Abstract
Available evidence on the bioactive, nutritional and putative detrimental properties of gut microbial metabolites has been evaluated to support a more integrated view of how prebiotics might affect host health throughout life. The present literature inventory targeted evidence for the physiological and nutritional effects of metabolites, for example, SCFA, the potential toxicity of other metabolites and attempted to determine normal concentration ranges. Furthermore, the biological relevance of more holistic approaches like faecal water toxicity assays and metabolomics and the limitations of faecal measurements were addressed. Existing literature indicates that protein fermentation metabolites (phenol, p-cresol, indole, ammonia), typically considered as potentially harmful, occur at concentration ranges in the colon such that no toxic effects are expected either locally or following systemic absorption. The endproducts of saccharolytic fermentation, SCFA, may have effects on colonic health, host physiology, immunity, lipid and protein metabolism and appetite control. However, measuring SCFA concentrations in faeces is insufficient to assess the dynamic processes of their nutrikinetics. Existing literature on the usefulness of faecal water toxicity measures as indicators of cancer risk seems limited. In conclusion, at present there is insufficient evidence to use changes in faecal bacterial metabolite concentrations as markers of prebiotic effectiveness. Integration of results from metabolomics and metagenomics holds promise for understanding the health implications of prebiotic microbiome modulation but adequate tools for data integration and interpretation are currently lacking. Similarly, studies measuring metabolite fluxes in different body compartments to provide a more accurate picture of their nutrikinetics are needed.
Keywords: Microbial metabolites, Prebiotic health benefits, Metagenome, Nutrikinetics
Introduction
For a long time, the colon was considered as an organ that merely absorbs water and electrolytes and converts undigested food residues to drive their excretion without having important physiological functions. Nowadays, it has been generally recognised that the microbial ecosystem inhabiting the gut profoundly affects human physiology and health. The gut bacteria can be considered as a highly active metabolic organ that provides metabolic traits that complement those encoded within our own genome. For instance, degradation of several structural polysaccharides in plant cell walls requires enzymes that are not encoded by the host but are available in specific bacteria( 1 ). The collective genetic information encoded in the intestinal micro-organisms is truly impressive and has been referred to as ‘our other genome’( 2 ).
The metabolites produced by the gut bacteria are accessible to the host's cells and in this way influence physiological processes both locally in the intestine and systemically. They contribute to the metabolic phenotype of the host and hence may influence the risk of disease( 3 ). Undigested carbohydrate and protein constitute the major substrates at the disposal of the microbiota for fermentation and result in the production of a range of well-established metabolites including SCFA, branched-chain fatty acids (BCFA), ammonia, amines, phenolic compounds and gases including hydrogen, methane and hydrogen sulfide. Other metabolic activities include the activation or inactivation of bioactive food components like isoflavanoids, flavanoids and plant lignans, the conversion of pro-drugs to drugs, the production of vitamins and the transformation of bile acids and xenobiotics( 4 , 5 ). Although the fermentative and metabolic activity in the human intestine has been studied for many decades, it remains difficult to evaluate bacterial metabolism in the colon in vivo. Most studies have relied on analysis of the composition of faeces, in vitro incubation studies using faecal inocula or experimental animal models.
An imbalance in the composition of the microbiota has been increasingly associated with the occurrence of chronic or lifestyle-related diseases such as inflammatory bowel disease (IBD), obesity and type 2 diabetes as well as with certain autoimmune diseases such as type 1 diabetes, coeliac disease or allergic asthma( 6 ). Therefore, manipulation of the microbiota has become a promising target for the improvement of host health. As diet is a major factor driving the composition and metabolism of the colonic microbiota, dietary interventions that modulate the supply of macronutrients (carbohydrates, proteins, fat) to the colon have been extensively investigated for this purpose. In particular prebiotics, defined as ‘selectively fermented food ingredients that allow specific changes in composition and/or activity of the microbiota that confer benefits upon host well-being and health’( 7 ), have been used in an attempt to improve gut health and by extension systemic health (http://www.worldgastroenterology.org/probiotics-prebiotics). Previously, the effect of prebiotic supplementation has been measured using the relative increase in Bifidobacterium and Lactobacillus species as markers( 8 ). However, increasing knowledge on the intestinal microbiota has shown that other genera or species may also confer health benefits, expanding the potential role of prebiotics. Emerging genera that may play a role in the maintenance of intestinal homeostasis and health include Eubacterium, Faecalibacterium, Roseburia and some species of Clostridia ( 7 ). Since the microbiota is characterised by a significant degree of functional redundancy( 9 ), meaning that different bacteria are able to perform similar functions, metabolise the same substrates and produce similar metabolites, analysing the activity of the microbiota rather than its composition and structure may be more relevant to assess the impact of prebiotic interventions.
In the present review, we have re-evaluated our current understanding of the role of bacterial metabolites in promoting or reducing health to estimate the potential applicability of those metabolites as markers of improved/reduced gut health. Moreover, we recognise that the contribution of the microbiota to the overall mammalian biochemistry may play distinctive roles at different developmental stages of the host. We have identified the limitations associated with analysis of single metabolites and have evaluated the usefulness of more holistic approaches including functional analysis of faecal water, metabolomics and metagenome analysis. The ultimate goal is to move towards a definition of ‘healthy metabolic signatures’ that might comprise integrated measures of metabolite patterns in different matrices.
Metabolites produced by microbial fermentation
The complex microbial ecosystem inhabiting the human intestinal tract produces a wide range of metabolites that interact with the host's cells and in this way influence the physiological processes in the colon. In addition, the metabolites may be absorbed and influence the overall mammalian biochemistry, thereby eliciting systemic effects. Table 1 provides an overview of the major bacterial compounds that can be found in the intestine.
Table 1.
List of bacterial metabolites that may be found in the intestine
| Type of metabolite | Metabolites |
|---|---|
| Metabolites derived from bacterial energy metabolism | ‘Terminal’ metabolites from carbohydrate fermentationSCFA: formate, acetate, propionate, butyrate, longer-chain fatty acidsBranched-chain fatty acids |
| ‘Intermediate’ metabolites from carbohydrate fermentationPartially degraded oligomeric carbohydrates (disaccharides, oligosaccharides, complex proteoglycans from mucins, etc.)Alcohols: methanol, ethanol, etc. | |
| Gaseous metabolitesFermentation gases: hydrogen, methane, carbon dioxideHighly volatile compounds: hydrogen sulfide | |
| Metabolites of fatty acid and lipid bioconversionLong-chain aldehydesFatty acids | |
| Metabolites from protein fermentationBranched-chain fatty acidsAmmonia and aminesAromatic derivatives of amino acids: phenols, cresols, indoles, etc. | |
| Metabolites derived from bioconversion of plant secondary compounds | Products of lignin/polyphenols bioconversion: equol, enterolactone, etc. |
| Metabolites from bacterial cytosolic compartment or secondary metabolism (spilled over by excess production, efflux or upon cell lysis) | Vitamins and cofactors (often in very small concentrations) Peptides (quorum-sensing signals of Gram-positive bacteria) Homoserine lactone (quorum-sensing signals of Gram-negative bacteria) Nucleic acids (free DNA, microRNA, etc.) Bacteriocins |
| Metabolites of the enterohepatic circulation | Bile acids Cholesterol, coprostanol Hormones and derivatives Glucuronide conjugates |
| Enzymes | Reductases Glucuronidases Glycohydrolases |
| Bacterial cell wall components |(of which several are immunoactive) | Lipopolysaccharide Polysaccharide A Peptidoglycan-derived structures Capsular polysaccharides (glycocalix) |
From this list of compounds, we selected a subset of metabolites that were considered relevant to improved or decreased health. Most of those metabolites are so-called primary metabolites which comprise products of metabolism that are essential for growth or that are the by-products of energy-yielding metabolism. Secondary metabolites (products which do not have an obvious role in cell metabolism such as vitamins) were not included for further analysis. The metabolites reviewed here include products of carbohydrate fermentation (acetic, propionic and butyric acid as well as lactic acid and succinic acid) and products of protein metabolism (ammonia, BCFA, phenol, amines, p-cresol, indole and hydrogen sulfide). In addition, metabolites of plant polyphenols have been included because of their putative health benefits and their bidirectional interaction with the intestinal microbiota.
Beneficial and harmful effects of relevant metabolites
Products of carbohydrate fermentation
SCFA
SCFA are mainly produced in the colon by bacterial fermentation of carbohydrates that escaped digestion in the small intestine. They are saturated aliphatic organic acids consisting of one to six carbons of which acetate (C2), propionate (C3) and butyrate (C4) are the most abundant ( ≥ 95 %)( 10 , 11 ). SCFA production mainly occurs in the proximal part of the colon where the availability of substrates is most abundant. The majority of SCFA (up to 95 %) are rapidly absorbed by the colonocytes resulting in decreasing concentrations from the proximal to distal colon. Only a minor fraction of SCFA (about 5 %) is excreted in faeces( 12 ).
Due to the inaccessibility of the human proximal colon for direct investigation and the rapid absorption of SCFA from the colonic lumen, it is extremely difficult to quantify SCFA production rates. Consequently, no systematic evaluation of normal ‘healthy’ production of SCFA is available. Assuming that 50–60 g carbohydrates reach the colon per d, the production of SCFA was estimated at 400–600 mmol/d( 10 ). Most studies measure faecal SCFA which are the resultant of their production and absorption. Therefore, faecal SCFA rather indicate losses and do not adequately reflect in situ production rates. Table 2 provides an overview of reported values in the literature for total and individual faecal SCFA in adults. Faecal excretion of total SCFA ranges from 60 to 90 μmol/g and might be slightly higher in obese subjects (80–100 μmol/g). SCFA are also detectable in urine, but are the remnant of gut, liver and systemic metabolism and do not reflect colonic generation either. In addition, acetate not only originates from the gut but also from endogenous metabolism, in particular fatty acid oxidation and glucose and/or amino acid metabolism( 13 , 14 ). Measurement of SCFA in plasma is similarly confounded. Stable isotope studies are required to reliably quantify colonic SCFA production as well as their metabolic fate in the host organism.
Table 2.
Faecal concentration of individual SCFA
| Subjects (n); age | Reported measure | Acetic acid | Propionic acid | Butyric acid | Total SCFA | Unit | Reference | |
|---|---|---|---|---|---|---|---|---|
| Healthy subjects | 10; 21–34 years | Mean (sd) | 218 (99) | 72 (37) | 58·7 (54·5) | 378 (188) | μmol/g dry weight | Whelan (2005)( 236 ) |
| 20; 20–40 years | Mean (sem) | 320·3 (24·9) | 97·3 (10·5) | 93·8 (9·13) | 511·4 (41·9) | μmol/g dry weight | Boler (2011)( 111 ) | |
| 13; 23–58 years | Median (IQR) | 52·2 | 23·2 (13·6–37·3) | 36·8 (5–128) | 119·3 (64·5–197·0) | μmol/g wet weight | Lewis (1997)( 237 ) | |
| 60; 18–24 years | Mean (sem) | 198·4 (14·2) | 55·2 (4·7) | 50·5 (4·9) | 304·1 | μmol/g dry weight | Lecerf (2012)( 109 ) | |
| 27; 18–55 years | Mean (sem) | 35·8 (2·4) | 11·4 (1·2) | 10·0 (1·1) | 61·1 (4·4) | μmol/g | Reimer (2012)( 238 ) | |
| 12; 18–65 years | 48 | 13·98 | 13·31 | 80·91 | μmol/g | Fernando (2010)( 239 ) | ||
| 46; 31–66 years | Mean (95 % CI) | 44·7 (39·7, 50·3) females 58·6 (49·8, 69·0) males | 12·3 (10·7, 14·0) females 16·1 (13·4, 19·5) males | 11·7 (9·8, 14·0) females 15·4 (12·1, 19·6) males | 69·5 (61·3, 78·7) females 90·5 (76·3, 108) males | μmol/g | McOrist (2011)( 240 ) | |
| 36 | Median (IQR) | 43·7 (34·0–52·2) | 13·1 (9·2–18·5) | 8·8 (5·2–11·5) | 91·8 (73·1–107·5) | μmol/g | Nemoto (2012)( 241 ) | |
| 20; 22–55 years | Mean (sem) | 42·13 (3·84) | 11·5 (1·19) | 11·28 (1·42) | 67·3 (6·22) | μmol/g | Tiihonen (2010)( 67 ) | |
| 8; 31–59 years | Mean (sd) | NR | NR | NR | 92·7 (33·9) | μmol/g | McOrist (2008)( 242 ) | |
| 20; 23–28 years | Mean (sem) | NR | NR | NR | 78·5 (6·4) | μmol/g | Hylla (1998)( 243 ) | |
| 30 | Mean (sd) | 50·5 (12·6) | 13·6 (5·2) | 14·1 (7·6) | 84·6 (22·9) | mmol/l | Schwiertz (2010)( 64 ) | |
| Obese subjects | 20; 22–55 years | Mean (sem) | 47·15 (3·80) | 13·64 (1·34) | 14·73 (1·47) | 78·79 (6·19) | μmol/g | Tiihonen (2010)( 67 ) |
| 91 | Mean (sd) | 58·5 (19·1) | 17·6 (7·6) | 18·3 (9·7) | 102 (33·5) | mmol/l | Brinkworth (2009)( 63 ) | |
| 32; 20–65 years | Mean (sem) | NR | NR | NR | 34 (6) | mmol/24 h | Benassi Evans (2010)( 244 ) | |
| 35 overweight 33 obese | Mean (sd) | 56·0 (18·2) 59·8 (18·3) | 18·3 (7·9) 19·3 (8·7) | 18·5 (10·1) 18·1 (10·0) | 98·7 (33·9) 103·9 (34·3) | mmol/l | Schwiertz (2010)( 64 ) |
IQR, interquartile range; NR, not reported.
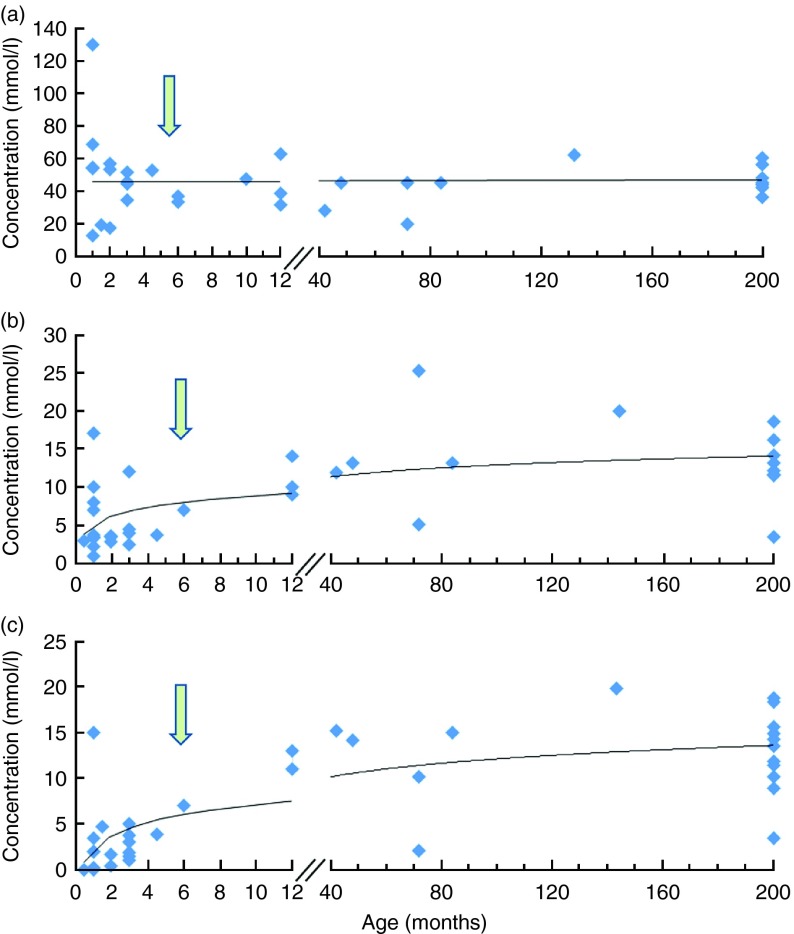
The pattern and amounts of faecal SCFA change through the different stages in life. In early infancy, the predominant SCFA are acetate and lactate in breast-fed infants and acetate and propionate in (unsupplemented) formula-fed infants( 15 ). In infants fed a formula supplemented with a mixture of galacto-oligosaccharides and fructo-oligosaccharides (9:1 ratio), faecal SCFA patterns were dominated by acetate, similarly as in breast-fed infants, with lower proportions of propionate and butyrate compared with the unsupplemented formula( 16 ). The levels of propionate have been reported to increase in the months before weaning. Butyrate production increases in the second part of the first year of life when faecal lactate levels fall to negligible values (CA Edwards, unpublished results). By the age of 2 years the pattern becomes more similar to that observed in adults( 17 ). Fig. 1 depicts the changes in SCFA from birth up to adulthood. In the elderly, the microbiota changes, with higher levels of Bacteroidetes( 18 ), which is likely to affect SCFA production. Nevertheless, no differences were detected in SCFA levels in a group of French 68- to 89-year-olds compared with a group of 30- to 46-year-olds( 19 ). In contrast, among participants in the pan-European project on the elderly gut microbiota (CROWNALIFE), elderly Europeans (76 ± 7·5 years; n 55) had lower concentrations of propionate, acetate and butyrate (by 30, 35 and 21 %, respectively) compared with younger adults (40 ± 9·7 years; n 53)( 20 ). With these apparently contradicting results obtained in different studies, it remains to be established what the normal patterns of SCFA in faecal material are during different stages of life.
