DNA replication has to be tightly regulated to ensure genome integrity such that DNA replication takes place only once per cell cycle. The Cdc6 sequential phosphorylation by GSK-3 and Cdk1 creates a binding site for Cdc4 ubiquitin ligase to promote Cdc6 degradation.
Abstract
To ensure genome integrity, DNA replication takes place only once per cell cycle and is tightly controlled by cyclin-dependent kinase (Cdk1). Cdc6p is part of the prereplicative complex, which is essential for DNA replication. Cdc6 is phosphorylated by cyclin-Cdk1 to promote its degradation after origin firing to prevent DNA rereplication. We previously showed that a yeast GSK-3 homologue, Mck1 kinase, promotes Cdc6 degradation in a SCFCdc4-dependent manner, therefore preventing rereplication. Here we present evidence that Mck1 directly phosphorylates a GSK-3 consensus site in the C-terminus of Cdc6. The Mck1-dependent Cdc6 phosphorylation required priming by cyclin/Cdk1 at an adjacent CDK consensus site. The sequential phosphorylation by Mck1 and Clb2/Cdk1 generated a Cdc4 E3 ubiquitin ligase–binding motif to promote Cdc6 degradation during mitosis. We further revealed that Cdc6 degradation triggered by Mck1 kinase was enhanced upon DNA damage caused by the alkylating agent methyl methanesulfonate and that the resulting degradation was mediated through Cdc4. Thus, Mck1 kinase ensures proper DNA replication, prevents DNA damage, and maintains genome integrity by inhibiting Cdc6.
INTRODUCTION
Initiation of DNA replication requires prior assembly of the prereplicative complex (pre-RC) by Cdc6- and Cdt1-dependent recruitment of the minichromosome maintenance complex (Mcm2–7) to the origin-bound origin recognition complex (Orc1-6) (Araki, 2010). Pre-RC assembly licenses the origin to further recruit the Dbf4-kinase/Cdc7 complex to form a bidirectional replication fork (Araki, 2010). DNA synthesis then occurs through the activity of S-phase/mitotic cyclin-Cdk complexes (Tanaka et al., 2007; Zegerman and Diffley, 2007). In Saccharomyces cerevisiae, this includes six B-type cyclins (Clb1–6) and one Cdk1 (Cdc28p) (Nasmyth, 1996).
Cells ensure that DNA replication occurs only once per cell cycle in order to maintain genome integrity. To achieve this, the pre-RC is rapidly disassembled after Cdk1-dependent phosphorylation, and reassembly is inhibited until the following cell cycle. The inhibition of pre-RC assembly is dependent on B-type cyclins (Dahmann et al., 1995; Ikui et al., 2007) and involves multiple overlapping mechanisms. Mcm2–7 is excluded from the nucleus after phosphorylation by the cyclin-Cdk complex (Labib et al., 1999; Nguyen et al., 2000). Orc2 and Orc6 are phosphorylated to inhibit pre-RC loading (Nguyen et al., 2001). To further prevent pre-RC assembly, Clb2 binds to the phosphorylated N-terminal domain of Cdc6 (Mimura et al., 2004), and Clb5p binds to the Arg-X-Leu (RXL) domain in Orc6 to sterically inhibit Cdt1/Mcm2–7 loading (Chen and Bell, 2011; Wilmes et al., 2004). In addition, Cdc6p levels are tightly regulated during the cell cycle. Cdc6 is transcribed in late mitosis and G1 and accumulates throughout G1; then its levels rapidly drop after passage through START (Piatti et al., 1995; Zwerschke et al., 1994). Cdc6 is also regulated through its localization (Honey and Futcher, 2007).
Multiple mutations affect rereplication control, including mutation of Orc2 and Orc6 phosphorylation sites (ORC2-ps and ORC6-ps) (Nguyen et al., 2001), forced nuclear localization of Mcm2–7 (MCM7-NLS) (Nguyen et al., 2000), and stabilization through N-terminal truncation of Cdc6p (CDC6ΔNT) (Drury et al., 1997). Combining these mutations is lethal and induces rereplication (Nguyen et al., 2001; Wilmes et al., 2004; Archambault et al., 2005). In addition, each of these individual mutations strongly synergizes with ORC6 mutation at the RXL domain (ORC6-rxl) (Wilmes et al., 2004). For instance, an ORC6-rxl CDC6ΔNT double mutant causes slow growth and increases the frequency of rereplication, with the functional outcome of causing DNA damage and activating the DNA damage checkpoint (Archambault et al., 2005; Ikui et al., 2007).
We previously found that the ORC6-rxl mutation is synthetically lethal with the deletion of the MCK1 gene, a yeast homologue of glycogen synthase kinase 3 (GSK-3) (Archambault et al., 2005; Ikui et al., 2012). Meiosis and centromere regulatory kinase (Mck1) is a serine/threonine kinase whose catalytic activity requires autophosphorylation on tyrosine residues (Lim et al., 1993; Brazill et al., 1997; Rayner et al., 2002). Mck1p was first found as a dosage suppressor of a centromere mutation that causes chromosomal missegregation (Shero and Hieter, 1991) and as an early regulator of genes required for meiosis and sporulation (Neigeborn and Mitchell, 1991). In addition, it is involved in the stress response by promoting the binding of transcription factors to the promoters of stress-responsive genes (Hirata et al., 2003). Accordingly, mck1-deletion cells are hot and cold sensitive and sensitive to the microtubule-destabilizing drug benomyl (Shero and Hieter, 1991; Andoh et al., 2000). Mck1 has a function in protein degradation; during heat stress, it promotes Rog1 degradation through the ubiquitin-ligase Rsp5 (Andoh et al., 2000). Mck1 also has a role in cell cycle regulation. It is a downstream regulator of calcineurin signaling that promotes ubiquitin-mediated Hsl1 degradation (Mizunuma et al., 2001) and Rcn1 degradation (Kishi et al., 2007). Mck1 also phosphorylates the sister cohesion protein Eco1, after Dbf4-kinase/Cdc7 and Cdk1-dependent priming phosphorylation, for ubiquitin-mediated degradation (Lyons et al., 2013).
There are eight CDK consensus sites in the N- and C-termini of Cdc6 that target Cdc6 for ubiquitin-mediated proteolysis (Drury et al., 1997, 2000; Elsasser et al., 1999; Perkins et al., 2001). In mitosis, Cdc6p is degraded through the SCFCdc4 ubiquitin ligase, which is dependent on Cdc6 T368 and S372 (Drury et al., 1997, 2000). Previously, we elucidated a novel function of Mck1 for DNA rereplication inhibition (Ikui et al., 2012). We found that Mck1 promotes Cdc6 degradation in a SCFCdc4-dependent manner through the C-terminus. The C-terminal residues 368TPTTSP372 in Cdc6 contain a GSK-3 consensus site in the form S/TxxxS/Tp. Theoretically, GSK-3 kinase phosphorylates the first S/T after the priming phosphorylation of the S/T residue at the fourth position by another kinase (Fiol et al., 1987). T368 and S372, which are important for Cdc6 degradation during mitosis, have been studied as putative Cdk1 phosphorylation sites (Drury et al., 2000).
In this article, we provide evidence in vitro and in vivo that Mck1, but not Cdk1, phosphorylates T368 only after priming phosphorylation of S372 by Clb2-Cdk1 to promote Cdc6 degradation during mitosis. The Cdc6 phosphodegron created by Cdk1 and Mck1 is crucial for Cdc4 binding to promote Cdc6 degradation. We also show that GSK3-dependent Cdc6 degradation is augmented after DNA damage stress in order to maintain genome integrity.
