Abstract
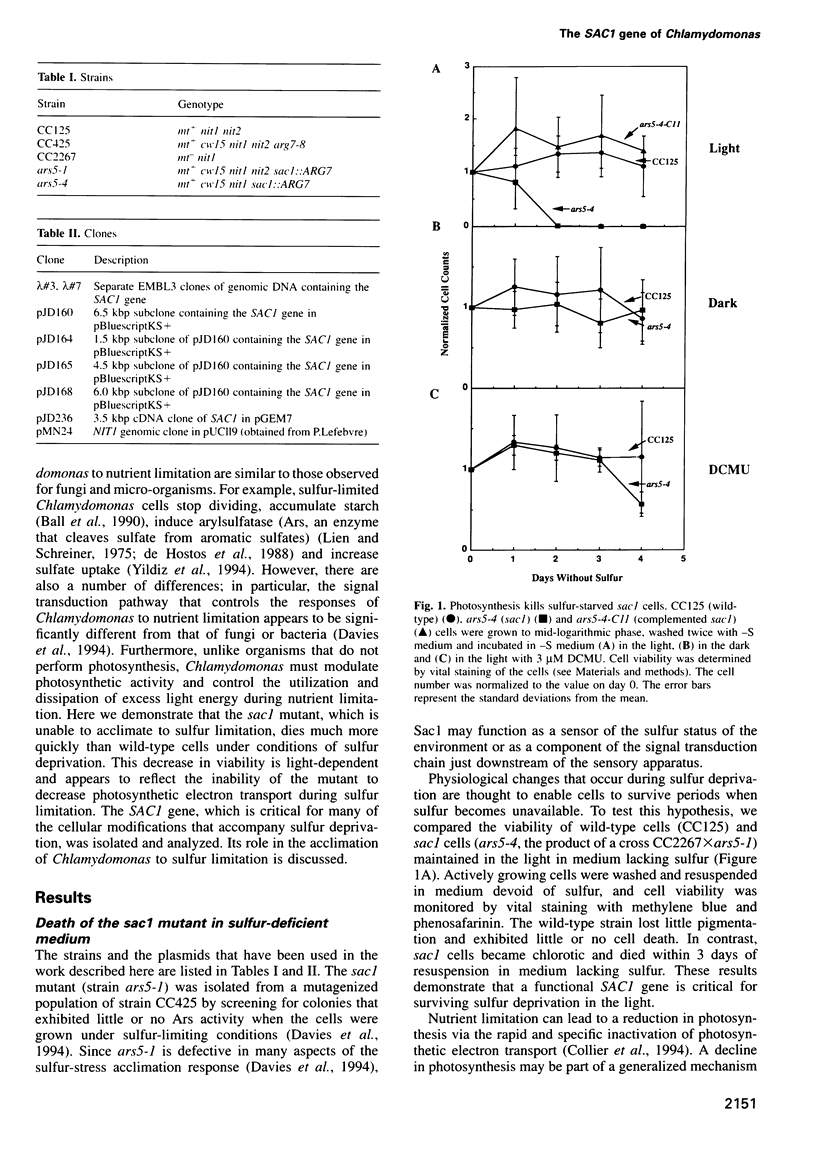
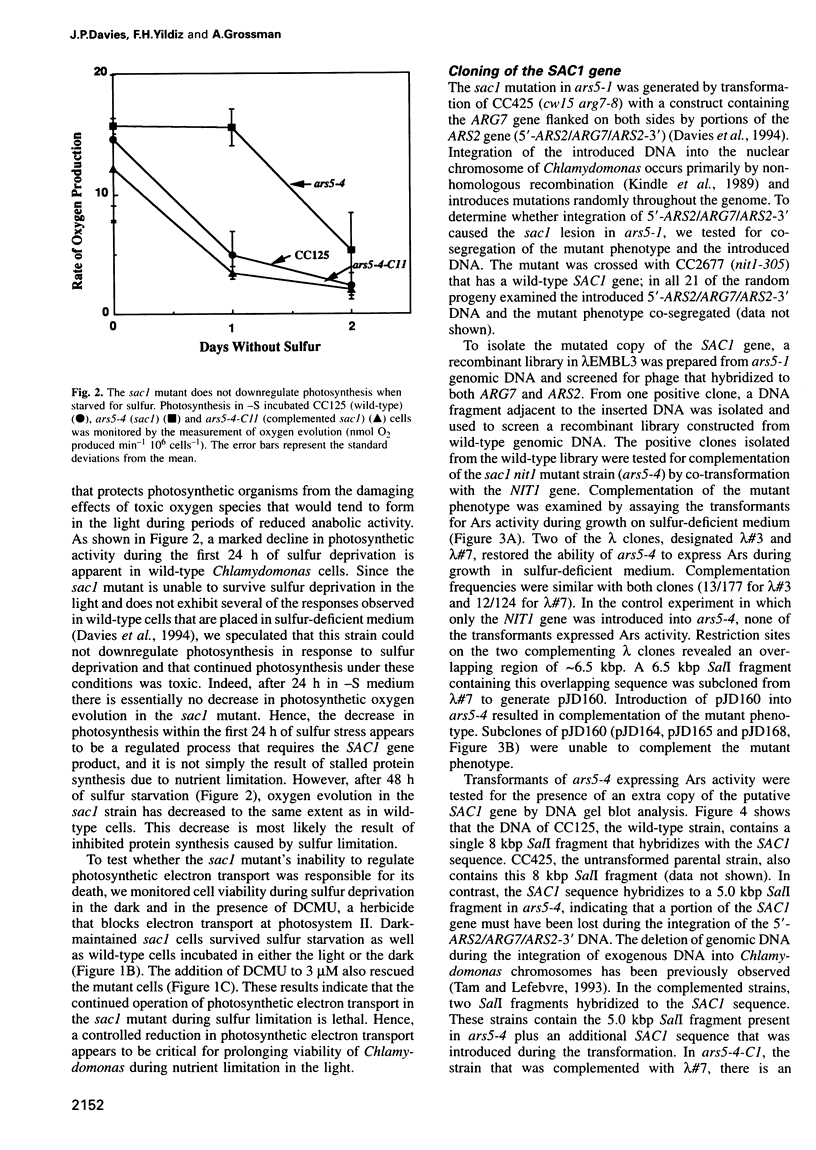
The sac1 mutant of Chlamydomonas reinhardtii is aberrant in most of the normal responses to sulfur limitation; it cannot synthesize arylsulfatase, does not take up sulfate as rapidly as wild-type cells, and does not synthesize periplasmic proteins that normally accumulate during sulfur-limited growth. Here, we show that the sac1 mutant dies much more rapidly than wild-type cells during sulfur deprivation; this emphasizes the vital role of the acclimation process. The loss of viability of the sac1 mutant during sulfur deprivation is only observed in the light and is mostly inhibited by DCMU. During sulfur-stress, wild-type cells, but not the sac1 mutant, downregulate photosynthesis. Thus, death of the sac1 mutant during sulfur deprivation is probably a consequence of its inability to downregulate photosynthesis. Furthermore, since SAC1 is necessary for the downregulation of photosynthesis, the process must be highly controlled and not simply the result of a general decrease in protein synthesis due to sulfur limitation. Genomic and cDNA copies of the SAC1 gene have been cloned. The deduced amino acid sequence of Sac1 is similar to an Escherichia coli gene that may involved in the response of E.coli to nutrient deprivation.