Fig. 1.
Evolution of faecal SCFA as a function of age: acetic acid (a); propionic acid (b); butyric acid (c). The arrows roughly indicate the change from breast-feeding to solid food with concurrent successional development of the gut microbiota away from one dominated by the bifidobacteria, which produce acetate and lactate during carbohydrate fermentation, to a more complex microbiota with higher relative abundance of Firmicutes, which produce acetate, propionate and butyrate as major SCFA endproducts of carbohydrate fermentation. The figures summarises the data reported in several studies( 15 , 17 , 71 , 84 , 126 , 228 – 235 ). A colour version of this figure can be found online at http://www.journals.cambridge.org/nrr
After uptake in the colonocytes, a considerable part of the SCFA is used as an energy source and is oxidised to carbon dioxide and ketone bodies( 21 ). The fraction that is not consumed by the colonocytes is transported across the basolateral membrane and reaches the liver via the portal bloodstream. Acetate is used by the liver as a precursor for the synthesis of cholesterol and long-chain fatty acids( 22 ). However, in individuals following a Western-type diet, high in refined carbohydrates, sugars and fatty acids and low in fibre, colonic acetate is likely to contribute only little to hepatic lipogenesis. A recent study in obese individuals even showed an inverse association between serum acetate levels and visceral adipose tissue( 23 ). In mice, it was shown that acetate derived from colonic fermentation of fermentable carbohydrates crosses the blood–brain barrier and directly suppresses appetite through central hypothalamic mechanisms( 24 ). Propionic acid is often the second most predominant SCFA and has received much attention for its potential roles in the reduction of lipogenesis, cholesterol synthesis inhibition, and more recently for its activation of G protein-coupled receptors (GPR) 41 and GPR43, release of satiety hormones and other metabolic and anti-inflammatory effects( 25 – 27 ). Butyric acid has been studied for its ability to promote colonic healing in colitis( 28 ) and its potential anti-cancer effects( 29 ), including apoptosis stimulation( 30 ), in part by inhibiting histone de-acetylase( 31 ). It has also been shown to inhibit oxidative damage in cultured cancer cells( 32 ) and it may improve gut barrier function( 33 ). Recent studies in mice have shown that oral administration of propionate and butyrate, but not acetate, facilitates the extra-thymic de novo generation of anti-inflammatory regulatory T (Treg) cells. In contrast, rectal administration of acetate and propionate, but not butyrate, promoted accumulation of Treg cells, suggesting that butyrate promotes de novo generation but not colonic accumulation of Treg cells, whereas acetate has an opposite activity and propionate is capable of both( 34 ). Furthermore, propionate and butyrate were shown to activate intestinal gluconeogenesis (IGN), which has beneficial effects on glucose and energy homeostasis, via complementary mechanisms. Whereas butyrate directly activates the IGN genes, propionate-mediated induction of IGN depends on a gut–brain communication axis involving the fatty acid receptor GPR41( 35 ). Those beneficial activities of SCFA produced in the intestine have been shown in different animal species including laboratory animals and production/farm animals( 36 – 38 ). However, in human subjects the relevant body of evidence is limited mainly because SCFA are traditionally only measured in faeces or fasting blood samples.
In vitro fermentation studies with prebiotics or dietary fibre have consistently resulted in increased levels of SCFA. In contrast, several human prebiotic intervention studies failed to demonstrate increased faecal SCFA, most likely due to the rapid colonic absorption of the SCFA, preventing them from being excreted in faeces( 39 – 44 ). The relative proportions of the SCFA vary between individuals and are particularly sensitive to the type of carbohydrate being fermented( 45 , 46 ). For example, the proportion of propionic acid production is increased during fermentation of guar gum, long-chain arabinoxylans, oats and oat fractions (oat bran and β-glucan), pectin, pulses, wheat dextrin and pyrodextrins( 47 – 56 ) whereas oligofructose predominantly yields acetate( 57 ). In contrast, the proportion of butyrate production increases with fermentation of starch and inulin-type fructans and often results from secondary fermentation of lactate and acetate, so-called cross-feeding between bacteria( 58 ). Indeed, this type of microbial cross-feeding could be viewed as an important physiological function supporting microbiota homeostasis and species richness, which have both been associated with gut health. In view of the different effects of acetic, propionic and butyric acids, the relative proportions of the acids produced are probably as relevant as their total levels.
Several animal and human studies have recently demonstrated an association between the gut microbiota composition and obesity. Initial studies found with a higher Firmicutes:Bacteroidetes ratio in the obese( 59 , 60 ). A greater fermentation capacity in both obese animals and human subjects compared with normal-weight subjects was suggested( 61 – 63 ) as well as increased concentrations of caecal or faecal SCFA. It was therefore hypothesised that the efficiency of the energy harvest from food was increased in obesity. However, later studies demonstrated that in human subjects, the relationship between the Firmicutes:Bacteroidetes ratio and obesity is less clear and human obesity may be associated with more subtle changes in the microbiota composition( 64 , 65 ). In a recent metagenomic study the typical ecological entity of microbiota ‘richness’ was highlighted as a strong determinant in body-weight control rather than its composition per se ( 66 ). Reported levels of faecal SCFA in obese subjects were higher than in normal-weight subjects (Table 2), although this has not been confirmed in all studies to date( 67 , 68 ). In addition, increased faecal SCFA levels do not necessarily indicate higher absorption rates and increased energy harvest by the host. Indeed, uptake of SCFA in colonocytes via the monocarboxylate transporter 1 (MCT-1) receptor is induced by fibre feeding (pectin) and butyrate, and moreover, inhibited by bile acids. Therefore, SCFA absorption might be reduced in the obese( 69 , 70 ). Fermentation capacity can be evaluated as in vitro production of SCFA from carbohydrates using faeces from healthy individuals and patients( 71 ). In pH-controlled faecal batch cultures, similar levels of SCFA were produced from α-gluco-oligosaccharides by microbiota from obese and lean subjects( 72 ). Overall, the relationships between the microbiota composition, intestinal SCFA levels and obesity are far from being elucidated. In a nice series of experiments in animals and human subjects, Cani and colleagues demonstrated how modulation of the microbiota by prebiotics controls endogenous glucagon-like peptide 2 production and the endocannabinoid system and contributes to the improvement in gut barrier function during obesity( 73 – 75 ). The direct involvement of specific gut bacteria and/or metabolites needs to be further investigated.
In recent studies, the intestinal bacteria have been implicated in the development of IBD and autoimmune diseases such as type 1 diabetes and coeliac disease. These conditions have been consistently characterised by a low abundance of butyrate-producing bacteria( 76 – 80 ). Functional analysis of the microbiota revealed remarkably lower levels of faecal SCFA in IBD( 81 – 83 ) whereas total SCFA and in particular acetate were found increased in coeliac disease( 84 – 86 ). Allergic children had lower faecal levels of propionate and butyrate than non-allergic children( 87 ). It remains to be explored to what extent these aberrant SCFA patterns are causative to the disease or can serve as markers of disease.
Lactate and succinate
Lactate and succinate are intermediates in the fermentation process of carbohydrates. In healthy conditions, they are further metabolised to acetate or butyrate and propionate, respectively, by cross-feeding species and do not substantially accumulate in the colonic lumen( 88 ). Recent evidence suggests that succinate acts as a signal for inflammation( 89 ). It stabilises the transcription factor hypoxia-inducible factor-1α (HIF-1α) in activated macrophages. When stabilised, HIF-1α up-regulates several genes including the inflammatory cytokine IL-1β, resulting in exacerbation of inflammation( 90 ). In addition, succinate acts as a ligand for GPR91, renamed SUNCR1. In the kidney, succinate-induced activation of GPR91 is reported to regulate the renin–angiotensin system and in dendritic cells, succinate signalling is required for enhanced antigen-presenting function. Increased levels of succinate have been linked to IBD as mice undergoing dextran sulfate sodium-induced colitis were shown to have more succinate in their caecum and faeces( 91 ) whereas in colonic tissue from dextran sulfate sodium-induced mice, succinate levels were lower than in control mice( 92 ). Therefore, succinate may be an ulcerogenic agent in the gut lumen, leading to mucosal damage and lower succinate levels in colonic tissues.
Lactate has two optical isomers, which are l-lactate and d-lactate. l-Lactate is produced from pyruvate by the enzyme lactate dehydrogenase during normal anaerobic metabolism whereas d-lactate is produced by many commensal bacteria in the colon. Increased levels of d-lactate in plasma and urine have been demonstrated in IBD( 81 , 93 ), intestinal ischaemia( 94 ), short bowel( 95 ) and appendicitis( 96 ) and are considered as a marker of dysbiosis and/or increased intestinal permeability. In faecal samples of IBD patients, mainly l-lactate levels are increased( 80 , 97 , 98 ), suggesting a mucosal origin of lactate( 99 ). As lactate is a potentially important co-substrate for many sulfate-reducing bacteria, increased colonic lactate levels may promote sulfide generation( 100 ) which is suspected of inhibiting the β-oxidation of butyrate in the colonocytes (see below).
Products of protein metabolism
Microbial products from protein metabolism include BCFA, ammonia, phenol, p-cresol, indole and hydrogen sulfide( 101 ). The toxic potential of these compounds is mainly derived from in vitro experiments, in which isolated cells or tissues are directly incubated with individual compounds, or from animal studies. The present review also encompasses the results of oral toxicity tests as a reasonable surrogate for assessing potential systemic effects. Minimally irritating concentrations were assessed as a marker of local effects and exceeded 1 % for all compounds, which is well above the concentrations occurring in the colon. In human studies, there is little evidence for adverse effects of protein fermentation metabolites( 102 ). In a recent study in healthy subjects, modulation of the degree of protein fermentation by changing dietary intake did not affect faecal water toxicity( 103 ).
An important determinant of the degree of proteolytic v. saccharolytic fermentation is the nutrient availability and in particular the ratio of available carbohydrate to nitrogen( 104 , 105 ). Therefore, the production of protein degradation products can generally be reduced by increasing the amount of fermentable carbohydrate reaching the colon in the form of resistant starch( 104 ) or prebiotic oligosaccharides( 106 – 112 ). In contrast, faecal ammonia, phenol and p-cresol were not affected after 4 weeks consumption of the polyol isomalt (30 g/d)( 113 ).
Phenol, p-cresol and indole
The phenolic compounds phenol, p-cresol and indole are the major metabolites of bacterial fermentation of the aromatic amino acids tyrosine, phenylalanine and tryptophan. These metabolites are largely and rapidly absorbed by the colonic mucosa cells and are excreted in urine after sulfate or glucuronide conjugation in the mucosa or the liver( 114 ). In healthy subjects, these compounds do not accumulate in the body. Therefore, their urinary elimination is often considered as a reliable estimate of their production in the colon( 105 ).
Many studies have reported significant interindividual variation in the urinary excretion of p-cresol and phenol in healthy adults. Data on ranges in other age groups (children, elderly) are scarce. Reported mean or median values vary between 10 and 55 mg/d for p-cresol (Table 3) and between 4 and 7·5 mg/d for phenol (Table 4). In obese individuals, urinary p-cresol and phenol levels at baseline were considerably higher than those reported in normal-weight adults (94·9 mg/d and 15·0 g/d for p-cresol and phenol, respectively) and decreased upon weight loss( 63 ). The levels of urinary p-cresol may increase in the very old( 115 ).
Table 3.
Reported excretion of p-cresol in urine and faeces
| Biofluid | Subjects (n); age | Reported measure | p-Cresol excretion | Unit | Reference | |
|---|---|---|---|---|---|---|
| Healthy subjects | Urine | 11; 35 ± 10 years | Mean (sem) | 454 (92)* | μmol/d | Birkett (1996)( 104 ) |
| 27; median 25 (IQR 23–29) years | Median (IQR) | 168 (93·3–304) and 208 (114–288) | μmol/d | Damen (2012)( 107 ) | ||
| 9; range 19–69 years | Mean (sem) | 408·3 (271·3) | μmol/d | Ling (1992)( 245 ) | ||
| 11; 39 ± 11 years | Median (IQR) | 532 (250–659)† p-cresyl sulfate | μmol × 1·73 m2 | Patel (2012)( 246 ) | ||
| 32; range 47–95 years | Mean (sd) | 510 (358)* | μmol/d | Renwick (1988)( 118 ) | ||
| 10; range 22–45 years 9; range 22–45 years | Mean (sd) | 248 (99)* 315 (206)* | μmol/d | De Preter (2004)( 247 ) | ||
| 15; 23 ± 1 years 15; 22 ± 1 years 15; 23 ± 1 years | Mean (sd) | 164 (101)* 186 (119)* 187 (96)* | μmol/d | De Preter (2007)( 248 ) | ||
| 10; 21 ± 1 years 9 | Median (IQR) | 196 (168–322)* 226 (141–368)* | μmol/d | De Preter (2007)( 249 ) | ||
| 20; median 23 (IQR 21–24) years | Median (IQR) | 297 (194–437) | μmol/d | Cloetens (2010)( 108 ) | ||
| 12; median 24 (IQR 21–28) years | Median (IQR) | 214 (107–315) | μmol/d | Cloetens (2008)( 250 ) | ||
| 20; range 19–41 years | Median (IQR) | 297 (239–349) | μmol/d | Windey (2012)( 103 ) | ||
| 19; range 21–53 years | Mean (sem) | 218 (58) | μmol/d | Gostner (2006)( 113 ) | ||
| Faeces | 11; range 3–11 years | Mean (sem) | 0·54 (0·29) | μmol/g faeces | Adams (1985)( 119 ) | |
| 11; 35 ± 10 years | Mean (sem) | 0·60 (0·07)* | μmol/g faeces | Birkett (1996)( 104 ) | ||
| 112; range 0–1 years | Mean (sd) | 0·14 (0·14) | μmol/g faeces | Heavey (2003)( 251 ) | ||
| 15; 23 ± 1 years 15; 22 ± 1 years 15; 23 ± 1 years | Mean (sd) | 124 (38)* 161 (53)* 101 (43)* | μmol/72 h | De Preter (2007)( 248 ) | ||
| 16; range 23–66 years | Mean (sem) | 58·86 (7·3) | μmol/g faeces | Clarke (2011)( 252 ) | ||
| 20; range 18–24 years | Mean (sem) | 0·52 (0·05)* | μmol/g dry weight | Lecerf (2012)( 109 ) | ||
| 19; range 21–53 years | Mean (sem) | 0·36 (0·04)* | μmol/g faeces | Gostner (2006)( 113 ) | ||
| 21; range 21–28 years | Mean (sem) | 1·5 (0·20) | μmol/g dry weight | Boler (2011)( 111 ) | ||
| Obese | Urine | 91, range 24–64 years | Mean (sd) | 879 (00) and 524 (259)* | μmol/d | Brinkworth (2009)( 63 ) |
| Faeces | 33, range 20–65 years | 0·54* | μmol/g faeces | Benassi-Evans (2010)( 244 ) |
IQR, interquartile range.
Calculated from reported values in mg/d using a molecular mass value for p-cresol of 108.
Calculated from reported values in mg/d using a molecular mass value for p-cresyl sulfate of 188.
Table 4.
Reported excretio of phenol in urine and faeces
| Biofluid | Subjects (n); age | Reported measure | Phenol excretion | Unit | Reference | |
|---|---|---|---|---|---|---|
| Healthy subjects | Urine | 11; 35 ± 10 years | Mean (sem) | 53 (0) | μmol/d | Birkett (1996)( 104 ) |
| 27; median 25 (IQR 23–29) years | Median (IQR) | 47 (37–66) and 43 (35–59) | μmol/d | Damen (2012)( 107 ) | ||
| 9; range 19–69 years | Mean (sem) | 73 (45) | μmol/d | Ling (1992)( 245 ) | ||
| 32; range 47–95 years | Mean (sd) | 67 (91) | μmol/d | Renwick (1988)( 118 ) | ||
| 15; 23 ± 1 years 15; 22 ± 1 years 15; 23 ± 1 years | Mean (sd) | 45 (21) 42 (25) 46 (22) | μmol/d | De Preter (2007)( 248 ) | ||
| 20; median 23 (IQR 21–24) years | Median (IQR) | 55 (31–76) | μmol/d | Cloetens (2010)( 108 ) | ||
| 19; range 21–53 years | Mean (sem) | 115 (32) | μmol/d | Gostner (2006)( 113 ) | ||
| Faeces | 11; range 3–11 years | Mean (sem) | 0·083 (0·034) | μmol/g faeces | Adams (1985)( 119 ) | |
| 11; 35 ± 10 years | Mean (sem) | 0·042 (0·006) | μmol/g faeces | Birkett (1996)( 104 ) | ||
| 112; range 0–1 years | Mean (sd) | 0·58 (0·84) | μmol/g faeces | Heavey (2003)( 251 ) | ||
| 15; 23 ± 1 years 15; 22 ± 1 years 15; 23 ± 1 years | Mean (sd) | 8·4 (7·1) 21·0 (16·5) 8·8 (6·9) | μmol/72 h | De Preter (2007)( 248 ) | ||
| 16; range 23–66 years | Mean (sem) | 0·7 (0·13) | μmol/g faeces | Clarke (2011)( 252 ) | ||
| 20; range 18–24 years | Mean (sem) | 0·0003 (0) | μmol/g dry weight | Lecerf (2012)( 109 ) | ||
| 19; range 21–53 years | Mean (sem) | 0·058 (0·015) | μmol/g faeces | Gostner (2006)( 113 ) | ||
| Obese | Urine | 91, range 24–64 years | Mean (sd) | 158 (119) and 159 (184) | μmol/d | Brinkworth (2009)( 63 ) |
| Faeces | 33, range 20–65 years | 0·037 | μmol/g faeces | Benassi-Evans (2010)( 244 ) |
IQR, interquartile range.