RESULTS
GSK-3 consensus site at T368-S372 in Cdc6 is responsible for Cdc6 degradation
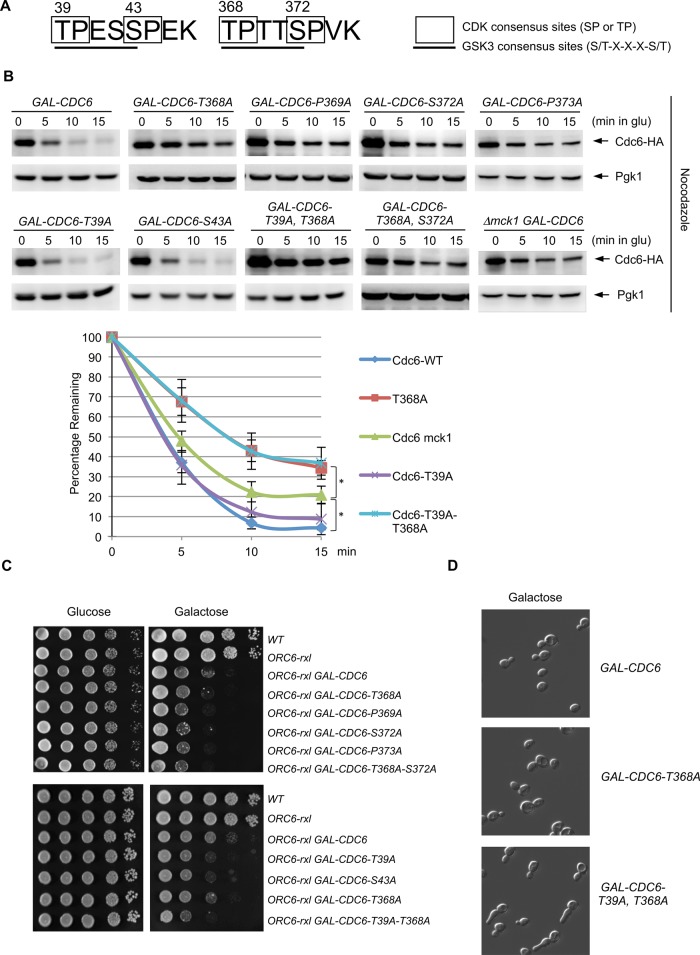
The GSK-3 consensus motif contains S/T-x-x-x-S/T. There are two GSK-3 consensus sites in Cdc6, one at each terminus (Figure 1A). The TPTTS GSK-3 consensus site (T368 to S372) in Cdc6 is overlapped with two CDK sites at TP (368 and 369) and SP (372 and 373) (Figure 1A). We mutated each amino acid from Thr, Ser, or Pro in the GSK-3 consensus site to Ala and examined the stability of Cdc6p. Wild-type Cdc6p was degraded after expression was shut off during mitosis (Figure 1B; Drury et al., 1997, 2000; Perkins et al., 2001). Cdc6-T368A, P369A, S372A, and P373A single mutants or T368A-S372A double-mutant proteins were stabilized, as was Cdc6p in mck1-deletion cells, indicating that the GSK consensus sequence from T368 to S372 is crucial for Cdc6 degradation (Figure 1B; Perkins et al., 2001). Cdc6 stabilization by P369A or P373A mutation indicates that T368 or S372 might be potential Cdk1 phosphorylation sites. A single mutation in Cdc6 at the N-terminus, T39A or S43A, did not alter Cdc6p stability during mitosis (Figure 1B). However, we observed high Cdc6p protein levels at time zero when the two mutations at the GSK-3 consensus sites T39A and T368A were combined (Figure 1B). Consistent with Cdc6 protein stability in CDC6-T39A, T368A double mutations (Figure 1B), we found that GAL-CDC6-T39A, T368A cells showed elongated bud and mitotic arrest (Figure 1D and Supplemental Figure S1). We also observed more stabilized Cdc6p in CDC6-T368A or CDC6-T39A, T368A cells than in Δmck1 cells (Figure 1B). This indicates that Cdc6-T368 site is phosphorylated by multiple kinases.
FIGURE 1:
Analysis of GSK-3 consensus sites in Cdc6. (A) Cdc6 contains two GSK-3 consensus sites, which overlap with two CDK consensus sites. (B) GAL-CDC6-HA strains with various mutations (T368A, P369A, S372A, P373A, T39A, S43A, T39A-T368A, and T368A-S372A) were expressed with galactose-containing medium for 2 h and then blocked with nocodazole for 2 h. Cdc6 expression was then suppressed by adding glucose. Protein extracts were collected every 5 min and subjected to Western blot analysis to observe Cdc6-HA. Pgk1 was used as a loading control. GAL-CDC6-HA in mck1-deletion cells was examined using the same method. Western blotting images for WT, Δmck1, CDC6-T39A, CDC6-T368A, and CDC6-T39A,T368A were quantified. Percentage of Cdc6 protein remaining relative to time zero is shown. Results are the average of three independent experiment, and error bars indicate SD. *p < 0.05. (C) Strains with the indicated genotypes were serially diluted 10-fold, plated on yeast extract/peptone/dextrose or yeast extract/peptone/galactose plates, and incubated at 30°C for 2 d. (D) GAL-CDC6, GAL-CDC6-T368A, or GAL-CDC6-T39A-T368A was grown in raffinose-containing medium first. Cdc6p was expressed with galactose for 3 h.
We previously found that mck1 deletion is synthetically lethal with an ORC6-rxl mutation that disrupts a control for DNA rereplication (Ikui et al., 2012). Given the Cdc6p stabilization in mck1-deletion cells (Figure 1B), we tested whether the CDC6 GSK-3 consensus-site mutations that stabilized Cdc6p also induce enhanced synthetic lethality in the ORC6-rxl mutant. Viability of yeast cells that contain various CDC6 mutations and the ORC6-rxl mutation were tested. The GAL-CDC6 ORC6-rxl mutant caused increased lethality on galactose-containing plates compared with controls (Figure 1C). The lethality was exacerbated when cells contained CDC6 mutations (T368A, P369A, S372A, and P373A single mutants or T368A and S372A double mutants) at the GSK-3 consensus sites (Figure 1C, top). ORC6-rxl GAL-CDC6-T39A, T368A cells showed enhanced lethality compared with ORC6-rxl GAL-CDC6-T39A or ORC6-rxl GAL-CDC6-T368A (Figure 1C, bottom). Next the CDC6-T368A mutation was integrated into the genome locus and was crossed with the ORC6-rxl and ORC6-rxl,ps mutants. The resulting double mutants, ORC6-rxl CDC6-T368A and ORC6-rxl,ps CDC6-T368A, showed slow growth (Supplemental Figure S2).
