The spindle checkpoint proteins Mad1 and Bub1 are dynamically recruited after induced de novo kinetochore assembly. Detached kinetochores compete with alternate binding sites in the nucleus to recruit Mad1 and Bub1 from very limited pools. Lateral kinetochore attachment to microtubules licenses Mad1 removal from kinetochores.
Abstract
We observed the dynamic recruitment of spindle checkpoint proteins Mad1 and Bub1 to detached kinetochores in budding yeast using real-time live-cell imaging and quantified recruitment in fixed cells. After induced de novo kinetochore assembly at one pair of sister centromeres, Mad1 appeared after the kinetochore protein Mtw1. Detached kinetochores were not associated with the nuclear envelope, so Mad1 does not anchor them to nuclear pore complexes (NPCs). Disrupting Mad1's NPC localization increased Mad1 recruitment to detached sister kinetochores. Conversely, increasing the number of detached kinetochores reduced the amount of Mad1 per detached kinetochore. Bub1 also relocalized completely from the spindle to detached sister centromeres after kinetochore assembly. After their capture by microtubules, Mad1 and Bub1 progressively disappeared from kinetochores. Sister chromatids that arrested with a lateral attachment to one microtubule exhibited half the Mad1 of fully detached sisters. We propose that detached kinetochores compete with alternate binding sites in the nucleus to recruit Mad1 and Bub1 from available pools that are small enough to be fully depleted by just one pair of detached kinetochores and that lateral attachment licenses Mad1 removal from kinetochores after a kinetic delay.
INTRODUCTION
Mechanisms to ensure that chromosomes are faithfully segregated are critical for maintaining hereditary continuity and avoiding aneuploidy-related diseases such as cancer in multicellular organisms. Chromosome missegregation is particularly harmful because it alters the dosages of numerous genes. A crucial cell cycle event therefore is initiation of chromosome segregation at the metaphase–anaphase boundary. The spindle checkpoint controls the timing of this transition by inhibiting the anaphase-promoting complex (APC) and its substrate specificity factor Cdc20 until all the chromosomes are properly organized on the spindle. Conditions that satisfy the spindle checkpoint relieve APCCdc20 inhibition, allowing APCCdc20 to trigger the precipitous and irreversible loss of cohesion between sister chromatids, thereby initiating anaphase chromosome segregation (reviewed in Musacchio and Salmon, 2007; Foley and Kapoor, 2013).
A key spindle checkpoint effector is the stable complex formed by Mad1 and Mad2 (Mad1/2), which localizes to kinetochores with defective attachments, at least in part through an interaction between Mad1 and Bub1 regulated by phosphorylation of Bub1 (Li and Benezra, 1996; Chen et al., 1998; Gillett et al., 2004; London and Biggins, 2014; Moyle et al., 2014). At kinetochores, Mad1/2 activates soluble copies of Mad2 to form inhibitory complexes with Cdc20 and APCCdc20 (Li et al., 1997; Fang et al., 1998; Hwang et al., 1998; Kim, 1998; De Antoni et al., 2005). Kinetochore localization of Mad1 is necessary for the checkpoint response to kinetochore detachment (Kastenmayer et al., 2005; Scott et al., 2005; Rodriguez-Bravo et al., 2014). Forced localization of Mad1 to kinetochores is sufficient to induce checkpoint arrest, even in the absence of attachment defects (Maldonado and Kapoor, 2011; Ballister et al., 2014; Kuijt et al., 2014). Thus Mad1 is a key marker of checkpoint activity at individual kinetochores, and its timely removal from kinetochores is an essential step toward checkpoint silencing.
Mad1/2 also localizes to nuclear pore complexes (NPCs; Chen et al., 1998; Campbell et al., 2001; Iouk et al., 2002). Evidence from human cells indicates that during interphase, NPC-localized Mad1/2 complexes generate a mitotic timing signal that imposes a minimum delay for anaphase initiation irrespective of progress toward biorientation (Rodriguez-Bravo et al., 2014). In yeast, NPC localization of Mad1 is required for the Kap121-transport inhibitory pathway (KTIP), a branch of the checkpoint that blocks nuclear import of Glc7, a protein phosphatase that opposes the checkpoint-activating kinases (Pinsky et al., 2009; Cairo et al., 2013). However, it is possible that the KTIP is not conserved in cells that undergo nuclear envelope breakdown (NEB) in mitosis, and Mad1 localization to NPCs may also have other, as-yet-undescribed functions. Here we investigate whether NPCs limit the availability of Mad1 to kinetochores.
A long-standing goal is to determine which events or conditions are directly monitored by the spindle checkpoint. A central challenge, therefore, is to understand how Mad1 is recruited to and removed from kinetochores and, in particular, to identify the modes of kinetochore–microtubule interaction that permit or exclude Mad1 binding to the kinetochore. Although this has been the subject of numerous experiments in many organisms (reviewed in Pinsky and Biggins, 2005; Khodjakov and Pines, 2010), it has been challenging to resolve the issue, in part because in most eukaryotic species, each kinetochore binds to multiple microtubules, allowing partial microtubule occupancy or simultaneous interactions with microtubules from both spindle poles. We avoided this complication by examining Saccharomyces cerevisiae cells, since each kinetochore binds to one microtubule in this yeast (Winey et al., 1995). Using both real-time fluorescence microscopy of living cells and quantitative fluorescence analysis of fixed cells, we examined the recruitment of Mad1 to a pair of detached sister centromeres after inducing de novo kinetochore assembly and observed its removal after kinetochore capture by microtubules. We also extended our analysis to examine Bub1 dynamics in the context of induced de novo kinetochore assembly. Our findings show the importance of lateral attachment and competition between different nuclear binding sites for Mad1 and Bub1 as key determinants of the spindle checkpoint response to unattached kinetochores.
RESULTS
Real-time analysis of kinetochore assembly and spindle checkpoint activation
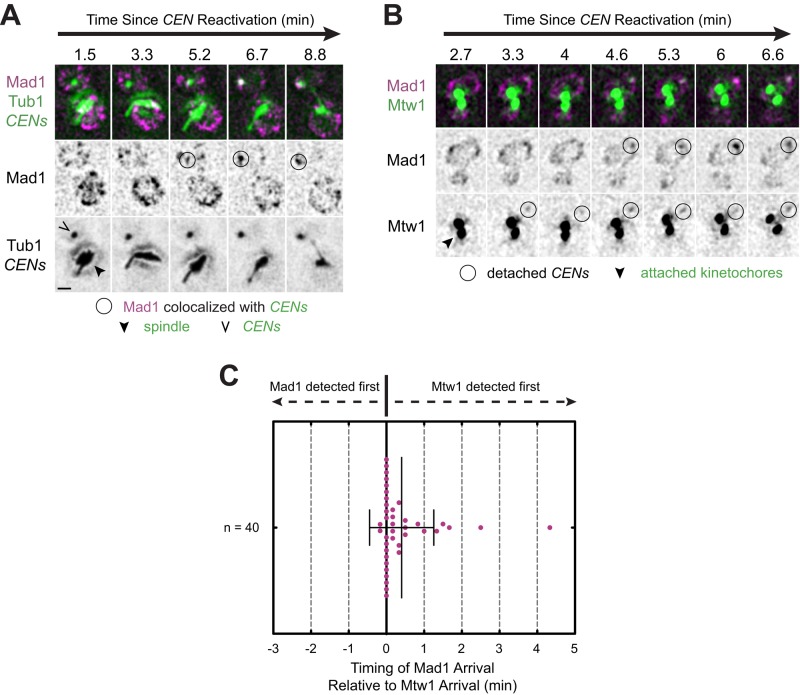
How rapidly the spindle checkpoint responds to kinetochore detachment after kinetochore assembly and in what manner Mad1 is initially recruited to kinetochores are not known. To observe Mad1 recruitment to detached kinetochores, we used a centromere reactivation assay (Tanaka et al., 2005). The assay allowed us to conditionally inactivate the centromeres of a single pair of sister chromatids (CEN3s on chromosome 3) in metaphase-arrested cells in order to remove the kinetochore proteins from the CEN3 centromeres and detach these chromosomes from the spindle. The centromeres could then be synchronously reactivated to assemble new kinetochores on the centromeric DNA (schematized in Supplemental Figure S1A). After centromere reactivation in cells expressing green fluorescent protein (GFP)–TUB1, marking the microtubules, and TetR-GFP, marking TetO-tagged centromeres, we observed capture events in approximately 42% of cells over 32 min of observation (Supplemental Figure S1B), with kinetics very similar to that published by Tanaka et al. (2005; Supplemental Figure S1C). Centromeres were captured on average 1300 nm (±650 SD, n = 85 centromeres) from the spindle and, after capture, translocated on microtubules at an average velocity of 970 nm/min (±610 SD, n = 84 centromeres), also in agreement with the results of Tanaka et al. (2005, 2007). Newly assembled kinetochores are expected to trigger the spindle checkpoint and recruit Mad1, since they are initially detached from the spindle (Chen et al., 1998; Gillett et al., 2004; Tanaka et al., 2005). To visualize the initial Mad1 recruitment to detached centromeres, we expressed a Mad1-3×mCherry fusion protein from the endogenous MAD1 locus (Supplemental Figure S1, D and E). After centromere reactivation, Mad1 accumulated at centromeres and then continuously colocalized with them as they moved within the nucleus (Figure 1A, and Supplemental Figure S2, A and B, and Supplemental Video S1).
FIGURE 1:
Mad1 recruitment to de novo assembled kinetochores. Cells bearing MET3pr-CDC20 and GAL1pr-CEN3-TetOs were grown for 3 h at 25°C in YP medium plus 2 mM methionine, 2% raffinose, and 2% galactose to synchronize cells in metaphase and inactivate CEN3. The cells were then released into synthetic medium plus 2 mM methionine containing 2% glucose at 25°C to reactivate CEN3 and then analyzed by epifluorescence microscopy, taking z-stacks every 10 s. Representative deconvolved, Gaussian-filtered MIPs are shown in A and B; scale bars, 1 μm. (A) Time series of Mad1-3×mCherry accumulation at a reactivated centromere in cells also expressing GFP-Tub1 and TetR-GFP (Supplemental Video S1). (B) Time series of Mad1-3×mCherry and Mtw1-3×GFP accumulation at a reactivated centromere (Supplemental Video S2). (C) Time lag between the first frame in which Mtw1 was detected and the first frame in which Mad1 was detected for at least five consecutive frames.
Mtw1 is a component of the microtubule-binding kinetochore protein network named KMN, after its key components in Caenorhabditis elegans, KNL-1 (Spc105 in yeast), the Mis12 complex (the Mtw1 or MIND complex in yeast), and the Ndc80 complex (Westermann et al., 2003; Cheeseman et al., 2006). Previous studies established that Mad1 requires outer kinetochore components for its localization to kinetochores (Gillett et al., 2004; Kim et al., 2012; London and Biggins, 2014; Moyle et al., 2014). Mtw1 requires DNA-binding kinetochore components, but not outer kinetochore components, for its own recruitment (Westermann et al., 2003; Cheeseman et al., 2004; Collins et al., 2005; Przewloka et al., 2007; Lampert et al., 2013). We therefore examined Mad1 recruitment with reference to the appearance of Mtw1 at kinetochores to estimate how rapidly Mad1 responds to detached kinetochores after their de novo assembly (Figure 1B, Supplemental Figure S2, C and D, and Supplemental Video S2). Mtw1-3×GFP foci were detectable 6 min, 35 s (±1 min, 53 s SD) after centromere reactivation, and Mad1 first appeared to continuously associate with detached centromeres 6 min, 55 s (±1 min, 53 s SD) after centromere reactivation (Supplemental Figure S2E). By comparing appearance times at individual centromeres, Mad1 was detected at the centromere after Mtw1 in 18 of 40 cells and before Mtw1 in only 2 of 40 cells (Figure 1C). In the remaining cells, Mad1 and Mtw1 were first detected in the same frame. On average, Mad1 appeared after Mtw1 with a delay of 24 s (±51 s SD; n = 40 centromeres; p < 0.01, Student's paired one-tailed t test; Figure 1C and Supplemental Figure S2E). Although we occasionally observed the intensities of both Mtw1 and Mad1 to increase concurrently, movement of the centromeres in the z-dimension might also account for concurrent changes in the fluorescence intensities of Mtw1 and Mad1. Thus we could not determine unambiguously whether individual Mad1 and Mtw1 proteins accumulate contemporaneously or whether Mad1 requires full incorporation of Mtw1 into the kinetochore before its own initial recruitment. Movement in the z-dimension, as well as the limited temporal resolution of our imaging scheme (10 s/frame) and incomplete fluorescent protein maturation, could also account for events in which Mtw1 appears at kinetochores earlier than, or simultaneously, with Mad1, which might lead to underestimation of the average delay between Mtw1 and Mad1 arrival. In any event, our measurements are consistent with the prediction that Mad1 responds to kinetochore detachment after Mtw1 assembles at the kinetochore.
