence of 39 conopeptides of M- and O-superfamilies with diverse cysteine patterns and divergent mature sequences clearly implies the affinity of these peptides for multiple ion channels. In addition, some conopeptide families such as con-ikot-ikot and conkunitzin that are shown to cause hyper-excitability (Bayrhuber et al. 2005; Walker et al. 2010) may be involved in inducing the excitotoxic shock during the prey capture by C. tribblei. However the function of one third of the identified conopeptide gene superfamilies in this study is still unknown.
Furthermore, conopeptides of a specific pharmacological family expressed in phylogenetically distant species have diverged in target specificity, and have different molecular targets as have been observed in α-conopeptides targeting different subtypes of nAChRs (Olivera and Teichert 2007). Therefore identification of diverse and divergent conopeptides and also new cysteine patterns in C.tribblei will provide a great source for discovery of peptides that may target different subtypes of ion channels and receptors making them valuable leads for drug development and biomedical applications.
Conclusion
This study is the first glimpse into the diverse conopeptide repertoire of C. tribblei as a representative of clade VIII (Espiritu et al. 2001) using high-throughput sequencing techniques. As expected, all of the identified conopeptides in the C. tribblei venom duct transcriptome were novel and some showed significantly divergent and different signal regions from other known conopeptides. Discovery of new conopeptides in C. tribblei will enhance our understanding of conopeptide diversity in this genus. In addition to conopeptides, other proteins contributing to post-translational modifications of conopeptides and also proteins involved in the efficient delivery of conopeptides in the prey body have been identified. Noticeably, C. tribblei is equipped with an arsenal of diverse conopeptides that may target different channels and receptors possibly working synergistically as ‘toxin cabals’ in order to enhance the efficiency of capturing prey and also to expand the range of its prey types.
Supplementary Material
Acknowledgments
The specimens used in this study were obtained in conjunction with a collection trip supported in part by ICBG grant 1U01TW008163. The Illumina sequencing performed in Oregon Health and Science University was funded by the Philippine PharmaSeas Drug Discovery Program. Roche 454 sequencing was supported by grant GM48677 (BMO) and performed in the Marine Science Institute, University of the Philippines. This study was partially supported by the Office of the Vice President for Academic Affairs, university of the Philippines through Philippine Genome Center. The data analysis was carried out using the High-Performance Computing Facility of the Advanced Science and Technology Institute and the Philippine e-Science Grid, Diliman, Quezon City. We would like to thank Joeriggo Reyes for the help in transcriptome analysis and Dr. Alexander Fedosov for the help in phylogenetic analysis. We would like to thank Maren Watkins for reviewing the conopeptide sequences and for her constructive comments.
Contributor Information
Neda Barghi, Marine Science Institute, University of the Philippines, Quezon City, Philippines.
Gisela P. Concepcion, Philippine Genome Center, Philippines, Marine Science Institute, University of the Philippines, Quezon City, Philippines
Baldomero M. Olivera, Department of Biology, University of Utah, Salt Lake City, UT, USA
Arturo O. Lluisma, Philippine Genome Center, Philippines, Marine Science Institute, University of the Philippines, Quezon City, Philippines
References
- Aguilar MB, Zugasti-Cruza A, Falcóna A, Batista CVF, Olivera BM, Heimer de la Coter EP. A novel arrangement of Cys residues in a paralytic peptide of Conus cancellatus (jr. syn.: Conus austini), a worm-hunting snail from the Gulf of Mexico. Peptides. 2013;41:38–44. doi: 10.1016/j.peptides.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH. Protein sequence databases. Curr Opin Chem Biol. 2004;8:76–80. doi: 10.1016/j.cbpa.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Bayrhuber M, Vijayan V, Ferber M, Graf R, Korukottu J, Imperial J, Garrett JE, Olivera BM, Terlau H, Zweckstetter M, Becker S. Conkunitzin-S1 is the first member of a new kunitz-type neurotoxin family, structural and functional characterization. J Biol Chem. 2005;280:23766–23770. doi: 10.1074/jbc.C500064200. [DOI] [PubMed] [Google Scholar]