Calculated from reported values in mg/d using a molecular mass value for phenol of 94.
Most studies on the urinary excretion of the indole metabolite indoxyl sulfate, also called indican, report values below 50 mg/d in healthy adults. In patients with liver cirrhosis( 116 ) and patients with diabetes( 117 ), excretion of indoxyl sulfate is higher (98·2 mg/d in cirrhosis, 65·7 mg/d in diabetics without neuropathy and 114·0 mg/d in diabetics with neuropathy) and correlates with steatorrhoea. In a study in patients with bladder cancer there was no evidence that phenolic microbial metabolites had promoting or co-carcinogenic activity for the human urinary bladder, as the urinary excretion of p-cresol, phenol and indoxyl sulfate was not different in the patients as compared with controls( 118 ).
Faecal excretion of phenolic compounds is not often reported, but in available studies amounts of 5–8 mg/d for p-cresol and 0·25–0·66 mg/d for phenol have been found. Interestingly, faecal excretion of p-cresol was 4-fold higher in a group of hyperactive children as compared with control children( 119 ).
The effects of phenolic compounds on intestinal cells have been determined mainly in in vitro incubation studies. Viability of colonic epithelial cells isolated from human biopsies was decreased after exposure to 1·25 mm-phenol, a physiologically relevant concentration, whereas higher phenol concentrations (20 mm) were required to reduce viability of HT-29 cells( 120 ). Notably, cell cultures from ulcerative colitis (UC) patients showed similar sensitivity to phenol exposure as cell cultures from control subjects at all concentrations tested.
Several papers have reported a concentration-dependent increase in paracellular permeability and reduced epithelial barrier function after incubation with phenol (1 μm to 21 mm) of Caco-2-monolayers or SK-CO15 intestinal cells( 121 , 122 ). Enhanced permeability was already apparent at concentrations of phenol that did not cause cell death. Similarly, p-cresol altered endothelial barrier function in human umbilical vein endothelial cells. In chronic kidney disease patients, p-cresol is considered a uraemic toxin. It accumulates in serum and might participate in the endothelial dysfunction that is observed in such patients( 123 ).
The European Food Safety Authority recently evaluated the toxicity of phenol following oral administration (http://www.efsa.europa.eu/en/efsajournal/doc/3189.pdf) and established a tolerable daily intake (TDI) of 0·5 mg/kg body weight per d. For a 75 kg individual, the TDI amounts to 37·5 mg/d, which is about 5-fold higher than the amount of phenol generated in the colon (7·5 mg/d) (assuming that urinary excretion rates reflect colonic generation rates). On repeat-dose administration to non-pregnant rats and mice, no consistent effects were seen at doses ≥ 250 mg/kg body weight per d.
The Joint FAO/WHO Expert Committee on Food Additives (JECFA) reviewed the oral toxicity of p-cresol in 2011. The systemic toxicity of p-cresol was evaluated in a 2-year study in rats following dietary administration as a 60:40 mixture of m-/p-cresol. (http://www.inchem.org/documents/jecfa/jecmono/v64je01.pdf). A no observed adverse effect level of 230 mg/kg body weight per d was identified, based on increased incidence of renal tubule adenomas in male rats at 720 mg/kg body weight per d. Effects seen in other studies (nasal sinuses, forestomach) were attributed to the local irritancy of p-cresol and did not reflect its systemic toxicity. Effects observed in a 13-week repeated exposure study in rats by oral administration were probably secondary to the local irritancy of p-cresol.
Data on the toxicity of indole are very limited. JECFA reviewed the effects after oral exposure to indole in 2006 (http://www.inchem.org/documents/jecfa/jecmono/v54je01.pdf). Following exposure of rats to indole at a dose of 100 mg/kg body weight per d in the diet for 460 d, signs of moderate reversible anaemia were apparent. No other adverse effects were observed. Rats fed a low-protein diet supplemented with indole (0·25–2 %) showed overall weight loss and growth retardation as well as haemolytic anaemia( 124 ).
Ammonia
Due to bacterial degradation of unabsorbed and endogenous nitrogenous compounds and endogenous nitrogen recycling, the colonic epithelium is constantly exposed to ammonia in millimolar concentrations( 125 ). Faecal ammonia excretion is comparable in overweight adults and normal-weight adults but is clearly lower in infants (Table 5). A recent study reported significantly higher faecal ammonia concentrations in children with autism spectrum disorders (ASD) (42·7 (SE 3·3) μmol/g faeces) compared with control children (32·3 (SE 1·9) μmol/g faeces)( 126 ). As elevated plasma ammonia concentrations have also been described in ASD, the authors suggested that higher faecal concentrations might translate into higher plasma ammonia concentrations.
Table 5.
Reported excretion of ammonia in faeces
| Population | Subjects (n); age | Reported measure | Ammonia excretion | Unit | Reference |
| Healthy subjects | 11; 35 ± 10 years | Mean (sem) | 23·3 (1·5) | μmol/g faeces | Birkett (1996)( 104 ) |
| 112; range 0–1 years | Mean (sd) | 7·7 (7·1) | μmol/g faeces | Heavey (2003)( 251 ) | |
| 16; range 23–66 years | Mean (sem) | 14·1 (1·7) | μmol/g faeces | Clarke (2011)( 252 ) | |
| 12; range 27–49 years | Mean (sd) | 5·1 (2·5) | μmol/g faeces | Slavin (2011)( 41 ) | |
| 9; 59 ± 3 years | Mean (sd) | 2·9 (0·5) | μmol/g faeces | Bianchi (2010)( 253 ) | |
| 46; 31–66 years | Mean (95 % CI) | 14·6 (12·6, 16·7) | μmol/kg faeces | McOrist (2011)( 240 ) | |
| 20; range 20–55 years | Mean (sem) | 22·6 (1·6) | μmol/g faeces | Tiihonen (2010)( 67 ) | |
| 8; range 21–60 years | Mean (sd) | 35·2 (7·9)* | μmol/g faeces | Shinohara (2010)( 110 ) | |
| 21; range 21–28 years | Mean (sem) | 137·5 (7·82) | μmol/g dry weight | Boler (2011)( 111 ) | |
| 36; range 22–67 years | Median (IQR) | 24·6 (16·5–30·0) | μmol/g faeces | Nemoto (2012)( 241 ) | |
| Obese | 19; range 20–55 years | Mean (sem) | 26·6 (2·2) | μmol/g faeces | Tiihonen (2010)( 67 ) |
| 121; 24–64 years | Mean (sd) | 26·5 (9·2) | μmol/g faeces | Brinkworth (2009)( 63 ) |
IQR, interquartile range.
Calculated from reported values in mg/d using a molecular mass value for ammonia of 17.
As early as the 1970s, reports on the effects of ammonia on epithelial cells have appeared. Visek (in 1978)( 127 ) was the first to report that ammonia alters nucleic acid synthesis, changes the morphology and intermediary metabolism of intestinal cells and reduces the lifespan of cells. After this initial report, several studies evaluated the impact of physiological concentrations of ammonia using isolated colonocytes, cell lines or animal colon tissue (for reviews, see Windey et al. ( 102 ) and Blachier et al. ( 128 )).
The European Food Safety Authority reviewed the effects of dietary administration of ammonia (as ammonium chloride) to rats for up to 30 months (http://www.efsa.europa.eu/en/efsajournal/doc/1925.pdf). It was not possible to identity a no observed adverse effect level from any of the studies, which in general used high doses to study the effects of metabolic acidosis, rather than the compound itself. Although a number of effects were observed, these were considered adaptive, and no adverse effects were observed at doses of 1100 mg/kg body weight per d for 30 months.
Hydrogen sulfide
Sulfate-reducing bacteria scavenge hydrogen as an electron donor and use sulfate as an oxidising agent for the dissimilation of organic matter. The major endproduct from this reaction is hydrogen sulfide. Luminal concentrations of sulfide are in the range of 1·0–2·4 mmol/l( 129 ) whereas faecal concentrations vary from 0·17 to 3·38 mmol/kg faeces( 130 , 131 ). It is probable that a large fraction of sulfide is bound to luminal compounds within the intestine.
The toxic potential of hydrogen sulfide on colonic cells has been extensively investigated. Sulfide influences oxidative metabolism of colonic epithelial cells by inhibiting cytochrome oxidase activity which catalyses the reduction of oxygen to water( 132 , 133 ). Several lines of evidence also suggest a role of hydrogen sulfide in the aetiology and/or risk of relapse of UC. In experimental animal models, a pathological condition similar to UC can be induced using undigestible sulfates in the form of dextran sulfate sodium or the sulfate-containing carrageenan. Whilst some studies found elevated faecal sulfide levels in patients with UC( 134 , 135 ), others did not( 136 ). However, detoxification of sulfide by the mucosal thiosulfate sulfurtransferase enzyme to the less toxic thiocyanate is impaired in UC patients( 137 ). In addition, a diet characterised by high meat intake as well as a high sulfur or sulfate intake was associated with increased likelihood of relapse in UC patients( 138 ). Exposure of non-transformed rat intestinal crypt cells (IEC-18 cells) to sodium hydrogen sulfide (50 μm) caused acute hypoxia and promoted early cell-cycle entry with an associated up-regulation of genes coding for proteins related to proliferative activity( 139 ). A series of in vitro experiments by Attene-Ramos et al. ( 140 ) revealed that hydrogen sulfide provokes genomic DNA damage in colonic cancer cells (HT-29 cells) at concentrations of 250 mm. No cellular metabolism was required for sulfide to induce genotoxicity and co-incubation with a radical scavenger reduced DNA damage induced by hydrogen sulfide, suggesting a radical-mediated mechanism( 141 ). In non-transformed human intestinal epithelial cells, the expression of genes involved in cell-cycle progression, inflammation and DNA repair response was modulated by sulfide. In particular, expression of the cyclo-oxygenase (COX)-2 gene, which is elevated in most human colorectal cancers (CRC), was significantly up-regulated( 142 ). Overexpression of COX-2 may play a decisive role in promoting CRC initiation or progression through the stimulation of angiogenesis, inhibition of apoptosis and increasing the proliferation in intestinal epithelial cells( 143 ).
In contrast to those reports on harmful effects, hydrogen sulfide is now also known to be a systemic signalling molecule. It is endogenously produced in micromolar concentrations from cysteine by the action of cystathionine γ-lyase and cystathionine β-synthase. At these low concentrations, it has been proposed that hydrogen sulfide is involved in neuromodulation of chloride secretion, in controlling ileum contractility and in nociception from the large intestine( 144 ). Blachier et al. proposed as a working hypothesis that any imbalance between levels of free sulfide in the large intestine and the capacity of epithelial cells to metabolise it will result in a loss of normal oxidative cell capacity( 144 ).
Information on the effects of oral exposure to hydrogen sulfide is very limited. The US Environmental Protection Agency (EPA) established a reference dose on the basis of effects observed in pigs, but subsequently withdrew this as it was concluded that the effect was irreproducible (http://www.epa.gov/iris/subst/0061.htm). Although the US EPA has established an inhalation reference concentration for hydrogen sulfide, this is based on local effects and hence is not suitable for assessing the systemic toxicity of the compound.
Branched-chain fatty acids
The BCFA isobutyrate, 2-methylbutyrate and isovalerate are produced by bacterial fermentation of valine, isoleucine and leucine, respectively. These BCFA constitute approximately 5–10 % of the total SCFA( 145 ). Similar to other protein fermentation metabolites, faecal BCFA concentrations are reduced after prebiotic intake( 106 , 146 , 147 ). In an in vitro incubation experiment with five different epithelial cell lines; minimal concentrations of isovalerate to induce cytotoxicity were lower than the concentrations produced by intestinal bacteria and both isovalerate and isobutyrate were able to induce apoptosis( 148 ). Several in vitro studies indicate that BCFA affect the exchange of ions in the colon and may act as a regulator of colonic Na+ absorption( 128 ) but little information is available regarding other effects of BCFA on colonic epithelial cells. Therefore, faecal concentrations of BCFA are considered only as markers for bacterial protein fermentation rather than markers of colonic health( 149 ).
Plant polyphenolic compounds/catabolites
While the physiological relevance of polyphenol catabolites derived from the gut microbiota is currently understudied, it is recognised that the majority of plant bioactive compounds must first be rendered biologically available, often through deglycation and hydrolysis by the gut microbiota before being absorbed by the host, and that microbial metabolic transformation can make an impact on polyphenol biological activity. While certain classes of phytochemical are broken down to unique catabolites, many different classes of polyphenol give rise to common small phenolic compounds. However, we currently do not know the physiological relevance of many of these compounds, or even the habitual or ‘normal’ concentration ranges of these phenolic acids, or how they respond to diet. For a few of these compounds, for example, the phyto-oestrogens equol, enterolactone and enterodiol, and the urolithins, specific health effects have been suggested( 150 ). Table 6 provides an overview of the microbial catabolites of common plant polyphenols and their putative health effects. For an up-to-date review on plant polyphenols catabolites and their putative health effects, see Del Rio et al. ( 150 ) and Dall'Asta et al. ( 151 ).
Table 6.
List of microbial catabolites of common plant polyphenols and their putative health effects( 155 )
| Plant polyphenol | Microbial catabolite | Possible health effects | References | |||||||||
| (–)-Epicatechin | 4-Hydroxyphenylacetic acid | Antimicrobial/antimycotic activity in vitro | Alakomi (2007)( 254 ) Ko (2009)( 255 ) Roowi (2010)( 256 ) | |||||||||
| 3-(3-Hydroxyphenyl)propionic acid | Antimicrobial activity against Gram-negative enterobacteria via outer membrane destabilisation | |||||||||||
| 5-(3,4-Dihydroxyphenyl)-γ-valeric acid | ? | |||||||||||
| (-)-5-(3′,4′-Dihydroxyphenyl)-γ-valerolactone | ? | |||||||||||
| (–)-Epigallocatechin | 4-Hydroxyphenylacetic acid | Antimicrobial/antimycotic activity in vitro | Roowi (2010)( 256 ) | |||||||||
| (-)-5-(3′,4′-Dihydroxyphenyl)-γ-valerolactone | ||||||||||||
| (–)-Epigallocatechin-3-O-gallate | Pyrocatechol | Ko (2009)( 255 ) Roowi (2010)( 256 ) Okello (2012)( 257 ) Ni (2008)( 258 ) Taguri (2006)( 259 ) | ||||||||||
| Pyrogallol | Antibacterial activity (especially against Gram-negative enterobacteria) An acetylcholinesterase inhibition greater than gallic acid parent Inhibition of Vibrio spp. quorum sensing | |||||||||||
| 4-Hydroxyphenylacetic acid | Antimicrobial/antimycotic activity in vitro | |||||||||||
| (-)-5-(3′,4′-Dihydroxyphenyl)-γ-valerolactone | ? | |||||||||||
| Daidzein | Equol | Phyto-oeotrogen important for heart and bone health, and possible colon cancer protectants | Jackman (2007)( 260 ) Ishimi (2009)( 261 ) Davis (2009)( 262 ) Selma (2009)( 263 ) | |||||||||
| Daidzein | O-demethylangolensin | Oestrogenic and/or anti-oestrogenic activity | Larrosa (2006)( 264 ) Selma (2009)( 263 ) | |||||||||
| Quercetin | 2-(3,4-Dihydroxyphenyl)acetic acid | Selma (2009)( 263 ) | ||||||||||
| 2-3-(3-Hydroxyphenyl)acetic acid | ||||||||||||
| 3,4-Dihydroxybenzoic acid | ||||||||||||
| Phloroglucinol | ||||||||||||
| 3-(3,4-Dihydroxyphenyl)propionic acid | ||||||||||||
| 3-(3-Hydroxyphenyl)propionic acid | ||||||||||||
| Kaempferol | 2-(4-Hydroxyphenyl)acetic acid | Selma (2009)( 263 ) | ||||||||||
| Naringenin | 3-(4-Hydroxyphenyl)propionic acid | Antimicrobial activity against Gram-negative enterobacteria via outer membrane destabilisation | Selma (2009)( 263 ) | |||||||||
| Phloroglucinol | ||||||||||||
| Isoxanthohumol | 8-Prenylnaringenin | ? | Selma (2009)( 263 ) | |||||||||
| Catechin and epicatechin | 3-(3-Hydroxyphenyl)propionic acid | Alakomi (2007)( 254 ) Selma (2009)( 263 ) | ||||||||||
| 5-(3′,4′-Dihydroxyphenyl)-γ-valerolactone | ||||||||||||
| 5-(3′-Hydroxyphenyl)-γ-valerolactone | ||||||||||||
| 3-Hydroxyhippuric acid pyrogallol | ||||||||||||
| 5-(3,4-Dihydroxyphenyl)valeric acid | ||||||||||||
| 5-(3-Hydroxyohenyl)valeric acid | ||||||||||||
| 3-(3,4-Dihydroxyphenyl)propionic acid | Antimicrobial activity against Gram-negative enterobacteria via outer membrane destabilisation | |||||||||||
| 5-(3-Methoxyohenyl)valeric acid | ||||||||||||
| 3-(3,4-Dihydroxyphenyl)propionic acid | ||||||||||||
| 5-(3-Methoxyohenyl)valeric acid | ||||||||||||
| 2,3-Dihydroxyphenoxyl 3-(3′,4′-dihydroxyphenyl)propionic acid | ||||||||||||
| Ellagitannins/ellagic acid | Urolithin-A | Oestrogenic and/or anti-oestrogenic activity, antimalarials | Del Rio (2010)( 265 ) Larrosa (2006)( 264 ) Dell'Agli (2010)( 266 ) | |||||||||
| Urolithin-B | Oestrogenic and/or anti-oestrogenic activity, antimalarials | |||||||||||
| Urolithin-C | Oestrogenic and/or anti-oestrogenic activity | |||||||||||
| Urolithin-D | Oestrogenic and/or anti-oestrogenic activity | |||||||||||
| Rutin | 3-Hydroxyphenylacetic acid | Rutin and catabolites inhibit advanced glycation endproduct formation in vitro Antimicrobial activity against Gram-negative enterobacteria via outer membrane destabilisation | Alakomi (2007)( 254 ) Jaganath (2009)( 154 ) Cervantes-Laurean (2006)( 267 ) | |||||||||
| 3,4-Dihydroxybenzoic acid | ||||||||||||
| 4-Hydroxybenzoic acid | ||||||||||||
| 3-(3-Hydroxyphenyl)propionic acid | ||||||||||||
| 3,4-Dihydroxyphenylacetic acid | ||||||||||||
| Lignans | Enterolactone | Phyto-oestrogen important for heart and bone health, and possible colon cancer protectants | Davis (2009)( 262 ) | |||||||||
| Enterodiol | Phyto-oestrogen important for heart and bone health, and possible colon cancer protectants | |||||||||||
Furthermore, the interaction between polyphenols and the microbiota is bi-directional. Recent evidence has shown that a number of polyphenols and their metabolites cause a selective stress or stimulus to some micro-organisms and influence other metabolic pathways like the production of SCFA( 152 , 153 ). In addition, in vitro incubation of faecal samples with quercetin-3-O-rutenoside (rutin) in the present of glucose as a carbon source showed a significant increase in deglycosylation of rutin and catabolism of quercetin, suggesting that prebiotic intervention might modify the bacterial metabolism of plant polyphenols( 154 ).