Detached kinetochores compete with NPCs and other detached kinetochores for Mad1
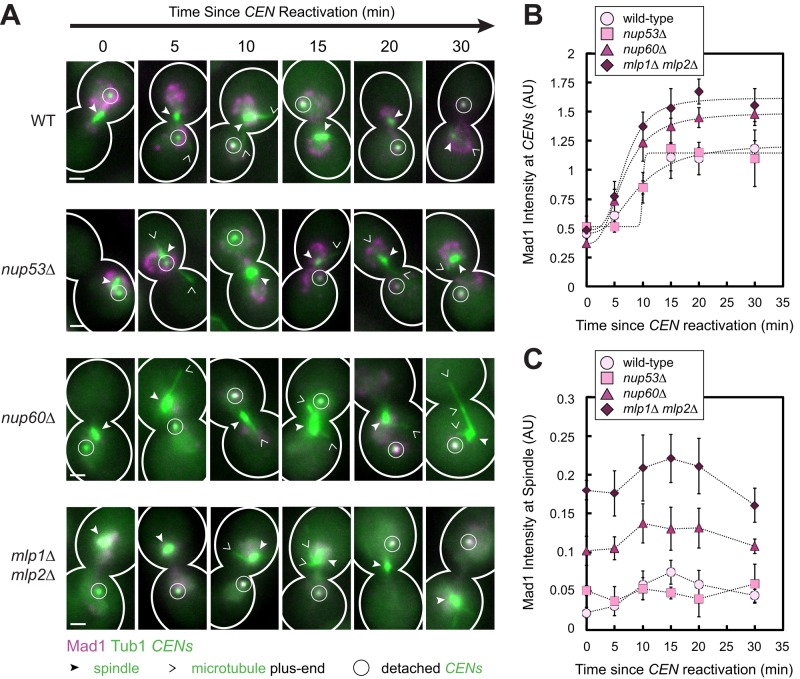
Because Mad1 localizes to NPCs and to detached kinetochores, these structures may compete to recruit Mad1 from the pool of unbound Mad1. Moreover, if NPCs sequester a sufficiently large fraction of the Mad1 molecules, they could limit the amount of free Mad1 available to bind to detached kinetochores. We therefore asked whether disrupting Mad1-binding sites at nuclear pores affects the amount of Mad1 localized to detached kinetochores. We found that it did. For this experiment, we measured the Mad1 intensity at detached sister centromeres in fixed cells at specified time points after centromere reactivation. In wild-type cells, Mad1 accumulated at reactivated sister centromeres until reaching a plateau after 15 min (Figure 2, A and B). Nup60 links Mlp1 and Mlp2 to the inner nuclear basket of the NPC. Deletion of Nup60 abolishes the NPC localization of the Mlps and Mad1/2, although a portion of the Mlps and Mad1/2 can remain localized to a spot on the nuclear envelope, called an Mlp focus, whose functional significance is unknown (Feuerbach et al., 2002; Scott et al., 2005). We found that deletion of NUP60 increased the total amount of Mad1 recruited to the newly assembled centromeres, resulting in a 35% increase in Mad1 intensity at centromeres at saturation compared with wild type (p < 0.01, extra sum-of-squares F test; Figure 2, A and B). Deletion of the genes encoding both the Mlps releases Mad1/2 from NPCs, as well as from Mlp foci (Scott et al., 2005). Cells lacking the Mlps exhibited a 55% increase in Mad1 intensity at centromeres compared with wild-type cells (p < 0.01, extra sum-of-squares F test; Figure 2, A and B). We also observed that significantly more Mad1 colocalized with spindles in cells lacking either Nup60 or the Mlps compared with wild-type cells at all time points, with a greater effect in mlp1Δ mlp2Δ cells than in nup60Δ cells (p < 0.01, Student's unpaired one-tailed t tests; Figure 2C). Attachment defects due to spindle abnormalities might account for the increased colocalization of Mad1 with spindles in our mlp1Δ mlp2Δ cells, since loss of Mlp2 is known to cause some spindle pole body (SPB) defects (Niepel et al., 2005). Nevertheless, the degree of Mad1 enrichment on reactivated sister centromeres correlated with the previously reported severity of disruption of Mad1-binding sites at the nuclear envelope in these different mutants (Scott et al., 2005), supporting the idea that both NPCs and Mlp foci sequester Mad1 from kinetochores.
FIGURE 2:
Effect of disrupting Mad1 binding to NPCs and Mlp foci on the amount of Mad1 recruited to a pair of detached sister centromeres. Wild-type, nup53Δ, nup60Δ, and mlp1Δ mlp2Δ cells expressing Mad1-3×mCherry, GFP-Tub1, and TetR-GFP and bearing MET3pr-CDC20 and GAL1pr-CEN3-TetOs were grown for 3 h at 25°C in YP medium plus 2 mM methionine, 2% raffinose, and 2% galactose to synchronize cells in metaphase and to inactivate CEN3. The cells were then released into synthetic medium plus 2 mM methionine containing 2% glucose at 25°C to reactivate CEN3. Cells were fixed after centromere reactivation at the indicated time points before imaging by epifluorescence microscopy. (A) Representative MIPs from each strain at each time point are shown. Scale bars, 1 μm. (B) Normalized Mad1 intensity at detached centromeres. Samples included at least 16 centromeres (mean of 108, median 113) for each strain at each time point. (C) Normalized Mad1 intensity at spindles. Samples included at least 153 spindles (mean of 454, median of 430) for each strain at each time point. Mean of three independent experiments. Error bars represent SEM. *p < 0.05.
In addition to causing a loss of the ability to recruit Mad1 to NPCs, deletion of Nup60 or the Mlps also disrupts the KTIP, a signaling pathway that inhibits the import of cargoes of the nuclear transport factor Kap121 through NPCs during mitosis and in response to nocodazole (Cairo et al., 2013). To determine whether the increased Mad1 recruitment to detached sister centromeres in these mutants was due to loss of the KTIP, we also examined the consequences of deleting Nup53, a Mad1-interacting nucleoporin that directly binds Kap121 and inhibits nuclear accumulation of Kap121 (Makhnevych et al., 2003). Of importance, in contrast to cells lacking Nup60 or the Mlps, Mad1 localization to NPCs is fundamentally undisturbed in the absence of Nup53 (Iouk et al., 2002; Scott et al., 2005; Figure 2A). We found that Mad1 recruitment to detached sister centromeres in cells lacking Nup53 was indistinguishable from recruitment in wild-type cells (Figure 2B). The increased Mad1 recruitment to detached centromeres in the absence of Nup60 or the Mlps, therefore, is not the result of losing the KTIP, because the pathway is defective in all three of the mutants. In addition, we further showed by immunoblotting that Mad1 is present in nup53Δ, nup60Δ, and mlp1Δ mlp2Δ cells at wild-type levels (Supplemental Figure S1E), indicating that our results are not due to differences in Mad1 expression or stability. Taken as a whole, our data suggest that NPCs sequester Mad1, preventing it from associating maximally with kinetochores or other Mad1-binding sites by limiting its availability.
The fact that releasing Mad1 from NPCs and Mlp foci increased Mad1 binding to centromeres in the reactivation assay implies that the concentration of free nuclear Mad1 is subsaturating for kinetochores, even when only one pair of chromosomes is detached from the spindle, as in the centromere reactivation assay. We reasoned that if this is true, kinetochores must also compete with one another for Mad1 binding. We tested this prediction in two ways. We repeated the analyses in cells bearing temperature-sensitive mutations that perturb kinetochore–microtubule attachments (Figure 3, A and C) and also in cells treated with the microtubule-destabilizing drug nocodazole to detach additional kinetochores (Figure 3, D and E).
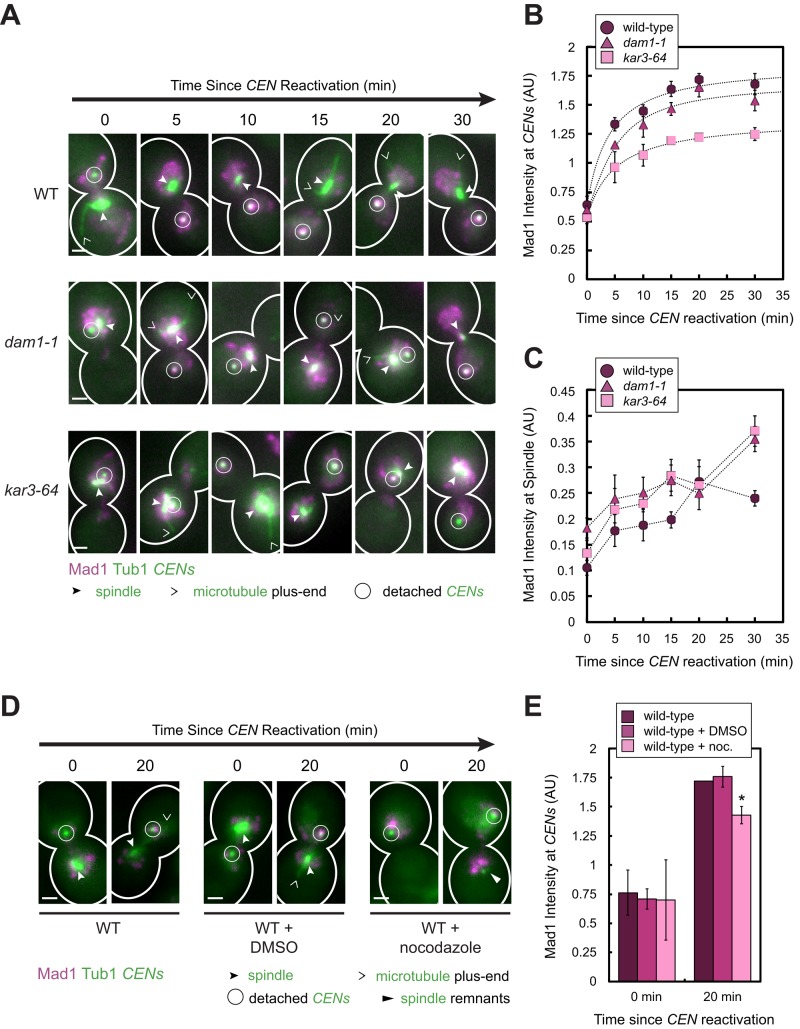
FIGURE 3:
Effect of increasing kinetochore attachment defects on the amount of Mad1 recruited to a single pair of detached sister centromeres. (A–C) Wild-type, dam1-1, and kar3-64 cells expressing Mad1-3×mCherry, GFP-Tub1, and TetR-GFP and bearing MET3pr-CDC20 and GAL1pr-CEN3-TetOs were grown for 2.25 h at 25°C in YP medium plus 2 mM methionine, 2% raffinose, and 2% galactose to synchronize cells in metaphase and to inactivate CEN3. The cells were shifted to 37°C, a restrictive temperature for both mutants, for 45 min and then released into synthetic medium plus 2 mM methionine containing 2% glucose at 37°C. Cells were fixed after centromere reactivation at the indicated time points before imaging by epifluorescence microscopy. (A) Representative MIPs from each strain at each time point. Scale bars, 1 μm. (B) Normalized Mad1 intensity at detached centromeres. Samples included at least 35 centromeres (mean of 94, median 95) for each strain at each time point. (C) Normalized Mad1 intensity at spindles. Samples included at least 174 spindle peaks (mean of 484, median of 458) for each strain at each time point. Mean of three independent experiments is shown in B and C. Error bars represent SEM. Results from these experiments also appear in Figure 6, E and F. (D, E) Wild-type cells were cultured as in A–C, except that the cells were synchronized in metaphase for 3 h at 25°C in the presence or absence of 15 μg/ml nocodazole and/or 1% DMSO. The cells were then released into synthetic medium plus 2 mM methionine containing 2% glucose at 25°C in the presence or absence of nocodazole and/or DMSO and then fixed and imaged as in A–C. (D) Representative MIPs from the strains at each time point. Scale bars, 1 μm. (E) Normalized Mad1 intensity at detached centromeres. Samples included at least 51 centromeres (mean of 125, median 95) for each strain at each time point. Error bars represent SEM. *p < 0.05.
Kar3 is a minus end–directed kinesin involved in lateral capture and kinetochore transport along the sides of microtubules, and Dam1 is the eponymous subunit of the Dam1 complex required for end-on attachment and microtubule depolymerization–driven kinetochore transport (Tanaka et al., 2005, 2007; Westermann et al., 2006). When either the kar3-64 and dam1-1 temperature-sensitive mutant was cultured for 45 min at 37°C, a restrictive temperature, we found that more Mad1 colocalized with spindles compared with wild-type cells at the time of centromere reactivation, consistent with an increased frequency of kinetochore–microtubule attachment defects in these mutants (p < 0.01, Student's unpaired one-tailed t tests; Figure 3, A and C). As predicted, less Mad1 accumulated at detached centromeres in these mutants, despite expressing Mad1 at a level indistinguishable from the level in the wild-type strain, as judged by immunoblotting (Supplemental Figure S1E). Centromeres in dam1-1 cells recruited 13% less Mad1 than wild-type cells, and centromeres in kar3-64 cells recruited 37% less Mad1 (p < 0.01, extra sum-of-squares F tests; Figure 3B).