Full text
PDF
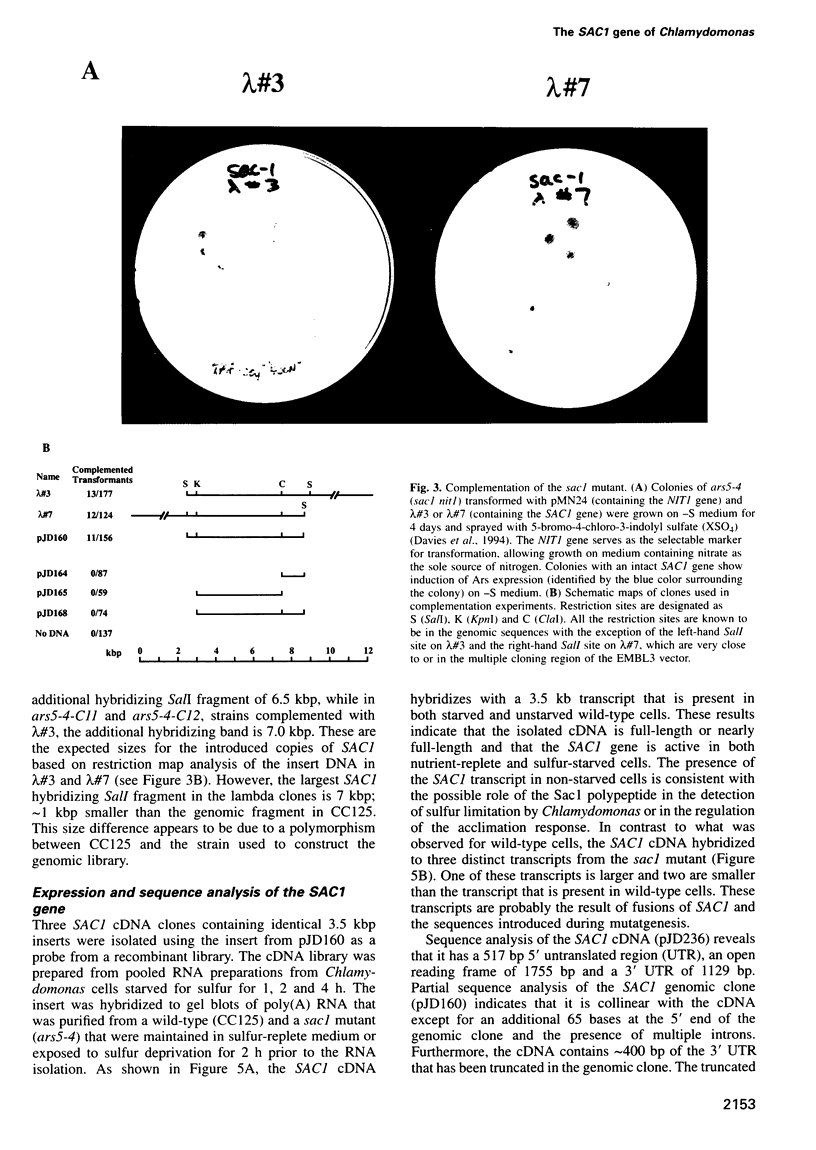

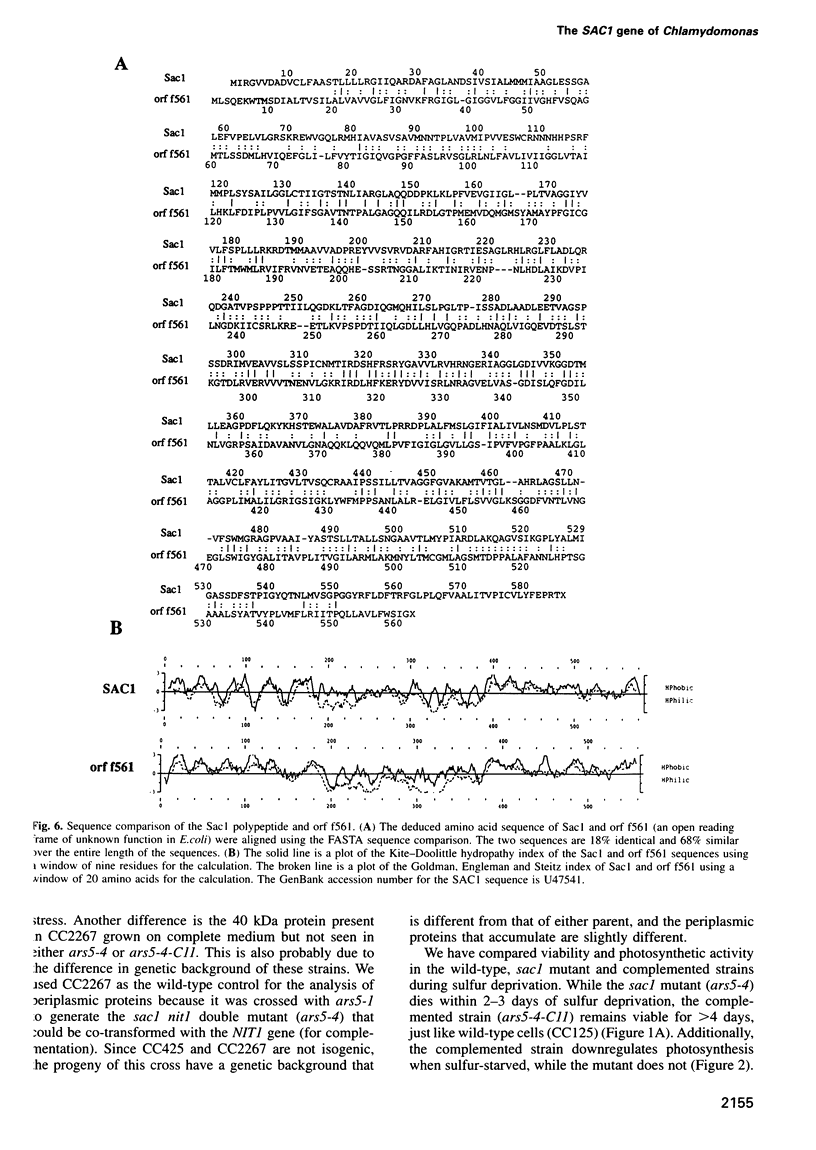
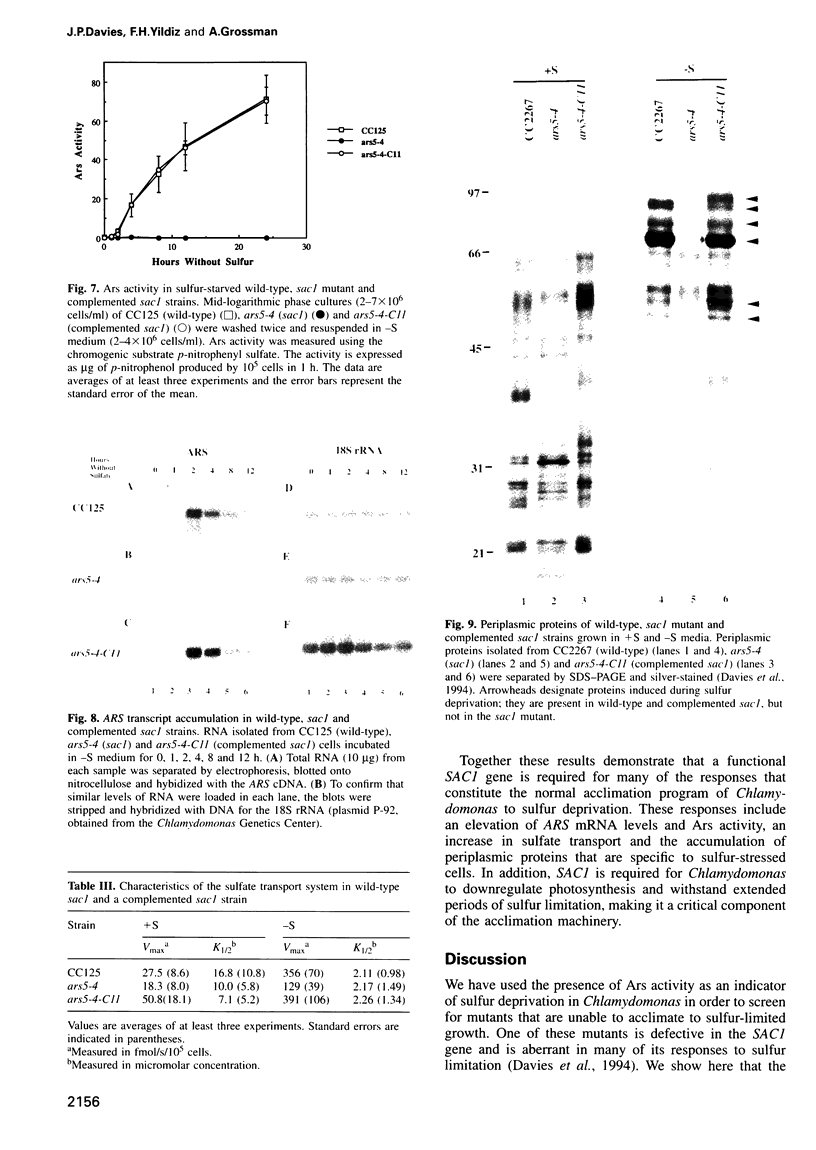



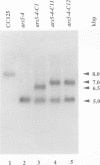
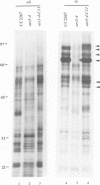
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi T., Murooka Y., Harada T. Regulation of arylsulfatase synthesis by sulfur compounds in Klebsiella aerogenes. J Bacteriol. 1975 Jan;121(1):29–35. doi: 10.1128/jb.121.1.29-35.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi T., Okamura H., Murooka Y., Harada T. Catabolite repression and derepression of arylsulfatase synthesis in Klebsiella aerogenes. J Bacteriol. 1974 Nov;120(2):880–885. doi: 10.1128/jb.120.2.880-885.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen S. P., Polazzi J. O., Gierse J. K., Easton A. M. Two novel heat shock genes encoding proteins produced in response to heterologous protein expression in Escherichia coli. J Bacteriol. 1992 Nov;174(21):6938–6947. doi: 10.1128/jb.174.21.6938-6947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames G. F. Bacterial periplasmic transport systems: structure, mechanism, and evolution. Annu Rev Biochem. 1986;55:397–425. doi: 10.1146/annurev.bi.55.070186.002145. [DOI] [PubMed] [Google Scholar]
- Apte B. N., Bhavsar P. N., Siddiqi O. The regulation of aryl sulphatase in Aspergillus nidulans. J Mol Biol. 1974 Jul 5;86(3):637–648. doi: 10.1016/0022-2836(74)90186-7. [DOI] [PubMed] [Google Scholar]
- Arst H. N., Jr Genetic analysis of the first steps of sulphate metabolism in Aspergillus nidulans. Nature. 1968 Jul 20;219(5151):268–270. doi: 10.1038/219268a0. [DOI] [PubMed] [Google Scholar]
- Breton A., Surdin-Kerjan Y. Sulfate uptake in Saccharomyces cerevisiae: biochemical and genetic study. J Bacteriol. 1977 Oct;132(1):224–232. doi: 10.1128/jb.132.1.224-232.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burland V., Plunkett G., 3rd, Daniels D. L., Blattner F. R. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics. 1993 Jun;16(3):551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- Caboche M., Rouzé P. Nitrate reductase: a target for molecular and cellular studies in higher plants. Trends Genet. 1990 Jun;6(6):187–192. doi: 10.1016/0168-9525(90)90175-6. [DOI] [PubMed] [Google Scholar]
