SUMMARY
Acetylation of histones at DNA regulatory elements plays a critical role in transcriptional activation. Histones are also modified by other acyl moieties, including crotonyl, yet the mechanisms that govern acetylation versus crotonylation and the functional consequences of this “choice” remain unclear. We show that the coactivator p300 has both crotonyltransferase and acetyltransferase activities and that p300-catalyzed histone crotonylation directly stimulates transcription to a greater degree than histone acetylation. Levels of histone crotonylation are regulated by the cellular concentration of crotonyl-CoA, which can be altered through genetic and environmental perturbations. In a cell-based model of transcriptional activation, increasing or decreasing the cellular concentration of crotonyl-CoA leads to enhanced or diminished gene expression, respectively, which correlates with the levels of histone crotonylation flanking the regulatory elements of activated genes. Our findings support a general principle wherein differential histone acylation (i.e. acetylation versus crotonylation) couples cellular metabolism to the regulation of gene expression.
INTRODUCTION
Histones are subject to a vast range of post-translational modifications with specific genomic localizations and well-documented functional roles in the regulation of transcription. Critical to our understanding of how histone modifications are regulated has been the identification and characterization of enzyme systems that catalyze the covalent modification of specific target residues. Histone lysine acetylation has been particularly well characterized within this paradigm, with the purification and identification of histone acetyltransferases prompting a breakthrough in our understanding of targeted lysine-acetylation’s direct role in gene regulation (Brownell et al., 1996). Despite this progress, little is known about the cellular regulation and functional relevance of a rapidly-expanding group of chemically related modifications known as histone acylations, of which acetylation is a well-studied member.
Histone lysine propionylation, butyrylation, malonylation, succinylation, 2-hydroxyisobutyrylation, and crotonylation have all been identified by tandem mass spectrometry (MS/MS) proteomic analysis over the past several years (Chen et al., 2007; Dai et al., 2014; Tan et al., 2011; Xie et al., 2012) (Figure S1A). These discoveries have increased the potential complexity of histone lysine modifications and have prompted interest in the functional consequence of differential acylation, both on histone proteins and non-histone proteins. Previous ChIP-seq analyses with “pan” acyl-specific antibodies have mapped histone crotonylation and histone 2-hydroxyisobutyrylation to regulatory elements of actively transcribed regions of the genome, generally coincident with the localization of histone acetylation (Dai et al., 2014; Tan et al., 2011). These studies demonstrate that histones flanking active regulatory elements are modified by a number of chemically distinct acylations, suggesting a role for these modifications in transcriptional regulation. Yet it remains unclear how a “choice” is made between various histone acylations and whether these modifications are functionally distinct from histone acetylation. Given that these modifications are derived from their respective charged acyl-CoAs, which populate various arms of intermediary metabolism, it has been proposed that differential histone acylation could be regulated metabolically and thereby act as a potential “integrator” of a cell’s metabolic state (Lin et al., 2012). However, this hypothesis has not been directly tested. By directly comparing histone crotonylation to histone acetylation, we sought to gain insights into the regulation of histone crotonylation, its role in transcription, and whether alterations in the balance between histone acetylation and crotonylation at regulatory elements have a functional consequence on gene expression.
Here, we show that the well-studied transcriptional co-activator and histone acetyltransferase (HAT) p300 also possesses histone crotonyltransferase (HCT) activity. Utilizing cell-free assays, we demonstrate that p300-catalyzed histone crotonylation directly stimulates transcription and does so to a greater degree than p300-catalyzed histone acetylation. As a result of p300’s dual enzymatic activities, the level of histone crotonylation in the cell is sensitive to changes in the cellular concentration of crotonyl-CoA. We implicate the cytoplasmic/nuclear metabolic enzyme acyl-CoA synthetase (ACSS2 or AceCS1) in the synthesis of crotonyl-CoA from the short-chain fatty acid (SCFA) crotonate in mammalian cells. Finally, by utilizing the lipopolysaccharide (LPS)-induced inflammatory response in the macrophage cell line RAW 264.7 as a model of transcriptional activation, we demonstrate that increasing or decreasing the cellular concentration of crotonyl-CoA prior to LPS-stimulation causes local changes in histone crotonylation, specifically at the histones flanking regulatory elements that show the greatest increase in p300 localization during the inflammatory response. This increase or decrease in crotonyl-CoA concentration also leads to enhanced or diminished levels, respectively, in expression of those specific activated genes.
RESULTS
HAT and HCT Activities Co-Purify with p300 from HeLa S3 Nuclear Extract
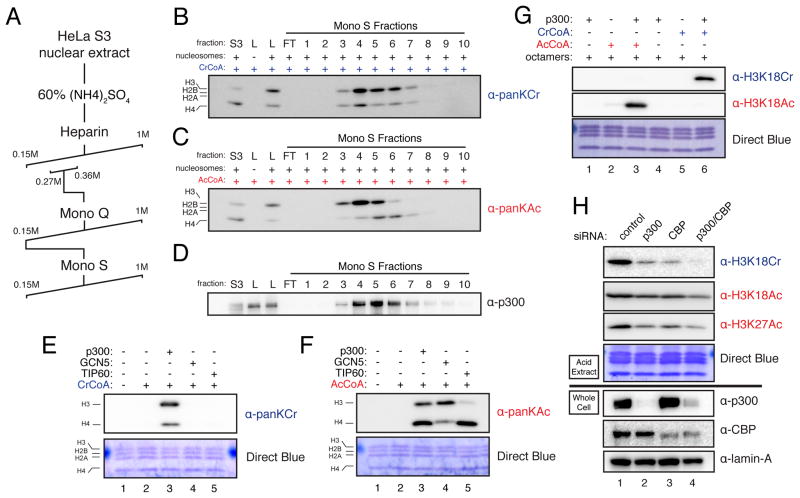
To identify enzyme(s) capable of catalyzing histone crotonylation, we sought to purify a histone crotonyltransferase (HCT) activity from HeLa S3 nuclear extracts. We employed an HCT assay that mirrors previously described histone acetyltransferase (HAT) assays (Mizzen et al., 1999), except that crotonyl-CoA, rather than acetyl-CoA was used as the high-energy acyl-donor, and immunoblotting of reaction products with a pan-crotonyl-lysine antibody (panKCr) was used as a measure of HCT activity. In parallel, HAT activity was determined by substituting acetyl-CoA in the reactions and immunoblotting reaction products with a pan-acetyl-lysine antibody (panKAc). Utilizing these assays, HCT activity was detected in crude HeLa S3 nuclear extracts (data not shown) and partially purified by the fractionation scheme outlined in Figure 1A and Figure S1B. Unexpectedly, HAT activity co-purified with HCT activity through multiple purification steps, including the final step in the partial purification scheme (Figure 1B and 1C). Co-elution of these two activities, albeit with subtle differences toward H3 and H4, suggested either that distinct HAT and HCT activities co-eluted throughout this purification or that a “HAT” might be responsible for the observed HCT activity.
Figure 1. p300 Co-Purifies With HCT Activity From Nuclear Extract and Has Intrinsic HCT Activity In Vitro and In Cells.