In metabolomic studies (see below) many of the compounds encountered relate to microbial catabolites of polyphenols that escape absorption in the small intestine( 155 , 156 ). These may become key markers of colonic bacterial activity.
Emerging metabolites
A range of amino acids and related molecules, including tryptophan, γ-aminobutyric acid, γ-hydroxybutyric acid, or biogenic amines and also host–microbiota co-metabolic metabolites including catecholamines like dopamine and noradrenaline, and bile acids have the potential to make an impact both in a beneficial or harmful way with the host depending on concentration and chemical profile( 157 ). Recent scientific interest has been focused on microbiota production of cell-signalling molecules and neurotransmitters which through the gut–liver–brain axis appear to regulate a number of diverse physiological functions including energy intake and expenditure, brain development and cognitive function and mood( 158 , 159 ). However, by and large, human data are scarce and it needs to be evaluated whether prebiotic intervention might affect these signalling pathways.
Factors that influence fermentation
The mechanisms controlling the metabolic activities of the colonic microbiota are only partly understood.
First of all, the type and quantity of dietary carbohydrate entering the colon have a dramatic impact on SCFA production. Many factors make an impact on the digestibility of carbohydrate in foods, not least their intrinsic chemical structure or biological availability in low-processed plant-derived foods. However, other food macromolecules ingested at the same meal, for example, red wine polyphenols or fat, and the load of complex carbohydrates like starch, can also determine the amount that reaches the colon( 160 , 161 ). Similarly, the extent of carbohydrate fermentation by the gut microbiota may be affected by other food components, for example, complex polyphenols, which may have antibacterial activity( 162 ).
Besides the diet, the specific phylogenetic and functional composition of the gut microbiota is influenced by a range of factors including host genetics, immunological factors and environmental factors including use of drugs such as antibiotics( 1 , 163 ). The large-intestinal microbiota has a strongly individual composition in human subjects and exhibits a remarkable compositional stability over time( 164 ), but is also amenable to dietary modulation( 165 ). Whereas some bacterial species are able to degrade a wide variety of substrates, other bacteria are nutritionally highly specialised( 1 ).
In addition, key geographic differences exist in bacteria and metabolites in different populations in different countries and regions, for example, in North and South China( 166 ), different countries in Europe( 163 ) and different populations (Afro-, Caucasian and Native-Americans) in the USA( 167 ). Some of these differences may be due to diet and lifestyle but genetic background may also be involved. These differences are important to take into account when investigating the relationship between bacterial metabolism and disease.
Finally, fermentation patterns are also determined by colonic transit times( 168 ). In a study by Cummings et al. a significant correlation was observed between colonic transit times and urinary excretion rates of phenols( 169 ). With shorter transit times, turnover rate is faster and microbial growth is more efficient, resulting in a greater mass of bacteria( 170 ). Similarly, in in vitro fermentation experiments with faecal inocula from volunteers with pharmacologically modified transit times, reduction of transit time was associated with increased production of SCFA and increased disappearance of substrate( 171 ). Although fermentation of carbohydrate occurs mainly in the proximal colon, a mixture of fermentable and less fermentable carbohydrates in the diet can push fermentation further around the colon and thus increase SCFA also in the more distal parts of the colon( 172 – 175 ). The European Food Safety Authority has recently approved two health claims for wheat bran in relation to two beneficial physiological effects, namely an increase in faecal bulk and a reduction of intestinal transit time. Whether increased SCFA production is responsible for the increased transit time in humans remains to be studied. In animal models, SCFA inhibit peristaltic contractile activity, however, only at concentrations above a physiological threshold( 176 ). In addition, the motor effects of SCFA may differ between species as intracolonic infusion of a 100 mm-SCFA solution did not modify transit in two healthy human subjects( 177 ). In a recent study in ten volunteers, infusion of a 100 mm-SCFA solution did not affect the phasic or tonic motor activity of the colon or the number of high-amplitude-propagated contractions( 178 ).
Gaps and limitations
Where to measure: choice of the biomatrix
A major obstacle in the evaluation of intestinal bacterial metabolism in vivo in human subjects is the inability to directly sample at the site of production. Therefore, much information has been obtained from analysis of faecal samples and supportive data from in vitro and experimental models. However, information on the activity of the intestinal microbiota can be derived from analysis of various biological samples including faecal samples, serum or plasma and urine samples. Zhao et al. ( 174 ) nicely showed that a modification of the intestinal microbiota in mice resulted in altered metabolite patterns in faeces. Administration of non-absorbable antibiotics resulted in increased levels of Bacteroides and Enterococcus species and was accompanied by a reduction in the overall fermentation of indigestible carbohydrates with lower levels of SCFA, lower levels of many amino acids and a disturbance of bile acid metabolism( 174 ). In contrast, the faecal metabolome in horses was shown not to be representative of the colonic metabolome( 179 ), which reflects the different patterns of bacterial metabolism and the absorption of products in different parts of the colon. Similarly, the impact of microbiota activity is reflected in the levels of several serum metabolites( 180 ). Interestingly, the host responds to many of those metabolites with phase II metabolism comparable with the response to drugs, as many metabolites are sulfated, glycine conjugated or glucuronidated to facilitate urinary excretion.
In recent years, many research efforts have focused on the mechanisms by which the SCFA acetate, propionate and butyrate affect host physiology, Nevertheless, reliable and quantifiable methodologies have rarely been employed to measure the relative SCFA production for different fibres in human subjects, or to quantify their relative contribution to circulating SCFA pools, for example, using stable isotope tracking or pharmaco- or nutri-kinetic approaches( 181 ). In addition, very few studies have examined the time course of SCFA production, absorption and utilisation after prebiotic intervention or examined the impact of other food components, underlying host disease or gut microbiota composition and genetic potential on these processes. Therefore it has been difficult to convincingly prove in human subjects a role for colonic SCFA produced from prebiotics in the key physiological processes proven to be regulated by SCFA in animal models. There is a critical need for multidisciplinary studies to address these questions and take that final mechanistic step from animal model to human physiology and health.
When to measure: snapshot analysis
Intestinal microbial fermentation is a dynamic process influenced by a wide range of factors (see above). Therefore, the nature and concentration of metabolites produced by the microbiota is context-dependent and the levels of each metabolite are a result of metabolic fluxes of highly variable rates, which are not adequately represented in steady-state metabolite profiles. The rapid uptake and conversion of metabolic intermediates, as well as their removal from the intestinal lumen through host absorption creates a highly dynamic system, which is strongly discrepant with the methodologies for analyses that can only provide a single time point quantification or ‘snapshot’. At present, very little is known about the short- or long-term variation in metabolite concentrations produced by the gut bacteria. The dynamic analysis of metabolic conversions within the microbiota may be further unravelled through the application of stable isotope-labelled nutrients( 182 ) that, in combination with metabolic modelling (for example, including the use of meta-transcriptome or meta-proteome datasets), may enable the determination of metabolic fluxes in the microbiota and the host mucosa.
More holistic approach
Functional analysis of faecal water
Prebiotics can affect the levels of many compounds in the gastrointestinal tract. These may be constitutively produced in the body, such as bile acids, or may be bacterial metabolites, such as SCFA. This complicates assessment of the potential effects of prebiotics on human health based on evaluation of individual substances. Hence, to supplement such assessment, a more holistic approach to the effects of prebiotics on the biological activity of the gastrointestinal milieu is required. One approach that might be useful for this purpose is the functional analysis of faecal water (for a review, see Osswald et al. ( 183 )). This should provide an integrated measure of the overall contribution of the compounds present to a defined biological endpoint, such as genotoxicity.
Faecal water has a number of potential advantages in such studies. It is non-invasive, it can reflect the effects of diet directly, it samples the initial target compartment, the gastrointestinal tract, it provides a measure of the total activity of what was present in the distal colon, it can provide repeated measurements over time, and subjects can serve as their own controls. Disadvantages include the practicalities of sample collection and the reluctance of some subjects to provide samples, the complexity of the biofluid may interfere in the assessment of some endpoints and it may not always be truly representative of the biological compartment of interest, because of modulation of intestinal contents before faecal excretion.
Perhaps the endpoint most widely assessed using faecal water is genotoxicity. A number of assays have been used for this purpose, including the Ames Salmonella test and SOS Chromo test for bacterial mutagenicity( 184 – 187 ). Over the past 15 years, those bacterial mutagenicity assays have been almost completely replaced by assays using mammalian cells as targets( 187 ). The Comet assay for DNA strand breaks in enterocyte cells (Caco-2, HT-29 and Hep G2 (liver derived)) has been commonly used to assess the genotoxic potential of faecal water. The considerable inter- and intra-individual variability between samples and individuals, possibly reflecting the effects of dietary variation, constitutes a major limitation. Use of a controlled dietary regimen for 9 d was of insufficient length to reduce interindividual variability, suggesting that changing the genotoxic potential of the gut microbiota may require much longer( 188 ). There is no ready means for normalisation of sample activity, equivalent, for example, to creatinine in urine. Efforts to use wet or dry weight of the stool have met with little success. However, the method of preparation of faecal water has little effect on the biological measurement that facilitates comparison across studies( 103 ).
The toxicity of faecal water has been assessed using a number of endpoints. Cytotoxicity has been measured by colorimetric cell viability assays based on the cleavage of a tetrazolium salt (for example, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) or water-soluble tetrazolium salt 1 (WST-1)) in the mitochondria of living cells to a coloured formazan derivative( 189 , 190 ). Changes in barrier function are assessed from impairment of tight junctions using transepithelial resistance( 191 , 192 ) or phenol red leakage( 193 , 194 ) in Caco-2 cells, and invasive potential of HTC116 cells using fluorescence-activated cell sorting (FACS) analysis( 191 ). In addition, the lytic activity of faecal water to erythrocytes has been quantified as a parameter of cytotoxicity and was significantly correlated to colonic cell proliferation( 195 ). Finally, the effects of faecal water on apoptosis rates in the human colon-derived cell lines HT-29 and FHC (fetal human cells) have been evaluated( 196 ). A number of apoptotic hallmarks were measured: changes in cell morphology, DNA fragmentation, FACS analysis of DNA strand breaks assessed using the terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) assay, and poly(ADP-ribose) polymerase cleavage.
In addition, several assays have been used to measure the effects of faecal water on cell proliferation. Using two human colon carcinoma cell lines, HT-29 and HCT 116, faecal water or their lipid extracts were found to activate activator protein-1 (AP-1), a transcription factor associated with the promotion of neoplastic transformation( 197 ), and to induce COX-2 promotor activity, which has been implicated in colon carcinogenesis( 198 ). Faecal water also inhibited cell cycle progression in HT-29 cells and down-regulated the gene expression of proliferating cell nuclear antigen, a protein essential for replication( 199 ).
Results on the effects of oligofructose and/or inulin treatment on the genotoxicity of faecal water in rats have been equivocal( 200 ). In a study in rats treated with azoxymethane, DNA damage assessed using the Comet assay was reduced with faecal samples obtained after 4 or more months of prebiotic treatment. Evidence from this study suggested that the antigenotoxic effects of prebiotics occur rapidly and that azoxymethane-induced tumour development increases the genotoxicity of faecal water( 201 ).
Relatively few studies to date have evaluated the effects of prebiotic administration on the biological effects of faecal water samples from human subjects. Feeding male smokers and non-smokers either plain sourdough bread, bread supplemented with prebiotics (inulin, linseed and soya flours) or bread additionally supplemented with antioxidants resulted, in the non-smoker group, in about 50 % reduction in DNA strand breaks induced by faecal water with both the control and test breads( 202 ). In smokers, the control and test breads reduced faecal water genotoxicity only in those with the glutathione S-transferase genotype GSTM1*0. Administration of polydextrose (8 g/d for 3 weeks) reduced faecal water genotoxicity using the Comet assay in thirty-three healthy subjects in a double-blind placebo controlled cross-over study( 44 ). Similarly, supplementation of the diet with konjac glucomannan (4·5 g/d) for 4 weeks in thirty healthy subjects significantly reduced faecal water genotoxicity( 203 ). In contrast, no changes in faecal water genotoxicity were observed after 4 weeks treatment with galacto-oligosaccharides (4 g/d) in a population above 50 years( 204 ).
Measurement of biological activity of faecal water samples is an attractive means of linking changes in the colonic contents with health outcomes. However, there are a number of potential limitations to this methodology. Standardisation of the assay protocols in terms of target cells and sample preparation is mandated to allow comparison of data between studies. To date there have been few studies using this approach in the evaluation of prebiotics.
Metabolomics
Metabolomics offers an alternative and holistic approach to understanding the interaction between the human gut microbiome and host metabolism as well as to identify possible biomarkers of gut health. Metabolomic studies allow simultaneous evaluation of a wide range of metabolites by a top-down approach bypassing the need for an a priori hypothesis. Several analytical platforms allow detection, identification and quantification of different ranges of molecules, including 1H-NMR, LC-MSMS and GC-MS( 205 , 206 ). However, due to the chemical diversity and different physico-chemical properties of the metabolites and the large dynamic range of metabolite concentrations in different biological samples, it is virtually impossible to measure the complete metabolome. In addition, the term ‘metabolome’ can refer to faeces( 205 ), urine or plasma( 207 ). By selecting a specific analytical platform and a biofluid in which metabolites will be measured, the metabolome will be reduced to those specific conditions. Metabolome approaches can be non-targeted so that any compounds are considered in a pattern, or targeted where more specific types of molecules are identified and sometimes quantified. In general, however, the strategy is to discover patterns of metabolites which are associated with disease states such as cancer( 208 – 210 ), the metabolic syndrome( 211 ), obesity( 212 ), CVD, diabetes( 213 ), gut disease such as UC( 205 ), irritable bowel syndrome( 214 ), peptic ulcer( 215 ) and the impact of changes in body weight or diet( 156 , 211 , 216 , 217 ).
Although broad-spectrum metabolomics is shedding new light on the metabolites derived from the gut microbiota at an unprecedented resolution, compound quantification has been found time-consuming and has not been a priority in many metabolomics protocols. However, there is still a place for accurate, reproducible and targeted analytical chemistry approaches to quantify selected panels of health-relevant metabolites present in various body biofluids including faeces. Accurate quantification is particularly important when determining subtle changes in metabolite levels in response to dietary interventions which rarely block or ‘turn-off’ pathways or metabolic activities as drugs do but rather modulate the production rate of metabolites. Advances in mass spectroscopy instrumentation and methodology have allowed the development of ‘targeted metabolomics’ approaches where there are panels of specific metabolites, often of related physiological relevance, as an example metabolites related to diabetes or dyslipidaemia, can be accurately and reproducibly quantified in blood and urine( 218 ).
Application of metabolomic analysis in the evaluation of putative health benefits of prebiotics might provide additional evidence or elucidate the molecular bases of their actions. Martin et al. used a model of mice colonised with a human baby's microbiota to evaluate the impact of prebiotic galactosyl-oligosaccharides on the metabolic changes in ten biofluids/compartments. Prebiotic administration significantly reduced the lipids in the liver and kidneys and altered the transmethylation metabolic pathways (homocysteine–betaine)( 219 ). So far, only a few intervention studies in human subjects applied metabolomics to evaluate the impact of a synbiotic( 220 – 222 ). Recently, a metabolomic approach was applied in a placebo-controlled trial in patients with Crohn's disease who received a dietary intervention with oligofructose-enriched inulin (OF-IN) (2 × 10 g/d) for 4 weeks( 223 ). Faecal butyrate levels were up-regulated to the levels found in faecal samples of healthy controls after OF-IN intake. In view of the immunomodulatory and anti-inflammatory properties of butyrate, these observations might encourage follow-up studies in Crohn's disease with prebiotics.