- Collier J. L., Grossman A. R. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: not all bleaching is the same. J Bacteriol. 1992 Jul;174(14):4718–4726. doi: 10.1128/jb.174.14.4718-4726.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford N. M., Arst H. N., Jr The molecular genetics of nitrate assimilation in fungi and plants. Annu Rev Genet. 1993;27:115–146. doi: 10.1146/annurev.ge.27.120193.000555. [DOI] [PubMed] [Google Scholar]
- DREYFUSS J. CHARACTERIZATION OF A SULFATE- AND THIOSULFATE-TRANSPORTING SYSTEM IN SALMONELLA TYPHIMURIUM. J Biol Chem. 1964 Jul;239:2292–2297. [PubMed] [Google Scholar]
- Davies J. P., Weeks D. P., Grossman A. R. Expression of the arylsulfatase gene from the beta 2-tubulin promoter in Chlamydomonas reinhardtii. Nucleic Acids Res. 1992 Jun 25;20(12):2959–2965. doi: 10.1093/nar/20.12.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. P., Yildiz F., Grossman A. R. Mutants of Chlamydomonas with Aberrant Responses to Sulfur Deprivation. Plant Cell. 1994 Jan;6(1):53–63. doi: 10.1105/tpc.6.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R. M., Teixeira A. R. Sulfur starvation in Lemna leads to degradation of ribulose-bisphosphate carboxylase without plant death. J Biol Chem. 1992 Apr 15;267(11):7253–7257. [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A. cys-3, the positive-acting sulfur regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. J Biol Chem. 1990 Jul 15;265(20):11942–11947. [PubMed] [Google Scholar]
- Goldstein A. H., Baertlein D. A., McDaniel R. G. Phosphate Starvation Inducible Metabolism in Lycopersicon esculentum: I. Excretion of Acid Phosphatase by Tomato Plants and Suspension-Cultured Cells. Plant Physiol. 1988 Jul;87(3):711–715. doi: 10.1104/pp.87.