(A) HCT partial purification scheme
(B–C) HCT activity co-elutes with HAT activity. HeLa S3 nuclear extracts were fractionated by the scheme detailed in (A). HCT and HAT activities were assayed by panKCr or panKAc immunoblot, respectively, at each round of fractionation and peak HCT activity was followed. The immublot readouts for HCT (B) and HAT activity (C) of the final Mono S fractions are shown here. S3 indicates crude unfractionated extract, L indicates the load/Mono S input, and FT indicates the column flow through. Equal volumes of each fraction were used in these assays. For MS/MS analysis of fractions with peak HCT activity (fractions 4, 5, and 6) see Figure S1C–E and Table S1.
(D) Immunoblot for p300 of the fractions assayed for activity in (B–C). Labels and loading are as in (B–C).
(E–F) HCT (E) and HAT (F) activities of purified recombinant p300, GCN5, and TIP60 were assayed using recombinant octamers as substrate. Reaction products were immunoblotted with the indicated antibodies.
(G) p300-mediated HCT and HAT reactions were performed as in (E–F) with the indicated reaction conditions. Reaction products were probed with the indicated site-specific antibodies.
(H) HeLa S3 cells were transfected with control or p300- and/or CBP-specific siRNA. 72 hours post-transfection, whole cell lysates and histones were prepared and immunoblotted with the indicated antibodies. Immunoblots of acid extracts are shown above the black line, while immunoblots of whole cell lysates are shown below the black line. See also Figure S1.
To identify proteins that were enriched during the purification of the HCT activity, we subjected peak Mono S fractions 4–6, and the crude nuclear extract (fraction “S3”) to MS/MS analysis (see Extended Experimental Procedures and Table S1). Interestingly, a number of previously described HATs were significantly enriched over crude nuclear extract, with the transcriptional coactivator p300 being the most enriched (Figures S1C–E). Based on Intensity Based Absolute Quantitation (iBAQ) values (Schwanhäusser et al., 2012), p300 went from being among the 10% least abundant of all detectible proteins in the crude extract to being among the 20% most abundant of all detectible proteins in the Mono S fraction 5 (Figures S1C–E). In support of the MS/MS data, immunoblot of the Mono S fractions for p300 show that the protein is markedly enriched from nuclear extracts through the purification of HCT activity (compare S3 to Mono S fraction 5 in Figure 1D).
p300 has HCT Activity In Vitro and In Cells
To directly test whether p300, or any of the other HATs enriched by our HCT purification, have intrinsic HCT activity, we purified recombinant p300, GCN5, TIP60, and MOF, and assayed their capacity to crotonylate and/or acetylate recombinant histone octamers by our previously described HCT and HAT assays. While all of these recombinant enzymes exhibited their well-characterized HAT activities, only p300 showed measurable HCT activity (Figures 1E–F and S1F–G).
We next sought to determine the principal sites of p300-catalyzed crotonylation and acetylation. Using semi-quantitative MS/MS analysis of in vitro reaction products by spectral counting (Carvalho et al., 2008), we found that H3K18 is the dominant site of both p300 crotonylation and acetylation, under these reaction conditions (Figure S1H). To confirm activity on H3K18, we utilized available site-specific antibodies against H3K18Cr or H3K18Ac to probe p300-driven HCT and HAT reaction products. Each antibody reacted with its intended target and failed to cross-react with the unintended target, lending support to both our MS/MS data and the acetyl vs. crotonyl specificities of the antibodies (Figure 1G). To confirm the site-specificity of these antibodies, recombinant H3 with a H3K18R mutation was used as a substrate in p300-driven HAT and HCT assays. Signals from both H3K18Cr and H3K18Ac antibodies were attenuated on the p300-modified H3K18R mutant as compared to the wild type H3, further confirming the antibody site-specificities (Figures S1I and S1J). These data confirm that p300 has intrinsic HCT activity in addition to its well-characterized HAT activity.
We next sought to determine whether p300 regulates H3K18Cr in cells. Knockdown of p300 or its paralog CBP by siRNA reduced the global levels of H3K18Cr, H3K18Ac, and H3K27Ac and double knockdown of p300 and CBP reduced the signals even further, as measured by immunoblot of acid-extracted histones (Figure 1H). In addition, overexpression of full-length p300 by transient transfection increased both H3K18Cr and H3K18Ac signals (Figure S1K). Taken together these data support the conclusion that p300 (and its paralog CBP) regulate both H3K18Cr and H3K18Ac in cells.
p300-Catalyzed Histone Crotonylation Stimulates Transcription in a Cell-Free System
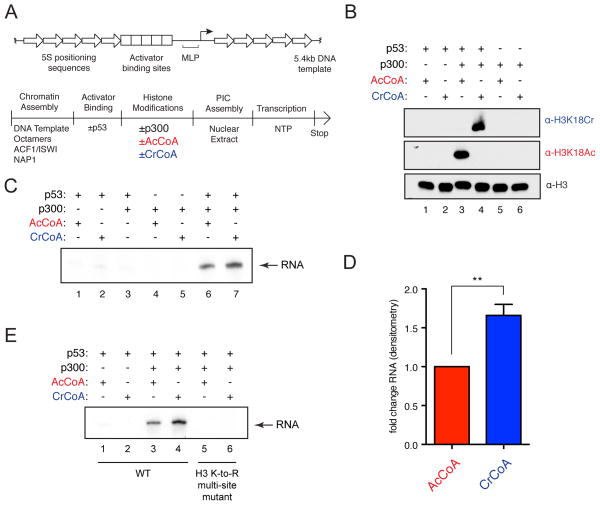
To directly test whether p300-catalyzed histone crotonylation plays a role in transcriptional regulation, we took advantage of a cell-free transcription assay wherein the presence of either acetyl-CoA or crotonyl-CoA could be experimentally controlled. p300’s HAT activity has long been implicated in activator-dependent stimulation of transcription from chromatinized templates in a reaction where maximal RNA synthesis is dependent upon p300 and acetyl-CoA (An et al., 2002). More recently, this assay has been modified to include natural activators, such as p53, and has been exploited to further our mechanistic understanding of transcriptional regulation in the context of chromatin (An et al., 2004; Tang et al., 2013). Similar experiments were conducted using a p53-dependent transcription assay, but with acetyl-CoA replaced by crotonyl-CoA (see schematic in Figure 2A). To ensure the integrity of these assays, the purities of acetyl-CoA and crotonyl-CoA were validated by HPLC analysis, which showed that both cofactors were pure and showed no traces of cross-contamination (Figure S2A). Using the H3K18Cr and H3K18Ac antibodies, we confirmed crotonylation of the recombinant chromatin substrate by p300 and its p53-dependence in this system (Figure 2B). We next performed the transcription assay and observed that p300-catalyzed crotonylation stimulated transcription in this system to a greater extent than did p300-catalyzed acetylation (Figure 2C and 2D). Under identical assay conditions, crotonyl-CoA containing reactions produced, on average, 1.66-fold greater transcript than did acetyl-CoA containing reactions, as measured by densitometry of autoradiograms from three independent experiments (Figures 2D and S2B).
Figure 2. Histone Crotonylation Stimulates Transcription In a Cell-Free System.