Link to metagenome analysis
Meta-omics technologies enable the simultaneous, ecosystem-wide readout of phylogenetic composition and function, providing a blueprint of the microbiota's functional potential. Metagenomics employs established technologies that are fuelled by the continuous advances made in sequencing technologies, allowing the cost- and time-effective creation of functional catalogues of the human intestinal microbiota in individuals( 2 ), which have confirmed a substantially distinct microbial community composition in the large v. the small intestine( 224 ). It is far from trivial to translate the genetic repertoire or the metagenome of the microbiota into its actual in situ activity, and there is a strong requirement for the further development of functional metagenomic approaches, including metatranscriptomics and metaproteomics. Finally, meta-metabolomics/metabonomics approaches (on faecal water, blood or urine) allow monitoring the pool of metabolites produced by the microbiota in the intestinal lumen (faecal water metabolomes), and their impact on the host's systemic biochemistry (urine and blood metabolomes).
A main bottleneck in these approaches is the biological interpretation of these data and their integration to decipher the underlying interactions. All the techniques mentioned above yield massive datasets with large proportions of unknown features (unannotated genes, unidentified spectra, etc.). In addition, all data are fragmented and include the assignment of incomplete spectra to fragmented gene sequences and partially sequenced species, which is a challenging problem even for meta-omics datasets that are generated from the same samples. Also the integration of metabolomic profiles and intestinal metagenome composition remains challenging, which is also a consequence of the ‘snapshot characteristics’ of the datasets generated.
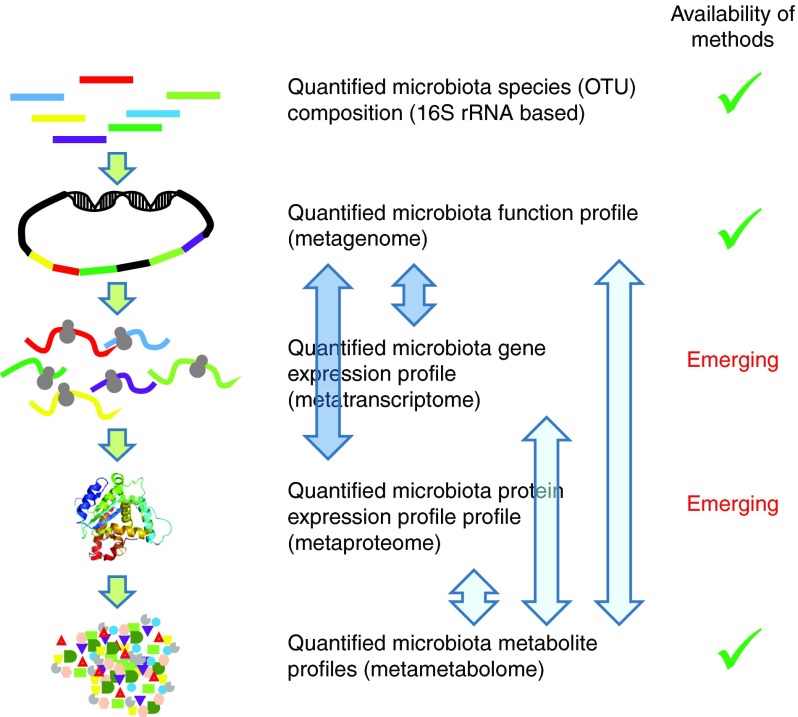
Fig. 2 shows a schematic presentation of the future needs to analyse the functional capacities of the microbiota.
Fig. 2.
Schematic presentation of the future needs for the functional analysis of the microbiota. Metagenome mapping of metatranscriptome and metaproteome data can rely on established methodologies (darker arrows), but the integration to these (functional) metagenome data with the meta-metabolome is far from trivial and in need of methodology development (lighter arrows). OTU, operational taxonomic units. A colour version of this figure can be found online at http://www.journals.cambridge.org/nrr
Bioinformatic processing and interpretation of large-scale metabolic datasets are hampered by the inherent broad scope of the current metabolic databases (for example, KEGG (Kyoto Encyclopedia of Genes and Genomes), etc.). These generic metabolic maps contain pathways that are not present in the gut ecosystem (for example, reactions requiring molecular oxygen as a substrate) and at the same time lack gut-specific pathways, which leads to a loss of both sensitivity and specificity in meta-omics data interpretation. This situation implies that there is a great need for gut-specific, specialised and curated databases, combined with dedicated software and visualisation tools to facilitate the effective interpretation of meta-omics datasets. These bioinformatic environments are under construction (J Raes, personal communication) and will be of great value for the metabolic deciphering of microbiota adaptations to dietary or pre-/pro-/symbiotic interventions. The detailed inference of metabolic potential and activity, when combined with species-function mapping and reconstruction of metabolic interactions between microbial groups, including the reconstruction of syntrophic chains and metabolite exchange( 225 ), will open the way towards novel strategies in mathematical modelling of the metabolic processes taking place in the intestine.
The challenges of data integration become even more pronounced when the intestine meta-omics data are to be connected to parameters that relate to the host's physiology. Some of these host analyses can encompass high-resolution measurements like blood or urine metabolite profiles, blood transcriptomes, or peripheral blood analyte patterns (cytokines, chemokines, hormones, etc.). Connecting these host measurements to meta-omics data is far from trivial, for example, because for many metabolites detected in blood or urine it is uncertain whether they are of intestinal origin. As a consequence, current studies usually limit themselves to the descriptive analysis of metabolic potential and/or activity across patient cohorts, in which the integration is commonly limited to the detection of correlated entities within the datasets that can be identified by multivariate statistics. Only in a few cases were the identified correlations explained through biological context and network biology reconstructions that explain the molecular relationships between the observed correlations. The latter process commonly requires a time-consuming and largely manual sifting of results combined with massive literature mining to decipher the biological context of the observed correlations. Systems biology mathematical frameworks that accelerate the conversion of correlation-based mining to comprehensive, hypothesis-generating biological interpretation, could accelerate the progress of the meta-omics field and its relevance in human health and disease. However, these computational frameworks are still in their infancy, and will require a substantial amount of validation before they can reliably be applied to effectively mine the complex multivariate datasets obtained through meta-omics and high-resolution host analyses.
Conclusions
Currently, there is insufficient evidence to use changes in levels of individual bacterial metabolites as markers in the assessment of prebiotic effectivity. Several in vitro and experimental animal studies indicate that protein fermentation metabolites including ammonia, phenol, p-cresol, indole or hydrogen sulfide intrinsically affect epithelial cellular metabolism and barrier function. However, there is no evidence from human studies that a reduction in faecal excretion of those metabolites contributes to health. Possibly, the impact of protein fermentation is overshadowed by other dietary or lifestyle factors. Although SCFA are generally recognised as markers of carbohydrate rather than protein fermentation in the colon and are therefore commonly considered as beneficial to health, a number of critical questions need to be answered before their concentrations can serve as biomarkers.
In particular, the lack of reliable concentration ranges defining the ‘normal’ or healthy state for these different metabolites in faeces and other biofluids, and the fact that steady-state metabolite concentrations or profiles do not take into account the rapid absorption and/or conversion of the metabolites, hampers the routine application of those techniques to human dietary interventions where microbiota modulation is an objective. There is an urgent need for dynamic, nutrikinetic-type studies, for example, with stable isotopes, to determine and quantify the pathway of microbial metabolites into the different body compartments. Functional analysis of faecal water toxicity has been proposed as a more holistic approach to link changes in colonic content to health outcomes but suffers from some practical considerations and the limited validation of this biomarker towards the end point of colorectal cancer.
Despite the challenges encountered in the integration of the different levels of quantitative analyses of the intestinal system through meta-omics and the corresponding host-specific parameters, the available meta-omics and other high-resolution analytical methods enable the determination of correlated multivariate signatures that can place potential metabolic or health markers in their context, thereby enhancing their value as markers in health and disease or in therapy efficacy evaluation. Of course, these meta-omics must first consider the prevailing ‘meta-data’ which govern nutrient concentrations within human biofluids, not least dietary intake, a difficult parameter to measure and control in free-living subjects. However, these developments may significantly refine our views of concepts like ‘the bandwidth of health’( 226 ) that postulate that multiple molecular solutions for a healthy functioning mucosa and/or microbiota exist. The multivariate signatures mentioned may enable appropriate population stratification for the more effective application of specific nutritional interventions in subpopulations that are predictably more responsive to a certain treatment. Meta-omic stratification of the human population is illustrated by the distinction of three ‘metagenomic enterotypes’ that are characterised by elevated community sizes of the Bacteroidetes, Prevotella and ruminococci( 227 ). Taken together, the deciphering of detailed and specific mechanisms of interaction in the host–microbe-metabolic interplay are a challenge for the future, but hold great promise for rationalised nutritional health improvement and/or even disease therapy in stratified population cohorts.
Acknowledgements
This work was conducted by an expert group of the European branch of the International Life Sciences Institute (ILSI Europe). The authors would like to thank Professor Joël Doré (Metagenomique et Écologie Intestinale, INRA, Jouy-en-Josas, Ile-de-France, France) and Dr Annick Bernalier (INRA, Clermont-Ferrand, France) for their contribution to the initial discussion sessions. The authors also thank Ms Agnès Méheust (formerly ILSI Europe) who coordinated the present study in its initial phase.
The expert group received funding from the ILSI Europe Prebiotics Task Force. Industry members of this task force are listed on the ILSI Europe website (www.ilsi.eu). For further information about ILSI Europe, please email info@ilsieurope.be or call +32 2 771 00 14. The opinions expressed and the conclusions of this publication are those of the authors and do not necessarily represent the views of ILSI Europe or those of its member companies.
All authors contributed to the discussion sessions, held to outline and delimit the content of the manuscript. K. A. V., A. R. B., C. A. E., M. K., A. N., J. R. and K. M. T. performed the literature search and contributed to the writing of the manuscript. All authors contributed to the discussion and interpretation of the literature data and approved the final manuscript.
None of the authors has any interests to declare that may conflict with the provision of their scientific input to this paper.
Abbreviations: BCFA, branched-chain fatty acid; COX, cyco-oxygenase; GPR, G protein-coupled receptor; IBD, inflammatory bowel disease; ILSI Europe, European branch of the International Life Sciences Institute; UC, ulcerative colitis
References
- 1. Scott KP, Gratz SW, Sheridan PO, et al. (2013) The influence of diet on the gut microbiota. Pharmacol Res 69, 52–60. [DOI] [PubMed] [Google Scholar]
- 2. Qin JJ, Li RQ, Raes J, et al. (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nicholson JK, Holmes E, Kinross J, et al. (2012) Host-gut microbiota metabolic interactions. Science 336, 1262–1267. [DOI] [PubMed] [Google Scholar]
- 4. Blaut M & Clavel T (2007) Metabolic diversity of the intestinal microbiota: implications for health and disease. J Nutr 137, 751S–755S. [DOI] [PubMed] [Google Scholar]
- 5. Marchesi J & Shanahan F (2007) The normal intestinal microbiota. Curr Opin Infect Dis 20, 508–513. [DOI] [PubMed] [Google Scholar]
- 6. Hooper LV, Littman DR & Macpherson AJ (2012) Interactions between the microbiota and the immune system. Science 336, 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roberfroid M, Gibson GR, Hoyles L, et al. (2010) Prebiotic effects: metabolic and health benefits. Br J Nutr 104, Suppl. 2, S1–S63. [DOI] [PubMed] [Google Scholar]
- 8. Gibson GR & Roberfroid MB (1995) Dietary modulation of the human colonic microbiota - introducing the concept of prebiotics. J Nutr 125, 1401–1412. [DOI] [PubMed] [Google Scholar]
- 9. Mahowald MA, Rey FE, Seedorf H, et al. (2009) Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A 106, 5859–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. den Besten G, van Eunen K, Groen AK, et al. (2013) The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54, 2325–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cook SI & Sellin JH (1998) Review Article: short chain fatty acids in health and disease. Aliment Pharmacol Ther 12, 499–507. [DOI] [PubMed] [Google Scholar]
- 12. Topping DL & Clifton PM (2001) Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev 81, 1031–1064. [DOI] [PubMed] [Google Scholar]
- 13. Freeland KR (2010) Wilson C & Wolever TMS Adaptation of colonic fermentation and glucagon-like peptide-1 secretion with increased wheat fibre intake for 1 year in hyperinsulinaemic human subjects. Br J Nutr 103, 82–90. [DOI] [PubMed] [Google Scholar]
- 14. Wolever TMS, Josse RG, Leiter LA, et al. (1997) Time of day and glucose tolerance status affect serum short-chain fatty acid concentrations in humans. Metabolism 46, 805–811. [DOI] [PubMed] [Google Scholar]
- 15. Edwards CA, Parrett AM, Balmer SE, et al. (1994) Faecal short chain fatty acids in breast-fed and formula-fed babies. Acta Paediatr 83, 459–462. [DOI] [PubMed] [Google Scholar]
- 16. Knol J, Scholtens P, Kafka C, et al. (2005) Colon microflora in infants fed formula with galacto- and fructo-oligosaccharides: more like breast-fed infants. J Pediatr Gastroenterol Nutr 40, 36–42. [DOI] [PubMed] [Google Scholar]
- 17. Midtvedt AC (1992) & Midtvedt T Production of short chain fatty-acids by the intestinal microflora during the 1st 2 years of human life. J Pediatr Gastroenterol Nutr 15, 395–403. [DOI] [PubMed] [Google Scholar]
- 18. Mariat D, Firmesse O, Levenez F, et al. (2009) The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 9, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Andrieux C, Membre JM, Cayuela C, et al. (2002) Metabolic characteristics of the faecal microflora in humans from three age groups. Scand J Gastroenterol 37, 792–798. [PubMed] [Google Scholar]
- 20. Gill CIR, Heavey P, McConville E, et al. (2007) Effect of fecal water on an in vitro model of colonic mucosal barrier function. Nutr Cancer 57, 59–65. [DOI] [PubMed] [Google Scholar]
- 21. Roediger WE (1982) Utilization of nutrients by isolated epithelial-cells of the rat colon. Gastroenterology 83, 424–429. [PubMed] [Google Scholar]
- 22. Wong JMW, de Souza R, Kendall CWC, et al. (2006) Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol 40, 235–243. [DOI] [PubMed] [Google Scholar]
- 23. Layden BT, Yalamanchi SK, Wolever TMS, et al. (2012) Negative association of acetate with visceral adipose tissue and insulin levels. Diabetes Metab Syndr Obes 5, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frost G, Sleeth ML, Sahuri-Arisoylu M, et al. (2014) The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat Commun 5, 3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kimura I, Inoue D, Maeda T, et al. (2011) Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41). Proc Natl Acad Sci U S A 108, 8030–8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Al-Lahham S, Roelofsen H, Rezaee F, et al. (2012) Propionic acid affects immune status and metabolism in adipose tissue from overweight subjects. Eur J Clin Invest 42, 357–364. [DOI] [PubMed] [Google Scholar]
- 27. Lin HV, Frassetto A, Kowalik EJ Jr, et al. (2012) Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLOS ONE 7, e35240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scheppach W, Sommer H, Kirchner T, et al. (1992) Effect of butyrate enemas on the colonic mucosa in distal ulcerative-colitis. Gastroenterology 103, 51–56. [DOI] [PubMed] [Google Scholar]
- 29. Fung KYC, Cosgrove L, Lockett T, et al. (2012) A review of the potential mechanisms for the lowering of colorectal oncogenesis by butyrate. Br J Nutr 108, 820–831. [DOI] [PubMed] [Google Scholar]
- 30. Fung KYC, Brierley GV, Henderson S, et al. (2011) Butyrate-induced apoptosis in HCT116 colorectal cancer cells includes induction of a cell stress response. J Proteome Res 10, 1860–1869. [DOI] [PubMed] [Google Scholar]
- 31. Shao YF, Gao ZH, Marks PA, et al. (2004) Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci U S A 101, 18030–18035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sauer J, Richter KK & Pool-Zobel BL (2007) Physiological concentrations of butyrate favorably modulate genes of oxidative and metabolic stress in primary human colon cells. J Nutr Biochem 18, 736–745. [DOI] [PubMed] [Google Scholar]