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green L. S., Grossman A. R. Changes in sulfate transport characteristics and protein composition of Anacystis nidulans R2 during sulfur deprivation. J Bacteriol. 1988 Feb;170(2):583–587. doi: 10.1128/jb.170.2.583-587.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann A., Sumper M. An inducible arylsulfatase of Volvox carteri with properties suitable for a reporter-gene system. Purification, characterization and molecular cloning. Eur J Biochem. 1994 Apr 1;221(1):143–150. doi: 10.1111/j.1432-1033.1994.tb18723.x. [DOI] [PubMed] [Google Scholar]
- Khamis S., Lamaze T., Lemoine Y., Foyer C. Adaptation of the Photosynthetic Apparatus in Maize Leaves as a Result of Nitrogen Limitation : Relationships between Electron Transport and Carbon Assimilation. Plant Physiol. 1990 Nov;94(3):1436–1443. doi: 10.1104/pp.94.3.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K. L. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K. L., Schnell R. A., Fernández E., Lefebvre P. A. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J Cell Biol. 1989 Dec;109(6 Pt 1):2589–2601. doi: 10.1083/jcb.109.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy H., Tal T., Shaish A., Zamir A. Cbr, an algal homolog of plant early light-induced proteins, is a putative zeaxanthin binding protein. J Biol Chem. 1993 Oct 5;268(28):20892–20896. [PubMed] [Google Scholar]
- Lien T., Schreiner O., Steine M. Purification of a derepressible arylsulfatase from Chlamydomonas reinhardti. Properties of the enzyme in intact cells and in purified state. Biochim Biophys Acta. 1975 Mar 28;384(1):168–179. doi: 10.1016/0005-2744(75)90106-0. [DOI] [PubMed] [Google Scholar]
- Lillie S. H., Pringle J. R. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol. 1980 Sep;143(3):1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluf G. A. Genetic and biochemical studies of distinct sulfate permease species in different developmental stages of Neurospora crassa. Arch Biochem Biophys. 1970 May;138(1):254–263. doi: 10.1016/0003-9861(70)90306-1. [DOI] [PubMed] [Google Scholar]