(A) Schematic of the transcription template (top) and the transcription reaction order of addition (bottom).
(B) p300 HAT and HCT reactions using recombinant chromatin with the indicated additions were analyzed by immunoblotting with the indicated antibodies.
(C) Transcription assays were performed under the indicated conditions. RNA products were visualized by autoradiography.
(D) Densitometry of autoradiographs comparing acetyl-CoA and crotonyl-CoA containing transcription reactions. Data represent mean fold change from three independent experiments ± standard deviation, p-value = 0.0013.
(E) In vitro transcription assays using chromatin reconstituted with recombinant wild-type H3 or with lysine residues 9, 14, 18, 23, 27, 36, and 56 mutated to arginine (K-to-R). RNA products were visualized by autoradiography. See also Figure S2.
To confirm that p300’s crotonyltransferase activity stimulates transcription by crotonylation of histone proteins and not of auxiliary factors from the added nuclear extract, we reconstituted recombinant chromatin with either wild-type histone H3 or a histone H3 with lysine residues 9, 14, 18, 23, 27, 36, and 56 mutated to arginine (K-to-R). For both acetylation and crotonylation, p300-driven RNA production was substantially inhibited when chromatin containing histone H3 K-to-R mutations was used as a template (Figure 2E), demonstrating that the acetylation or crotonylation of the mutated histone lysine residues is necessary for the production of transcript in this cell-free system. The modest, yet reproducible and statistically significant, difference between histone acetylation and histone crotonylation in the stimulation of transcription provides evidence that histone acetylation and crotonylation are functionally distinct.
The Cellular Concentration of Crotonyl-CoA Regulates Global Histone Crotonylation
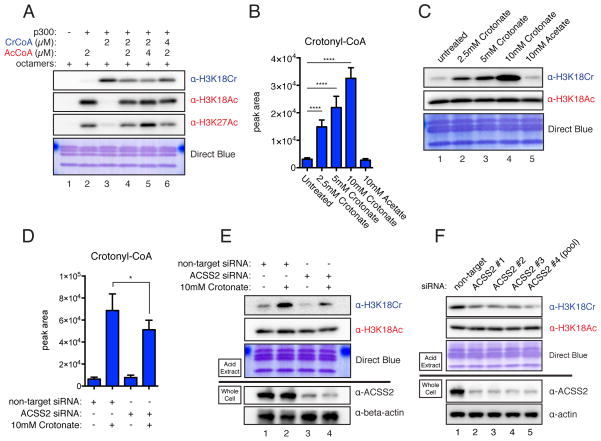
To test whether changes in histone crotonylation would have an effect on gene expression in cells, we next sought to gain insights into how differential acylation is regulated in mammalian cells. Given that p300 can catalyze both histone acetylation and crotonylation, we hypothesized that a differentially modified substrate would be the direct result of the cellular concentrations of either acetyl-CoA or crotonyl-CoA that would, in turn, be regulated by extracellular and intracellular sources of these cofactors. To provide a preliminary test of this hypothesis, p300 reactions were performed in which acetyl-CoA and crotonyl-CoA were mixed in varying ratios. Indeed, altering the relative concentrations of these two acyl-CoAs dictated how much of each modification was present in the final reaction product (Figure 3A). We conclude that crotonyl-CoA can compete with acetyl-CoA for p300’s activity toward its substrate and, consequentially, the relative concentrations of crotonyl-CoA to acetyl-CoA will determine the enzyme’s reaction products. While H3K18 is not the exclusive site of either p300-catalyzed acetylation or crotonylation, the levels of H3K18Cr and H3K18Ac can be used as a surrogate for p300’s differential activity in vitro and in cell-based assays.
Figure 3. Histone Crotonylation is Regulated Metabolically by the Concentration of Crotonyl-CoA.
(A) In vitro p300 HAT/HCT reactions were performed with indicated concentrations of crotonyl-CoA and acetyl-CoA. Reaction products were immunoblotted with the indicated antibodies.
(B) LC-MS analysis of cellular crotonyl-CoA levels extracted from HeLa S3 cells cultured with the indicated concentration of sodium crotonate (pH 7.4) or sodium acetate (pH 7.4) added to full media for 12 hours. The data represent mean peak area ± standard deviation of four independent experiments. Summary of p-value is as follows: ns (p>0.05), * (p≤0.05), ** (p≤0.01), *** (p≤0.001), **** (p≤0.0001).
(C) Histones were acid extracted from HeLa S3 cells treated as in (B) and immunoblotted by the indicated antibodies.
(D) HeLa S3 cells were transfected with control or ACSS2-specific siRNA (pool of 5). 12 hours prior to harvest 10 mM crotonate was added to the media as indicated. 72 hours post-transfection cells were harvested and subject to LC-MS analysis as in (B). The data represent mean peak area ± standard deviation of four independent experiments. p-value summary as in (B).
(E) Same as (D), except 72 hours post-transfection, histones and whole cell lysates were prepared, and immunoblotted with the indicated antibodies. Immunoblots of acid extracts are shown above the black line, while immunoblots of whole cell lysates are shown bellow the black line.
(F) HeLa S3 cells were transfected with control or specific siRNAs to ACSS2. ACSS2 #1-3 are unique single RNAs, and ACSS2 #4 is the pool of 5 used in (D–E). Analysis and labeling as in (C) and (E). See also Figures S3 and S4.
To investigate the relative cellular concentrations of acetyl-CoA and crotonyl-CoA, we established a targeted LC-MS approach that allowed us to extract CoA species from whole cells in culture and measure the abundance of acetyl-CoA and crotonyl-CoA (see Experimental Procedures). We assayed HeLa S3 cells growing in full media under exponential growth conditions and observed a small yet measurable pool of crotonyl-CoA, which was approximately 1000x less abundant than acetyl-CoA (Figure 3B and S3A, first column). The pool of crotonyl-CoA is small in comparison to acetyl-CoA, the most abundant CoA-species measured and the product of several major biosynthetic pathways in multiple organelles. The low basal abundance of crotonyl-CoA places it well below the Kd of p300 (Meier, 2013), which should allow any fluctuation in its concentration to affect downstream crotonyltransferase reactions. To test this prediction, we experimentally increased the concentration of crotonyl-CoA in HeLa S3 cells by adding sodium crotonate to the culture media, and observed a dose-dependent increase in both the cellular concentration of crotonyl-CoA and the global levels of H3K18Cr (Figures 3B and 3C). The relatively low abundance of cellular crotonyl-CoA allows for this dramatic fold change in its availability. In contrast, addition of sodium acetate to cells does not dramatically increase the abundance of cellular acetyl-CoA and has little to no effect on global levels of H3K18Ac (Figures S3A and 3C). Here again, we utilize the site-specific and acyl-specific antibodies (H3K18Ac and H3K18Cr) as a tenable surrogate measure for the differential acylation state of histone residues modified by p300/CBP in the cell. Moreover, the increase in histone crotonylation is not specific to H3K18. A dose-dependent signal increase was also observed with the pan-crotonyl-lysine antibody, not only in H3, but also in the other core histones, upon crotonate addition (Figure S3B). We also observed a similar dose-dependent increase in global histone crotonylation in HEK293T cells, mouse embryonic stem cells, and RAW 264.7 cells, a mouse macrophage cell line, demonstrating that this is not a HeLa S3-specific phenomenon (Figure S3C–E). To rule out the possibility that crotonate is acting as a histone deacetylase (HDAC) inhibitor, we carried out a fluorometric-based HDAC assay with HeLa S3 nuclear extracts as a general source of HDAC activity in the presence of increasing concentrations of crotonate, acetate, or butyrate. While butyrate showed the expected inhibition of HDAC activity, crotonate and acetate showed minimal inhibition (Figure S3F).