- 33. Ploger S, Stumpff F, Penner GB, et al. (2012) Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann N Y Acad Sci 1258, 52–59. [DOI] [PubMed] [Google Scholar]
- 34. Arpaia N, Campbell C, Fan X, et al. (2013) Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. De Vadder F, Kovatcheva-Datchary P, Goncalves D, et al. (2014) Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 156, 84–96. [DOI] [PubMed] [Google Scholar]
- 36. Faber TA, Hopkins AC, Middelbos IS, et al. (2011) Galactoglucomannan oligosaccharide supplementation affects nutrient digestibility, fermentation end-product production, and large bowel microbiota of the dog. J Anim Sci 89, 103–112. [DOI] [PubMed] [Google Scholar]
- 37. Theil PK, Jorgensen H, Serena A, et al. (2011) Products deriving from microbial fermentation are linked to insulinaemic response in pigs fed breads prepared from whole-wheat grain and wheat and rye ingredients. Br J Nutr 105, 373–383. [DOI] [PubMed] [Google Scholar]
- 38. Van Immerseel F, Russell JB, Flythe MD, et al. (2006) The use of organic acids to combat Salmonella in poultry: a mechanistic explanation of the efficacy. Avian Pathol 35, 182–188. [DOI] [PubMed] [Google Scholar]
- 39. Petry N, Egli I, Chassard C, et al. (2012) Inulin modifies the bifidobacteria population, fecal lactate concentration, and fecal pH but does not influence iron absorption in women with low iron status. Am J Clin Nutr 96, 325–331. [DOI] [PubMed] [Google Scholar]
- 40. Linetzky Waitzberg D, Alves Pereira CC, Logullo L, et al. (2012) Microbiota benefits after inulin and partially hydrolized guar gum supplementation: a randomized clinical trial in constipated women. Nutr Hosp 27, 123–129. [DOI] [PubMed] [Google Scholar]
- 41. Slavin J & Feirtag J (2011) Chicory inulin does not increase stool weight or speed up intestinal transit time in healthy male subjects. Food Funct 2, 72–77. [DOI] [PubMed] [Google Scholar]
- 42. Worthley DL, Le Leu RK, Whitehall VL, et al. (2009) A human, double-blind, placebo-controlled, crossover trial of prebiotic, probiotic, and synbiotic supplementation: effects on luminal, inflammatory, epigenetic, and epithelial biomarkers of colorectal cancer. Am J Clin Nutr 90, 578–586. [DOI] [PubMed] [Google Scholar]
- 43. Costabile A, Klinder A, Fava F, et al. (2007) Whole-grain wheat breakfast cereal has a prebiotic effect on the human gut microbiota: a double-blind, placebo-controlled, crossover study. Br J Nutr 99, 110–120. [DOI] [PubMed] [Google Scholar]
- 44. Costabile A, Fava F, Roytio H, et al. (2012) Impact of polydextrose on the faecal microbiota: a double-blind, crossover, placebo-controlled feeding study in healthy human subjects. Br J Nutr 108, 471–481. [DOI] [PubMed] [Google Scholar]
- 45. Timm DA, Stewart ML, Hospattankar A, et al. (2010) Wheat dextrin, psyllium, and inulin produce distinct fermentation patterns, gas volumes, and short-chain fatty acid profiles in vitro . J Med Food 13, 961–966. [DOI] [PubMed] [Google Scholar]
- 46. Pylkas AM, Juneja LR & Slavin JL (2005) Comparison of different fibers for in vitro production of short chain fatty acids by intestinal microflora. J Med Food 8, 113–116. [DOI] [PubMed] [Google Scholar]
- 47. Connolly ML, Lovegrove JA & Tuohy KM (2010) In vitro evaluation of the microbiota modulation abilities of different sized whole oat grain flakes. Anaerobe 16, 483–488. [DOI] [PubMed] [Google Scholar]
- 48. Cuervo A, Salazar N, Ruas-Madiedo P, et al. (2013) Fiber from a regular diet is directly associated with fecal short-chain fatty acid concentrations in the elderly. Nutr Res 33, 811–816. [DOI] [PubMed] [Google Scholar]
- 49. Kedia G, Vazquez JA, Charalampopoulos D, et al. (2009) In vitro fermentation of oat bran obtained by debranning with a mixed culture of human fecal bacteria. Curr Microbiol 58, 338–342. [DOI] [PubMed] [Google Scholar]
- 50. Hughes SA, Shewry PR, Gibson GR, et al. (2008) In vitro fermentation of oat and barley derived β-glucans by human faecal microbiota. FEMS Microbiol Ecol 64, 482–493. [DOI] [PubMed] [Google Scholar]
- 51. Laurentin A & Edwards CA (2004) Differential fermentation of glucose-based carbohydrates in vitro by human faecal bacteria – a study of pyrodextrinised starches from different sources. Eur J Nutr 43, 183–189. [DOI] [PubMed] [Google Scholar]
- 52. Nordlund E, Aura AM, Mattila I, et al. (2012) Formation of phenolic microbial metabolites and short-chain fatty acids from rye, wheat, and oat bran and their fractions in the metabolical in vitro colon model. J Agric Food Chem 60, 8134–8145. [DOI] [PubMed] [Google Scholar]
- 53. Nilsson A, Johansson E, Ekstrom L, et al. (2013) Effects of a brown beans evening meal on metabolic risk markers and appetite regulating hormones at a subsequent standardized breakfast: a randomized cross-over study. PLOS ONE 8, e59985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Van den Abbeele P, Venema K, van de Wiele T, et al. (2013) Different human gut models reveal the distinct fermentation patterns of arabinoxylan versus inulin. J Agric Food Chem 61, 9819–9827. [DOI] [PubMed] [Google Scholar]
- 55. Hobden MR, Martin-Morales A, Guerin-Deremaux L, et al. (2013) In vitro fermentation of Nutriose® FB06, a wheat dextrin soluble fibre, in a continuous culture human colonic model system. PLOS ONE 8, e77128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Noack J, Timm D, Hospattankar A, et al. (2013) Fermentation profiles of wheat dextrin, inulin and partially hydrolyzed guar gum using an in vitro digestion pretreatment and in vitro batch fermentation system model. Nutrients 5, 1500–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang X & Gibson GR (1993) Effects of the in vitro fermentation of oligofructose and inulin by bacteria growing in the human large intestine. J Appl Bacteriol 75, 373–380. [DOI] [PubMed] [Google Scholar]
- 58. Morrison DJ, Mackay WG, Edwards CA, et al. (2006) Butyrate production from oligofructose fermentation by the human faecal flora: what is the contribution of extracellular acetate and lactate? Br J Nutr 96, 570–577. [PubMed] [Google Scholar]
- 59. Lyra A, Lahtinen S, Tiihonen K, et al. (2010) Intestinal microbiota and overweight. Benef Microbes 1, 407–421. [DOI] [PubMed] [Google Scholar]
- 60. Ley RE, Turnbaugh PJ, Klein S, et al. (2006) Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023. [DOI] [PubMed] [Google Scholar]
- 61. Turnbaugh PJ, Ley RE, Mahowald MA, et al. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. [DOI] [PubMed] [Google Scholar]
- 62. Payne AN, Chassard C, Zimmermann M, et al. (2011) The metabolic activity of gut microbiota in obese children is increased compared with normal-weight children and exhibits more exhaustive substrate utilization. Nutr Diabetes 1, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Brinkworth GD, Noakes M, Clifton PM, et al. (2009) Comparative effects of very low-carbohydrate, high-fat and high-carbohydrate, low-fat weight-loss diets on bowel habit and faecal short-chain fatty acids and bacterial populations. Br J Nutr 101, 1493–1502. [DOI] [PubMed] [Google Scholar]
- 64. Schwiertz A, Taras D, Schafer K, et al. (2010) Microbiota and SCFA in lean and overweight healthy subjects. Obesity 18, 190–195. [DOI] [PubMed] [Google Scholar]
- 65. Tims S, Derom C, Jonkers DM, et al. (2013) Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J 7, 707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Le Chatelier E, Nielsen T, Qin J, et al. (2013) Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. [DOI] [PubMed] [Google Scholar]
- 67. Tiihonen K, Ouwehand AC & Rautonen N (2010) Effect of overweight on gastrointestinal microbiology and immunology: correlation with blood biomarkers. Br J Nutr 103, 1070–1078. [DOI] [PubMed] [Google Scholar]
- 68. Fleissner CK, Huebel N, Abd El-Bary MM, et al. (2010) Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br J Nutr 104, 919–929. [DOI] [PubMed] [Google Scholar]
- 69. Borthakur A, Priyamvada S, Kumar A, et al. (2012) A novel nutrient sensing mechanism underlies substrate-induced regulation of monocarboxylate transporter-1. Am J Physiol Gastrointest Liver Physiol 303, G1126–G1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Goncalves P, Catarino T, Gregorio I, et al. (2012) Inhibition of butyrate uptake by the primary bile salt chenodeoxycholic acid in intestinal epithelial cells. J Cell Biochem 113, 2937–2947. [DOI] [PubMed] [Google Scholar]
- 71. Parrett AM, Edwards CA & Lokerse E (1997) Colonic fermentation capacity in vitro. Development during weaning in breast-fed infants is slower for complex carbohydrates than for sugars. Am J Clin Nutr 65, 927–933. [DOI] [PubMed] [Google Scholar]
- 72. Sarbini SR, Kolida S, Gibson GR, et al. (2013) In vitro fermentation of commercial α-gluco-oligosaccharide by faecal microbiota from lean and obese human subjects. Br J Nutr 109, 1980–1989. [DOI] [PubMed] [Google Scholar]
- 73. Cani PD, Possemiers S, van de Wiele T, et al. (2009) Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 58, 1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Geurts L, Lazarevic V, Derrien M, et al. (2011) Altered gut microbiota and endocannabinoid system tone in obese and diabetic leptin-resistant mice: impact on apelin regulation in adipose tissue. Front Microbiol 2, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Geurts L, Neyrinck AM, Delzenne NM, et al. (2014) Gut microbiota controls adipose tissue expansion, gut barrier and glucose metabolism: novel insights into molecular targets and interventions using prebiotics. Benef Microbes 5, 3–17. [DOI] [PubMed] [Google Scholar]
- 76. de Goffau MC, Fuentes S, van den Bogert B, et al. (2014) Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia 57, 1569–1577. [DOI] [PubMed] [Google Scholar]
- 77. de Goffau MC, Luopajarvi K, Knip M, et al. (2013) Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes 62, 1238–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Brown CT, Davis-Richardson AG, Giongo A, et al. (2011) Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS ONE 6, e25792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang W, Chen L, Zhou R, et al. (2014) Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol 52, 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Machiels K, Joossens M, Sabino J, et al. (2014) A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63, 1275–1283. [DOI] [PubMed] [Google Scholar]
- 81. Huda-Faujan N, Abdulamir AS, Fatimah AB, et al. (2010) The impact of the level of the intestinal short chain fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem J 4, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Treem WR, Ahsan N, Shoup M, et al. (1994) Fecal short-chain fatty acids in children with inflammatory bowel disease. J Pediatr Gastroenterol Nutr 18, 159–164. [DOI] [PubMed] [Google Scholar]
- 83. Tjellström B, Högberg L, Stenhammar L, et al. (2012) Effect of exclusive enteral nutrition on gut microflora function in children with Crohn's disease. Scand J Gastroenterol 47, 1454–1459. [DOI] [PubMed] [Google Scholar]
- 84. Tjellström B, Högberg L, Stenhammar L, et al. (2013) Faecal short-chain fatty acid pattern in childhood coeliac disease is normalised after more than one year's gluten-free diet. Microb Ecol Health Dis 2013, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tjellström B, Stenhammar L, Högberg L, et al. (2010) Screening-detected and symptomatic untreated celiac children show similar gut microflora-associated characteristics. Scand J Gastroenterol 45, 1059–1062. [DOI] [PubMed] [Google Scholar]
- 86. Tjellström B, Stenhammar L, Högberg L, et al. (2005) Gut microflora associated characteristics in children with celiac disease. Am J Gastroenterol 100, 2784–2788. [DOI] [PubMed] [Google Scholar]
- 87. Bottcher MF, Nordin EK, Sandin A, et al. (2000) Microflora-associated characteristics in faeces from allergic and nonallergic infants. Clin Exp Allergy 30, 1590–1596. [DOI] [PubMed] [Google Scholar]
- 88. Macfarlane S & Macfarlane GT (2003) Regulation of short-chain fatty acid production. Proc Nutr Soc 62, 67–72. [DOI] [PubMed] [Google Scholar]
- 89. Mills E & O'Neill LA (2014) Succinate: a metabolic signal in inflammation. Trends Cell Biol 24, 313–320. [DOI] [PubMed] [Google Scholar]
- 90. Tannahill GM, Curtis AM, Adamik J, et al. (2013) Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ariake K, Ohkusa T, Sakurazawa T, et al. (2000) Roles of mucosal bacteria and succinic acid in colitis caused by dextran sulfate sodium in mice. J Med Dent Sci 47, 233–241. [PubMed] [Google Scholar]
- 92. Shiomi Y, Nishiumi S, Ooi M, et al. (2011) GCMS-based metabolomic study in mice with colitis induced by dextran sulfate sodium. Inflamm Bowel Dis 17, 2261–2274. [DOI] [PubMed] [Google Scholar]
- 93. Song WB, Lv YH, Zhang ZS, et al. (2009) Soluble intercellular adhesion molecule-1, d-lactate and diamine oxidase in patients with inflammatory bowel disease. World J Gastroenterol 15, 3916–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Murray MJ, Gonze MD, Nowak LR, et al. (1994) Serum d( − )-lactate levels as an aid to diagnosing acute intestinal ischemia. Am J Surg 167, 575–578. [DOI] [PubMed] [Google Scholar]
- 95. Halperin ML & Kamel KS (1996) d-Lactic acidosis: turning sugar into acids in the gastrointestinal tract. Kidney Int 49, 1–8. [DOI] [PubMed] [Google Scholar]
- 96. Duzgun AP, Bugdayci G, Sayin B, et al. (2007) Serum d-lactate: a useful diagnostic marker for acute appendicitis. Hepatogastroenterology 54, 1483–1486. [PubMed] [Google Scholar]
- 97. Vernia P, Caprilli R, Latella G, et al. (1988) Fecal lactate and ulcerative colitis. Gastroenterology 95, 1564–1568. [DOI] [PubMed] [Google Scholar]
- 98. Hove H & Mortensen PB (1995) Influence of intestinal inflammation (IBD) and small and large-bowel length on fecal short-chain fatty-acids and lactate. Dig Dis Sci 40, 1372–1380. [DOI] [PubMed] [Google Scholar]
- 99. Hove H, Holtug K, Jeppesen PB, et al. (1995) Butyrate absorption and lactate secretion in ulcerative colitis. Dis Colon Rectum 38, 519–525. [DOI] [PubMed] [Google Scholar]
- 100. Marquet P, Duncan SH, Chassard C, et al. (2009) Lactate has the potential to promote hydrogen sulphide formation in the human colon. FEMS Microbiol Lett 299, 128–134. [DOI] [PubMed] [Google Scholar]
- 101. Nyangale EP, Mottram DS & Gibson GR (2012) Gut microbial activity, implications for health and disease: the potential role of metabolite analysis. J Proteome Res 11, 5573–5585. [DOI] [PubMed] [Google Scholar]
- 102. Windey K, De Preter V & Verbeke K (2012) Relevance of protein fermentation to gut health. Mol Nutr Food Res 56, 184–196. [DOI] [PubMed] [Google Scholar]
- 103. Windey K, De Preter V, Louat T, et al. (2012) Modulation of protein fermentation does not affect fecal water toxicity: a randomized cross-over study in healthy subjects. PLOS ONE 7, e52387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Birkett A, Muir J, Phillips J, et al. (1996) Resistant starch lowers fecal concentrations of ammonia and phenols in humans. Am J Clin Nutr 63, 766–772. [DOI] [PubMed] [Google Scholar]
- 105. Smith EA & Macfarlane GT (1996) Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol 81, 288–302. [DOI] [PubMed] [Google Scholar]
- 106. Swanson KS, Grieshop CM, Flickinger EA, et al. (2002) Fructooligosaccharides and lactobacillus acidophilus modify bowel function and protein catabolites excreted by healthy humans. J Nutr 132, 3042–3050. [DOI] [PubMed] [Google Scholar]
- 107. Damen B, Cloetens L, Broekaert WF, et al. (2012) Consumption of breads containing in situ-produced arabinoxylan oligosaccharides alters gastrointestinal effects in healthy volunteers. J Nutr 142, 470–477. [DOI] [PubMed] [Google Scholar]
- 108. Cloetens L, Broekaert WF, Delaedt Y, et al. (2010) Tolerance of arabinoxylan-oligosaccharides and their prebiotic activity in healthy subjects: a randomised, placebo-controlled cross-over study. Br J Nutr 103, 703–713. [DOI] [PubMed] [Google Scholar]
- 109. Lecerf JM, Depeint F, Clerc E, et al. (2012) Xylo-oligosaccharide (XOS) in combination with inulin modulates both the intestinal environment and immune status in healthy subjects, while XOS alone only shows prebiotic properties. Br J Nutr 108, 1847–1858. [DOI] [PubMed] [Google Scholar]
- 110. Shinohara K, Ohashi Y, Kawasumi K, et al. (2010) Effect of apple intake on fecal microbiota and metabolites in humans. Anaerobe 16, 510–515. [DOI] [PubMed] [Google Scholar]