Mck1 phosphorylates Thr-368 in Cdc6 after priming by Clb2/Cdk1
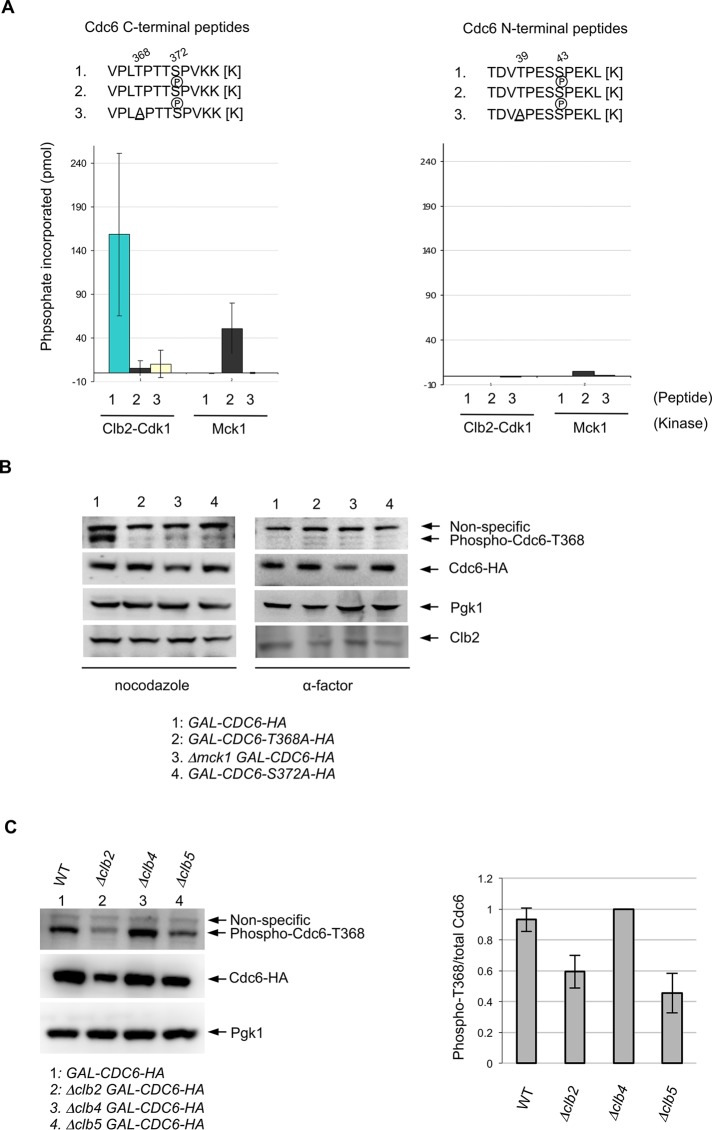
To identify the Cdk1 or Mck1 phosphorylation sites on Cdc6, we performed an in vitro kinase assay using various Cdc6 synthetic peptides with Mck1p kinase or Cdk1p kinase purified from yeast. Clb2/Cdk1 phosphorylated the wild-type Cdc6 synthetic peptide containing the C-terminal GSK-3 consensus site; however, the phosphorylation was abolished when the Cdc6 peptide contained a phosphate at S372 (Figure 2A, left). This indicates that Clb2/Cdk1 targets the S372 site but not the T368 site. On the other hand, Mck1 did not phosphorylate the wild-type Cdc6 peptide unless the peptide contained a phosphate at S372 (Figure 2A, left). The Mck1-dependent Cdc6 phosphorylation was abolished when we used Cdc6 peptides that contained an alanine mutation at T368 (Figure 2A, left). This result proved that Cdc6-T368 is phosphorylated by Mck1 in vitro. Mck1 weakly phosphorylated the Cdc6 peptide that contained the N-terminal GSK-3 consensus site at T39. However, the phosphorylation efficiency was ∼20-fold less than that at the C-terminal T368 (Figure 2A, right). Clb2/Cdk1 did not phosphorylate the Cdc6 peptide that contained the Cdk1 consensus site at T39 and S43 (Figure 2A, right).
FIGURE 2:
Mck1 phosphorylates the T368 site in Cdc6 after priming by Clb2-Cdk1. (A) To measure Clb2-Cdk1 and Mck1 kinase activities on Cdc6 phosphopeptides, various synthetic peptides of Cdc6 (residues 36–47 or 365–376; shown above) were incubated with purified kinases from asynchronous yeast cultures and [γ-32P]ATP. For each kinase, phosphate incorporation was normalized to a control reaction without peptides. Values for the C-terminal peptides represent the average from three independent experiments. Error bars represent SD. (B) Indicated strains were grown in raffinose-containing medium first. Cdc6 expression was induced with galactose for 2 h. Cells were blocked with nocodazole or α-factor for 2 h. Western blotting was performed using anti–phosphoT368 of Cdc6, anti-HA, anti-Pgk1, and anti-Clb2 antibodies, respectively. (C) Indicated strains were grown in raffinose-containing medium first, and then galactose was added to induce Cdc6 expression for 2 h. Cells were blocked with nocodazole for 2 h. Western blotting was performed using anti–phosphoT368 of Cdc6, anti-HA, and anti-Pgk1 antibodies, respectively (left). Band intensity for the phospho-Cdc6-T368 was quantified and normalized to the total amount of Cdc6 (right). The ratio phospho-T368/total Cdc6 in Δclb4 was set as 1. Results are the average of three independent experiments; error bars indicate SD.
Next we tested whether Mck1 kinase phosphorylates Cdc6-T368 in vivo, using a phosphospecific antibody against the T368 site. Cdc6 was phosphorylated at the T368 site in wild-type cells when the cell cycle was arrested during mitosis using nocodazole (Figure 2B, left). The Cdc6-T368 phosphorylation was abolished in the Cdc6-T368A mutant as well as in Δmck1 cells. We did not detect T368 phosphorylation when the priming site at S372 was mutated to alanine, indicating that Mck1 phosphorylates T368 only when the S372 priming site is phosphorylated (Figure 2B, left). Furthermore, the T368 site was not phosphorylated in wild-type cells arrested in G1 phase with α-factor (Figure 2B, right). These results support our priming model (Ikui et al., 2012) that Cdc6 requires Cdk1-dependent priming at S372 in order to be phosphorylated by Mck1 at T368 during mitosis.
We next tested the cyclin dependence of Mck1-induced Cdc6 phosphorylation at T368. The phosphorylation status of Cdc6-T368 was analyzed in vivo in GAL-CDC6, Δclb2 GAL-CDC6, Δclb4 GAL-CDC6 or Δclb5 GAL-CDC6 cells during mitosis. Cdc6-T368 phosphorylation was dependent on the mitotic cyclin Clb2 (Figure 2C), which is consistent with our in vitro kinase assay results. The phosphorylation was also dependent on the S-phase cyclin Clb5. However, the phosphorylation was still observed in clb4-deletion cells (Figure 2C).
Cdc6 phosphorylation by Mck1 and Cdk1 creates a Cdc4 phosphodegron
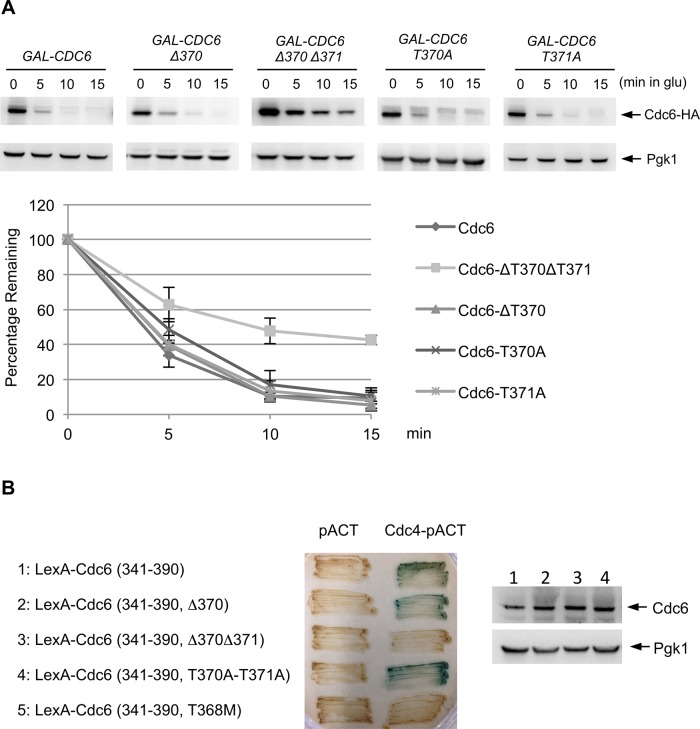
We previously proposed a model in which double phosphorylation of Cdc6 by Mck1 and Cdk1 may create a phosphodegron that is recognized by Cdc4 (Ikui et al., 2012). To test this, we created a GAL-CDC6-Δ370 mutant strain lacking one amino acid between the Mck1 and Cdk1 phosphorylation sites and whose expression is controlled under the GAL promoter. Similar to wild-type Cdc6p, the Cdc6-Δ370 protein was still degradable after glucose addition, whereas the Cdc6-Δ370Δ371 protein, lacking two of the three amino acids of the phosphodegron, was more stable than both single mutants (Figure 3A). Cdc6-T370A or Cdc6-T371A proteins were also degraded, signifying that spacing and not the specific amino acid sequence of 370 and 371 is critical for recognition, potentially by Cdc4 (Figure 3A). To determine whether Cdc4 binds to Cdc6 with specific spacing between the two phosphorylations at T368 and S372, we tested the binding efficiency between Cdc4 and Cdc6 by a yeast two-hybrid assay. Consistent with the stabilization data, Cdc4 did not bind to Cdc6-Δ370Δ371 (Figure 3B). However, Cdc4 recognized wild-type Cdc6 and Cdc6-Δ370 (Figure 3B). Each LexA-Cdc6 fusion protein was expressed at comparative levels, and therefore the protein-binding defect between Cdc4 and Cdc6 in the Cdc6-Δ370Δ371 mutant is not due to poor protein expression levels (Figure 3B, bottom). We conclude that Cdc4 binds to the Cdc6 phosphodegron at T368 and S372 when there are two or three amino acids between phosphorylations. Only one amino acid between phosphorylations (Cdc6-Δ370Δ371) causes stability because Cdc4 does not recognize it. T368M mutant was used as a negative control (Perkins et al., 2001).