To further demonstrate that addition of crotonate is increasing histone crotonylation through the direct production of crotonyl-CoA and not through an indirect mechanism, we asked whether knockdown of the Acyl-CoA Synthetase, ACSS2, would attenuate the increase in histone crotonylation. ACSS2 is known to charge acetate with free CoA-SH to form acetyl-CoA in mammalian cells, is known to localize to the cytosol and the nucleus (Wellen et al., 2009), and has been shown to charge longer chain SCFAs in vitro (Frenkel and Kitchens, 1977), making it an excellent candidate for the conversion of crotonate to crotonyl-CoA. Supporting this notion, knockdown of ACSS2 prior to addition of crotonate to HeLa S3 cells reduced the amount of crotonyl-CoA produced and the levels of H3K18Cr (Figure 3D and 3E, compare columns/lanes 2 and 4). Intriguingly, we also observed that ACSS2 knockdown reduced the global levels of H3K18Cr in untreated cells (Figure 3E, compare lanes 1 and 3). This suggests that basal concentrations of crotonyl-CoA used for histone crotonylation are synthesized from endogenous sources of crotonate. To further support this observation and to rule out potential complications due to siRNA off-target effects, we tested four separate ACSS2-specific RNAs that all showed a global reduction in H3K18Cr (Figure 3F). These data suggest that ACSS2 is responsible, at least in part, for the production of crotonyl-CoA to be used for histone crotonylation.
We next asked whether impairing acetyl-CoA production would have an effect on histone crotonylation. Our in vitro reactions predict that lowering the concentration of acetyl-CoA will lead to an increase in H3K18Cr by reducing the competition for p300/CBP (Figure 3A). A growing literature has shown that the cytoplasmic and nuclear enzyme ATP-Citrate Lyase (ACL) is responsible for producing acetyl-CoA used for histone acetylation in mammalian cells (Lee et al., 2014; Wellen et al., 2009). Knockdowns of ACL by three unique siRNAs resulted in the expected reduction in global levels of H3K18Ac and, as predicted by our in vitro experiments, an increase in global levels of H3K18Cr (Figure S4A). Furthermore, the increase in H3K18Cr can be reversed by the addition of acetate, which has previously been shown to restore histone acetylation levels in the absence of ACL (Figure S4B) (Wellen et al., 2009). Recently, the pyruvate dehydrogenase (PDH) complex, which synthesizes acetyl-CoA for the TCA cycle in the mitochondria, has been shown to translocate to the nucleus where it synthesizes acetyl-CoA used for histone acetylation (Sutendra et al., 2014). Upon knockdown of PDHE1α, a critical subunit of the PDH complex, we observed both the previously reported reduction in H3K18Ac and the predicted increase in H3K18Cr (Figure S4C). Given the relatively low steady-state levels of crotonyl-CoA to acetyl-CoA in the cell and the decrease in acetyl-CoA observed with ACL knockdown, we do not suggest that crotonyl-CoA is completely replacing acetyl-CoA, but rather that the crotonyl-CoA that is present has less competition for p300 and is therefore more often used for p300-catalyzed histone acylation.
H3K18Cr is Associated with Active Chromatin and Correlates with p300 Localization
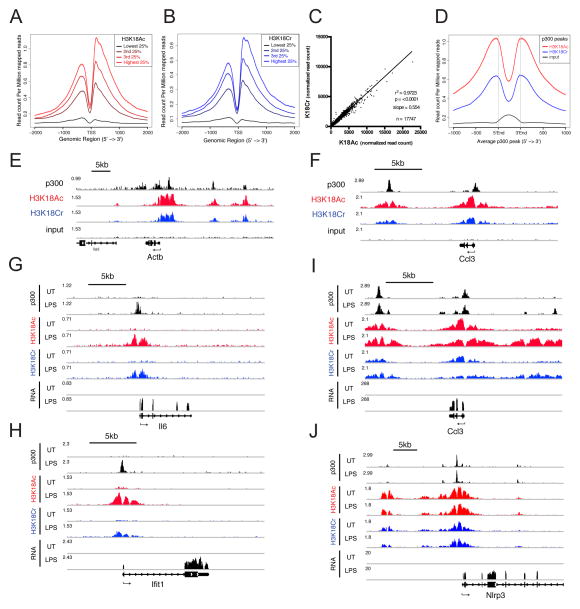
To investigate the role of differential acylation in transcriptional activation within a cellular context, we turned to the LPS-induced inflammatory response in the macrophage cell line RAW 264.7. The macrophage LPS response involves a well-characterized transcriptional program that requires p300 recruitment to many sites of downstream gene activation to facilitate histone acetylation and chromatin remodeling that are critical for the inflammatory response (Ghisletti et al., 2010; Ramirez-Carrozzi et al., 2009). We performed RNA-seq and ChIP-seq for H3K18Ac and H3K18Cr on cells pre- and post- LPS stimulation (120′). We also made use of published p300 ChIP-seq from macrophages under similar stimulation conditions (Ghisletti et al., 2010).
We first assessed the ChIP-seq data for H3K18Ac and H3K18Cr under unstimulated conditions and observed that both histone modifications map to the transcription start site (TSS) and other regulatory elements (Figure 4). Both exhibit strong correlations with gene expression (Figures 4A and 4B) and with each other (Figure 4C), and are significantly enriched at peaks of p300 across the genome (Figure 4D), as predicted by our findings that both modifications are catalyzed by p300. The trends observed by average profiles of H3K18Ac and H3K18Cr reads are also clearly observed at individual genes (Figures 4E and 4F). These data point to a role for p300-catalyzed crotonylation in general transcriptional activation and reaffirm the notion that active regulatory elements are modified by a number of histone acylations.
Figure 4. H3K18Cr is Associated With Active Chromatin, Correlates with p300 Peaks, and is Induced at “De Novo-Activated” Regulatory Elements Upon LPS Stimulation.
(A–B) All genes with FPKM >1 were split into 4 equal groups based on their expression levels calculated from RNA-seq of unstimulated RAW 264.7 cells. The average profile of H3K18Ac (A) and H3K18Cr (B) ChIP-seq data from unstimulated RAW 264.7 cells are plotted for each group at TSS +/− 2kb.
(C) Correlation between H3K18Ac and H3K18Cr ChIP-seq read counts within all H3K18Cr peaks (17,747). Plotted as normalized read counts (read counts per million mapped reads).
(D) Average profile of H3K18Ac, H3K18Cr, and input ChIP-seq data from unstimulated RAW 264.7 around all annotated p300 peaks from unstimulated macrophages.