- Biggs JS, Olivera BM, Kantor YI. α-conopeptides specifically expressed in the salivary gland of Conus pulicarius. Toxicon. 2008;52:101–105. doi: 10.1016/j.toxicon.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs JS, Watkins M, Puillandre N, Ownby J, Christensen S, Moreno, Lopez-Vera E, Christensen S, Moreno KJ, Navarro AL, Corneli PS, Olivera BM. Evolution of conus peptide toxins: analysis of Conus californicus Reeve, 1844. Mol Phylogenet Evol. 2010;56:1–12. doi: 10.1016/j.ympev.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanella JJ, Bitincka L, Smalley J. MatGAT: An application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics. 2003;4:29. doi: 10.1186/1471-2105-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Garrett JE, Watkins M, Olivera BM. Purification and characterization of a novel excitatory peptide from Conus distans venom that defines a novel gene superfamily of conotoxins. Toxicon. 2008;52:139–145. doi: 10.1016/j.toxicon.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Gilad Y, Avidan N, Ben-Asher E, Levy Z, Fainzilber M. Mechanisms for evolving hypervariability: the case of conopeptides. Mol Biol Evol. 2001;18:120–131. doi: 10.1093/oxfordjournals.molbev.a003786. [DOI] [PubMed] [Google Scholar]
- Duckert P, Brunak S, Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel. 2004;17:107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- Duda TF, Chang D, Lewis BD, Lee T. Geographic variation in venom allelic composition and diets of the widespread predatory marine gastropod Conus ebraeus. PloS One. 2009;4:e6245. doi: 10.1371/journal.pone.0006245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda TF, Palumbi SR. Gene expression and feeding ecology: evolution of piscivory in the venomous gastropod genus Conus. Proc R Soc London B. 2004;271:1165–1174. doi: 10.1098/rspb.2004.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S, Biass D, Stöcklin R, Favreau P. Dramatic intraspecimen variations within the injected venom of Conus consors: an unsuspected contribution to venom diversity. Toxicon. 2010;55:1453–1462. doi: 10.1016/j.toxicon.2010.02.025. [DOI] [PubMed] [Google Scholar]
- Dutertre S, Jin A, Kaas Q, Jones A, Alewood PF, Lewis RJ. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol Cell Proteomics. 2013;12:312–329. doi: 10.1074/mcp.M112.021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliger CA, Richmond TA, Lenaric ZN, Pierce NT, Sweedler JV, Gilly WF. Diversity of conotoxin types from Conus californicus reflects a diversity of prey types and a novel evolutionary history. Toxicon. 2011;57:311–322. doi: 10.1016/j.toxicon.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England LJ, Imperial J, Jacobsen R, Craig AG, Gulyas J, Akhtar M, Rivier J, Julius D, Olivera BM. Inactivation of a serotonin-gated ion channel by a polypeptide toxin from marine snails. Science. 1998;281:575–578. doi: 10.1126/science.281.5376.575. [DOI] [PubMed] [Google Scholar]
- Espiritu DJ, Watkins M, Dia-Monje V, Cartier GE, Cruz LJ, Olivera BM. Venomous cone snails: molecular phylogeny and the generation of toxin diversity. Toxicon. 2001;39:1899–1916. doi: 10.1016/s0041-0101(01)00175-1. [DOI] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology. 1994;3:294–299. [PubMed] [Google Scholar]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29:644–654. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit, a user-friendly biological sequence alignment editor and analysis program for Windows 95, 98, NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Huelsenbeck JP, Ronquist F, Hall B. MrBayes: bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hu H, Bandyopadhyay PK, Olivera BM, Yandell M. Characterization of the Conus bullatus genome and its venom-duct transcriptome. BMC Genomics. 2011;12:60. doi: 10.1186/1471-2164-12-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Bandyopadhyay PK, Olivera BM, Yandell M. Elucidation of the molecular envenomation strategy of the cone snail Conus geographus through transcriptome sequencing of its venom duct. BMC Genomics. 