FIGURE 3:
Cdc6 phosphodegron for Cdc4. (A) GAL-CDC6-HA strains with various mutations (Δ370Δ371, Δ370, T370A, or T371A) were incubated in raffinose-containing medium and transferred to galactose-containing medium for 2 h to induce Cdc6 expression, and then cells were blocked with nocodazole. Cdc6 expression was suppressed by adding glucose. Protein samples were collected every 5 min and subjected to Western blot analysis to observe Cdc6-HA. Pgk1 was used as a loading control. Western blotting images were quantified. Results represent the average of three experiments, with error bars indicating SD. (B) Yeast two-hybrid analysis was performed to determine the binding between Cdc4p and the Cdc6p C-terminus. Full-length CDC4 in the pACT plasmid, containing the GAL4 activation domain (GAD), was cotransformed into L40 yeast strains along with the various CDC6 mutants in the pBTM116 plasmid fused to LexA. Transformants were assayed for β-galactosidase activity, as visualized in blue. Proteins were extracted from each strain (1–4), and Cdc6 protein levels were examined by Western blotting.
Cdc6 degradation upon DNA damage is mediated by Mck1
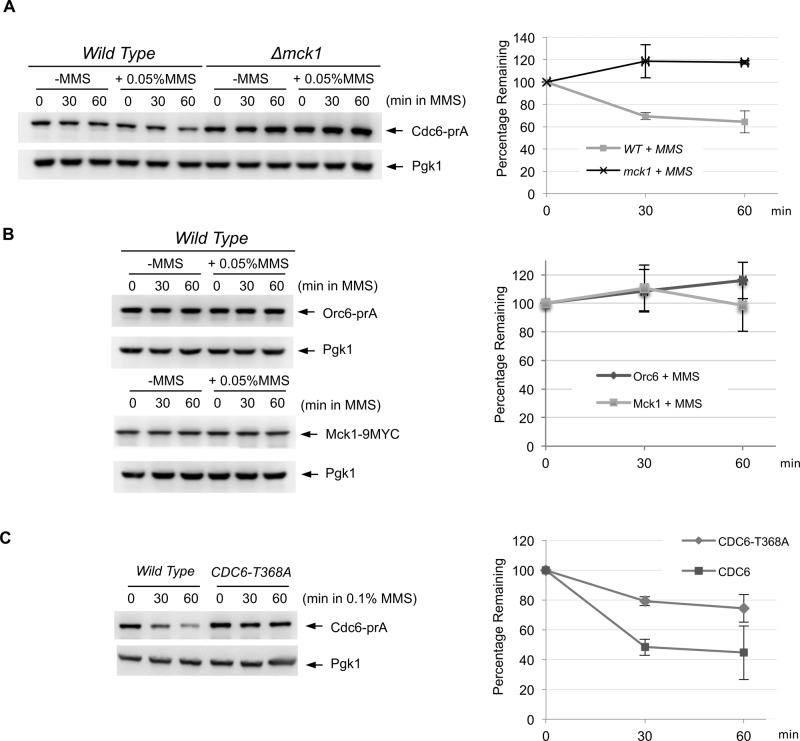
It has been shown that the DNA alkylating agent methyl methanesulfonate (MMS) triggers Cdc6 degradation in S. cerevisiae (Hall et al., 2007). We tested the possibility that DNA damage–induced Cdc6 degradation is dependent on Mck1 kinase. Cdc6p from the endogenous promoter was degraded after MMS treatment, as previously reported (Figure 4A; Hall et al., 2007), whereas Orc6 and Mck1 protein levels remained stable (Figure 4B). This suggests that MMS-induced protein degradation is specific to Cdc6p and is not indicative of an overall reduction in replication protein or general protein levels. The MMS-dependent Cdc6 degradation was suppressed in Δmck1 cells (Figure 4A), suggesting that Cdc6 degradation after DNA damage was mediated by Mck1 kinase. We also observed suppression of Cdc6 degradation in the CDC6-T368A mutant, in which T368A mutation was integrated at the endogenous locus (Figure 4C). We conclude that DNA damage triggers Mck1-dependent Cdc6 degradation.
FIGURE 4:
DNA damage triggers Cdc6 degradation in Mck1-dependent manner. (A) CDC6-prA or Δmck1 CDC6-prA cells were incubated in yeast extract/peptone/dextrose (YEPD) medium to log phase, and then MMS was added (0.05% final). Protein extracts were made at 0-, 30-, or 60-min incubation and subjected to Western blotting to visualize endogenous Cdc6-prA. Pgk1 was used as a loading control. The same experiment was repeated three times to quantify Cdc6 protein levels. Average percentage of Cdc6p remaining is shown. Bars represent SD. (B) Protein degradation of Orc6-prA or Mck1-9MYC was examined by the same method using IgG or anti-MYC antibodies, respectively. (C) CDC6-prA or CDC6-T368A-prA cells were grown in YEPD to log phase. MMS at 0.1% concentration was added, and protein samples were collected after 0, 30, and 60 min. The protein extracts were subjected to Western blotting to visualize Cdc6-prA. The same experiment was performed three times. Cdc6 protein levels were quantified, and average percentage of Cdc6p remaining is shown. Bars represent SD.
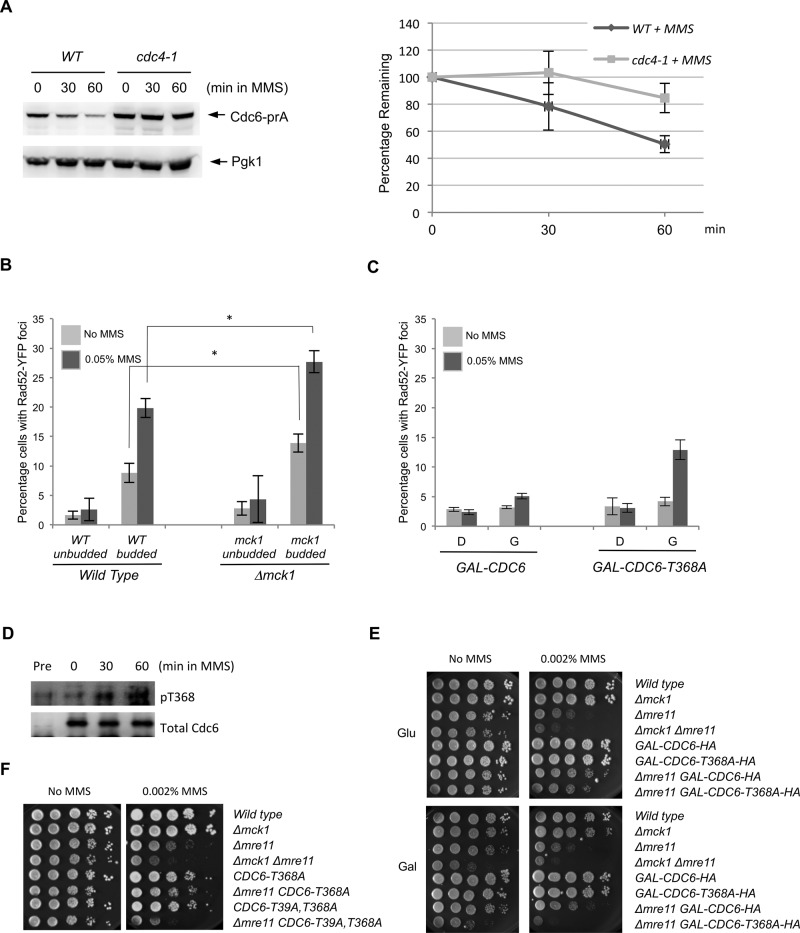
Cdc6 degradation is mediated by the SCFCdc4 complex (Drury et al., 1997; Elsasser et al., 1999; Perkins et al., 2001). We found that MMS-induced Cdc6 degradation was suppressed when Cdc4 was defective (Figure 5A). Most likely, Cdc6 phosphorylations upon DNA damage are recognized by the SCFCdc4 complex for subsequent Cdc6p ubiquitination. It is known that Cdc6 degradation is also mediated through Tom1 and Dia2 (Hall et al., 2007; Kim et al, 2012). We further tested whether Cdc6 degradation upon MMS treatment is mediated by the Tom1 or Dia2 ubiquitin ligases. We did not observe significant difference in Cdc6 degradation rate between wild-type, Δdia2, or Δtom1 deletion cells after MMS treatment (Supplemental Figure S3).