(E–F) Genome browser representation of normalized ChIP-seq reads for p300, H3K18Ac, H3K18Cr, and input from unstimulated macrophages at a “housekeeping” gene (Actb) (E) and a lineage specific constitutively expressed (“pre-activated”) gene (Ccl3) (F). Normalized to total mapped reads. The y-axis maximum is given at the far left of each track. Arrow below refseq gene track indicates directionality of transcription.
(G–J) Genome browser representation of RNA-seq reads and ChIP-seq reads for p300, H3K18Ac, and H3K18Cr from unstimulated (UT) and 120′ LPS-stimulated (LPS) macrophages at two “de novo-activated” genes (Il6 and Ifit1) (G and H, respectively) and two “pre-activated” genes (Ccl3 and Nlrp3) (I and J). Data presented as in (E–F). See also Figure S5.
H3K18Cr and H3K18Ac are Induced at “De Novo-Activated” Regulatory Elements Upon LPS Stimulation
Stimulation of macrophages by LPS initiates a well-characterized, rapid, and robust transcription program (Bhatt et al., 2012; Ramirez-Carrozzi et al., 2009). For our interests in investigating differential acylation by p300, we focused on two unique classes of LPS-induced genes: I) those showing increased p300 recruitment upon LPS stimulation (fold (reads) > 4), which we will term “de novo-activated”, and II) those showing pre-localized levels of p300 (prior to LPS stimulation) that are not significantly changed upon stimulation (−1.3 < fold (reads) < 1.3), which we term “pre-activated.” The ChIP-seq traces for p300, H3K18Ac, H3K18Cr, and RNA-seq of genes from both classes ± LPS stimulation (120′) demonstrate how the chromatin landscape at these two different classes of genes responds to LPS stimulation. While “de novo-activated” genes (Figures 4G–H and S5A–C) show an induction in both modifications only upon LPS stimulation, “pre-activated” genes (Figure 4I and 4J) have peaks of both H3K18Ac and H3K18Cr prior to LPS stimulation. These trends are also observed by average profiles of H3K18Ac and H3K18Cr centered on the TSS of all “pre-activated” and all “de novo-activated” genes (Figures S5D–G).
To test whether altering the levels of histone crotonylation would affect gene activation, we asked whether increasing or decreasing the concentration of crotonyl-CoA in RAW 264.7 cells prior to LPS stimulation would alter the differential acylation state at the regulatory elements of “de novo-activated” genes and affect the gene’s expression upon LPS-stimulation and subsequent p300 recruitment. We focused on the “de novo-activated” genes because we sought to examine genes in which the p300-catalyzed acylation of histones is a rate-limiting step in the transcription reaction. We hypothesize that “de novo-activated” genes will be more responsive to changes in the concentrations of crotonyl-CoA than the “pre-activated” genes, which are both acetylated and crotonylated prior to LPS stimulation.
Increasing Crotonyl-CoA Concentrations by Crotonate Addition Increases H3K18Cr at the Promoters of “De Novo-Activated” Genes and Enhances Gene Expression
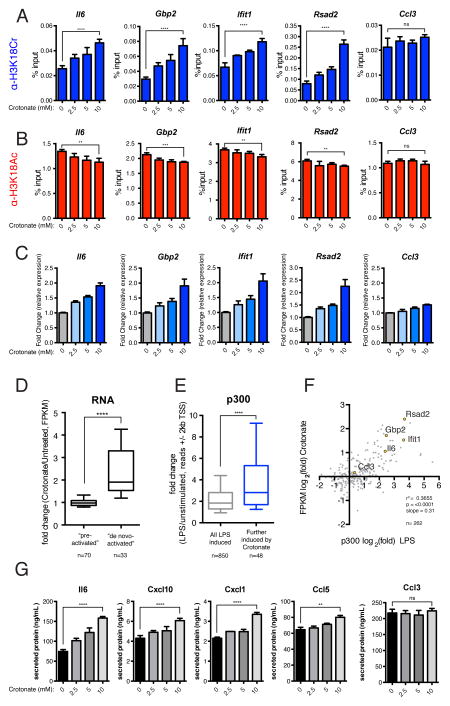
To experimentally alter the cellular concentration of crotonyl-CoA available for p300 to use upon LPS-stimulation, we first pretreated RAW 264.7 cells with varying concentrations of sodium crotonate for 6hrs prior to LPS stimulation, which increases the cellular concentration of crotonyl-CoA in a dose-dependent manner (Figure S6A). After 2 hours of LPS stimulation, we performed ChIP for H3K18Cr and H3K18Ac. In support of our hypothesis, we observe a dose-dependent increase in H3K18Cr at the promoters of four “de novo-activated” genes (Il6, Gbp2, Ifit1, and Rsad2) and no significant change in H3K18Cr at a “pre-activated” (pre-acylated) gene (Ccl3) (Figure 5A). Furthermore, we see an associated dose-dependent decrease in H3K18Ac at the promoters of the “de novo-activated” genes tested and no significant change in H3K18Ac at the “pre-activated” gene (Figure 5B). This same trend is observed in an independent replicate using a single concentration of sodium crotonate (Figure S6B).
Figure 5. Increasing the Concentration of Crotonyl-CoA Prior to LPS Stimulation Leads to a Greater Induction of H3K18Cr at, and Enhanced Stimulation of, “De Novo-Activated” Inflammatory Genes Upon LPS Stimulation.
(A) qPCR analysis of H3K18Cr ChIP products from RAW 264.7 cells pre-treated with the indicated concentration of sodium crotonate (pH 7.4) (mM) for 6 hours prior to a 2-hour LPS stimulation. Primers were designed for TSS-proximal ChIP-seq peaks of H3K18Ac and H3K18Cr (see Experimental Procedures). ChIP-qPCR results for four “de novo-activated” genes (Il6, Gbp2, Ifit1, and Rsad2) and one “pre-activated” gene (Ccl3) are shown. Data are represented as mean of % input ± standard deviation of technical replicates. Summary of p-value is as follows: ns (p>0.05), * (p≤0.05), ** (p≤0.01), *** (p≤0.001), **** (p≤0.0001).
(B) Experiment and analysis are same as (A), except ChIP was performed with H3K18Ac antibody.
(C) RT-qPCR analysis of LPS-stimulated (120′) RAW 264.7 cells pre-treated with the indicated concentration of sodium crotonate (pH 7.4) (mM). Relative expression is normalized to Gapdh. RT-qPCR data for the same set of genes as in (A–B). Data are represented as the mean fold-change in relative expression due to crotonate addition from three independent experiments ± standard deviation.
(D) The fold changes in FPKM over LPS-stimulation due to crotonate pre-treatment, as measured by RNAseq, are represented as a box and whisker plot (10th – 90th percentile) for both “pre-activated” and “de novo-activated” genes. P-value summaries are as in (A).
(E) Box and whisker plot representation (10th – 90th percentile) of the fold change in p300 read counts, due to LPS stimulation, +/− 2kb from the TSS of two sets of genes, (1) all LPS induced genes (fold change (FPKM) ≥2 upon LPS stimulation, n=850) and (2) a subset of (1) whose expression was further induced ≥2-fold due to pre-treatment with crotonate (n=48). P-value summaries are as in (A).