2012;13:284. doi: 10.1186/1471-2164-13-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob RB, McDougal OM. The M-superfamily of conotoxins: a review. Cell Mol Life Sci. 2010;67:17–27. doi: 10.1007/s00018-009-0125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas Q, Westermann J, Craik DJ. Conopeptide characterization and classifications: an analysis using ConoServer. Toxicon. 2010;55:1491–1509. doi: 10.1016/j.toxicon.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Kaas Q, Yu R, Jin A, Dutertre S, Craik DJ. ConoServer: updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 2012;40:D325–D330. doi: 10.1093/nar/gkr886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordiŝ D, Gubenŝek F. Adaptive evolution of animal toxin multigene families. Gene. 2000;261:43–52. doi: 10.1016/s0378-1119(00)00490-x. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–360. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, Mcwilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lavergne V, Dutertre S, Jin A, Lewis RJ, Taft RJ, Alewood PF. Systematic interrogation of the Conus marmoreus venom duct transcriptome with ConoSorter reveals 158 novel conotoxins and 13 new gene superfamilies. BMC Genomics. 2013;14:708. doi: 10.1186/1471-2164-14-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lirazan MB, Hooper D, Corpuz GP, Ramilo CA, Bandyopadhyay P, Cruz LJ, Olivera BM. The spasmodic peptide defines a new conotoxin superfamily. Biochemistry. 2000;39:1583–1588. doi: 10.1021/bi9923712. [DOI] [PubMed] [Google Scholar]
- Lirazan MB, Jimenez EC, Craig AG, Olivera BM, Cruz LJ. Conophysin-R, a Conus radiatus venom peptide belonging to the neurophysin family. Toxicon. 2002;40:901–908. doi: 10.1016/s0041-0101(02)00079-x. [DOI] [PubMed] [Google Scholar]
- Lluisma AO, Milash BA, Moore M, Olivera BM, Bandyopadhyay PK. Novel venom peptides from the cone snail Conus pulicarius discovered through next-generation sequencing of its venom duct transcriptome. Mar Genomics. 2012;5:43–51. doi: 10.1016/j.margen.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu S, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam T, Wang J. SOAPdenovo2: an empirically improved memory-efficient short-read de Novo assembler. GigaScience. 2012;1(18):1–6. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim J, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–381. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricq AV, Jensen S, Walker C, Madsen D, Olivera BM, Ellison M. Conus polypeptides. US2010/0197567 A1 United States Patent. 2009
- McIntosh JM, Ghomashchi F, Gelb MH, Dooley DJ, Stoehr SJ, Giordani AB, Naisbitt SR, Olivera BM. Conodipine-M, a novel phospholipase A2 isolated from the venom of the marine snail Conus magus. J Biol Chem. 1995;270:3518–3526. doi: 10.1074/jbc.270.8.3518. [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Liu L, Shao X, Chi C, Wang C. Identification of a novel class of conotoxins defined as V-conotoxins with a unique cysteine pattern and signal peptide sequence. Peptides. 2008;29:985–991. doi: 10.1016/j.peptides.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Olivera BM. Conus peptides: biodiversity-based discovery and exogenomics. J Biol Chem. 2006;281:31173–31177. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Corneli PS, Watkins M, Fedosov A. Biodiversity of cone snails and other venomous marine gastropods: evolutionary success through neuropharmacology. Annu Rev Anim Biosci. 2014;2:487–513. doi: 10.1146/annurev-animal-022513-114124. [DOI] [PubMed] [Google Scholar]
- Olivera BM, McIntosh JM, Cruz LJ, Luque FA, Gray WR. Purification and sequence of a presynaptic peptide toxin from Conus geographus venom. biochemistry. 1984;22:5078–5090. doi: 10.1021/bi00317a001. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Teichert RW. Diversity of the neurotoxin Conus peptides, a model for concerted pharmacological discovery. Mol Interventions. 2007;7:251–260. doi: 10.1124/mi.7.5.7. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Wood T, Zhang Z, Miller W. Comparison of DNA sequences with protein sequences. Genomics. 1997;46:24–36. doi: 10.1006/geno.1997.4995. [DOI] [PubMed] [Google Scholar]