Next we asked whether disruption of additional kinetochore attachments at a normal growth temperature by treatment with nocodazole also affected the amount of Mad1 recruited to reactivated centromeres. As expected, in the majority of the cells exposed to nocodazole for 3 h at 25°C before centromere reactivation, the detached centromere was the only visible green spot (Figure 3D, nocodazole-treated cells at 0 min), although in some cells, spindle remnants could be seen (Figure 3D, nocodazole-treated cells at 20 min). Mad1 also accumulated at one or a few bright spots in these cells, which likely correspond to unlabeled detached kinetochores (Figure 3D, nocodazole-treated cells; see the next section and Figure 4, A and B, and Supplemental Figure S3A). As with the temperature-sensitive mutants described earlier, we found that the amount of Mad1 recruited to the labeled centromeres 20 min after their reactivation was reduced by 17% in the cells treated with nocodazole compared with untreated cells (p < 0.01, Student's paired one-tailed t test), whereas dimethyl sulfoxide (DMSO) had no effect (Figure 3E). Taken together, these findings suggest that Mad1-binding sites at unattached kinetochores compete with one another, in addition to competing with NPCs, to recruit Mad1 from a limiting nucleoplasmic pool. We later discuss the implications of these results with respect to checkpoint sensitivity.
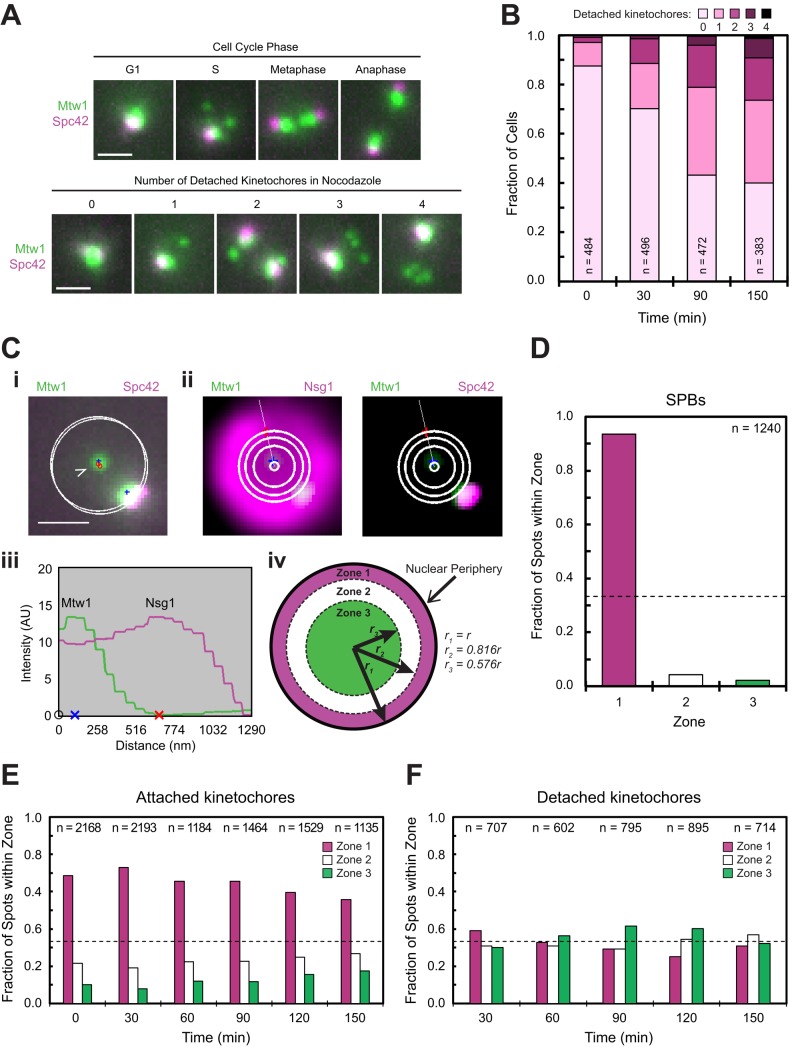
FIGURE 4:
Analysis of the subnuclear localization distribution of kinetochores detached using nocodazole. Asynchronously dividing cells were exposed to nocodazole for 0 to 150 min before fixation and imaging by epifluorescence microscopy. Scale bars, 1 μm. (A) Top, cells expressing Mtw1-3×GFP and Spc42-RFP before treatment with nocodazole (time zero). Bottom, same strain after treatment with nocodazole for 150 min. Representative MIPs. (B) Number of detached Mtw1 spots per cell at the time points indicated from the experiment in A. (C) Localization of spots in cells expressing Mtw1-3×GFP, Spc42-CFP, and Nsg1-mCherry. (i) Representative MIP of a cell containing two Mtw1 spots (blue pluses), one detached and one attached (open and closed arrows, respectively). White circles depict the automated fit to the Nsg1 fluorescence in same planes as the Mtw1 spots. (ii) Fit to Nsg1 fluorescence (left) and localization of the detached spot (right). Images are the sum of fluorescence in the peak-intensity plane of the spot of interest (blue plus) and the two adjacent planes after Gaussian filtering and background subtraction. White lines depict the trace used to measure distance of the spot from the center and the periphery. The three larger concentric circles delineate three subnuclear zones of equal area (see iv also). (iii) Fluorescence intensity profiles across the lines shown in ii. Blue x marks the Mtw1 centroid, and red x marks the nuclear boundary. (iv) Cartoon illustrating the mathematical relationship between the nuclear radius (r) and the boundaries of the three concentric subnuclear zones of equal area. (D–F) Subnuclear localization distributions of Spc42 spots in cells expressing Spc42-GFP and Nsg1-mCherry at time zero, before nocodazole addition (D) and Mtw1 spots that colocalized with Spc42 (E) or not (F) in cells expressing Mtw1-3×GFP, Spc42-CFP, and Nsg1-mCherry. Dotted lines at 33.3% in D–F represent the expected fraction of spots that should appear in each zone if the spots are randomly localized.
Mad1 does not physically link detached kinetochores to nuclear pore complexes
Because Mad1's NPC and kinetochore localization domains are encoded by domains on opposite ends of the primary amino acid sequence (Kastenmayer et al., 2005; Scott et al., 2005), Mad1 might potentially interact with these structures simultaneously. During interphase in fission yeast, kinetochores are anchored to the nuclear envelope independent of microtubules (Funabiki, 1993; Hou et al., 2012), and in budding yeast, Mlp1 and Mlp2 tether a subset of stress-inducible gene loci to NPCs (Tan-Wong et al., 2009; Bermejo et al., 2012). We therefore investigated whether the Mlps might similarly position budding yeast kinetochores that are detached from microtubules to the nuclear envelope by binding to Mad1.
During the normal cell cycle in budding yeast, detached kinetochores appear only briefly during S phase, after centromere replication and de novo kinetochore assembly (Kitamura et al., 2007). Consistent with this observation, when we fluorescently labeled Mtw1 and the SPB component Spc42 to use as references for assessing kinetochore attachment, we observed detached kinetochores in only 12% of asynchronously dividing cells, and then exclusively in cells with unduplicated SPBs (Figure 4, A and B, and Supplemental Figure S3A). However, detached kinetochores accumulated after depolymerization of microtubules with nocodazole, even in cells with duplicated SPBs (Figure 4, A and B, and Supplemental Figure S3A). To quantitatively analyze the spatial distribution of detached and attached kinetochores, we developed an algorithm to assign fluorescent foci to one of three concentric subnuclear zones of equal area, using Nsg1 as a marker for the nuclear periphery (Figure 4C and Supplemental Figure S3B; Materials and Methods; Dieppois et al., 2006; Taddei et al., 2006). We postulated that if Mad1 links detached kinetochores to NPCs, then the localization of detached kinetochores should be biased toward the nuclear periphery. However, using our algorithm and analyzing more than at least 600 kinetochores per condition, we found at all time points examined after nocodazole treatment that detached kinetochores were randomly distributed in the nucleus (Figure 4F), in contrast to attached kinetochores and SPBs, which were concentrated at the nuclear periphery (Figure 4, D and E). Moreover, we obtained concordant results when we measured the distribution of detached kinetochores manually (Supplemental Figure S3C), and also when we used Mtw1 fluorescence intensity, instead of Spc42 as a means of classifying kinetochores as detached (Supplemental Figure S4). Thus we found no evidence that Mad1 tethers detached kinetochores to the nuclear periphery, in agreement with our conclusion that Mad1-binding sites at the nuclear envelope compete with detached kinetochores to recruit Mad1 instead of simultaneously binding the same Mad1 molecules.
Mad1 disappears from sister centromeres upon their retrieval to the spindle, and its timely removal depends on Nup60
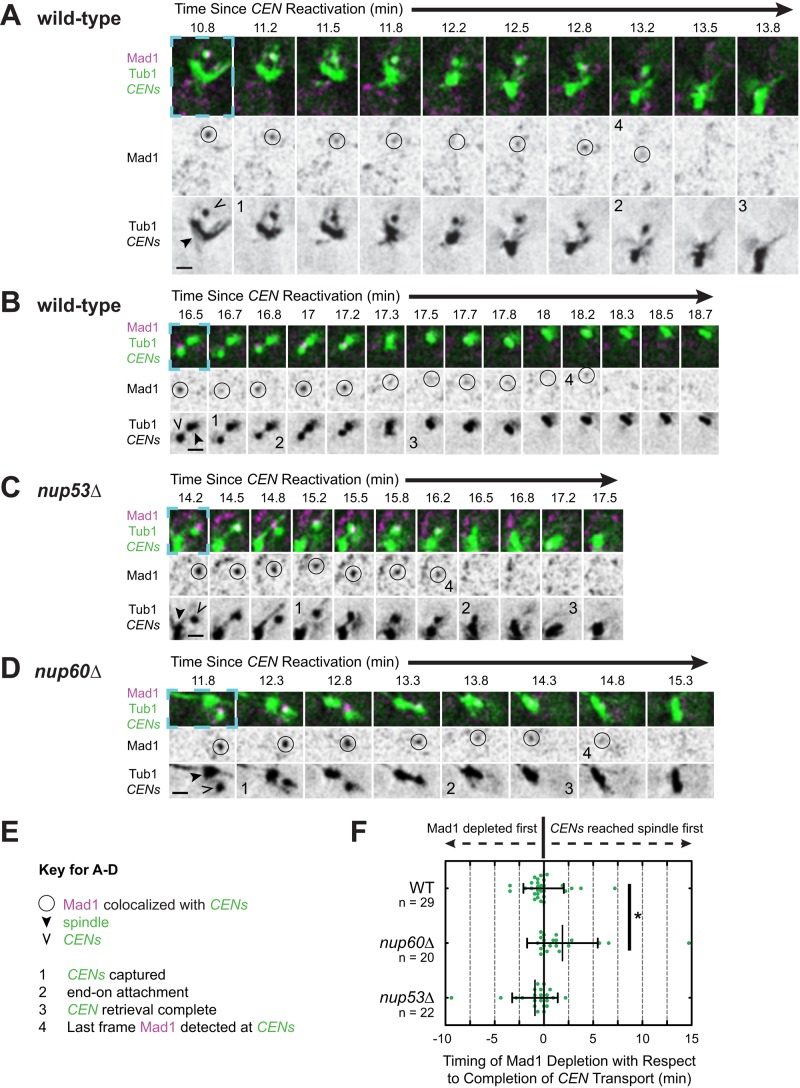
It is unclear whether the checkpoint distinguishes between kinetochore associations with the sides of microtubules (lateral attachments) and interactions with the microtubule plus end (end-on attachments). To determine which mode(s) of attachment satisfy the spindle checkpoint, we used live-cell imaging and evaluated the timing and manner of Mad1 removal after kinetochore capture. For this purpose, we observed cells expressing Mad1-3×mCherry, TetR-GFP, and GFP-Tub1 using real-time microscopy (Figure 5 and Supplemental Figure S5). Centromere capture was defined as when sister centromeres initiated a processive directional movement toward the spindle. Completion of capture was defined as when the fluorescence of the centromere could no longer be distinguished from the spindle fluorescence. In all cells examined, Mad1 was detected at the centromere before the capture event. Similarly, Mad1 disappeared from the centromere after the capture event, although the last frame at which Mad1 could be detected at sister centromeres varied from cell to cell (Figure 5F). In the majority of cells (18 of 29 cells), Mad1 disappeared from the centromere before it arrived at the spindle (Figure 5F). Because centromeres and microtubules were both tagged using GFP, we could not always observe the individual capturing microtubules and therefore could not discern the mode of attachment. However, on occasion, we could clearly resolve the capturing microtubule (Figure 5). We were also aided by kymograph and fluorescence intensity analysis (Supplemental Figures S5 and S6). In kymographs, the conversion from a lateral to an end-on attachment is accompanied by a change in the slope of the centromeres' changing position over time (events marked “2” in Figure 5 and Supplemental Figure S5) resulting from the different average velocities of Kar3-mediated lateral transport and Dam1-mediated end-on transport (600 and 1500 nm/min, respectively; Tanaka et al., 2007).