(F) Scatter plot representation of all LPS-stimulated genes with annotated p300 peaks ± 500bp from TSS (n=262), where x = fold change in p300 read counts due to LPS, and y = fold change in expression (FPKM) over LPS-induction due to crotonate pre-treatment; both are plotted as log2 values. Four “de novo-activated” gene (Il6, Gbp2, Ifit1, and Rsad2) and one “pre-activated” gene (Ccl3) are highlighted in yellow.
(G) Chemokine and cytokine protein abundance in supernatants from LPS-stimulated (16hr) RAW 264.7 cells pre-treated with the indicated concentration of sodium crotonate. Data for four “de novo-activated” chemo/cytokines (Il6, Cxcl10, Cxcl1, and Ccl5) and one “pre-activated” chemokine (Ccl3) are represented here as the mean of two independent experiments ± standard deviation. P-value summaries are as in (A). See also Figure S6.
With the ability to fine tune the amount of H3K18Cr present on the promoters of “de novo-activated” genes, we next measured mRNA by RT-qPCR from cells treated with varying concentrations of sodium crotonate prior to a 2 hour LPS stimulation. Gene expression of the “de novo-activated” genes was increased in a dose-dependent manner, while the “pre-activated” gene was only minimally affected (Figure 5C). Additional tested genes (“de novo-activated:” Cmpk2, Cxcl10, Ifnb, and Ccl5, and “pre-activated:” Pim1) followed the same trend (Figure S6C). These data provide evidence correlating the abundance of H3K18Cr at the proximal regulatory elements of a gene to its expression, supporting the notion that the balance between histone crotonylation and histone acetylation (i.e. differential acylation) has a functional consequence on gene expression.
To explore the scope of LPS-induced genes that are further induced due to crotonate pre-treatment, we next performed RNA-seq on LPS stimulated (120′) cells with and without pre-treatment of sodium crotonate (10mM). Consistent with our RT-qPCR data of select genes, “de novo-activated” genes were on average further stimulated by crotonate pre-treatment with a mean fold increase of 2.4 over LPS-induction, whereas “pre-activated” genes were on average unaffected with a mean fold change of 1.0 (Figure 5D and S6D–E). We next asked whether all crotonate-responsive genes (>2-fold increase over LPS stimulation) also show a greater than average recruitment of p300 upon LPS stimulation. To do this, we compared the fold-change of p300 reads (+/− 2kb from TSS) of a list of all genes whose expression is induced >2-fold (“All LPS-induced”, n=850) to a subset of that list of genes that are further induced >2-fold over LPS-stimulation due to crotonate pre-treatment (“Further induced by crotonate”, n=48). Notably, we observed a significantly greater than average fold-change in p300 recruitment to crotonate-responsive genes (Figure 5E). We also observed a statistically significant positive correlation between the fold-change in p300-localization upon LPS and the fold-change in mRNA abundance due to crotonate treatment (Figure 5F), further establishing the link between transcriptional activation and differential histone acylation.
Increasing Cellular Concentrations of Crotonyl-CoA Prior to LPS Stimulation Enhances Cytokine and Chemokine Secretion Upon LPS Stimulation
To test whether the effects observed in gene expression lead to changes in the functional arm of the inflammatory response, we measured how pre-treating cells with crotonate effected cytokine and chemokine secretion upon LPS stimulation. We first performed an LPS time-course with or without crotonate treatment (10mM), collected supernatants 0, 3, 6, and 16 hours post LPS-stimulation, and observed a greater concentration of secreted Il6 in cells pre-treated with crotonate at every time point post-LPS-stimulation, as measured by a standard ELISA (Figure S6F). We next employed a multiplex bead-based immunoassay (see Extended Experimental Procedures) to more broadly analyze the effect of crotonate treatment on cytokine and chemokine secretion. We collected supernatant from cells pre-treated with increasing concentrations of sodium crotonate 16hrs post LPS-stimulation and measured the concentrations of a variety of LPS-induced chemokines and cytokines (Il6, Cxcl10, Cxcl1, Ccl5, Ccl3, Il10, Ccl22, and Tnfα). In agreement with the standard ELISA assay, we observed a dose-dependent increase in Il6 secretion due to sodium crotonate addition (Figure 5G). We also observed a dose-dependent increase in Cxcl10, Cxcl1, Ccl5, Il10, and Ccl22 secretion, all of which are products of “de novo activated” genes (Figure 5G and S6E). Furthermore, the secreted factors expressed from “pre-activated” genes (Ccl3 and Tnfα) did not show a significant change in protein secretion due to addition of sodium crotonate (Figure 5G and S6G).
Knockdown of ACSS2 Reduces H3K18Cr at the Promoters of “De Novo-activated” Genes and Attenuates Gene Expression
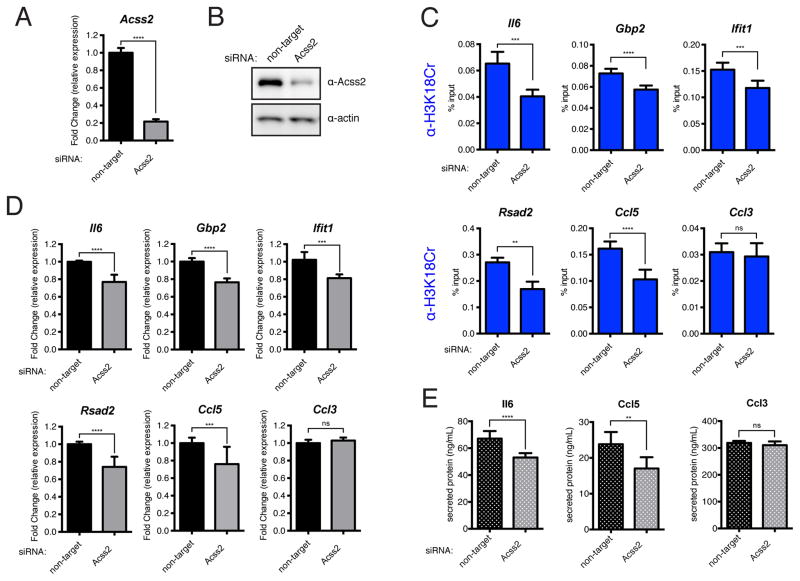
To test whether decreasing crotonyl-CoA available for histone crotonylation prior to LPS-stimulation would have a negative effect on “de novo-activated” genes, we performed siRNA knockdown of ACSS2 in RAW 264.7 cells prior to a 120′ LPS stimulation. We confirmed knockdown of ACSS2 both at the RNA level by RT-qPCR (Figure 6A) and at the protein level by immunoblot (Figure 6B). We next performed ChIP for H3K18Cr in LPS-stimulated (120′) cells that had been transfected with either non-target or ACSS2-specific siRNA. As expected, we observed significant decreases in H3K18Cr due to ACSS2 knockdown in five “de novo-activated” genes (Il6, Gbp2, Ifit1, Rsad2, and Ccl5) and no significant change in the “pre-activated” gene (Ccl3) (Figure 6C). We next performed RT-qPCR under the same experimental conditions with three independent replicates and observed a reduction in mRNA abundance for the five “de novo-activated” genes tested (Il6, Gbp2, Ifit1, Rsad2, and Ccl5) and no change for the “pre-activated” gene (Ccl3) (Figure 6D). Furthermore, under similar experimental conditions we observed a reduction in the LPS-induced secretion of Il6 and Ccl5, but no significant change in Ccl3 secretion, upon knockdown of ACSS2 (Figure 6E).