- Marzluf G. A. Genetic and metabolic controls for sulfate metabolism in Neurospora crassa: isolation and study of chromate-resistant and sulfate transport-negative mutants. J Bacteriol. 1970 Jun;102(3):716–721. doi: 10.1128/jb.102.3.716-721.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluf G. A., Metzenberg R. L. Positive control by the cys-3 locus in regulation of sulfur metabolism in Neurospora. J Mol Biol. 1968 Apr 28;33(2):423–437. doi: 10.1016/0022-2836(68)90199-x. [DOI] [PubMed] [Google Scholar]
- Metzenberg R. L., Ahlgren S. K. Mutants of Neurospora deficient in aryl sulfatase. Genetics. 1970 Mar-Apr;64(3):409–422. [PMC free article] [PubMed] [Google Scholar]
- Paietta J. V. Molecular cloning and analysis of the scon-2 negative regulatory gene of Neurospora crassa. Mol Cell Biol. 1990 Oct;10(10):5207–5214. doi: 10.1128/mcb.10.10.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paietta J. V. Molecular cloning and regulatory analysis of the arylsulfatase structural gene of Neurospora crassa. Mol Cell Biol. 1989 Sep;9(9):3630–3637. doi: 10.1128/mcb.9.9.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paietta J. V. Production of the CYS3 regulator, a bZIP DNA-binding protein, is sufficient to induce sulfur gene expression in Neurospora crassa. Mol Cell Biol. 1992 Apr;12(4):1568–1577. doi: 10.1128/mcb.12.4.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier G., Schmidt G. W. Chlororespiration: an adaptation to nitrogen deficiency in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4791–4795. doi: 10.1073/pnas.88.11.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumley F. G., Schmidt G. W. Nitrogen-dependent regulation of photosynthetic gene expression. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2678–2682. doi: 10.1073/pnas.86.8.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saux C., Lemoine Y., Marion-Poll A., Valadier M. H., Deng M., Morot-Gaudry J. F. Consequence of Absence of Nitrate Reductase Activity on Photosynthesis in Nicotiana plumbaginifolia Plants. Plant Physiol. 