FIGURE 5:
Mad1 removal from sister centromeres after capture. Wild-type, nup53Δ, and nup60Δ cells expressing Mad1-3×mCherry, GFP-Tub1, and TetR-GFP and bearing MET3pr-CDC20 and GAL1pr-CEN3-TetOs were grown for 3 h at 25°C in YP medium plus 2 mM methionine, 2% raffinose, and 2% galactose to synchronize cells in metaphase and inactivate CEN3. The cells were then released into synthetic medium plus 2 mM methionine containing 2% glucose at 25°C to reactivate CEN3 and examined by live-cell epifluorescence microscopy, taking z-stacks every 10 s after centromere reactivation. Representative deconvolved, Gaussian-filtered MIPs are shown in A–D; scale bars, 1 μm. (A–D) Time series showing Mad1 disappearance from sister centromeres after capture. Note that the time intervals depicted vary across the strains in order to show key events. The blue dashed boxes surrounding the first frame of each time series highlight the corresponding regions of the expanded views shown in Supplemental Figure S5 and Supplemental Videos S3–S6. (E) Key for A–D. (F) Timing of Mad1's final disappearance from centromeres relative to the completion of capture. The completion of capture was defined as the first frame in which the centromeres could no longer be resolved from the spindle. Error bars represent SD. *p < 0.05.
In some cases, Mad1 was no longer detectable at the centromere soon after end-on transport began (Figure 5A, Supplemental Figures S5, A and B, and S6A, and Supplemental Video S3). In other cells, however, Mad1 clearly remained colocalized with the centromere until after the centromere arrived at the spindle (Figure 5B, Supplemental Figures S5, C and D, and S6B, and Supplemental Video S4). The mean velocity of sister centromere transport increased in 75.4% of cells after Mad1 depletion (n = 57 centromeres). The velocity increased from 530 (±620 SD, n = 62 centromeres) to 1280 nm/min (±1060 SD, n = 72 centromeres), which are similar to the velocities of lateral and end-on transport, respectively (Tanaka et al., 2007). This coincidence suggested that perhaps the establishment of an end-on attachment might be sufficient to license Mad1 removal from both sister centromeres. On average, however, Mad1's disappearance coincided with the completion of centromere capture (2 s before, n = 29 centromeres; Figure 3F). Because biorientation occurs rapidly after the completion of centromere retrieval (Tanaka et al., 2005), the average is more consistent with the idea that both sisters need to interact with microtubules before Mad1 is fully removed from them. However, there may be a kinetic delay between the event that licenses Mad1 removal (e.g., the establishment of a lateral or an end-on attachment) and the timing of Mad1 disappearance due to a requirement for modifications of the kinetochore that reduce Mad1 affinity.
If Mad1 is eliminated from captured kinetochores by a combination of exchange and modification of its binding sites, the concentration of free nuclear Mad1 could influence the kinetics of Mad1 disappearance. We asked, therefore, whether releasing Mad1 from NPCs influences the timing of Mad1 disappearance from sister centromeres after their capture by microtubules. Because of the increased localization of Mad1 to spindles in mlp1Δ mlp2Δ cells (Figure 2C), we limited our analysis to nup53Δ and nup60Δ cells and considered only the centromeres of cells in which Mad1 did not colocalize with the spindle before centromere capture. We found that the timing and manner of Mad1 disappearance in nup53Δ-negative control cells were indistinguishable from those in wild-type cells (Figure 5, C and F, Supplemental Figures S5, E and F, and S6C, and Supplemental Video S5). In contrast, Mad1's disappearance was delayed 1 min, 50 s on average in nup60Δ cells such that Mad1 continued to colocalize with centromeres until after they arrived at the spindle in the majority of cells (70%, n = 20 cells; Figure 5, D and F, Supplemental Figures S5, G and H, and S6D, and Supplemental Video S6). The consequences of NUP60 deletion in the reactivation assay were largely specific to Mad1 localization, since we failed to detect differences relative to wild-type cells in several other parameters that we measured after centromere reactivation, including the timing of capture after reactivation, the distance of centromeres from the spindle at the time of capture, and the duration and velocity of transport. Consistent with the delay in Mad1 removal, however, the mean distance from the spindle at the time of Mad1 disappearance was reduced in nup60Δ cells to 250 ± 370 nm (SD, n = 20 centromeres), compared with wild-type cells, in which Mad1 disappeared at a mean distance of 740 ± 790 nm (SD, n = 26 centromeres). Together these observations are consistent with our conclusion that NPCs are a sink for Mad1 localization that controls Mad1 association and dissociation from kinetochores.
Mad1 localization to sister centromeres is reduced by half after lateral attachment
Sister centromeres are retrieved to the spindle within 1 min, 49 s (±1 min, 21 s; n = 87 cells) of capture. During this time, they can switch from lateral to end-on attachment (Tanaka et al., 2005). Finally, they biorient soon after arrival at the spindle (Tanaka et al., 2005). We began our live-cell investigations assuming that Mad1 binding to the kinetochore reaches a new steady state within seconds of establishing an attachment. However, the variability in the events and timing associated with Mad1's disappearance, and the fact that Mad1 removal could be delayed by deletion of NUP60, indicated that this assumption might not be valid (Figure 5 and Supplemental Figures S5 and S6). Our results are consistent with the possibility that either a lateral or an end-on attachment might be a sufficient condition to license Mad1 removal from sisters after a kinetic delay. We were intrigued, however, that the intensity of Mad1 at sister centromeres diminished after their initial association with an individual microtubule (e.g., Figure 5A, Supplemental Figures S5, A and B, and S5, A and B, and Supplemental Video S3), a result expected if Mad1 is partially released after lateral attachment.
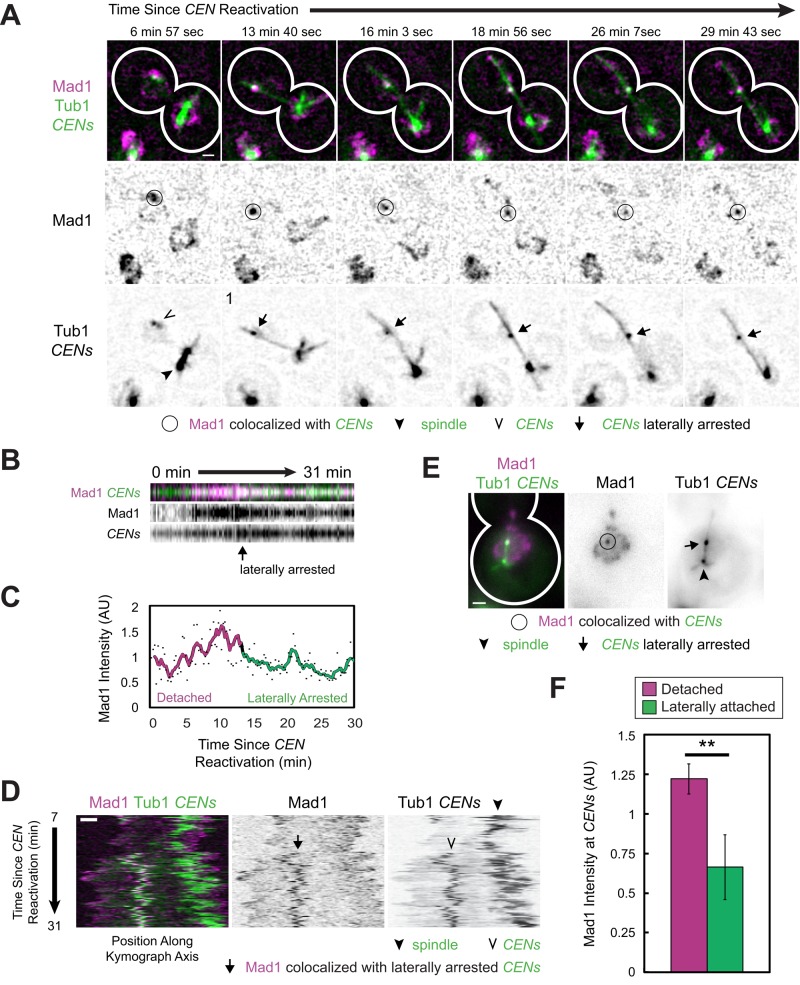
Minus end–directed lateral transport of kinetochores along microtubules is defective in kar3-64 cells at the restrictive temperature, and a majority of centromeres are transported toward the spindle pole via end-on attachment to the depolymerizing microtubule (Tanaka et al., 2007). However, a small subset of centromeres (<5%) associate laterally with microtubules but fail to subsequently form end-on attachments and translocate poleward, resulting in a laterally attached standstill phenotype (see Figure 8A of Tanaka et al., 2007). We exploited this phenotype in order to quantify the intensity of Mad1 at laterally attached kinetochores. We first observed the localization of Mad1 in kar3-64 cells at the restrictive temperature of 37°C using real-time microscopy in live cells. As expected, a minority of kinetochores arrested with lateral attachments to microtubules after capture in the absence of Kar3 function (Figure 6, A–C, and Supplemental Video S7). In such cells, the plus end of the associated microtubule underwent successive switches between polymerization and depolymerization, with the plus end depolymerizing back to the centromeres several times but failing to establish a stable end-on association (Figure 6D and Supplemental Video S7). Meanwhile, Mad1 remained localized to laterally attached centromeres throughout the observation period (Figure 6, A and D, and Supplemental Video S7), although we again noted diminution of Mad1 intensity after lateral attachment (Figure 6, B and C). We exploited our fixed-cell assay to quantitatively analyze Mad1 intensity in kar3-64 cells. At 15, 20, or 30 min after centromere reactivation, we classified centromeres as detached or associated with microtubules laterally (Figure 6E). Strikingly, we found that Mad1 at sister centromeres that were laterally associated with microtubules was reduced by 46% compared with fully detached sisters (p < 0.01, Student's paired one-tailed t test; Figure 6F). Thus, in kar3-64 cells, lateral attachment is sufficient to release about half of the bound Mad1 from sister centromeres.
FIGURE 6:
Mad1 localization to sister centromeres associated laterally with microtubules. (A–D) kar3-64 cells expressing Mad1-3×mCherry, GFP-Tub1, and TetR-GFP and bearing MET3pr-CDC20 and GAL1pr-CEN3-TetOs were grown for 2.25 h at 25°C in YP medium plus 2 mM methionine, 2% raffinose, and 2% galactose to synchronize cells in metaphase and inactivate CEN3. The cells were shifted to 37°C, a restrictive temperature for both mutants, for 45 min, and then released into synthetic medium plus 2 mM methionine containing 2% glucose at 37°C. Cells were then examined by live-cell epifluorescence microscopy at 37°C, taking z-stacks every 14.4 s after centromere reactivation. Deconvolved, Gaussian-filtered MIPs are shown in A (Supplemental Video S7). (A) Time series of a representative cell in which the sister centromeres became laterally arrested. (B) Kymograph of a series of images approximately centered on the centromeres in A. (C) Mad1 intensity at the centromeres in A and B normalized by taking the ratio of the mCherry signal to the associated GFP intensity (dots). A color-coded sliding window average of five frames is also shown. (D) Kymograph along the axis connecting the spindle to the centromere after capture. (E, F) Cells of the same strain as in A–D were fixed after centromere reactivation at 15, 20, and 30 min before imaging by epifluorescence microscopy. (Data are from the same experiments as in Figure 3.) (E) Representative MIPs of laterally attached centromeres identified in fixed kar3-64 cells. (F) Normalized Mad1 intensity at detached and laterally attached sister centromeres. Mean of three independent experiments. Samples were 222, 292, and 315 detached centromeres and 16, 20, and 29 laterally attached centromeres, respectively. Error bars represent SEM. **p < 0.01. Scale bars, 1 μm.
Behavior of Bub1 at spindles and detached, newly assembled kinetochores
In this study, we used Mad1 as a marker for spindle checkpoint activity at pairs of reactivated sister centromeres because it is a key downstream component of the spindle checkpoint signaling pathway. Because Bub1 and Mad2 act immediately upstream and downstream of Mad1, respectively, in the recruitment pathway that links spindle checkpoint proteins to kinetochores (Li and Benezra, 1996; Chen et al., 1998; Gillett et al., 2004; London and Biggins, 2014; Moyle et al., 2014), we also initiated studies of their dynamics. However, we were unable to study Mad2 using the centromere reactivation system because when we fused the 3×mCherry tag to the Mad2 N- or C-terminus using linkers of different lengths, cells exhibited sensitivity to benomyl, the fusion protein did not localize to NPCs in dividing cells or to kinetochores in nocodazole, and they showed a checkpoint defect (Hoyt et al., 1991; Li and Murray, 1991; Gillett et al., 2004). In contrast, we were able to generate a Bub1-3×mCherry fusion protein that retained spindle checkpoint function (Supplemental Figure S7A).