Figure 6. Knockdown of ACSS2 Prior to LPS Stimulation Leads to a Decreased Induction of H3K18Cr at, and Decreased Stimulation of, “De Novo-Activated” Inflammatory Genes Upon LPS Stimulation.
(A–B) RAW 264.7 cells were transfected with control or Acss2-specific siRNAs, 72 hours post-transfection cells were harvested for either RT-qPCR analysis (A) or immunoblot analysis (B) to assess knockdown efficiency. For (A) data is presented as mean fold change due to ACSS2 knockdown of technical replicates ± standard deviation. Summary of p-value is as follows: ns (p>0.05), * (p≤0.05), ** (p≤0.01), *** (p≤0.001), **** (p≤0.0001).
(C) qPCR analysis of H3K18Cr ChIP products from LPS-stimulated (120′) RAW 264.7 cells that had been transfected with either control siRNA or siRNAs specific for Acss2 72 hours prior to stimulation. ChIP-qPCR results for five “de novo-activated” genes (Il6, Gbp2, Ifit1, Rsad2, and Ccl5) and one “pre-activated” gene (Ccl3) are shown here. Data are represented as mean of % input of technical replicates ± standard deviation. P-value summaries are as in (A).
(D) RT-qPCR analysis of LPS-stimulated (120′) RAW 264.7 cells previously transfected with either control or Acss2-specific RNAs, as in (C). Relative expression is normalized to Gapdh. RT-qPCR data are shown for the same set of genes as in (C). Data are represented as the mean fold-change in relative expression due to Acss2 knockdown from three independent experiments ± standard deviation. P-value summaries are as in (A).
(E) Chemokine and cytokine protein abundance in supernatants from LPS-stimulated (16hr) RAW 264.7 cells transfected with the indicated RNAs, as in (C). Data for two “de novo-activated” chemo/cytokines (Il6 and Ccl5) and one “pre-activated” chemokine (Ccl3) are represented here as the mean of two independent experiments ± standard deviation. P-value summaries are as in (A).
Based on our findings, we conclude that differential histone acylation (crotonylation vs. acetylation) at specific lysine residues (H3K18, in this study, and likely other histone sites) is regulated metabolically by a previously unappreciated balance in cellular levels of crotonyl-CoA and acetyl-CoA. We favor the view that the concentrations of crotonyl-CoA and acetyl-CoA (and likely other short-chain acylations) are “translated” by the co-activators p300 and CBP into the differential acylation states of local chromatin influencing transcriptional activation by mechanisms that remain unclear (Figure 7). Our findings lend support to emerging studies linking metabolism to the alteration of chromatin landscapes and thereby the regulation of gene expression.
Figure 7. Enzymatic and Metabolic Regulation of Differential Histone Acylation.
Schematic diagram of the pathways involved in the enzymatic and metabolic regulation of histone crotonylation and histone acetylation (differential acylation). The differential acylation state of chromatin is regulated by the relative concentrations of acetyl-CoA and crotonyl-CoA, which are synthesized through distinct metabolic pathways, diagramed here. Branches of the diagram that are still unknown are marked by question marks. The PDH (pyruvate dehydrogenase), ACSS2 (Acyl-CoA synthetase), and ACL (ATP Citrate Lyase) reactions occur in both the cytosol and nuclear compartments. We favor the model that DNA-sequence-specific transcription factors (TF) recruit p300/CBP to specific genomic loci where they will “translate” the nuclear/cytosolic acyl-CoA levels by differentially acylating histones, thereby facilitating transcription to varying degrees.
DISCUSSION
Although a variety of histone lysine acylations, diverse in both chemical structure and site of modification, have recently been identified, it remains unclear whether they are functionally distinct from one another or from acetylation, the archetypal histone acylation. Focusing on histone crotonylation, we show that the modification is dynamically regulated both enzymatically, by p300/CBP, and metabolically, by the relative concentration of crotonyl-CoA and acetyl-CoA, in mammalian cells. Furthermore, we establish a role for histone crotonylation in both cell-free and cell-based models of transcriptional activation. We demonstrate that by experimentally altering the cellular concentration of crotonyl-CoA the histones that flank specific regulatory elements exhibit an increased level of H3K18Cr and that the expression of that gene is positively effected.
Regulation of histone modifications, and thereby regulation of chromatin structure, by metabolite availability has been proposed as a potential mechanism for gene regulation under the premise that many chromatin-modifying enzymes utilize products of intermediary metabolism as cofactors essential for their activities (Gut and Verdin, 2013; Kaelin and McKnight, 2013; Meier, 2013). Our study introduces a new dimension to this paradigm in that fluctuations in both acetyl-CoA and crotonyl-CoA are shown to affect global and local chromatin landscapes leading to distinct functional outputs. While our study clearly establishes that the concentration of crotonyl-CoA influences transcriptional activation through its effects on differential acylation, it remains unclear whether the physiological concentrations of crotonyl-CoA are as dynamic as they are in our cell culture experiments where crotonate addition to media was used to experimentally alter the cellular concentration of crotonyl-CoA.
By utilizing a recombinant chromatin template, we show that p300-catalyzed histone crotonylation directly stimulated transcription to a greater degree than p300-catalyzed histone acetylation. We propose that fluctuations in acyl-CoA metabolism are “translated” to chromatin through p300 or other, yet uncovered, acyl-transferases. Our study focused largely on H3K18 because of its identification as the dominant site of p300-driven crotonylation in our in vitro assays, the clear regulation of its global levels in mammalian cells by p300/CBP, and the availability of acyl-specific antibodies for this modification. However, our MS/MS analysis also detected other sites of crotonylation on H3. Since the multi-site H3 K-to-R mutations ablate this transcriptional response, we conclude that histone crotonylation per se is required for this response; but exactly how that response is mediated and how it may differ mechanistically from the stimulation associated with acetylation remains unclear.
Lysine crotonylation, like acetylation, will neutralize the positive charge of the ε-amino group of lysine, leading to the possibility of charge-based cis-effects on the chromatin fiber where the increase bulk and rigidity of crotonyl may lead to an enhanced effect. It is currently unknown whether there are specific “readers” for crotonyl-lysine that take advantage of this increased bulk and rigidity, which would add functionality to these modifications through trans-effects. The surprising discoveries that methyl-lysine “readers” can distinguish mono-, di-, and tri-methylated lysines (Taverna et al., 2007), a comparably small chemical difference compared to the diversity of acylations, provides precedent for such discrimination.