FIGURE 5:
Cdc6 degradation upon DNA damage is mediated through the ubiquitin pathway. (A) CDC6-prA or cdc4-1 CDC6-prA cells under the endogenous promoter were grown to log phase at 26°C first and then the temperature was increased to 36°C for 1.5 h. Then MMS was added (0.1% final). Samples were collected after 0, 30, or 60 min. The same experiment was performed three times, and Cdc6 protein levels were quantified. Error bars represent SD. (B) RAD52-YFP or Δmck1 RAD52-YFP cells were grown to log phase in low-fluorescence medium. MMS was added (0.05% final) for 90 min. Rad52-YFP foci were visualized under a fluorescence microscope to assay for double-stranded DNA breaks before and after treatment with MMS. Percentage of cells containing Rad52 foci was determined in unbudded or budded cells. One hundred cells were counted for each sample. Percentage is average from three independent experiments, and error bars represent the SD. *p < 0.05. (C) GAL-CDC6 or GAL-CDC6-T368A cells were grown to log phase in raffinose-containing medium first, and then either glucose or galactose was added for 3 h of incubation. Finally, MMS was then added (0.05% final) for 1 h. Rad52-YFP foci formation was determined with or without MMS treatment. One hundred cells were counted for each sample. Percentage is average from three independent experiments, and error bars represent SD. (D) cdc4-1 GAL-CDC6-HA cells were incubated in raffinose-containing medium first. Galactose was then added for 1 h of incubation, followed by nocodazole for 1.5 h. The temperature was then raised to 36°C to inactivate Cdc4 function for 1.5 h. Finally, cells were treated with MMS (0.1% final) and sampled every 30 min. Western blotting was performed using anti-phosphoT368 of Cdc6 and anti-HA. Control experiment was performed in raffinose-containing medium, indicated as “pre.” (E, F) Cells with indicated genotypes were serially diluted 10-fold on yeast extract/peptone/dextrose or yeast extract/peptone/galactose plates containing 0.002% MMS. The plates were incubated at 30°C for 2 d.
Cells with stabilized Cdc6 are susceptible to DNA damage
The homologous recombination protein Rad52 localizes to sites of double-stranded DNA breaks (DSBs) (Lisby et al., 2001). MMS increases the frequency of DSBs in budded cells, as detected by the formation of Rad52-YFP foci (Lisby et al., 2003), likely due to an increased frequency of replication fork collapse (Tercero and Diffley, 2001). To test the functional significance of Mck1-mediated Cdc6 degradation during DNA damage stress, we counted the frequency of Rad52-YFP foci in wild-type cells or Δmck1 cells after MMS treatment. In unbudded cells during G1 phase, we did not observe a significant difference in the frequency of Rad52-YFP foci between wild-type and Δmck1 cells after MMS treatment (Figure 5B). Consistent with previous work, however, more Rad52-YFP foci were formed in wild-type budded cells, both with and without MMS treatment (Figure 5B; Lisby et al., 2003). In untreated Δmck1 cells, we observed a statistically higher rate of Rad52–yellow fluorescent protein (YFP) foci formation (Figure 5B). The Rad52-YFP foci frequency was increased after MMS treatment in both wild-type and Δmck1 cells, but more strikingly in Δmck1 cells (Figure 5B). Furthermore, cells expressing GAL-CDC6-T368A, which contains a mutation at the Mck1 phosphorylation site, had an increase in the frequency of Rad52-YFP foci formation after MMS treatment when grown in galactose-containing but not glucose-containing media compared with GAL-CDC6 cells (Figure 5C).
We further obtained evidence that the Cdc6-T368 site is more phosphorylated after MMS treatment (Figure 5D). This result supports our conclusion that Cdc6 is degraded after phosphorylation at the T368 site through Mck1 kinase. To test whether Mck1 has a role in preventing lethality in response to DNA damage, we tested cell viability of Δmck1 cells in MMS. The Δmck1 cells were not sensitive to MMS, indicating that cell viability of Δmck1 was maintained due to DNA damage checkpoint activation in the presence of MMS (Figure 5E). We therefore tested whether deletion of a DNA damage checkpoint component may enhance lethality in Δmck1 cells. Deleting both MRE11 (a part of the MRX complex) and MCK1 genes enhanced lethality in response to MMS (Figure 5E). Furthermore, a combination of Δmre11 and the GAL-CDC6-T368A mutation also enhanced the cell lethality to MMS (Figure 5E), supporting the idea that DNA damage in Δmck1 or GAL-CDC6-T368A cells is augmented when a DNA checkpoint component is eliminated.
We next tested whether the endogenous CDC6-T368A mutation behaves in the same way as GAL-CDC6-T368A. The CDC6-T368A cells did not show significant MMS sensitivity, similar to Δmck1 (Figure 5F). Furthermore, Δmre11 CDC6-T368A double mutant did not have a significant difference in viability in response to MMS compared with Δmre11 (Figure 5F). We created CDC6-T39A,T368A, in which both mutations were integrated into the endogenous locus. CDC6-T39A,T368A cells did not show MMS sensitivity (Figure 5F). However, deleting MRE11 in CDC6-T39A,T368A cells enhanced lethality, similar to Δmck1 Δmre11 (Figure 5F). Furthermore, there was increased Rad52 foci formation in budded cells with endogenous CDC6-T39A,T368A after MMS exposure (Supplemental Figure S4).
DISCUSSION
The mechanism of Cdc6 protein degradation is complex. Cdk1-dependent Cdc6 phosphorylations and degradations have been extensively studied in order to understand the molecular mechanism of Cdc6 control. The Cdc6 phosphorylation sites have been studied based on the CDK consensus motif ST or TP (Drury et al., 2000; Perkins et al., 2001; Boronat and Campbell, 2007; Honey and Futcher, 2007). There are two GSK-3 consensus sites in Cdc6 (Figure 1A). Both of the GSK-3 sites in Cdc6 overlap with two CDK consensus sites; the second of each CDK site, at S43 or S372, is conserved, as it contains a lysine at the fourth position, K46 and K375, respectively (Figure 1A). In this study, we found that the phosphorylation site at T368 in Cdc6, which was previously studied as a CDK site, is actually a GSK-3 phosphorylation site (Figure 2). GSK-3 kinase requires priming phosphorylation (Lee et al., 2012; Lyons et al., 2013). Therefore we hypothesized that within the GSK-3 consensus site in Cdc6p (368TPTTS372), S372 would serve as the priming site for T368 phosphorylation by Mck1 (Ikui et al., 2012). Because S372 occurs within a conserved Cdk1 site (T/SPxR/K), we predicted that Cdk1 would provide this priming phosphorylation (Ikui et al., 2012). We showed that T368 is phosphorylated by Mck1 (Figure 2A). The Cdc6-S372 site, followed by a basic residue at the fourth position, was efficiently phosphorylated by Clb2/Cdk to serve as the priming phosphorylation site. Mck1 also phosphorylated the Cdc6 N-terminal GSK-3 consensus site at T39, but with less efficiency (Figure 2A). We also tested the possibility that Clb2/Cdk1 phosphorylates S43, the other conserved CDK site, but we determined that it did not (unpublished data). It is possible that G1 cyclins or S-phase cyclins coupled with Cdk1 may phosphorylate the S43 site. This would allow cell cycle–dependent Cdc6 degradation whose timing relies on Cdk1-priming.