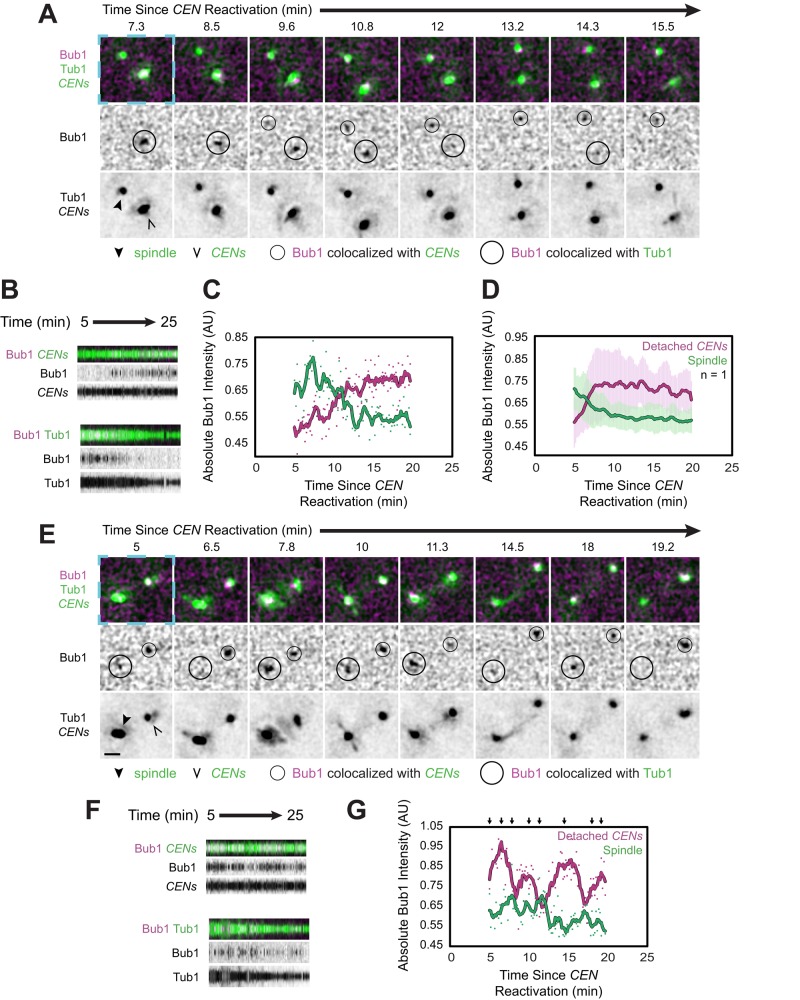
When we examined Bub1-3×mCherry localization using the centromere reactivation assay, we observed that some Bub1 already colocalized with the spindle at the start of the observation period (5 min after centromere reactivation) in 52% of cells (n = 185 cells; Figure 7, A–D, Supplemental Figure S7B, and Supplemental Video S8). This early localization is consistent with the observation that Bub1 is detected at spindles in metaphase during the normal cell cycle (Gillett et al., 2004). Bub1 was eventually detected at the reactivated centromere in 74% of cells that exhibited detached centromeres (n = 119 cells; Figure 7, A–D, and Supplemental Video S8). Along the same lines, among the cells in which Bub1 colocalized with the spindle at the start of our observation and was later detected at the centromeres (n = 42 cells), Bub1's accumulation at the centromeres was accompanied by its disappearance from the spindle in at least 43% of the cells (Figure 7, A–D, and Supplemental Video S8). Indeed, the amount of Bub1 lost from the spindle was approximately the same as that recruited to the centromeres on average, and the changes occurred with similar kinetics at both sites (Figure 7, A–D, and Supplemental Video S8). Although photobleaching might partially contribute to Bub1's disappearance from spindles, we also noted that the intensity of Bub1 at the spindle and the centromeres fluctuated over time, and these intensities varied inversely (Figure 7, E–G, Supplemental Figure S7, C and D, and Supplemental Video S9). Thus, as Bub1 accumulated at centromeres, it disappeared from the spindle, but, conversely, the amount of Bub1 that colocalized with the spindle occasionally increased, and this increase was accompanied by a reduction of Bub1 at the detached centromeres (Figure 7, E–G, Supplemental Figure S7, C and D, and Supplemental Video S9). Together these observations indicate that Bub1-binding sites associated with the spindle and the detached centromeres directly compete to recruit Bub1. Moreover, as with Mad1, the entire pool of available Bub1 is small enough to be completely bound by its binding sites at a single pair of sister centromeres.
FIGURE 7:
Bub1 localization to spindles and de novo assembled kinetochores. Cells expressing Bub1-3×mCherry, GFP-Tub1, and TetR-GFP and bearing MET3pr-CDC20 and GAL1pr-CEN3-TetOs were grown for 3 h at 25°C in YP medium plus 2 mM methionine, 2% raffinose, and 2% galactose to synchronize cells in metaphase and inactivate CEN3. The cells were then released into synthetic medium plus 2 mM methionine containing 2% glucose at 25°C to reactivate CEN3 and then analyzed by epifluorescence microscopy beginning 5 min later, taking z-stacks every 10 s. Representative deconvolved, Gaussian-filtered MIPs are shown in A and E. Scale bars, 1 μm. The blue dashed boxes surrounding the first frames of A and E highlight the corresponding regions of the expanded views shown in Supplemental Figure S7, B and C, and Supplemental Videos S8 and S9. (A) Time series showing the accumulation of Bub1-3×mCherry at the reactivated centromeres and its disappearance from the spindle, marked by GFP-Tub1 (Supplemental Video S8). (B) Kymographs from the cell in A, made from 15 × 15 boxes approximately centered on the centromeres (top) and 19 × 19 boxes approximately centered on the spindle (bottom). (C) Absolute (i.e., not normalized) intensity of Bub1 at the centromeres and spindle shown in A and B. (D) Absolute intensity of Bub1 at the centromeres and spindle, averaged across the cells in which Bub1 recruitment to the centromeres was accompanied by its disappearance from the spindle. Error bars represent SD. (E) Time series of a cell in which Bub1 alternately accumulated at the centromeres or the spindle and its accumulation at one site was accompanied by its disappearance from the other site (Supplemental Video S9). (F) Kymographs from the cell in E, made as in B. (G) Absolute intensity of Bub1 (not normalized) at the centromeres and spindle shown in D and E. Arrows above the graph refer mark the frames depicted in E. Color-coded sliding window averages of five frames are also shown.
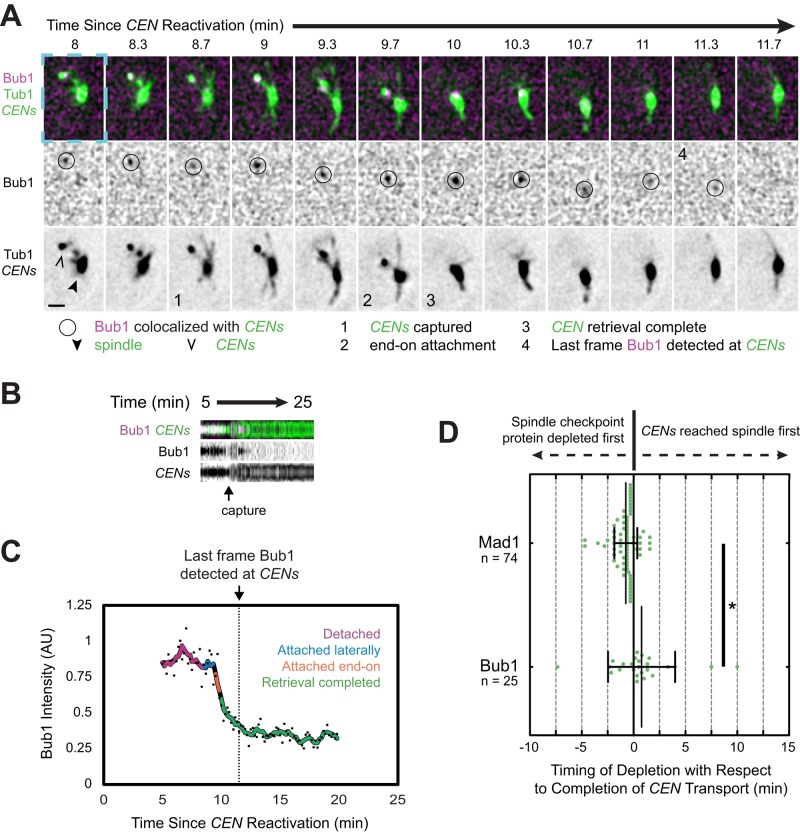
We sought to determine whether Bub1 expression is required for Mad1 localization in the context of de novo kinetochore assembly. We expressed Bub1 fused to an auxin-inducible degron tag (AID*) and the FLAG tag (Bub1-AID*-6×FLAG) at the endogenous BUB1 locus in a centromere reactivation strain also expressing OsTIR1-9×MYC and Mad1-3×mCherry (Nishimura et al., 2009; Morawska and Ulrich, 2013). As expected, Bub1 was degraded after treatment of the cells with 250 μM auxin indole-3-acetic acid (IAA), decreasing to 18% of the normal level after 60 min (Supplemental Figure S8A). When we reactivated centromeres in the cells after 60 min of treatment with 250 μM IAA, we found that although centromere capture and retrieval occurred normally, Mad1 recruitment to the centromeres was greatly diminished (Supplemental Figure S8, B–E, and Supplemental Video S11), consistent with the placement of Bub1 upstream of Mad1 in the kinetochore recruitment pathway. Moreover, when we examined the timing of the final disappearance of Bub1-3×mCherry from centromeres, we noted that Bub1 began to disappear after their capture and was no longer detectable on average 46 s after retrieval was complete, 1 min, 30 s later than the timing of Mad1's disappearance (Figure 8, Supplemental Figure S7D, and Supplemental Video S10). Indeed, Bub1 continued to colocalize with centromeres until after they were fully retrieved to the spindle in the majority of cells, in contrast to Mad1 (17 of 29 centromeres; Figure 8D). Together these observations provide further support the emerging model that Bub1 is the receptor for Mad1 at kinetochores (Brady and Hardwick, 2000; Sharp-Baker and Chen, 2001; Gillett et al., 2004; Kim et al., 2012; Yamagishi et al., 2012; London and Biggins, 2014; Moyle et al., 2014).
FIGURE 8:
Bub1 removal from sister centromeres after capture. Cells expressing Bub1-3×mCherry, GFP-Tub1, and TetR-GFP and bearing MET3pr-CDC20 and GAL1pr-CEN3-TetOs were treated as described in Figure 7. Representative deconvolved, Gaussian-filtered MIPs are shown in A. Scale bars, 1 μm. (A) Time series showing Bub1 disappearance from sister centromeres after capture. The blue dashed boxes surrounding the first frame highlights the corresponding regions of the expanded views shown in Supplemental Figure S7D and Supplemental Video S10. (B) Kymograph from the cell in A, made from 15 by 15 boxes approximately centered on the centromeres (top). (C) Bub1 intensity at the centromeres in A and B normalized by taking the ratio of the mCherry signal to the associated GFP intensity (dots). A color-coded sliding window average of five frames is also shown. (D) Timing of the final disappearance of Bub1-3×mCherry and Mad1-3×mCherry from centromeres relative to the completion of capture. The completion of capture was defined as the first frame in which the centromeres could no longer be resolved from the spindle. Error bars represent SD. *p < 0.05.
DISCUSSION
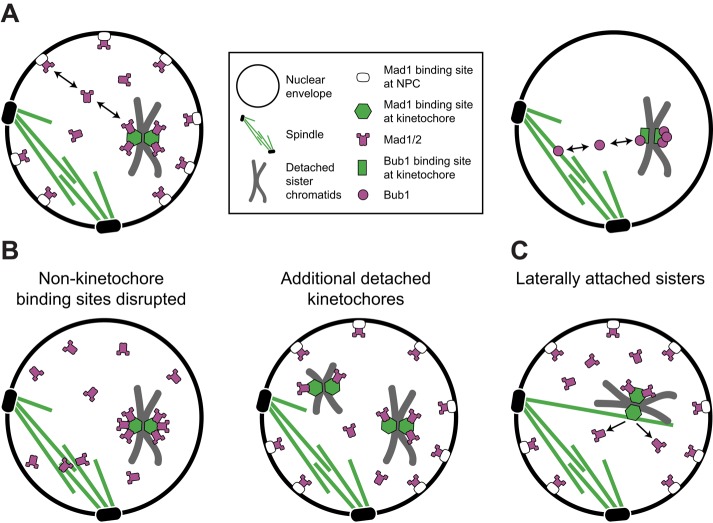
Here we used fluorescence imaging–based experiments to identify factors that control Mad1's kinetochore association and dissociation. Due to the binding of Mad1 to both kinetochores and NPCs, we considered that Mad1 might mediate a direct physical interaction between the two structures. However, we found no evidence that Mad1 mediates such an interaction (Figure 4 and Supplemental Figures S3 and S4). In addition, Mad1's recruitment to kinetochores does not require Mad1 localization to NPCs (Figures 2 and 5D and Supplemental Video S6; Scott et al., 2005, 2009; Cairo et al., 2013). To the contrary, more Mad1 accumulates at kinetochores when its binding partners at NPCs are eliminated (Figures 2 and 5D and Supplemental Video S6). In light of these observations, we favor a model in which Mad1 is recruited to detached kinetochores from a nucleoplasmic pool of free Mad1 such that NPCs and kinetochores compete to bind Mad1 (Figure 9A, left). Indeed, we found evidence that available Mad1 is limiting, which has important implications for the characteristics of the checkpoint response to kinetochore detachment.