Our studies illustrate that the intra-and inter-cellular metabolic state of the cell can directly influence the local chromatin landscape by causing fluctuations in metabolite/cofactor concentrations required for histone crotonylation and acetylation. We put forward a proof-of-concept for the functional connection between acyl-CoA metabolism and transcriptional activation, operating through the differential acylation of histones. Future work as to the physiological range of acyl-CoA concentrations in a variety of cell-types, environments, and stress conditions will be critical to understand specifically how and for what biological process nature exploits this hitherto unexplored mechanism of gene regulation.
EXPERIMENTAL PROCEDURES
Nuclear Extract Preparation and Fractionation
For nuclear extract, HeLa S3 cells were grown in suspension culture in DMEM supplemented with 10% FBS. Soluble nuclear extract was prepared by an ammonium sulfate extraction protocol detailed in extended experimental procedures. Column chromatography was performed using the AKTA Purifier system with the following columns: in-house packed POROS Heparin 50 uM (Invitrogen), Mono Q 5/50 GL (GE Healthcare), and Mono S 5/50 GL (GE Healthcare). Fractions were collected over a linear potassium chloride gradient from 0.15 M to 1 M.
Immunoblot Analysis
Histones were purified from cells using a standard acid extraction protocol (Shechter et al., 2007). Whole cell lysates were prepared by boiling cell pellets in 2x Laemmli sample buffer for 5 minutes followed by brief vortexing. The following antibodies were used in this study: Pan-KCr (PTM-Biolabs 501), H3K18Cr (PTM-Biolabs 517), pan-KAc (PTM-Biolabs 105), H3K18Ac (Abcam 1191), H3K27Ac (Active Motif 39685), H3K56Ac (Abcam 76307), H3 (Abcam 1791), p300 (Santa Cruz 584), CBP (Santa Cruz 7300), ACSS2 (Cell Signaling 3658), alpha-actin (Sigma A2066), beta-actin (Abcam 8224), and lamin-A (Abcam 26300). The ACL antibody was a gift from the Thompson Lab.
Cell Culture and Transfections
HeLa S3, HEK 293T, and RAW 264.7 cells were cultured in DMEM, supplemented with 10% FBS, and 1% GlutaMAX (Gibco) at 37°C under 5% CO2. Sodium crotonate was prepared by dissolving solid crotonic acid (MP 207938) in water, followed by titration with sodium hydroxide to pH 7.4. Sodium acetate was prepared similarly with acetic acid. siRNA transfections were performed using Dharmafect 1 (Dharmacon) as per manufacturer’s instruction. The following siRNAs were used at the following concentrations: non-target control (Dharmacon D-001810-10, 25nM), p300 (Dharmacon L-003486, 25nM), CBP (Dharmacon L-003477, 25nM), ACSS2 #1 (Ambion s31745, 20nM), ACSS2 #2 (Ambion s31746, 20nM), ACSS2 #3 (Ambion s31747, 20nM), ACSS2 #4 (Dharmacon L-010396, 20nM), ACL #1 (Ambion s915, 10nM), ACL #2 (Ambion s916, 10nM), ACL #3 (Ambion s917, 10nM), PDHE1α (Ambion s10245, 20nM), mouse ACSS2 (Dharmacon L-065412, 25nM). DNA transfections were performed using lipofectomine 2000 as per manufacturer’s instruction. For LPS stimulation, RAW 264.7 cells were plated at ~25% density >24 hours prior to stimulation. LPS from Salmonella typhosa (Sigma L2387) was added at 100 ng/mL and samples were collected at indicated time points.
Supplementary Material
Acknowledgments
We would like to thank P.W. Lewis, S.Z. Josefowicz, B.W. Carey, and C. Lu for valuable discussion and scientific input. This work was funded by institutional support from The Rockefeller University to C.D.A and R.G.R., NIH grants GM105933, CA160036, and RR020839 to Y.Z., and a National Science Foundation Graduate Research Fellowship Program award (DGE-1325261) to B.R.S. Y.Z. is a member of the scientific advisory board of PTM Biolab, Co. Ltd. (Chicago, IL).
Footnotes
ACCESSION NUMBERS
The Gene Expression Omnibus (GEO) accession number for the data sets reported in this paper is GSE63889.
Supplementary information includes Extended Experimental Procedures, six figures, and one table.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- An W, Palhan VB, Karymov MA, Leuba SH, Roeder RG. Selective requirements for histone H3 and H4 N termini in p300-dependent transcriptional activation from chromatin. Molecular Cell. 2002;9:811–821. doi: 10.1016/s1097-2765(02)00497-5. [DOI] [PubMed] [Google Scholar]
- Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript Dynamics of Proinflammatory Genes Revealed by Sequence Analysis of Subcellular RNA Fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Carvalho PC, Hewel J, Barbosa VC, Yates JR. Identifying differences in protein expression levels by spectral counting and feature selection. Genet Mol Res. 2008;7:342–356. doi: 10.4238/vol7-2gmr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Peng C, Montellier E, Lu Z, Chen Y, Ishii H, Debernardi A, Buchou T, Rousseaux S, Jin F, et al. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nature Chemical Biology. 2014;10:365–370. doi: 10.1038/nchembio.1497. [DOI] [PubMed] [Google Scholar]
- Frenkel EP, Kitchens RL. Purification and properties of acetyl coenzyme A synthetase from bakers’ yeast. J Biol Chem. 1977;252:504–507. [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and Characterization of Enhancers Controlling the Inflammatory Gene Expression Program in Macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Gut P, Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nature. 2013;502:489–498. doi: 10.1038/nature12752. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Jr, McKnight SL. Influence of Metabolism on Epigenetics and Disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JV, Carrer A, Shah S, Snyder NW, Wei S, Venneti S, Worth AJ, Yuan ZF, Lim HW, Liu S, et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metabolism. 2014;20:306–319. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Su X, He B. Protein Lysine Acylation and Cysteine Succination by Intermediates of Energy Metabolism. ACS Chem Biol. 2012;7:947–960. doi: 10.1021/cb3001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier JL. Metabolic Mechanisms of Epigenetic Regulation. ACS Chem Biol. 2013;8:2607–2621. doi: 10.1021/cb400689r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen CA, Brownell JE, Cook RG, Allis CD. Histone acetyltransferases: preparation of substrates and assay procedures. Meth Enzymol. 1999;304:675–696. doi: 10.1016/s0076-6879(99)04041-0. [DOI] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A Unifying Model for the Selective Regulation of Inducible Transcription by CpG Islands and Nucleosome Remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2012;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E, Michelakis ED. A Nuclear Pyruvate Dehydrogenase Complex Is Important for the Generation of Acetyl-CoA and Histone Acetylation. Cell. 2014;158:84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. Identification of 67 Histone Marks and Histone Lysine Crotonylation as a New Type of Histone Modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Chen WY, Shimada M, Nguyen UTT, Kim J, Sun XJ, Sengoku T, McGinty RK, Fernandez JP, Muir TW, et al. SET1 and p300 Act Synergistically, through Coupled Histone Modifications, in Transcriptional Activation by p53. Cell. 2013;154:297–310. doi: 10.1016/j.cell.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-Citrate Lyase Links Cellular Metabolism to Histone Acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, Boeke JD, Zhao Y. Lysine succinylation and lysine malonylation in histones. Molecular & Cellular Proteomics. 2012;11:100–107. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.