The GSK-3 consensus sequence, S/T-x-x-x-S/T, contains three amino acids between the priming and phosphorylation sites. Together, they create a phosphodegron for SCFCdc4-dependent ubiquitination (Ikui et al., 2012; Lyons et al., 2013). The GSK-3 sites at the N-terminus and C-terminus of Cdc6 coincide with a binding site for the Cdc4 E3 ubiquitin ligase (Perkins et al., 2001). It was shown that the protein binding between Cdc4 and Cdc6 is abolished when there is a mutation at T368 or S372 (Perkins et al., 2001). Here we show that the double phosphorylations created by Cdk1 at S372 and Mck1 at T368 serve as a phosphodegron for Cdc4 binding. The spacing of two or three, and not one, amino acids between the two phosphorylations is important for Cdc4 recognition (Figure 3B). This is consistent with a previous report for Eco1p (Lyons et al., 2013). Mck1 requires a priming phosphorylation. For example, Eco1 must be primed by Dbf4 and Cdk1 for cohesion function regulation (Lyons et al., 2013) and Rpc53 must be primed by Kns1 in the TOR signaling pathway before Mck1 can phosphorylate either substrate (Lee et al., 2012). Possibly, Mck1 substrates are favored targets of Cdc4 (Mizunuma et al., 2001; Kishi et al., 2007; Ikui et al., 2012; Lyons et al., 2013; Edenberg et al., 2014), because the GSK-3 consensus motif and the priming model can determine precise Cdc4 specificity. These reports, together with our findings, strongly suggest that there might be more GSK-3 substrates that are targeted in a Cdc4-dependent manner.
The presence of a separate Mck1-dependent degradation mechanism of Cdc6 raised the question of why such a mechanism is needed. First, we tested whether Mck1 is regulated in a cell cycle–dependent manner. Mck1 protein was stable throughout the cell cycle, as were its localization and kinase activities (Supplemental Figure S5). Mck1 was shown to promote protein degradation in response to stress (Andoh et al., 2000). We therefore reasoned that the Mck1-dependent Cdc6p degradation could be involved in the cell's response to stress. The Cdc6 degradation in response to MMS was dependent on Mck1 kinase (Figure 4, A and C). Furthermore, MMS-induced Cdc6 degradation was mediated through SCF F-box protein (Cdc4) (Figure 5A). The continuous Mck1 activity throughout the cell cycle may help to phosphorylate and degrade Cdc6p when cells are exposed to DNA damage agent.
We also tested whether hydroxyurea (HU) or benomyl promotes Cdc6 degradation, since Δmck1-deletion cells are sensitive to these reagents (Shero and Hieter, 1991). However, Cdc6 was stable after HU or benomyl treatment (Supplemental Figure S6). These results indicate that Mck1-dependent Cdc6 degradation is triggered by specific DNA damage through alkylating agents. It has been reported that Sld3 is phosphorylated and inhibits late origin firing when cells are treated with MMS (Lopez-Mosqueda et al., 2010; Zegerman and Diffley, 2010). The inhibition of late origin firing is dependent on Rad53 (Lopez-Mosqueda et al., 2010; Zegerman and Diffley, 2010). Mck1-dependent Cdc6 degradation may serve as an additional mechanism to block origin firing, and it would be of interest to know whether the Mck1-dependent mechanism inhibits early and/or late origin firing. It remains to be investigated whether Mck1 is activated upon DNA damage directly. We observed that Cdc6 is more stabilized in CDC6-T368A than in Δmck1 (Figure 1B). This result raised the possibility that Cdc6p may be phosphorylated through another kinase, such as Rad53 or Mec1.
Cells undergoing S phase are more prone to DNA damage by MMS (Lisby et al., 2003). We also observed that Δmck1 cells accumulate more DNA damage in budded cells, which includes S-phase cells, than in wild-type cells (Figure 5C). We conclude that DNA replication has to be controlled and inhibited in early S phase by inhibiting Cdc6p so that cells do not proceed through S phase when cells are exposed to DNA damage agents. Continuous initiation of DNA replication during S phase under DNA damage stress will lead to accumulation of DNA damage and lethality.
It will be also interesting to identify novel Mck1 substrates. We searched for proteins that contain a GSK-3 consensus motif similar to that in Cdc6, S/T-P-X-X-S/T-P-X-R/K. Using the MOTIF search database (www.genome.jp/tools/motif/MOTIF2.html), we identified 17 candidate proteins that contain a similar GSK-3 consensus sequence, including three DNA replication proteins, Cdc6, Mcm3, and Sld2. Although we do not yet have evidence that Mck1 targets these proteins, it would be interesting to study whether Mck1 targets other proteins besides Cdc6p.
The Δmck1 cells are not sensitive to MMS (Figure 5E), which suggests that the DNA damage checkpoint might be activated and protects Δmck1 cells even after the accumulation of DNA damage. Deletion of a DNA damage checkpoint gene enhanced Δmck1 sensitivity to MMS, supporting the hypothesis (Figure 5E). Desany et al. (1998) reported that overexpression of MCK1 suppresses lethality in rad53 mutants, indicating that Mck1p has a role in DNA damage repair. We propose a model in which Cdc6 inhibition by Mck1 under DNA damage stress ensures genome integrity and maintenance of proper cell proliferation. We also observed that overexpression of CDC6 but not the CDC6-T368A mutant partially rescues MMS sensitivity in the mre11-deletion strain (Figure 5E, bottom). Mcm complex plays a role in the DNA damage–induced signaling that controls DNA replication (Cortez et al., 2004). Overexpression of Cdc6 helps Mcm complex to be loaded on DNA (Frigola et al., 2013), which may enhance the DNA damage signaling pathway to rescue the Δmre11 lethality to MMS.
In humans, deregulation of replication factors, such as Cdc6, has been observed in many cancers. For instance, Cdc6 overexpression has been associated with brain tumors (Ohta et al., 2001), cervical cancer (Murphy et al., 2005), and lung carcinomas (Karakaidos et al., 2004). Given the implication of Cdc6 up-regulation in human cancers (Borlado and Mendez, 2008), it would be of considerable interest to study whether mammalian GSK-3 kinase plays a role in Cdc6 degradation. Cdc6 degradation during alkylating DNA damage is conserved between humans and yeasts (Hall et al., 2007). Therefore it is possible that human GSK-3 kinase, similar to Mck1, might promote Cdc6 degradation. A link between mammalian GSK-3 kinase and the DNA damage response has been reported. GSK-3 kinase phosphorylates the oncogenic metazoan transcription factor c-Myc after DNA damage triggered by ultraviolet light. The phosphorylated c-Myc is targeted for ubiquitination by SCFFbw7, the human homologue of Cdc4 (Popov et al., 2007). Of interest, c-Myc was shown to have a nontranscriptional role in the initiation of DNA replication (Dominguez-Sola et al., 2007). Although it is not clear whether the c-Myc degradation during DNA damage is associated with the replication function of c-Myc, it would support the general idea that DNA replication is inhibited during DNA damage through degradation of replication proteins.