1987 May;84(1):67–72. doi: 10.1104/pp.84.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott W. A., Metzenberg R. L. Location of Aryl Sulfatase in Conidia and Young Mycelia of Neurospora crassa. J Bacteriol. 1970 Dec;104(3):1254–1265. doi: 10.1128/jb.104.3.1254-1265.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam L. W., Lefebvre P. A. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics. 1993 Oct;135(2):375–384. doi: 10.1093/genetics/135.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Jacquemin I., Surdin-Kerjan Y. MET4, a leucine zipper protein, and centromere-binding factor 1 are both required for transcriptional activation of sulfur metabolism in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Apr;12(4):1719–1727. doi: 10.1128/mcb.12.4.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Scott J. D., McMullen B., Hurwitz M., Krebs E. G., Wigler M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Apr;7(4):1371–1377. doi: 10.1128/mcb.7.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. B., Tatchell K. SRA5 encodes the low-Km cyclic AMP phosphodiesterase of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jan;8(1):505–510. doi: 10.1128/mcb.8.1.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz F. H., Davies J. P., Grossman A. R. Characterization of Sulfate Transport in Chlamydomonas reinhardtii during Sulfur-Limited and Sulfur-Sufficient Growth. Plant Physiol. 1994 Mar;104(3):981–987. doi: 10.1104/pp.104.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos E. L., Schilling J., Grossman A. R. Structure and expression of the gene encoding the periplasmic arylsulfatase of Chlamydomonas reinhardtii. Mol Gen Genet. 1989 Aug;218(2):229–239. doi: 10.1007/BF00331273. [DOI] [PubMed] [Google Scholar]
- de Hostos E. L., Togasaki R. K., Grossman A. Purification and biosynthesis of a derepressible periplasmic arylsulfatase from Chlamydomonas reinhardtii. J Cell Biol. 1988 Jan;106(1):29–37. doi: 10.1083/jcb.106.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]