FIGURE 9:
Model for spindle checkpoint surveillance of kinetochore–microtubule attachment. (A) After de novo kinetochore assembly, Mad1 is recruited to kinetochores from the nucleoplasmic pool of free Mad1 (left). The concentration of this unbound pool at steady state is limited by the abundance and affinities of Mad1 receptors at kinetochores and nonkinetochore binding sites, including NPCs and Mlp foci in budding yeast, or the spindle matrix in metazoan cells (left). Bub1-binding sites at detached kinetochores directly compete with Bub1-binding sites at the spindle for a limited amount of Bub1. The entire available pool can be recruited to a single pair of kinetochores (right). (B) Perturbations that disrupt Mad1 binding to nonkinetochore sites increase the pool of free Mad1, allowing Mad1 binding sites at detached kinetochores to be more fully occupied (left). Normally, however, sequestration of Mad1 to nonkinetochore binding sites restricts its availability, so detached kinetochores compete for a subsaturating pool of Mad1 (right). The number of Mad1 molecules bound to each detached kinetochore decreases as the number of detached kinetochores increases, making Mad1's kinetochore localization disproportionately affected by the first or last detached kinetochore. (C) Mad1 interprets the status of each sister kinetochore independently. Thus lateral attachment of sisters to a microtubule licenses Mad1 removal from the laterally attached sister. The precise timing of Mad1 disappearance, however, is variable, likely due to the kinetics of enzymatic modifications of the kinetochore that reduce Mad1 affinity after attachment.
Mad1 localization responds disproportionately to the first or last detached kinetochore
Because one detached chromosome is sufficient to delay anaphase in a variety of organisms, a long-standing view is that the spindle checkpoint response is “all or nothing” (Spencer and Hieter, 1992; Rieder, 1994; Li and Nicklas, 1995; Pangilinan and Spencer, 1996). How can such sensitivity be reconciled with quantitative studies indicating that the strength of the spindle checkpoint response varies with the number of detached kinetochores (Collin et al., 2013; Dick and Gerlich, 2013)? Here we discovered in budding yeast that the amount of Mad1 recruited to detached kinetochores is strongly affected by perturbations that alter the abundance of kinetochore and nonkinetochore binding sites for Mad1 (Figures 2 and 3). The concentration of free Mad1 is limited by its sequestration to NPCs to such a degree that the amount available is less than saturating for even a single pair of detached kinetochores (Figure 2). We also discovered that detached kinetochores compete with Bub1-binding sites associated with the spindle to recruit Bub1 and that the total pool of Bub1 is so small that it can be fully depleted by just two detached kinetochores (Figure 7 and Supplemental Figures S7, C and D, and 9A, right). If Bub1 is the sole receptor for Mad1/2 at the kinetochore (London and Biggins, 2014; Moyle et al., 2014; Supplemental Figure S8), then the low abundance of Bub1 would also impose a strict limit on the amount of Mad1/2 that can be recruited to kinetochores, on top of the limit imposed by sequestration of Mad1 to NPCs. Because detached kinetochores compete for access to Mad1 and Bub1, cells with more than one chromosome detached recruit less Mad1 per kinetochore than cells with only one detached chromosome (Figures 3 and 7B). This may explain why the strength of the kinetochore-derived checkpoint signal saturates at a low number of detached kinetochores per cell (see Figure 5G of Dick and Gerlich, 2013).
Although NPCs are disassembled during nuclear envelope breakdown (NEB) in cells that undergo open mitosis, the Mlp orthologues in Drosophila melanogaster and Aspergillus nidulans become enriched in the vicinity of the spindle during mitosis, and it has been suggested that this “spindle matrix” of Mlps corrals Mad1 to maintain a high local concentration of Mad1 near kinetochores, facilitating its recruitment to kinetochores (Qi et al., 2004; Lee et al., 2008; De Souza et al., 2009; Lince-Faria et al., 2009; Cairo et al., 2013). In contrast, we propose that in budding yeast, the Mlps limit the availability of Mad1/2 to kinetochores by sequestering Mad1/2 at NPCs.
The concentration of Mad1 and Mad2 strongly affects spindle checkpoint signaling and cell physiology. Mitotic duration, a measure of checkpoint strength, scales with the amount of Mad2 recruited to kinetochores, and the checkpoint is strongly inhibited by reductions of Mad1, and especially Mad2, expression (Barnhart et al., 2011; Collin et al., 2013; Heinrich et al., 2013). Their expression is reduced in many human tumors, and reducing expression of either or overexpressing Mad2 is sufficient to drive aneuploidy and tumorigenesis in animal models (reviewed in Schuyler et al., 2012). These observations are in accord with our proposal that the amount of Mad1/2 that can bind kinetochores is minimal, even under normal conditions (Figure 9, A and B). The distribution of Mad1/2 is disproportionately affected by the number of detached kinetochores in the context of one or only a few detachments precisely because of this limited availability (Figure 9, A and B). As a consequence, the spindle checkpoint will respond disproportionately to the first, or, even more vitally, the final detached chromosome as a cell approaches anaphase. Rather than being “all or nothing,” the spindle checkpoint of budding yeast is “nearly all or nothing.”
Lateral attachments satisfy the spindle checkpoint
Mad1 recruitment to kinetochores is key to controlling the metaphase–anaphase transition, but it is only part of the story. Failure to silence the checkpoint after biorientation results in chronic cell cycle arrest and loss of viability (Hardwick et al., 1996; Farr and Hoyt, 1998; Pinsky et al., 2009; Meadows, 2013). Because Mad1's kinetochore localization is sufficient to induce metaphase arrest (Maldonado and Kapoor, 2011; Ballister et al., 2014; Kuijt et al., 2014), we also investigated which event or condition triggers Mad1 removal from kinetochores. In metazoan cells, Mad1/2 may be removed in part by cytoplasmic dynein, a microtubule motor believed to move Mad1/2 from kinetochores toward spindle poles after kinetochores attach to microtubules (Howell et al., 2000, 2001; Chan et al., 2009). Budding yeast do not undergo NEB, however, and yeast dynein appears to be excluded from the nucleus and to not contribute to kinetochore attachment (Sheeman et al., 2003; Tanaka et al., 2007). A possible budding yeast protein that could provide this activity of metazoan dynein is Kar3, the only minus end–directed motor in the nucleus, which supports lateral attachment and poleward transport, like metazoan dynein (Rieder and Alexander, 1990; Tanaka et al., 2005, 2007; Yang et al., 2007). Yet Kar3 colocalizes with centromeres during retrieval (Tanaka et al., 2007), and we never observed Mad1 moving toward the spindle poles ahead of centromere transport (Figure 5). Thus it seems unlikely that Kar3 strips Mad1 from kinetochores.
In our real-time imaging studies, we noted an unanticipated variation in the timing of Mad1 removal from centromeres consistent with a kinetic delay (Figure 5). This was also true of Bub1 (Figure 8). Because removal of Mad1 (and Bub1) might require enzymatic removal of checkpoint-activating phosphorylations by protein phosphatase I (Lesage et al., 2011; Funabiki and Wynne, 2013; Primorac et al., 2013; London and Biggins, 2014), we postulated that the disappearance of Mad1 from kinetochores after the establishment of a satisfactory attachment might depend on both the turnover of Mad1 at kinetochores and the kinetics of such enzymatic modifications that reduce the affinity of Mad1 for kinetochores. Bearing this in mind, we sought to disrupt mechanical transitions after capture, reasoning that by arresting centromeres at an intermediate stage of retrieval, enzymatic alterations of the kinetochore might reach a steady state. In that case, the amount of Mad1 detected at the centromeres should more closely report on the suitability of the attachment with respect to the spindle checkpoint. To this end, we were able to observe kinetochores arrested with lateral attachments, in an organism in which each kinetochore only interacts with one microtubule, for as long as 17 min by exploiting the kar3-64 mutant (Figure 6; Tanaka et al., 2007). This mutant afforded us the ability to directly observe that lateral attachments cause Mad1 to dissociate from kinetochores.
Many studies have examined whether the spindle checkpoint monitors attachment to microtubules, tension between sister bioriented kinetochores, or both (McIntosh, 1991; Li and Nicklas, 1995; Rieder et al., 1995; Pinsky and Biggins, 2005) but little attention has been paid to whether the checkpoint distinguishes lateral attachments, which are the initial mode of kinetochore–microtubule association, from end-on attachments (Merdes and De Mey, 1990; Rieder and Alexander, 1990; Tanaka et al., 2005; Gachet et al., 2008). An unusual budding yeast mutant, dam1-765, fails to delay anaphase or recruit Mad1 to spindles despite indirect evidence that a subset of its kinetochores may be uncoupled from microtubule ends during metaphase (Shimogawa et al., 2006, 2010). However, whether kinetochore attachments in dam1-765 yeast are functionally equivalent to the lateral attachments formed initially upon capture is not clear (Kitamura et al., 2007; Shimogawa et al., 2010). Another study in fission yeast examined whether sister kinetochores are relieved of Mad2 upon lateral capture but failed to detect its disappearance from kinetochores until after retrieval to the spindle was complete (Saitoh et al., 2008). However, our observations suggest that Mad1/2 removal might be kinetically delayed after attachment (Figure 5). Moreover, because three to four microtubules bind to each fission yeast kinetochore (Ding et al., 1993), attachment of sister kinetochores to just one microtubule in this organism may result in an effect too subtle to detect.
In our studies, we were able to measure Mad1 levels at two sister kinetochores bound to one microtubule in the kar3-64 mutant and found that amount of Mad1 at laterally arrested centromeres was half that of fully detached sister centromeres (Figure 6E). Although we cannot exclude the possibility that the Mad1 we detected was distributed proportionally between both sisters due to the subdiffraction colocalization of sister kinetochores under these conditions, we suggest that a likely possibility is that Mad1 is eliminated specifically from the one sister centromere that interacts directly with the capturing microtubule. Our data support a model in which the spindle checkpoint interprets the attachment status of each sister kinetochore independently and lateral association to a microtubule suffices to license Mad1 removal from the attached sister (Figure 9C). Moreover, because laterally attached centromeres lack tension, the data strongly support the possibility that the primary defect monitored by Mad1 in budding yeast is attachment.
MATERIALS AND METHODS
Strain culture and construction
All yeast strains were W303 derivatives (Supplemental Table S1). We created haploid strains bearing multiple genomic modifications by mating and sporulation. To construct an integrating vector for tagging Mad1 with 3×mCherry, we amplified a 3′ fragment of the MAD1 open reading frame (ORF) corresponding to nucleotides 1593–2250 from yeast genomic DNA and ligated the fragment into pRS305::3×mCherry (Markus et al., 2009) between the SalI and BamHI restriction sites, creating pDD2405 (pRS305::MAD1(1593-2250)-3×mCherry). We integrated the plasmid by linearizing it with SnaBI restriction enzyme. To tag Bub1 with 3×mCherry, we amplified a 3′ fragment of the BUB1 ORF (nucleotides 255–3066) from yeast genomic DNA and inserted it into pRS305::3×mCherry, as in the foregoing, creating pDD2447 (pRS305::BUB1(255-3066)-3×mCherry). We integrated the plasmid by linearizing it with HindIII. We confirmed that Mad1-3×mCherry and Bub1-3×mCherry retain spindle checkpoint function by comparing the growth of cells expressing the fusion protein to wild-type cells and mad1Δ yeast (Supplemental Figures S1D and S7A). Whereas mad1Δ and bub1Δ yeast grow poorly in the presence of benomyl due to a failure to arrest in metaphase despite microtubule destabilization, the growth of MAD1-3×mCherry and BUB1-3×mCherry yeast is indistinguishable from that of wild type (Supplemental Figures S1D and S7A; Hoyt et al., 1991; Li and Murray, 1991). We constructed other fluorescent protein fusions and gene deletions in diploid yeast using standard methods of PCR product-mediated recombination (Longtine et al., 1998). All other genetic constructs were derived from strains described previously by others.
We routinely cultured strains bearing GAL1pr-CEN3-TetR and MET3pr-CDC20 in synthetic minimal medium lacking methionine supplemented with 2% dextrose and essential nutrients at 25°C. To prevent accumulation of the red pigment P-ribosylamino imidazole, strains bearing pRS412 were cultured using synthetic medium lacking adenine supplemented with 2% dextrose and essential nutrients at 25°C. Otherwise, yeast were routinely grown using yeast extract/peptone (YP) medium with 2% dextrose at 25°C. To prepare benomyl plates, we made fresh benomyl stocks at 10 mg/ml in DMSO. The final concentration of DMSO in benomyl and negative control plates was 0.2% in YP agarose plates with 2% dextrose. For serial dilution assays, we grew yeast overnight to saturation, adjusted each strain to OD600 = 1, and then made 10-fold serial dilutions, spotting 4 μl of each onto agar plates.
Epifluorescence microscopy
For cell fixation, we suspended cell pellets in 4% paraformaldehyde plus 3.4% sucrose, fixing for 10 min at room temperature, before washing with three times with 1 ml of 100 mM potassium phosphate, pH 7.5, plus 1.2 M sorbitol. We stored fixed cells in phosphate/sorbitol buffer at 4°C and imaged within 48 h. For both live- and fixed-cell imaging, we allowed the cells to settle and adhere to concanavalin A (ConA)–coated, 25-mm round #1.5 glass coverslips (Warner Instruments, Hamden, CT) for 7 min before washing four times with 1 ml of wash medium while observing the cells in bright field in order to adjust the density of cells. For fixed cells, we used phosphate-buffered saline, pH 7.4, at room temperature for washes and imaging. For live-cell imaging, washes are described later.