MATERIALS AND METHODS
Plasmids and strains
Standard methods were used for mating, tetrad dissection, and transformation. All strains listed in Supplemental Table S1 are congenic with W303. p305-based GAL-CDC6-HA plasmid was a gift from Stephen P. Bell (Massachusetts Institute of Technology, Cambridge, MA). GAL-CDC6-HA strains were made as described previously (Wilmes et al., 2004). Briefly, GAL-CDC6-HA plasmid was linearized with StuI and integrated at the URA locus in the wild-type strain. The copy number of the integrated GAL-CDC6 was determined by real-time PCR. GAL-CDC6 mutations were made by site-directed mutagenesis (QuikChange II XL mutagenesis kit; Agilent, Santa Clara, CA) using the GAL-CDC6-HA plasmid as a template. The mutagenized GAL-CDC6 plasmid was integrated at the URA locus in wild-type cells, and the copy number was determined as described. MCK1-9MYC strain was constructed as described previously (Knop et al., 1999). CDC6-T368A and CDC6-T39A,T368A strains were generated by two-step PCR in order to integrate the mutation into the genomic locus (Figures 4C and 5F; Toulmay and Schneiter, 2006).
Cell cycle block experiments
Mitotic cell cycle arrest was achieved using 15 μg/ml nocodazole for 2 h at 30°C. GAL-CDC6–HA strains were grown to log phase in medium containing 3% raffinose. Galactose was then added to induce CDC6-HA expression for 2 h, followed by incubation in nocodazole for another 2 h. Glucose was then added to shut off the expression. For the α-factor arrest experiments, log-phase cells were arrested in G1 using 100 nM α-factor for 2 h at 30°C.
Western blotting
Cells were lysed by agitation in SDS sample buffer with glass beads using FastPrep (MP Biomedicals, Santa Ana, CA) for 20 s, twice, at speed 6. Proteins were separated by SDS–PAGE with 10% polyacrylamide gel, except for the samples in Figure 2, which were separated using SDS–PAGE with Novex 4–20% Tris-glycine polyacrylamide gel (Invitrogen, Life Technologies, Carlsbad, CA). Western blot analysis was performed using anti-hemagglutinin (HA) antibody 3F10 (Roche, Penzberg, Germany) at 1:2000 dilution, anti-Clb2 antibody (a generous gift from Frederick Cross, Rockefeller University, New York, NY), anti–phospho Cdc6-T368 antibody at 1:1000 dilution (custom-made antibody by 21st Century Bio, Marlboro, MA), anti-cMYC antibody 9E10 (Sigma-Aldrich, St. Louis, MO) at 1:2000 dilution, and anti-Pgk1 (Life Technologies, Carlsbad, CA) at 1:2000 as a loading control. Protein A–tagged proteins were probed using horseradish peroxidase–conjugated anti-rabbit immunoglobulin G (Sigma-Aldrich) at 1:5000.
To detect phospho–Cdc6-T368, CDC6 was immunoprecipitated by HA-affinity purification first in order to enhance the signal (Ikui et al., 2012).
Kinase assay
Clb2-Cdk1 and Mck1 proteins were purified from yeast cells expressing TAP-tagged proteins as previously described (Lyons and Morgan, 2011). The TAP-tagged plasmid for Clb2 and Mck1 were kindly provided by David O Morgan (University of California, San Francisco, San Francisco, CA; Lyons et al., 2013). Peptide kinase assay was performed as described (Lyons et al., 2013) at room temperature for 30 min using purified Mck1 or Clb2-Cdk1 with 5 μM peptides (synthesized by NeoBioLab, Woburn, MA) in a 15-μl total reaction, with 0.6 μCi of [γ-32P]ATP (3000 μCi mmol−1; PerkinElmer, Melville, NY). After stopping the reaction and washing, radioactivity was measured using a scintillation counter (Tri-Carb 2800; PerkinElmer; Puig et al., 2001).
Microscope
Cells were grown in low-fluorescence medium, which was prepared as described previously (Sheff and Thorn, 2004). Cells were imaged on an Eclipse 90i microscope (Nikon, Tokyo, Japan) with a total internal reflection fluorescence 60×/1.45 numerical aperture Plan Apochromatic objective lens (Nikon) fitted with a cooled Clara interline charge-coupled device camera (Andor, Belfast, United Kingdom) and using an Intensilight Ultra High Pressure 130-W mercury lamp (Nikon) as an illumination source. Images were acquired using the NIS Element BR software (Nikon). Image acquisition times for green fluorescent protein, YFP, 4′,6-diamidino-2-phenylindole (DAPI), and differential interference contrast (DIC) were 300, 200, 50, and 50 ms, respectively. All images were processed using ImageJ software (National Institutes of Health, Bethesda, MD; Schneider et al., 2012). To visualize RAD52-YFP foci, the DIC image was acquired, followed by seven YFP images at 0.5-μm intervals along the z-axis. YFP image stacks were combined through maximum intensity projection in order to count Rad52-YFP foci along the entire z-stack for each field. The corresponding DIC images were used as a reference to determine the budding of the cells. Log-phase MCK1-GFP cells were lightly fixed in 4% paraformaldehyde and then stained with DAPI. The fixed cells were imaged using the relevant wavelengths, and the images were pseudocolored and processed using ImageJ.
Yeast two-hybrid assay
The pBTM116 constructs containing Cdc6 is a generous gift from J. Diffley's lab (Francis Crick Institute, London, UK; Perkins et al., 2001). The CDC6-pBTM116 plasmid was subjected to site-directed mutagenesis to create the various Cdc6 mutants (QuikChange Lightning Kit; Agilent). The CDC4/pACT plasmid and each of the various CDC6/pBMT116 plasmids were cotransformed into L40 strain and plated on synthetic defined-Leu/Trp plates, and β-galactosidase activity was measured as described previously (Ikui et al., 2012).
Supplementary Material
Acknowledgments
We gratefully acknowledge David Morgan for plasmids and yeast strains used in Figure 2. We also thank John Diffley and Stephen Bell for the BTM116-CDC6 and GAL-CDC6 constructs, respectively. CDC6-T368A and CDC6-T39A,T368A mutants were created by Shoily Khondker. A.E.I. was supported by National Institutes of Health Grant 5SC3GM105498 and a Professional Staff Congress–City University of New York Enhanced Award.
Abbreviation used:
- MMS
methyl methanesulfonate.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-07-1213) on May 20, 2015.