- 111. Boler BM, Serao MC, Bauer LL, et al. (2011) Digestive physiological outcomes related to polydextrose and soluble maize fibre consumption by healthy adult men. Br J Nutr 106, 1864–1871. [DOI] [PubMed] [Google Scholar]
- 112. Wutzke KD, Lotz M & Zipprich C (2010) The effect of pre- and probiotics on the colonic ammonia metabolism in humans as measured by lactose-[15N2]ureide. Eur J Clin Nutr 64, 1215–1221. [DOI] [PubMed] [Google Scholar]
- 113. Gostner A, Blaut M, Schäffer V, et al. (2006) Effect of isomalt consumption on faecal microflora and colonic metabolism in healthy volunteers. Br J Nutr 95, 40–50. [DOI] [PubMed] [Google Scholar]
- 114. Roediger W & Babidge W (1997) Human colonocyte detoxification. Gut 41, 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Collino S, Montoliu I, Martin FPJ, et al. (2013) Metabolic signatures of extreme longevity in northern Italian centenarians reveal a complex remodeling of lipids, amino acids, and gut microbiota metabolism. PLOS ONE 8, e56564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Patney NL, Mehrotra MP, Khanna HK, et al. (1976) Urinary indican excretion in cirrhosis of liver. J Assoc Physicians India 24, 291–295. [PubMed] [Google Scholar]
- 117. Patney NL, Saxena SK, Mehrotra MP, et al. (1977) Urinary indican in diabetes mellitus. J Indian Med Assoc 68, 94–97. [PubMed] [Google Scholar]
- 118. Renwick AG, Thakrar A, Lawrie CA, et al. (1988) Microbial amino-acid metabolites and bladder cancer: no evidence of promoting activity in man. Hum Toxicol 7, 267–272. [DOI] [PubMed] [Google Scholar]
- 119. Adams RF, Murray KE & Earl JW (1985) High levels of fecal para-cresol in a group of hyperactive children. Lancet ii, 1313–1313. [DOI] [PubMed] [Google Scholar]
- 120. Pedersen G, Brynskov J & Saermark T (2002) Phenol toxicity and conjugation in human colonic epithelial cells. Scand J Gastroenterol 37, 74–79. [DOI] [PubMed] [Google Scholar]
- 121. Mccall IC, Betanzos A, Weber DA, et al. (2009) Effects of phenol on barrier function of a human intestinal epithelial cell line correlate with altered tight junction protein localization. Toxicol Appl Pharmacol 241, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hughes R, Kurth MJ, McGilligan V, et al. (2008) Effect of colonic bacterial metabolites on Caco-2 cell paracellular permeability in vitro . Nutr Cancer 60, 259–266. [DOI] [PubMed] [Google Scholar]
- 123. Meijers BK, Van Kerckhoven S, Verbeke K, et al. (2009) The uremic retention solute p-cresyl sulfate and markers of endothelial damage. Am J Kidney Dis 54, 891–901. [DOI] [PubMed] [Google Scholar]
- 124. Roe DA (1971) Effects of methionine and inorganic sulfate on indole toxicity and indican excretion in rats. J Nutr 101, 645–654. [DOI] [PubMed] [Google Scholar]
- 125. Smith EA & Macfarlane GT (1997) Dissimilatory amino acid metabolism in human colonic bacteria. Anaerobe 3, 327–337. [DOI] [PubMed] [Google Scholar]
- 126. Wang L, Christophersen CT, Sorich MJ, et al. (2012) Elevated fecal short chain fatty acid and ammonia concentrations in children with autism spectrum disorder. Dig Dis Sci 57, 2096–2102. [DOI] [PubMed] [Google Scholar]
- 127. Visek WJ (1978) Diet and cell-growth modulation by ammonia. Am J Clin Nutr 31, S216–S220. [DOI] [PubMed] [Google Scholar]
- 128. Blachier F, Mariotti F, Huneau JF, et al. (2007) Effects of amino acid-derived luminal metabolites on the colonic epithelium and physiopathological consequences. Amino Acids 33, 547–562. [DOI] [PubMed] [Google Scholar]
- 129. Macfarlane GT, Gibson GR & Cummings JH (1992) Comparison of fermentation reactions in different regions of the human colon. J Appl Bacteriol 72, 57–64. [DOI] [PubMed] [Google Scholar]
- 130. Florin T, Neale G, Gibson GR, et al. (1991) Metabolism of dietary sulfate: absorption and excretion in humans. Gut 32, 766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Magee EA, Richardson CJ, Hughes R, et al. (2000) Contribution of dietary protein to sulfide production in the large intestine: an in vitro and a controlled feeding study in humans. Am J Clin Nutr 72, 1488–1494. [DOI] [PubMed] [Google Scholar]
- 132. Leschelle X, Goubern M, Andriamihaja M, et al. (2005) Adaptative metabolic response of human colonic epithelial cells to the adverse effects of the luminal compound sulfide. Biochim Biophys Acta 1725, 201–212. [DOI] [PubMed] [Google Scholar]
- 133. Medani M, Collins D, Docherty NG, et al. (2011) Emerging role of hydrogen sulfide in colonic physiology and pathophysiology. Inflamm Bowel Dis 17, 1620–1625. [DOI] [PubMed] [Google Scholar]
- 134. Pitcher MCL, Beatty ER & Cummings JH (2000) The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut 46, 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Levine J, Ellis CJ, Furne JK, et al. (1998) Fecal hydrogen sulfide production in ulcerative colitis. Am J Gastroenterol 93, 83–87. [DOI] [PubMed] [Google Scholar]
- 136. Moore J, Babidge W, Millard S, et al. (1998) Colonic luminal hydrogen sulfide is not elevated in ulcerative colitis. Dig Dis Sci 43, 162–165. [DOI] [PubMed] [Google Scholar]
- 137. De Preter V, Arijs I, Windey K, et al. (2012) Decreased mucosal sulfide detoxification is related to an impaired butyrate oxidation in ulcerative colitis. Inflamm Bowel Dis 18, 2371–2380. [DOI] [PubMed] [Google Scholar]
- 138. Jowett SL, Seal CJ, Pearce MS, et al. (2004) Influence of dietary factors on the clinical course of ulcerative colitis: a prospective cohort study. Gut 53, 1479–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Deplancke B & Gaskins HR (2003) Hydrogen sulfide induces serum-independent cell cycle entry in nontransformed rat intestinal epithelial cells. FASEB J 17, 1310–1312. [DOI] [PubMed] [Google Scholar]
- 140. Attene-Ramos MS, Wagner ED & Plewa MJ (2006) Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res 4, 9–14. [DOI] [PubMed] [Google Scholar]
- 141. Attene-Ramos MS, Wagner ED, Gaskins HR, et al. (2007) Hydrogen sulfide induces direct radical-associated DNA damage. Mol Cancer Res 5, 455–459. [DOI] [PubMed] [Google Scholar]
- 142. Attene-Ramos MS, Nava GM, Muellner MG, et al. (2010) DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ Mol Mutagen 51, 304–314. [DOI] [PubMed] [Google Scholar]
- 143. Koehne CH & Dubois RN (2004) Cox-2 inhibition and colorectal cancer. Semin Oncol 31, 12–21. [DOI] [PubMed] [Google Scholar]
- 144. Blachier F, Davila AM, Mimoun S, et al. (2010) Luminal sulfide and large intestine mucosa: friend or foe? Amino Acids 39, 335–347. [DOI] [PubMed] [Google Scholar]
- 145. Mortensen PB & Clausen MR (1996) Short-chain fatty acids in the human colon: relation to gastrointestinal health and disease. Scand J Gastroenterol 31, 132–148. [DOI] [PubMed] [Google Scholar]
- 146. Makelainen HS, Makivuokko HA, Salminen SJ, et al. (2007) The effects of polydextrose and xylitol on microbial community and activity in a 4-stage colon simulator. J Food Sci 72, M153–M159. [DOI] [PubMed] [Google Scholar]
- 147. Alles MS, Katan MB, Salemans JMJI, et al. (1997) Bacterial fermentation of fructooligosaccharides and resistant starch in patients with an ileal pouch anal anastomosis. Am J Clin Nutr 66, 1286–1292. [DOI] [PubMed] [Google Scholar]
- 148. Sakurazawa T & Ohkusa T (2005) Cytotoxicity of organic acids produced by anaerobic intestinal bacteria on cultured epithelial cells. J Gastroenterol 40, 600–609. [DOI] [PubMed] [Google Scholar]
- 149. Kim E, Coelho D & Blachier F (2013) Review of the association between meat consumption and risk of colorectal cancer. Nutr Res 33, 983–994. [DOI] [PubMed] [Google Scholar]
- 150. Del Rio D, Rodriguez-Mateos A, Spencer JPE, et al. (2013) Dietary (poly)phenolics in human health: structures, bioavailability, and evidence of protective effects against chronic diseases. Antioxid Redox Signal 18, 1818–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Dall'Asta M, Calani L, Tedeschi M, et al. (2012) Identification of microbial metabolites derived from in vitro fecal fermentation of different polyphenolic food sources. Nutrition 28, 197–203. [DOI] [PubMed] [Google Scholar]
- 152. Bolca S, Van de Wiele T & Possemiers S (2013) Gut metabotypes govern health effects of dietary polyphenols. Curr Opin Biotechnol 24, 220–225. [DOI] [PubMed] [Google Scholar]
- 153. Tzounis X, Rodriguez-Mateos A, Vulevic J, et al. (2011) Prebiotic evaluation of cocoa-derived flavanols in healthy humans by using a randomized, controlled, double-blind, crossover intervention study. Am J Clin Nutr 93, 62–72. [DOI] [PubMed] [Google Scholar]
- 154. Jaganath IB, Mullen W, Lean ME, et al. (2009) In vitro catabolism of rutin by human fecal bacteria and the antioxidant capacity of its catabolites. Free Radic Biol Med 47, 1180–1189. [DOI] [PubMed] [Google Scholar]
- 155. Tuohy KM, Conterno L, Gasperotti M, et al. (2012) Up-regulating the human intestinal microbiome using whole plant foods, polyphenols, and/or fiber. J Agric Food Chem 60, 8776–8782. [DOI] [PubMed] [Google Scholar]
- 156. Moco S, Martin FPJ & Rezzi S (2012) Metabolomics view on gut microbiome modulation by polyphenol-rich foods. J Proteome Res 11, 4781–4790. [DOI] [PubMed] [Google Scholar]
- 157. Asano Y, Hiramoto T, Nishino R, et al. (2012) Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am J Physiol Gastrointest Liver Physiol 303, G1288–G1295. [DOI] [PubMed] [Google Scholar]
- 158. Foster JA & McVey Neufeld K-A (2013) Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci 36, 305–312. [DOI] [PubMed] [Google Scholar]
- 159. Rhee SH, Pothoulakis C & Mayer EA (2009) Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat Rev Gastroenterol Hepatol 6, 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Gin H, Rigalleau V, Caubet O, et al. (1999) Effects of red wine, tannic acid, or ethanol on glucose tolerance in non-insulin-dependent diabetic patients and on starch digestibility in vitro . Metabolism 48, 1179–1183. [DOI] [PubMed] [Google Scholar]
- 161. Henry CJ, Lightowler HJ, Newens KJ, et al. (2008) The influence of adding fats of varying saturation on the glycaemic response of white bread. Int J Food Sci Nutr 59, 61–69. [DOI] [PubMed] [Google Scholar]
- 162. Bazzocco S, Mattila I, Guyot S, et al. (2008) Factors affecting the conversion of apple polyphenols to phenolic acids and fruit matrix to short-chain fatty acids by human faecal microbiota in vitro . Eur J Nutr 47, 442–452. [DOI] [PubMed] [Google Scholar]
- 163. Fallani M, Amarri S, Uusijarvi A, et al. (2011) Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology 157, 1385–1392. [DOI] [PubMed] [Google Scholar]
- 164. Zoetendal EG, Rajilic-Stojanovic M & De Vos WM (2008) High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57, 1605–1615. [DOI] [PubMed] [Google Scholar]
- 165. Albenberg L & Wu GD (2014) Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology 146, 1564–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Yap IKS, Brown IJ, Chan Q, et al. (2010) Metabolome-wide association study identifies multiple biomarkers that discriminate north and south Chinese populations at differing risks of cardiovascular disease: INTERMAP study. J Proteome Res 9, 6647–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Ou JH, Delany JP, Zhang M, et al. (2012) Association between low colonic short-chain fatty acids and high bile acids in high colon cancer risk populations. Nutr Cancer 64, 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Macfarlane GT & Macfarlane S (2011) Fermentation in the human large intestine: its physiologic consequences and the potential contribution of prebiotics. J Clin Gastroenterol 45, suppl., S120–S127. [DOI] [PubMed] [Google Scholar]
- 169. Cummings JH, Hill MJ, Bone ES, et al. (1979) Effect of meat protein and dietary fiber on colonic function and metabolism. II. Bacterial metabolites in feces and urine. Am J Clin Nutr 32, 2094–2101. [DOI] [PubMed] [Google Scholar]
- 170. Stephen AM, Wiggins HS & Cummings JH (1987) Effect of changing transit-time on colonic microbial-metabolism in man. Gut 28, 601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. El Oufir L, Barry JL, Flourie B, et al. (2000) Relationships between transit time in man and in vitro fermentation of dietary fiber by fecal bacteria. Eur J Clin Nutr 54, 603–609. [DOI] [PubMed] [Google Scholar]
- 172. Govers MJ, Gannon NJ, Dunshea FR, et al. (1999) Wheat bran affects the site of fermentation of resistant starch and luminal indexes related to colon cancer risk: a study in pigs. Gut 45, 840–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Morita T, Kasaoka S, Hase K, et al. (1999) Psyllium shifts the fermentation site of high-amylose cornstarch toward the distal colon and increases fecal butyrate concentration in rats. J Nutr 129, 2081–2087. [DOI] [PubMed] [Google Scholar]
- 174. Zhao Y, Wu J, Li JV, et al. (2013) Gut microbiota composition modifies fecal metabolic profiles in mice. J Proteome Res 12, 2987–2999. [DOI] [PubMed] [Google Scholar]
- 175. Washington N, Harris M, Mussellwhite A, et al. (1998) Moderation of lactulose-induced diarrhea by psyllium: effects on motility and fermentation. Am J Clin Nutr 67, 317–321. [DOI] [PubMed] [Google Scholar]
- 176. Cherbut C (2003) Motor effects of short-chain fatty acids and lactate in the gastrointestinal tract. Proc Nutr Soc 62, 95–99. [DOI] [PubMed] [Google Scholar]
- 177. Kamath PS, Phillips SF, O'Connor MK, et al. (1990) Colonic capacitance and transit in man: modulation by luminal contents and drugs. Gut 31, 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Jouet P, Moussata D, Duboc H, et al. (2013) Effect of short-chain fatty acids and acidification on the phasic and tonic motor activity of the human colon. Neurogastroenterol Motil 25, 943–949. [DOI] [PubMed] [Google Scholar]
- 179. Dougal K, Harris PA, Edwards A, et al. (2012) A comparison of the microbiome and the metabolome of different regions of the equine hindgut. FEMS Microbiol Ecol 82, 642–652. [DOI] [PubMed] [Google Scholar]
- 180. Wikoff WR, Anfora AT, Liu J, et al. (2009) Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 106, 3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Verbeke K, Ferchaud-Roucher V, Preston T, et al. (2010) Influence of the type of indigestible carbohydrate on plasma and urine short-chain fatty acid profiles in healthy human volunteers. Eur J Clin Nutr 64, 678–684. [DOI] [PubMed] [Google Scholar]
- 182. de Graaf AA & Venema K (2007) Gaining insight into microbial physiology in the large intestine: a special role for stable isotopes. Adv Microb Physiol 53, 73–168, 313–314. [DOI] [PubMed] [Google Scholar]
- 183. Osswald K, Becker TW, Grimm M, et al. (2000) Inter- and intra-individual variation of faecal water - genotoxicity in human colon cells. Mutat Res 472, 59–70. [DOI] [PubMed] [Google Scholar]
- 184. Bosworth D & Venitt S (1986) Testing human fecal extracts for genotoxic activity with the SOS chromotest: the importance of controlling for fecal enzyme activity. Mutagenesis 1, 143–149. [DOI] [PubMed] [Google Scholar]
- 185. De Kok TMCM & van Maanen JMS (2000) Evaluation of fecal mutagenicity and colorectal cancer risk. Mutat Res 463, 53–101. [DOI] [PubMed] [Google Scholar]
- 186. Mower HF, Ichinotsubo D, Wang LW, et al. (1982) Fecal mutagens in two Japanese populations with different colon cancer risks. Cancer Res 42, 1164–1169. [PubMed] [Google Scholar]
- 187. Gratz SW, Wallace RJ & El-Nezami HS (2011) Recent perspectives on the relations between fecal mutagenicity, genotoxicity and diet. Front Pharmacol 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Klinder A, Karlsson PC, Clune Y, et al. (2007) Fecal water as a non-invasive biomarker in nutritional intervention: comparison of preparation methods and refinement of different endpoints. Nutr Cancer 57, 158–167. [DOI] [PubMed] [Google Scholar]
- 189. Karlsson PC, Huss U, Jenner A, et al. (2005) Human fecal water inhibits COX-2 in colonic HT-29 cells: role of phenolic compounds. J Nutr 135, 2343–2349. [DOI] [PubMed] [Google Scholar]
- 190. Nordling MM, Glinghammar B, Karlsson PC, et al. (2003) Effects on cell proliferation, activator protein-1 and genotoxicity by fecal water from patients with colorectal adenomas. Scand J Gastroenterol 38, 549–555. [DOI] [PubMed] [Google Scholar]
- 191. Morrow DMP, Ryan MP & McGlynn H (1996) An in vitro model to assess tumour promotor activity. Br J Cancer 74, 68. [Google Scholar]
- 192. Gill CIR, McDougall GJ, Glidewell S, et al. (2010) Profiling of phenols in human fecal water after raspberry supplementation. J Agric Food Chem 58, 10389–10395. [DOI] [PubMed] [Google Scholar]