For 25°C and fixed-cell imaging, we used a Nikon Eclipse Ti microscope (Nikon Instruments, Melville, NY) controlled by MetaMorph (Molecular Devices, Sunnyvale, CA) equipped with a Plan Apo VC 100×/1.4 Oil OFN25 DIC N2 objective (with Type NF immersion oil, Nikon), a MOV-2000 piezo stage (Applied Scientific Instrumentation, Eugene, OR), a Perfect Focus System (Nikon), a temperature-controlled enclosure system (InVivo Scientific, St. Louis, MO), and a Neo sCMOS camera (Andor Technology, South Windsor, CT; 65-nm effective pixel size). We used the SPECTRA X Light Engine (Lumencor, Beaverton, OR) for excitation with a 470/22-nm single-band bandpass excitation filter for GFP, a 575/25-nm single-band bandpass excitation filter for mCherry, and a 524/628-nm dual-band bandpass filter for GFP/mCherry emission (Brightline; Semrock, Lake Forest, IL).
For 37°C imaging, we used an Olympus IX81 microscope (Olympus America, Center Valley, PA) controlled by MetaMorph equipped with a 100×/1.40 PlanApo Oil ∞/0.17 objective, a WeatherStation temperature controller (PrecisionControl, Farmington Hills, MI), and an ORCA-R2 camera (Hamamatsu Photonics, Hamamatsu, Japan; 64.5-nm effective pixel size). We used Type 37LDF immersion oil (Cargille Labs, Cedar Grove, NJ). Excitation light was attenuated using neutral density filters. For GFP, we used a 488-nm argon-ion laser (Melles Griot, Albuquerque, NM) with an HQ 480/40-nm excitation filter and Q 505 long-pass and HQ 535/50-nm emission filters (Chroma Technology, Bellows Falls, VT). For mCherry, we used a 561-nm diode-pumped solid-state laser (Melles Griot) with HQ 565 nm long-pass and HQ 620/60-nm emission filters (Chroma Technology).
Centromere reactivation assay
The centromere reactivation assay was described previously (Supplemental Figure S1; Tanaka et al., 2005). Specifically, we grew strains to mid logarithmic growth phase in synthetic methionine-dropout medium plus 2% raffinose before inoculating 2-ml cultures of cells to 0.2 OD600 unit/ml of methionine- and galactose-containing medium (YP, 2% raffinose, 2% galactose, and 2 mM methionine) to arrest cells in metaphase and detach CEN3s. We allowed the strains to grow for 3 h at 25°C, or for 2.25 h at 25°C and then for 45 min at 37°C, before resuspending them in synthetic tryptophan-dropout medium, 2% raffinose, 2% galactose, and 2 mM methionine, and then prepared cells for imaging. For live imaging, we concentrated the cells to 100 μl and transferred 50 μl of the cells to ConA-coated coverglass to adhere (described earlier). We washed the cells using synthetic tryptophan-dropout medium plus 2 mM methionine before reactivating the centromeres with 1 ml of synthetic tryptophan-dropout medium and 2% dextrose plus 2 mM methionine. Movies were acquired by taking three images at 0.7-μm spacing centered on a medial focal plane, alternating between red and green acquisitions. For experiments at 25°C, stacks were acquired every 10 s. For experiments at 37°C, stacks were acquired every 14.4 s. For the auxin-inducible degron system, we freshly prepared a 500 mM stock of IAA in DMSO. After culturing cells in methionine- and galactose-containing medium for 2.5 h at 25°C, we treated them with 250 μM IAA for the times specified in the text, and we included 250 μM IAA in the wash and centromere reactivation media.
For fixed-cell imaging after centromere reactivation, we washed live cells twice with 500 μl of synthetic tryptophan-dropout medium plus 2 mM methionine and then reactivated the centromeres by resuspending the cells in 1 ml of synthetic tryptophan-dropout medium and 2% dextrose plus 2 mM methionine before pelleting the cells and fixing them at the appropriate time point. For time zero, cells were fixed immediately after washing. The cells were maintained at 25 or 37°C using a water bath, as appropriate, until fixation. Images of fixed cells were acquired by taking 27–29 images at 0.2-μm spacing centered on a medial focal plane.
Image processing
We used ImageJ (National Institute of Health, Bethesda, MD) for image cropping, setting intensity ranges, maximum intensity projections (MIPs), kymographs, time stamps, and scale bars. We used Illustrator CS6 (Adobe Systems, San Jose, CA) for all other image annotation. We used MATLAB (MathWorks, Natick, MA) for programmatic image analysis. For automated spot and nucleus identification, we applied Laplacian of Gaussian filters and thresholding. For automated centroid calculation, intensity measurements, and localization of nuclear boundaries, we applied a Gaussian filter to the raw image data. These transformations were applied to obtain the data presented in Figures 2, B and C, 3, B and C, 4, C and F, and 6E and Supplemental Figures S3B and S4. We created a custom MetaMorph macro to perform nearest-neighbors deconvolution on the z-stack from each time point (settings: filter size, 9; scaling factor, 0.97; result scale, 25; suppress noise; λ = 507 for GFP and λ = 610 for mCherry) and then convert the resulting stacks into a MIP series. We smoothed the resulting MIPs in ImageJ with a Gaussian filter (radius, 1 pixel) and normalized the series to adjust for photobleaching. These transformations were applied to all real-time imaging data (images in Figures 1 and 5–8, Supplemental Figures S2, S5, S7, and S8, and all videos). All kymographs were generated in ImageJ using the straight-line and reslice stack tools (settings: output spacing, 1 pixel; slice count, 15; and avoid interpolation). We present SD projections of the resulting 15-kymograph stacks to distinguish pixels with large variations in intensity (Figures 6, B and D, 7, B and F, and 8B and Supplemental Figures S2, B, D, and S5, B, D, F, and H). We made kymographs of aligned centromere spots symmetrical in the space dimension by averaging them with their mirror images (Figures 6B, 7, B and F, and 8B and Supplemental Figure S2, B and D). All nondeconvolved z-stack images are shown as MIPs, except Figure 4Cii (Figures 2A, 3, A and D, 4A, and 6E). Minimum and maximum intensities were set after all other transformations and adjusted to the same values for all images within a given experiment before finally converting the 16-bit images to 8-bit.
Quantitative analysis of subnuclear localization and fluorescence intensities
To detach kinetochores by microtubule depolymerization in liquid cultures, we treated asynchronously dividing cells with 15 μg/ml nocodazole (final) from a 100× stock dissolved in DMSO for the indicated times. Cells at time zero were not treated with nocodazole. For comparison of GAL gene cluster enrichment at the nuclear periphery across experimental conditions (Supplemental Figure S3B), asynchronously dividing cells bearing GAL-LacOs and expressing LacI-GFP and Nsg1-mCherry were cultured in medium containing raffinose until log phase and then switched to medium containing the indicated carbon sources for 2 h before fixation and imaging by epifluorescence microscopy, as in Green et al. (2012).
To analyze the subnuclear distribution of fluorescent spots in fixed cells in a semiautomated manner, we created custom routines using MATLAB. The program identifies all fluorescent spots in all planes, and then, for each spot, the program 1) localizes the center of mass of the spot within its plane of maximal intensity, 2) calculates the center of the nucleus in the same plane with reference to a marker for the nuclear periphery (Nsg1-mCherry), 3) identifies the location of the nuclear boundary relative to the spot using the same marker, and 4) assigns the spot to one of three concentric subnuclear zones of equal area, as in Taddei et al. (2006) and Dieppois et al. (2006) (Figure 4D and Supplemental Figure S3B). We tested our localization accuracy by localizing nuclear features with known spatial distributions as controls. When we quantified the distribution Spc42, which is at the outer plane of the nuclear envelope, nearly 94% of Spc42 spots were appropriately assigned to outermost nuclear zone (Figure 4D; Jaspersen and Winey, 2004). In the presence of nocodazole, attached kinetochores cluster in close proximity to SPBs via interactions with short microtubules that persist in the presence of the drug. As expected, attached kinetochores were also disproportionately localized to the outermost zone, even after nocodazole treatment (Figure 4E). Finally, we examined the localization of the GAL gene cluster under various conditions using our algorithm and obtained results in close agreement with those of Cabal et al. (2006) and Green et al. (2012) (Supplemental Figure S3B). We concluded that the algorithm can accurately detect the enrichment of objects near the nuclear envelope and is sensitive to localization changes resulting from experimental perturbations.
For manual localization of spots (Supplemental Figure S3C), raw image stacks were analyzed in ImageJ. We first determined the coordinates of the peak-intensity pixel of a spot, then assigned the spot to a subnuclear zone by approximating the distance to the nearest nuclear boundary and the diameter of the nucleus along the same line. The nuclear boundaries were defined as the Nsg1 peak-intensity pixels at either extreme of the nucleus along the line.
We created variants of our localization program to allow user-based classification of spots (e.g., classification of kinetochores as attached or detached by reference to Spc42; Figures 2, B and C, 3, B and C, 4, C, E, and F, and 6F) and/or to calculate the intensities of spots (Figures 2, B and C, 3, B and C, and 6 and Supplemental Figure S4). These programs are available in the Supplemental Material. For quantification of Mad1 intensities at detached CEN3s, spindles, or laterally attached CEN3s (Figures 2, B and C, 3, B and C, and 6F), we measured the mCherry and GFP intensities in the region surrounding the GFP peaks in the maximum intensity plane using the signal and background masks depicted in Supplemental Figure S3D. After background subtraction, we calculated the normalized Mad1 intensity by dividing the mCherry signal by the GFP signal.
To classify Mtw1 spots as detached based on their fluorescence intensity (Supplemental Figure S4), we estimated an appropriate intensity threshold. Mtw1 spots at time zero exhibited a mean intensity of 871 AU (±553 SD, n = 2611 spots), and fitting the data to a Gaussian function gave a distribution centered on 719 AU (±576 SD, R2 = 0.93; Supplemental Figure S4D). Treatment with nocodazole broadened the distribution over time (Supplemental Figure S4D), as microtubule instability causes spindles to collapse or separate into halves containing a random subset of kinetochores (Figure 4A). A distinct population of dim Mtw1 spots accumulated in response to nocodazole that is likely correspond to individual detached kinetochores of unreplicated chromosomes and pairs of detached sister kinetochores (Supplemental Figure S4D). We fitted the intensity histograms in Supplemental Figure S4D for times after nocodazole treatment to the sum of an exponential function and a Gaussian function in order to approximate the center of the peak at the left (not depicted). We averaged the Gaussian-fit centers across the five time points after treatment with nocodazole and obtained a mean of 82.4 AU (±36.2 SD) for the peak. Using this information, we selected 150 AU, approximately 2 SDs above the mean, as the threshold for “dim” kinetochores (Supplemental Figure S4, E and F).
Statistical analysis
We fitted data for Mad1 accumulation at centromeres to the sigmoidal binding equation, (Bmin − Bmax)/(1 + e(time-halftime)/rate) + Bmax, and made statistical comparisons of the curve-fitting results using the extra sum-of-squares F test in GraphPad Prism (GraphPad Software, San Diego, CA; Figures 2B and 3B). For comparisons of Mad1 intensities on spindles in mutants versus wild type at individual time points, we used the Student's t test (unpaired, one-tailed) for each of three independent experiments (Figures 2C and 3C) and combined p values across independent experiments using Fisher's method. For comparison of GAL gene cluster enrichment at the nuclear periphery across experimental conditions (Supplemental Figure S3B), we used the one-tailed Fisher's exact test for each comparison from each of three independent experiments and combined p values across independent experiments using Fisher's method. For comparison of the timing of Mad1 removal from centromeres in different strains, we used the Student's t test (unpaired, one-tailed; Figure 5F). To compare the timing of Bub1 removal to Mad1 removal, we used the Student's t test (one-tailed) with Welch's correction (Figure 8D). For comparison of Mad1 intensity at detached kinetochores to laterally attached kinetochores, we used the Student's t test (paired, one-tailed; Figure 6F). For all experiments subjected to multiple-hypothesis testing, we corrected the p values with the Holm–Bonferroni method using the baseline significance threshold (α) of 0.05. We report significant results as either p < 0.05 or p < 0.01.
Immunoblotting
We arrested strains in metaphase with their centromeres detached as in the centromere reactivation experiments at 25 or 37°C, except for an Nsg1-mCherry control strain that we grew to mid log phase in YP/dextrose. We then prepared whole-cell extracts by trichloroacetic acid precipitation, loaded extracts onto a 10% SDS–PAGE gel, and transferred them to a 0.45-μm nitrocellulose membrane that we blocked for 30 min with 2% milk Tris-buffered saline (pH 7.5) plus 0.05% Tween 20 (TBS-T). To obtain a specific signal with polyclonal anti-RFP (Rockland Immunochemicals, Gibertsville, PA), it was necessary to pre-deplete the antibody using an acetone powder of yeast from an isogenic strain lacking Mad1-3×mCherry. We flash froze a cell pellet in liquid N2, pulverized the cells using a 6870 Freezer/Mill (SPEX SamplePrep, Metuchen, NJ), and resuspended the powder in 10 volumes of −20°C acetone, precipitating on ice for 1 h before recovering the powder by centrifugation at 3000 × g for 5 min. The powder was dried by evaporation and stored in the dark at room temperature. We diluted anti-RFP 1:200 into 2% milk TBS-T containing 10 mg/ml yeast acetone powder, adsorbing the nonspecific antibodies overnight at 4°C on a rotating agitator. We then recovered the supernatant after centrifugation at 3000 × g for 10 min. We first incubated the nitrocellulose membrane with the pre-depleted anti-RFP supernatant at 4°C overnight and then incubated the membrane with 1:15,000 anti-Pgk1 (Molecular Probes, Carlsbad, CA) in 2% milk TBS-T for 1 h at room temperature. Between antibodies, we washed the membrane three times for 10 min each using TBS-T at room temperature. Detection was performed using fluorescent secondary antibodies and imaged using the Odyssey system (LI-COR Biosciences, Lincoln, NE).