REFERENCES
- Andoh T, Hirata Y, Kikuchi A. Yeast glycogen synthase kinase 3 is involved in protein degradation in cooperation with Bul1, Bul2, and Rsp5. Mol Cell Biol. 2000;20:6712–6720. doi: 10.1128/mcb.20.18.6712-6720.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr Opin Cell Biol. 2010;22:766–771. doi: 10.1016/j.ceb.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Archambault V, Ikui AE, Drapkin BJ, Cross FR. Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol Cell Biol. 2005;25:6707–6721. doi: 10.1128/MCB.25.15.6707-6721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlado LR, Mendez J. CDC6: from DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis. 2008;29:237–243. doi: 10.1093/carcin/bgm268. [DOI] [PubMed] [Google Scholar]
- Boronat S, Campbell JL. Mitotic Cdc6 stabilizes anaphase-promoting complex substrates by a partially Cdc28-independent mechanism, and this stabilization is suppressed by deletion of Cdc55. Mol Cell Biol. 2007;27:1158–1171. doi: 10.1128/MCB.01745-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazill DT, Thorner J, Martin GS. Mck1, a member of the glycogen synthase kinase 3 family of protein kinases, is a negative regulator of pyruvate kinase in the yeast Saccharomyces cerevisiae. J Bacteriol. 1997;179:4415–4418. doi: 10.1128/jb.179.13.4415-4418.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Bell SP. CDK prevents Mcm2-7 helicase loading by inhibiting Cdt1 interaction with Orc6. Genes Dev. 2011;25:363–372. doi: 10.1101/gad.2011511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Glick G, Elledge SJ. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Natl Acad Sci USA. 2004;101:10078–10083. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmann C, Diffley JF, Nasmyth KA. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JF. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JF. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr Biol. 2000;10:231–240. doi: 10.1016/s0960-9822(00)00355-9. [DOI] [PubMed] [Google Scholar]
- Edenberg ER, Vashisht AA, Topacio BR, Wohlschlegel JA, Toczyski DP. Hst3 is turned over by a replication stress-responsive SCFCdc4 phospho-degron. Proc Natl Acad Sci USA. 2014;111:5962–5967. doi: 10.1073/pnas.1315325111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S, Chi Y, Yang P, Campbell JL. Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Mol Biol Cell. 1999;10:3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiol CJ, Mahrenholz AM, Wang Y, Roeske RW, Roach PJ. Formation of protein kinase recognition sites by covalent modification of the substrate. Molecular mechanism for the synergistic action of casein kinase II and glycogen synthase kinase 3. J Biol Chem. 1987;262:14042–14048. [PubMed] [Google Scholar]
- Frigola J, Remus D, Mehanna A, Diffley JF. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495:339–343. doi: 10.1038/nature11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JR, Kow E, Nevis KR, Lu CK, Luce KS, Zhong Q, Cook JG. Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol Biol Cell. 2007;18:3340–3350. doi: 10.1091/mbc.E07-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata Y, Andoh T, Asahara T, Kikuchi A. Yeast glycogen synthase kinase-3 activates Msn2p-dependent transcription of stress responsive genes. Mol Biol Cell. 2003;14:302–312. doi: 10.1091/mbc.E02-05-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey S, Futcher B. Roles of the CDK phosphorylation sites of yeast Cdc6 in chromatin binding and rereplication. Mol Biol Cell. 2007;18:1324–1336. doi: 10.1091/mbc.E06-06-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikui AE, Archambault V, Drapkin BJ, Campbell V, Cross FR. Cyclin and cyclin-dependent kinase substrate requirements for preventing rereplication reveal the need for concomitant activation and inhibition. Genetics. 2007;175:1011–1022. doi: 10.1534/genetics.106.068213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikui AE, Rossio V, Schroeder L, Yoshida S. A yeast GSK-3 kinase Mck1 promotes Cdc6 degradation to Inhibit DNA re-replication. PLoS Genet. 2012;8:e1003099. doi: 10.1371/journal.pgen.1003099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakaidos P, Taraviras S, Vassiliou LV, Zacharatos P, Kastrinakis NG, Kougiou D, Kouloukoussa M, Nishitani H, Papavassiliou AG, Lygerou Z, et al. Overexpression of the replication licensing regulators hCdt1 and hCdc6 characterizes a subset of non-small-cell lung carcinomas: synergistic effect with mutant p53 on tumor growth and chromosomal instability—evidence of E2F-1 transcriptional control over hCdt1. Am J Pathol. 2004;165:1351–1365. doi: 10.1016/S0002-9440(10)63393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Zhang W, Koepp DM. The Hect domain E3 ligase Tom1 and the F-box protein Dia2 control Cdc6 degradation in G1 phase. J Biol Chem. 2012;287:44212–44220. doi: 10.1074/jbc.M112.401778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi T, Ikeda A, Nagao R, Koyama N. The SCFCdc4 ubiquitin ligase regulates calcineurin signaling through degradation of phosphorylated Rcn1, an inhibitor of calcineurin. Proc Natl Acad Sci USA. 2007;104:17418–17423. doi: 10.1073/pnas.0704951104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Labib K, Diffley JF, Kearsey SE. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat Cell Biol. 1999;1:415–422. doi: 10.1038/15649. [DOI] [PubMed] [Google Scholar]
- Lee J, Moir RD, McIntosh KB, Willis IM. TOR signaling regulates ribosome and tRNA synthesis via LAMMER/Clk and GSK-3 family kinases. Mol Cell. 2012;45:836–843. doi: 10.1016/j.molcel.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MY, Dailey D, Martin GS, Thorner J. Yeast MCK1 protein kinase autophosphorylates at tyrosine and serine but phosphorylates exogenous substrates at serine and threonine. J Biol Chem. 1993;268:21155–21164. [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Mosqueda J, Maas NL, Jonsson ZO, Defazio-Eli LG, Wohlschlegel J, Toczyski DP. Damage-induced phosphorylation of Sld3 is important to block late origin firing. Nature. 2010;467:479–483. doi: 10.1038/nature09377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons NA, Fonslow BR, Diedrich JK, Yates JR, 3rd, Morgan DO. Sequential primed kinases create a damage-responsive phosphodegron on Eco1. Nat Struct Mol Biol. 2013;20:194–201. doi: 10.1038/nsmb.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons NA, Morgan DO. Cdk1-dependent destruction of Eco1 prevents cohesion establishment after S phase. Mol Cell. 2011;42:378–389. doi: 10.1016/j.molcel.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S, Seki T, Tanaka S, Diffley JF. Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature. 2004;431:1118–1123. doi: 10.1038/nature03024. [DOI] [PubMed] [Google Scholar]
- Mizunuma M, Hirata D, Miyaoka R, Miyakawa T. GSK-3 kinase Mck1 and calcineurin coordinately mediate Hsl1 down-regulation by Ca2+ in budding yeast. EMBO J. 2001;20:1074–1085. doi: 10.1093/emboj/20.5.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy N, Ring M, Heffron CC, King B, Killalea AG, Hughes C, Martin CM, McGuinness E, Sheils O, O'Leary JJ. p16INK4A, CDC6, and MCM5: predictive biomarkers in cervical preinvasive neoplasia and cervical cancer. J Clin Pathol. 2005;58:525–534. doi: 10.1136/jcp.2004.018895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. At the heart of the budding yeast cell cycle. Trends Genet. 1996;12:405–412. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- Neigeborn L, Mitchell AP. The yeast MCK1 gene encodes a protein kinase homolog that activates early meiotic gene expression. Genes Dev. 1991;5:533–548. doi: 10.1101/gad.5.4.533. [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Irie K, Li JJ. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr Biol. 2000;10:195–205. doi: 10.1016/s0960-9822(00)00337-7. [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- Ohta S, Koide M, Tokuyama T, Yokota N, Nishizawa S, Namba H. Cdc6 expression as a marker of proliferative activity in brain tumors. Oncol Rep. 2001;8:1063–1066. doi: 10.3892/or.8.5.1063. [DOI] [PubMed] [Google Scholar]
- Perkins G, Drury LS, Diffley JF. Separate SCF(CDC4) recognition elements target Cdc6 for proteolysis in S phase and mitosis. EMBO J. 2001;20:4836–4845. doi: 10.1093/emboj/20.17.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a 'reductional' anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov N, Herold S, Llamazares M, Schulein C, Eilers M. Fbw7 and Usp28 regulate myc protein stability in response to DNA damage. Cell Cycle. 2007;6:2327–2331. doi: 10.4161/cc.6.19.4804. [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Rayner TF, Gray JV, Thorner JW. Direct and novel regulation of cAMP-dependent protein kinase by Mck1p, a yeast glycogen synthase kinase-3. J Biol Chem. 2002;277:16814–16822. doi: 10.1074/jbc.M112349200. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- Shero JH, Hieter P. A suppressor of a centromere DNA mutation encodes a putative protein kinase (MCK1) Genes Dev. 1991;5:549–560. doi: 10.1101/gad.5.4.549. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- Toulmay A, Schneiter R. A two-step method for the introduction of single or multiple defined point mutations into the genome of Saccharomyces cerevisiae. Yeast. 2006;23:825–831. doi: 10.1002/yea.1397. [DOI] [PubMed] [Google Scholar]
- Wilmes GM, Archambault V, Austin RJ, Jacobson MD, Bell SP, Cross FR. Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 2004;18:981–991. doi: 10.1101/gad.1202304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature. 2010;467:474–478. doi: 10.1038/nature09373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerschke W, Rottjakob HW, Kuntzel H. The Saccharomyces cerevisiae CDC6 gene is transcribed at late mitosis and encodes a ATP/GTPase controlling S phase initiation. J Biol Chem. 1994;269:23351–23356. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.