- 193. Jovov B, Wills NK & Lewis SAA (1991) spectroscopic method for assessing confluence of epithelial-cell cultures. Am J Physiol 261, C1196–C1203. [DOI] [PubMed] [Google Scholar]
- 194. Ramakers JD, Volman JJ, Biorklund M, et al. (2007) Fecal water from ileostomic patients consuming oat β-glucan enhances immune responses in enterocytes. Mol Nutr Food Res 51, 211–220. [DOI] [PubMed] [Google Scholar]
- 195. Lapre JA & VanderMeer R (1992) Diet-induced increase of colonic bile-acids stimulates lytic activity of fecal water and proliferation of colonic cells. Carcinogenesis 13, 41–44. [DOI] [PubMed] [Google Scholar]
- 196. Haza AI, Glinghammar B, Grandien A, et al. (2000) Effect of colonic luminal components on induction of apoptosis in human colonic cell lines. Nutr Cancer 36, 79–89. [DOI] [PubMed] [Google Scholar]
- 197. Glinghammar B, Holmberg K & Rafter J (1999) Effects of colonic lumenal components on AP-1-dependent gene transcription in cultured human colon carcinoma cells. Carcinogenesis 20, 969–976. [DOI] [PubMed] [Google Scholar]
- 198. Glinghammar B & Rafter J (2001) Colonic luminal contents induce cyclooxygenase 2 transcription in human colon carcinoma cells. Gastroenterology 120, 401–410. [DOI] [PubMed] [Google Scholar]
- 199. Zeng HW & Davis CD (2003) Down-regulation of proliferating cell nuclear antigen gene expression occurs during cell cycle arrest induced by human fecal water in colonic HT-29 cells. J Nutr 133, 2682–2687. [DOI] [PubMed] [Google Scholar]
- 200. Pool-Zobel BL (2005) Inulin-type fructans and reduction in colon cancer risk: review of experimental and human data. Br J Nutr 93, S73–S90. [DOI] [PubMed] [Google Scholar]
- 201. Klinder A, Forster A, Caderni G, et al. (2004) Fecal water genotoxicity is predictive of tumor-preventive activities by inulin-like oligofructoses, probiotics (Lactobacillus rhamnosus and Bifidobacterium lactis), and their synbiotic combination. Nutr Cancer 49, 144–155. [DOI] [PubMed] [Google Scholar]
- 202. Glei M, Habermann N, Osswald K, et al. (2005) Assessment of DNA damage and its modulation by dietary and genetic factors in smokers using the comet assay: a biomarker model. Biomarkers 10, 203–217. [DOI] [PubMed] [Google Scholar]
- 203. Wu WT, Cheng HC & Chen HL (2011) Ameliorative effects of konjac glucomannan on human faecal β-glucuronidase activity, secondary bile acid levels and faecal water toxicity towards Caco-2 cells. Br J Nutr 105, 593–600. [DOI] [PubMed] [Google Scholar]
- 204. Walton GE, van den Heuvel EGHM, Kosters MHW, et al. (2012) A randomised crossover study investigating the effects of galacto-oligosaccharides on the faecal microbiota in men and women over 50 years of age. Br J Nutr 107, 1466–1475. [DOI] [PubMed] [Google Scholar]
- 205. Le Gall G, Noor SO, Ridgway K, et al. (2011) Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res 10, 4208–4218. [DOI] [PubMed] [Google Scholar]
- 206. Martin FPJ, Collino S & Rezzi S (2011) 1H NMR-based metabonomic applications to decipher gut microbial metabolic influence on mammalian health. Magn Reson Chem 49, Suppl. S1, S47–S54. [DOI] [PubMed] [Google Scholar]
- 207. Martin FPJ, Rezzi S, Pere-Trepat E, et al. (2009) Metabolic effects of dark chocolate consumption on energy, gut microbiota, and stress-related metabolism in free-living subjects. J Proteome Res 8, 5568–5579. [DOI] [PubMed] [Google Scholar]
- 208. Cheng Y, Xie GX, Chen TL, et al. (2012) Distinct urinary metabolic profile of human colorectal cancer. J Proteome Res 11, 1354–1363. [DOI] [PubMed] [Google Scholar]
- 209. Qiu YP, Cai GX, Su MM, et al. (2010) Urinary metabonomic study on colorectal cancer. J Proteome Res 9, 1627–1634. [DOI] [PubMed] [Google Scholar]
- 210. Qiu Y, Cai G, Su M, et al. (2009) Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J Proteome Res 8, 4844–4850. [DOI] [PubMed] [Google Scholar]
- 211. Tulipani S, Llorach R, Jauregui O, et al. (2011) Metabolomics unveils urinary changes in subjects with metabolic syndrome following 12-week nut consumption. J Proteome Res 10, 5047–5058. [DOI] [PubMed] [Google Scholar]
- 212. Calvani R, Miccheli A, Capuani G, et al. (2010) Gut microbiome-derived metabolites characterize a peculiar obese urinary metabotype. Int J Obes 34, 1095–1098. [DOI] [PubMed] [Google Scholar]
- 213. Suhre K, Meisinger C, Doring A, et al. (2010) Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS ONE 5, e13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Ponnusamy K, Choi JN, Kim J, et al. (2011) Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J Med Microbiol 60, 817–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Ktsoyan ZA, Beloborodova NV, Sedrakyan AM, et al. (2011) Profiles of microbial fatty acids in the human metabolome are disease-specific. Front Microbiol 1, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Martin FPJ, Sprenger N, Montoliu I, et al. (2010) Dietary modulation of gut functional ecology studied by fecal metabonomics. J Proteome Res 9, 5284–5295. [DOI] [PubMed] [Google Scholar]
- 217. Fava F, Gitau R, Griffin BA, et al. (2013) The type and quantity of dietary fat and carbohydrate alter faecal microbiome and short-chain fatty acid excretion in a metabolic syndrome ‘at-risk’ population. Int J Obes 37, 216–223. [DOI] [PubMed] [Google Scholar]
- 218. Seiber JN, Molyneux RJ & Schieberle P (2014) Targeted metabolomics: a new section in the Journal of Agricultural and Food Chemistry . J Agric Food Chem 62, 22–23. [DOI] [PubMed] [Google Scholar]
- 219. Martin FPJ, Sprenger N, Yap IKS, et al. (2009) Panorganismal gut microbiome-host metabolic crosstalk. J Proteome Res 8, 2090–2105. [DOI] [PubMed] [Google Scholar]
- 220. Ndagijimana M, Laghi L, Vitali B, et al. (2009) Effect of a synbiotic food consumption on human gut metabolic profiles evaluated by 1H nuclear magnetic resonance spectroscopy. Int J Food Microbiol 134, 147–153. [DOI] [PubMed] [Google Scholar]
- 221. Vitali B, Ndagijimana M, Cruciani F, et al. (2010) Impact of a synbiotic food on the gut microbial ecology and metabolic profiles. BMC Microbiol 10, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. De Preter V, Ghebretinsae AH, Abrahantes JC, et al. (2011) Impact of the synbiotic combination of Lactobacillus casei Shirota and oligofructose-enriched inulin on the fecal volatile metabolite profile in healthy subjects. Mol Nutr Food Res 55, 714–722. [DOI] [PubMed] [Google Scholar]
- 223. De Preter V, Joossens M, Ballet V, et al. (2013) Metabolic profiling of the impact of oligofructose-enriched inulin in Crohn's disease patients: a double-blinded randomized controlled trial. Clin Transl Gastroenterol 4, e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Zoetendal EG, Raes J, van den Bogert B, et al. (2012) The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J 6, 1415–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Faust K & Raes J (2012) Microbial interactions: from networks to models. Nat Rev Microbiol 10, 538–550. [DOI] [PubMed] [Google Scholar]
- 226. Bron PA, van Baarlen P & Kleerebezem M (2012) Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat Rev Microbiol 10, 66–78. [DOI] [PubMed] [Google Scholar]
- 227. Arumugam M, Raes J, Pelletier E, et al. (2011) Enterotypes of the human gut microbiome. Nature 473, 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. De Filippo C, Cavalieri D, Di Paola M, et al. (2010) Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107, 14691–14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Guerin-Danan C, Andrieux C, Popot F, et al. (1997) Pattern of metabolism and composition of the fecal microflora in infants 10 to 18 months old from day care centers. J Pediatr Gastroenterol Nutr 25, 281–289. [DOI] [PubMed] [Google Scholar]
- 230. Siigur U, Ormisson A & Tamm A (1993) Faecal short-chain fatty acids in breast-fed and bottle-fed infants. Acta Paediatr 82, 536–538. [DOI] [PubMed] [Google Scholar]
- 231. Holscher HD, Faust KL, Czerkies LA, et al. (2012) Effects of prebiotic-containing infant formula on gastrointestinal tolerance and fecal microbiota in a randomized controlled trial. JPEN J Parenter Enteral Nutr 36, 95S–105S. [DOI] [PubMed] [Google Scholar]
- 232. Norin E, Midtvedt T & Bjorksten B (2004) Development of faecal short-chain fatty acid pattern during the first year of life in Estonian and Swedish infants. Microb Ecol Health Dis 16, 8–12. [Google Scholar]
- 233. Bakker-Zierikzee AM, Alles MS, Knol J, et al. (2005) Effects of infant formula containing a mixture of galacto- and fructo-oligosaccharides or viable Bifidobacterium animalis on the intestinal microflora during the first 4 months of life. Br J Nutr 94, 783–790. [DOI] [PubMed] [Google Scholar]
- 234. Samuelsson U & Ludvigsson J (2004) The concentrations of short-chain fatty acids and other microflora-associated characteristics in faeces from children with newly diagnosed type 1 diabetes and control children and their family members. Diabet Med 21, 64–67. [DOI] [PubMed] [Google Scholar]
- 235. Parrett AM & Edwards CA (1997) In vitro fermentation of carbohydrate by breast fed and formula fed infants. Arch Dis Child 76, 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Whelan K, Judd PA, Preedy VR, et al. (2005) Fructooligosaccharides and fiber partially prevent the alterations in fecal microbiota and short-chain fatty acid concentrations caused by standard enteral formula in healthy humans. J Nutr 135, 1896–1902. [DOI] [PubMed] [Google Scholar]
- 237. Lewis SJ & Heaton KW (1997) Increasing butyrate concentration in the distal colon by accelerating intestinal transit. Gut 41, 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Reimer RA, Pelletier X, Carabin IG, et al. (2012) Faecal short chain fatty acids in healthy subjects participating in a randomised controlled trial examining a soluble highly viscous polysaccharide versus control. J Hum Nutr Diet 25, 373–377. [DOI] [PubMed] [Google Scholar]
- 239. Fernando WMU, Hill JE, Zello GA, et al. (2010) Diets supplemented with chickpea or its main oligosaccharide component raffinose modify faecal microbial composition in healthy adults. Beneficial Microbes 1, 197–207. [DOI] [PubMed] [Google Scholar]
- 240. McOrist AL, Miller RB, Bird AR, et al. (2011) Fecal butyrate levels vary widely among individuals but are usually increased by a diet high in resistant starch. J Nutr 141, 883–889. [DOI] [PubMed] [Google Scholar]
- 241. Nemoto H, Kataoka K, Ishikawa H, et al. (2012) Reduced diversity and imbalance of fecal microbiota in patients with ulcerative colitis. Dig Dis Sci 57, 2955–2964. [DOI] [PubMed] [Google Scholar]
- 242. McOrist AL, Abell GC, Cooke C, et al. (2008) Bacterial population dynamics and faecal short-chain fatty acid (SCFA) concentrations in healthy humans. Br J Nutr 100, 138–146. [DOI] [PubMed] [Google Scholar]
- 243. Hylla S, Gostner A, Dusel G, et al. (1998) Effects of resistant starch on the colon in healthy volunteers: possible implications for cancer prevention. Am J Clin Nutr 67, 136–142. [DOI] [PubMed] [Google Scholar]
- 244. Benassi-Evans B, Clifton P, Noakes M, et al. (2010) High-protein/high red meat and high-carbohydrate weight-loss diets do not differ in their effect on faecal water genotoxicity tested by use of the WIL2-NS cell line and with other biomarkers of bowel health. Mutat Res 703, 130–136. [DOI] [PubMed] [Google Scholar]
- 245. Ling WH & Hanninen O (1992) Shifting from a conventional diet to an uncooked vegan diet reversibly alters fecal hydrolytic activities in humans. J Nutr 122, 924–930. [DOI] [PubMed] [Google Scholar]
- 246. Patel KP, Luo FJG, Plummer NS, et al. (2012) The production of p-cresol sulfate and indoxyl sulfate in vegetarians versus omnivores. Clin J Am Soc Nephrol 7, 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. De Preter V, Geboes K, Verbrugghe K, et al. (2004) The in vivo use of the stable isotope-labelled biomarkers lactose-[15N]ureide and [2H4]tyrosine to assess the effects of pro- and prebiotics on the intestinal flora of healthy human volunteers. Br J Nutr 92, 439–446. [DOI] [PubMed] [Google Scholar]
- 248. De Preter V, Coopmans T, Rutgeerts P, et al. (2007) Influence of long-term administration of lactulose and Saccharomyces boulardii on the colonic generation of phenolic compounds and on different gastro-intestinal parameters. J Am Coll Nutr 25, 541–549. [DOI] [PubMed] [Google Scholar]
- 249. De Preter V, Vanhoutte T, Huys G, et al. (2007) Effects of Lactobacillus casei Shirota, Bifidobacterium breve, and oligofructose-enriched inulin on colonic nitrogen-protein metabolism in healthy humans. Am J Physiol Gastrointest Liver Physiol 292, G358–G368. [DOI] [PubMed] [Google Scholar]
- 250. Cloetens L, De Preter V, Swennen K, et al. (2008) Dose-response effect of arabinoxylooligosaccharides on gastrointestinal motility and on colonic bacterial metabolism in healthy volunteers. J Am Coll Nutr 27, 512–518. [DOI] [PubMed] [Google Scholar]
- 251. Heavey PM, Savage SAH, Parrett A, et al. (2003) Protein-degradation products and bacterial enzyme activities in faeces of breast-fed and formula-fed infants. Br J Nutr 89, 509–515. [DOI] [PubMed] [Google Scholar]
- 252. Clarke JM, Topping DL, Christophersen CT, et al. (2011) Butyrate esterified to starch is released in the human gastrointestinal tract. Am J Clin Nutr 94, 1276–1283. [DOI] [PubMed] [Google Scholar]
- 253. Bianchi MA, Scazzina F, Del Rio D, et al. (2010) Ability of a high-total antioxidant capacity diet to increase stool weight and bowel antioxidant status in human subjects. Br J Nutr 104, 1500–1507. [DOI] [PubMed] [Google Scholar]
- 254. Alakomi HL, Puupponen-Pimia R, Aura AM, et al. (2007) Weakening of Salmonella with selected microbial metabolites of berry-derived phenolic compounds and organic acids. J Agric Food Chem 55, 3905–3912. [DOI] [PubMed] [Google Scholar]
- 255. Ko HS, Jin RD, Krishnan HB, et al. (2009) Biocontrol ability of Lysobacter antibioticus HS124 against phytophthora blight is mediated by the production of 4-hydroxyphenylacetic acid and several lytic enzymes. Curr Microbiol 59, 608–615. [DOI] [PubMed] [Google Scholar]
- 256. Roowi S, Stalmach A, Mullen W, et al. (2010) Green tea flavan-3-ols: colonic degradation and urinary excretion of catabolites by humans. J Agric Food Chem 58, 1296–1304. [DOI] [PubMed] [Google Scholar]
- 257. Okello EJ, Leylabi R & McDougall GJ (2012) Inhibition of acetylcholinesterase by green and white tea and their simulated intestinal metabolites. Food Funct 3, 651–661. [DOI] [PubMed] [Google Scholar]
- 258. Ni N, Choudhary G, Li M, et al. (2008) Pyrogallol and its analogs can antagonize bacterial quorum sensing in Vibrio harveyi . Bioorg Med Chem Lett 18, 1567–1572. [DOI] [PubMed] [Google Scholar]
- 259. Taguri T, Tanaka T & Kouno I (2006) Antibacterial spectrum of plant polyphenols and extracts depending upon hydroxyphenyl structure. Biol Pharm Bull 29, 2226–2235. [DOI] [PubMed] [Google Scholar]
- 260. Jackman KA, Woodman OL & Sobey CG (2007) Isoflavones, equol and cardiovascular disease: pharmacological and therapeutic insights. Curr Med Chem 14, 2824–2830. [DOI] [PubMed] [Google Scholar]
- 261. Ishimi Y (2009) Soybean isoflavones in bone health. Forum Nutr 61, 104–116. [DOI] [PubMed] [Google Scholar]
- 262. Davis CD & Milner JA (2009) Gastrointestinal microflora, food components and colon cancer prevention. J Nutr Biochem 20, 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Selma MV, Espin JC & Tomas-Barberan FA (2009) Interaction between phenolics and gut microbiota: role in human health. J Agric Food Chem 57, 6485–6501. [DOI] [PubMed] [Google Scholar]
- 264. Larrosa M, Gonzalez-Sarrias A, Garcia-Conesa MT, et al. (2006) Urolithins, ellagic acid-derived metabolites produced by human colonic microflora, exhibit estrogenic and antiestrogenic activities. J Agric Food Chem 54, 1611–1620. [DOI] [PubMed] [Google Scholar]
- 265. Del Rio D, Borges G & Crozier A (2010) Berry flavonoids and phenolics: bioavailability and evidence of protective effects. Br J Nutr 104, S67–S90. [DOI] [PubMed] [Google Scholar]
- 266. Dell'Agli M, Galli GV, Bulgari M, et al. (2010) Ellagitannins of the fruit rind of pomegranate (Punica granatum) antagonize in vitro the host inflammatory response mechanisms involved in the onset of malaria. Malar J 9, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Cervantes-Laurean D, Schramm DD, Jacobson EL, et al. (2006) Inhibition of advanced glycation end product formation on collagen by rutin and its metabolites. J Nutr Biochem 17, 531–540. [DOI] [PubMed] [Google Scholar]