Supplementary Material
Acknowledgments
We thank Joshua Johnson for assistance with unpublished experiments related to this study; Connie Peng, Jeff Woodruff, Ryan Joyner, Aaron Welch, Adrianne Pigula, Itziar Ibarlucea-Benitez, and all the members of the Barnes and Drubin labs for support and helpful discussions; Elisa Dutz for MATLAB code; the labs of Karsten Weis, Tomo Tanaka, Tim Huffaker, Sue Biggins, and David Lydall for yeast strains; and Wei-Lih Lee and Doug Koshland for plasmids. We also thank our reviewers for suggestions that helped to improve the manuscript. Funding was provided by the National Science Foundation Graduate Research Fellowship Program (N.I.K.) and the National Institutes of Health (Grant GM 47842 to G.B.).
Abbreviations used:
- AID
auxin-inducible degron
- APC
anaphase-promoting complex
- IAA
indole-3-acetic acid
- KTIP
Kap121-transport inhibitory pathway
- MIP
maximum intensity projection
- NEB
nuclear envelope breakdown
- NPC
nuclear pore complex
- SPB
spindle pole body
- TBS-T
Tris-buffered saline (pH 7.5) plus 0.05% Tween 20
- YP
yeast extract/peptone.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-05-0276) on May 28, 2015.
REFERENCES
- Ballister ER, Riegman M, Lampson MA. Recruitment of Mad1 to metaphase kinetochores is sufficient to reactivate the mitotic checkpoint. J Cell Biol. 2014;204:901–908. doi: 10.1083/jcb.201311113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart EL, Dorer RK, Murray AW, Schuyler SC. Reduced Mad2 expression keeps relaxed kinetochores from arresting budding yeast in mitosis. Mol Biol Cell. 2011;22:2448–2457. doi: 10.1091/mbc.E09-01-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Kumar A, Foiani M. Preserving the genome by regulating chromatin association with the nuclear envelope. Trends Cell Biol. 2012;22:465–473. doi: 10.1016/j.tcb.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Brady DM, Hardwick KG. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr Biol. 2000;10:675–678. doi: 10.1016/s0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- Cairo L V, Ptak C, Wozniak RW. Mitosis-specific regulation of nuclear transport by the spindle assembly checkpoint protein Mad1p. Mol Cell. 2013;49:109–120. doi: 10.1016/j.molcel.2012.10.017. [DOI] [PubMed] [Google Scholar]
- Campbell M, Chan G, Yen T. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J Cell Sci. 2001;114:953–963. doi: 10.1242/jcs.114.5.953. [DOI] [PubMed] [Google Scholar]
- Chan YW, Fava LL, Uldschmid A, Schmitz MHA, Gerlich DW, Nigg EA, Santamaria A. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J Cell Biol. 2009;185:859–874. doi: 10.1083/jcb.200812167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin P, Nashchekina O, Walker R, Pines J. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat Cell Biol. 2013;15:1378–1385. doi: 10.1038/ncb2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins KA, Castillo AR, Tatsutani SY, Biggins S. De novo kinetochore assembly requires the centromeric histone H3 variant. Mol Biol Cell. 2005;16:5649–5660. doi: 10.1091/mbc.E05-08-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- De Souza CP, Hashmi SB, Nayak T, Oakley B, Osmani SA. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol Biol Cell. 2009;20:2146–2159. doi: 10.1091/mbc.E08-08-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick AE, Gerlich DW. Kinetic framework of spindle assembly checkpoint signalling. Nat Cell Biol. 2013;15:1370–1377. doi: 10.1038/ncb2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois G, Iglesias N, Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr KA, Hoyt MA. Bub1p kinase activates the Saccharomyces cerevisiae spindle assembly checkpoint. Mol Cell Biol. 1998;18:2738–2747. doi: 10.1128/mcb.18.5.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerbach F, Galy V, Trelles-Sticken E, Fromont-Racine M, Jacquier A, Gilson E, Olivo-Marin J-C, Scherthan H, Nehrbass U. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat Cell Biol. 2002;4:214–221. doi: 10.1038/ncb756. [DOI] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Wynne DJ. Making an effective switch at the kinetochore by phosphorylation and dephosphorylation. Chromosoma. 2013;122:135–158. doi: 10.1007/s00412-013-0401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet Y, Reyes C, Courthéoux T, Goldstone S, Gay G, Serrurier C, Tournier S. Sister kinetochore recapture in fission yeast occurs by two distinct mechanisms, both requiring Dam1 and Klp2. Mol Biol Cell. 2008;19:1646–1662. doi: 10.1091/mbc.E07-09-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillett ES, Espelin CW, Sorger PK. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J Cell Biol. 2004;164:535–546. doi: 10.1083/jcb.200308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EM, Jiang Y, Joyner R, Weis K. A negative feedback loop at the nuclear periphery regulates GAL gene expression. Mol Biol Cell. 2012;23:1367–1375. doi: 10.1091/mbc.E11-06-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Heinrich S, Geissen E-M, Kamenz J, Trautmann S, Widmer C, Drewe P, Knop M, Radde N, Hasenauer J, Hauf S. Determinants of robustness in spindle assembly checkpoint signalling. Nat Cell Biol. 2013;15:1328–1339. doi: 10.1038/ncb2864. [DOI] [PubMed] [Google Scholar]
- Hou H, Zhou Z, Wang Y, Wang J, Kallgren SP, Kurchuk T, Miller EA, Chang F, Jia S. Csi1 links centromeres to the nuclear envelope for centromere clustering. J Cell Biol. 2012;199:735–744. doi: 10.1083/jcb.201208001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol. 2000;150:1233–1250. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol. 2002;159:807–819. doi: 10.1083/jcb.200205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Kastenmayer JP, Lee MS, Hong AL, Spencer FA, Basrai MA. The C-terminal half of Saccharomyces cerevisiae Mad1p mediates spindle checkpoint function, chromosome transmission fidelity and CEN association. Genetics. 2005;170:509–517. doi: 10.1534/genetics.105.041426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Pines J. Centromere tension: a divisive issue. Nat Cell Biol. 2010;12:919–923. doi: 10.1038/ncb1010-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- Kim S, Sun H, Tomchick DR, Yu H, Luo X. Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting. Proc Natl Acad Sci USA. 2012;109:6549–6554. doi: 10.1073/pnas.1118210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijt TEF, Omerzu M, Saurin AT, Kops GJPL. Conditional targeting of MAD1 to kinetochores is sufficient to reactivate the spindle assembly checkpoint in metaphase. Chromosoma. 2014;123:471–480. doi: 10.1007/s00412-014-0458-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F, Mieck C, Alushin GM, Nogales E, Westermann S. Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. J Cell Biol. 2013;200:21–30. doi: 10.1083/jcb.201210091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Sterling H, Burlingame A, McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008;22:2926–2931. doi: 10.1101/gad.1677208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage B, Qian J, Bollen M. Spindle checkpoint silencing: PP1 tips the balance. Curr Biol. 2011;21:R898–R903. doi: 10.1016/j.cub.2011.08.063. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Li Y, Gorbea C, Mahaffey D, Rechsteiner M, Benezra R. MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc Natl Acad Sci USA. 1997;94:12431–12436. doi: 10.1073/pnas.94.23.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lince-Faria M, Maffini S, Orr B, Ding Y, Cláudia Florindo, Sunkel CE, Tavares A, Johansen J, Johansen KM, Maiato H. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J Cell Biol. 2009;184:647–657. doi: 10.1083/jcb.200811012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N, Biggins S. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev. 2014:140–152. doi: 10.1101/gad.233700.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Makhnevych T, Lusk CP, Anderson AM, Aitchison JD, Wozniak RW. Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell. 2003;115:813–823. doi: 10.1016/s0092-8674(03)00986-3. [DOI] [PubMed] [Google Scholar]
- Maldonado M, Kapoor TM. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat Cell Biol. 2011;13:475–482. doi: 10.1038/ncb2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus SM, Punch JJ, Lee W-L. Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr Biol. 2009;19:196–205. doi: 10.1016/j.cub.2008.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR. Structural and mechanical control of mitotic progression. Cold Spring Harb Symp Quant Biol. 1991;56:613–619. doi: 10.1101/sqb.1991.056.01.070. [DOI] [PubMed] [Google Scholar]
- Meadows JC. Interplay between mitotic kinesins and the Aurora kinase-PP1 (protein phosphatase 1) axis. Biochem Soc Trans. 2013;41:1761–1765. doi: 10.1042/BST20130191. [DOI] [PubMed] [Google Scholar]
- Merdes A, De Mey J. The mechanism of kinetochore-spindle attachment and polewards movement analyzed in PtK2 cells at the prophase-prometaphase transition. Eur J Cell Biol. 1990;53:313–325. [PubMed] [Google Scholar]
- Morawska M, Ulrich HD. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast. 2013;30:341–351. doi: 10.1002/yea.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle MW, Kim T, Hattersley N, Espeut J, Cheerambathur DK, Oegema K, Desai A. A Bub1-Mad1 interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J Cell Biol. 2014;204:647–657. doi: 10.1083/jcb.201311015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Niepel M, Strambio-de-Castillia C, Fasolo J, Chait BT, Rout MP. The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J Cell Biol. 2005;170:225–235. doi: 10.1083/jcb.200504140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Pangilinan F, Spencer F. Abnormal kinetochore structure activates the spindle assembly checkpoint in budding yeast. Mol Biol Cell. 1996;7:1195–1208. doi: 10.1091/mbc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Biggins S. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Pinsky BA, Nelson CR, Biggins S. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr Biol. 2009;19:1182–1187. doi: 10.1016/j.cub.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primorac I, Weir JR, Chiroli E, Gross F, Hoffmann I, van Gerwen S, Ciliberto A, Musacchio A. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. Elife. 2013;2:e01030. doi: 10.7554/eLife.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka MR, Zhang W, Costa P, Archambault V, D'Avino PP, Lilley KS, Laue ED, McAinsh AD, Glover DM. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS One. 2007;2:e478. doi: 10.1371/journal.pone.0000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Rath U, Wang D, Xu YZ, Ding Y, Zhang W, Blacketer MJ, Paddy MR, Girton J, Johansen J, et al. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol Biol Cell. 2004;15:4854–4865. doi: 10.1091/mbc.E04-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Bravo V, Maciejowski J, Corona J, Buch HK, Collin P, Kanemaki MT, Shah JV, Jallepalli PV. Nuclear pores protect genome integrity by assembling a premitotic and mad1-dependent anaphase inhibitor. Cell. 2014;156:1017–1031. doi: 10.1016/j.cell.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S, Kobayashi Y, Ogiyama Y, Takahashi K. Dual regulation of Mad2 localization on kinetochores by Bub1 and Dam1/DASH that ensure proper spindle interaction. Mol Biol Cell. 2008;19:3885–3897. doi: 10.1091/mbc.E08-03-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler SC, Wu Y-F, Kuan VJ-W. The Mad1-Mad2 balancing act—a damaged spindle checkpoint in chromosome instability and cancer. J Cell Sci. 2012;125:4197–4206. doi: 10.1242/jcs.107037. [DOI] [PubMed] [Google Scholar]
- Scott RJ, Cairo L V, Van de Vosse DW, Wozniak RW. The nuclear export factor Xpo1p targets Mad1p to kinetochores in yeast. J Cell Biol. 2009;184:21–29. doi: 10.1083/jcb.200804098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Lusk CP, Dilworth DJ, Aitchison JD, Wozniak RW. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:4362–4374. doi: 10.1091/mbc.E05-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp-Baker H, Chen R-H. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and Cenp-E, independently of its kinase activity. J Cell Biol. 2001;153:1239–1250. doi: 10.1083/jcb.153.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D. Determinants of S. cerevisiae dynein localization and activation. Curr Biol. 2003;13:364–372. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- Shimogawa MM, Graczyk B, Gardner MK, Francis SE, White EA, Ess M, Molk JN, Ruse C, Niessen S, Yates JR 3rd, et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr Biol. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogawa MM, Wargacki MM, Muller EG, Davis TN. Laterally attached kinetochores recruit the checkpoint protein Bub1, but satisfy the spindle checkpoint. Cell Cycle. 2010;9:3619–3628. doi: 10.4161/cc.9.17.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F, Hieter P. Centromere DNA mutations induce a mitotic delay in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1992;89:8908–8912. doi: 10.1073/pnas.89.19.8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- Westermann S, Cheeseman IM, Anderson S, Yates JR, Drubin DG, Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Wang H-W, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Yang C-H, Tanno Y, Watanabe Y. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat Cell Biol. 2012;14:746–752. doi: 10.1038/ncb2515. [DOI] [PubMed] [Google Scholar]
- Yang Z, Tulu US, Wadsworth P, Rieder CL. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr Biol. 2007;17:973–980. doi: 10.1016/j.cub.2007.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.