- Puillandre N, Koua D, Favreau P, Olivera BM, Stöcklin R. Molecular phylogeny, classification and evolution of conopeptides. J Mol Evol. 2012;74:297–309. doi: 10.1007/s00239-012-9507-2. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Tolle DP, Barrett AJ. Evolutionary families of peptidase inhibitors. Biochem J. 2004;378:705–716. doi: 10.1042/BJ20031825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remigio EA, Duda TF. Evolution of ecological specialization and venom of a predatory marine gastropod. Mol Ecol. 2008;17:1156–1162. doi: 10.1111/j.1365-294X.2007.03627.x. [DOI] [PubMed] [Google Scholar]
- Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, Griffith M, Raymond A, Thiessen N, Cezard T, Butterfield YS, Newsome R, Chan SK, She R, Varhol R, Kamoh B, Prabhu A, Tam A, Zhao Y, Moore RA, Hirst M, Marra MA, Jones SJM, Hoodless PA, Birol I. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7:909–915. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- Safavi-Hemami H, Siero WA, Kuang Z, Williamson NA, Karas JA, Page LR, MacMillan D, Callaghan B, Kompella SN, Adams DJ, Norton RS, Purcell AW. Embryonic toxin expression in the cone snail Conus victoriae: primed to kill or divergent function? J Biol Chem. 2011;286:22546–22557. doi: 10.1074/jbc.M110.217703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavi-hemami H, Gorasia DG, Steiner AM, Williamson NA, Karas JA, Gajewiak J, Olivera BM, Bulaj G, Purcell AW. Modulation of conotoxin structure and function is achieved through a multienzyme complex in the venom ducts of cone snails. J Biol Chem. 2012;287:34288–34303. doi: 10.1074/jbc.M112.366781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: Robust de Novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AB, van Marle A, van Elk R, Bogerd J, van Heerikhuizen H, Geraerts WP. Evolutionary conservation of the insulin gene structure in invertebrates: cloning of the gene encoding molluscan insulin-related peptide III from Lymnaea stagnalis. J Mol Endocrinol. 1993;11:103–113. doi: 10.1677/jme.0.0110103. [DOI] [PubMed] [Google Scholar]
- Terrat Y, Biass D, Dutertre S, Favreau P, Remm M, Stöcklin R, Piquemal D, Ducancel F. High-resolution picture of a venom duct transcriptome: case study with the marine snail Conus consors. Toxicon. 2012;59:34–46. doi: 10.1016/j.toxicon.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Van Kesteren RE, Smit AB, De Lange RP, Kits KS, Van Golen FA, Van Der Schors RC, De With ND, Burke JF, Geraerts WP. Structural and functional evolution of the vasopressin/oxytocin superfamily: vasopressin-related conopressin is the only member present in Lymnaea, and is involved in the control of sexual behavior. J Neurosci. 1995;15:5989–5998. doi: 10.1523/JNEUROSCI.15-09-05989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CS, Jensen S, Ellison M, Matta JA, Lee WY, Imperial SJ, Duclos N, Brockie PJ, Madsen DM, Isaac JTR, Olivera BM, Maricq AV. A novel Conus snail polypeptide causes excitotoxicity by blocking desensitization of AMPA receptors. Curr Biol. 2010;19:900–908. doi: 10.1016/j.cub.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Han Y, Shao X, Chi C, Guo Z. Molecular cloning, expression and characterization of protein disulfide isomerase from Conus marmoreus. FEBS J. 2007;274:4778–4787. doi: 10.1111/j.1742-4658.2007.06003.x. [DOI] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Boloud L, Wang J. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34(Web service issue):293–297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H, Xiong Z, Que H, Xie Y, Holland PWH, Paps J, Zhu Y, Wu F, Chen Y, Wang J, Peng C, Meng J, Yang L, Liu J, Wen B, Zhang N, Huang Z, Zhu Q, Feng Y, Mount A, Hedgecock D, Xu Z, Liu Y, Domazet-Lošo T, Du Y, Sun X, Zhang S, Liu B, Cheng P, Jiang X, Li J, Fan D, Wang W, Fu W, Wang T, Wang B, Zhang J, Peng Z, Li Y, Li N, Wang J, Chen M, He Y, Tan F, Song X, Zheng Q, Huang R, Yang H, Du X, Chen L, Yang M, Gaffney PM, Wang S, Luo L, She Z, Ming Y, Huang W, Zhang S, Huang B, Zhang Y, Qu T, Ni P, Miao G, Wang J, Wang Q, Steinberg CEW, Wang H, Li N, Qian L, Zhang G, Li Y, Yang H, Liu X, Wang J, Yin Y, Wang J. The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 2012;490:49–54. doi: 10.1038/nature11413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.