Summary
The hippocampus has become the focus of research in several neurodegenerative disorders. Automatic segmentation of this structure from magnetic resonance (MR) imaging scans of the brain facilitates this work. Segmentation techniques must be evaluated using a dataset of MR images with accurate hippocampal outlines generated manually. Manual segmentation is not a trivial task. Lack of a unique segmentation protocol and poor image quality are only two factors that have confounded the consistency required for comparative study. We have developed a publicly available dataset of T1-weighted (T1W) MR images of epileptic and nonepileptic subjects along with their hippocampal outlines to provide a means of evaluation of segmentation techniques. This dataset contains 50 T1W MR images, 40 epileptic and 10 nonepileptic. All images were manually segmented by a widely used protocol. Twenty five images were selected for training and were provided with hippocampal labels. Twenty five other images were provided without labels for testing algorithms. The users are allowed to evaluate their generated labels for the test images using 11 segmentation similarity metrics. Using this dataset, we evaluated two segmentation algorithms, Brain Parser and Classifier Fusion and Labeling (CFL), trained by the training set. For Brain Parser, an average Dice coefficient of 0.64 was obtained with the testing set. For CFL, this value was 0.75. Such findings indicate a need for further improvement of segmentation algorithms in order to enhance reliability.
Keywords: segmentation algorithm, hippocampus, magnetic resonance imaging, validation
Introduction
Both the natural process of aging and several neurodegenerative and lesional conditions affect the brain. The study of the shape and tissue characteristics of brain structures provides us with the means to monitor changes over time and to shed light on the nature of clinical progression. Magnetic resonance (MR) imaging is widely used as the medium of choice because of its resolution and the ability to assess the intrinsic nature of defined components. Select areas may be outlined on sequential images and studied by 3D segmentation. Manual segmentation, however, is both tedious and time-consuming; hence, automatic segmentation algorithms are preferred.
Numerous automated segmentation techniques have been proposed. Comparison is difficult as each proposed technique uses its own unique algorithm upon a different set of images. On the other hand, the creation of individual sets of training and testing samples would burden the development of automatic techniques as it would require the outline of select structures and, moreover, consideration of the appearance of the site as it might be defined by contrast, signal-to-noise ratio (SNR) and head position. Commonly, analyses are performed on relatively few subjects and may be biased towards those upon whom it may be more suited.
A publicly available online dataset of MR images with a sequential outline of a select structure such as the hippocampus would provide a validation tool for all segmentation algorithms for comparison. In this work, we have provided such a dataset of images and have used it to compare two published algorithms, Brain Parser (Tu et al., 2008) and Classifier Fusion and Labeling (CFL) (Aljabar et al., 2007).
Variations in the volume and architecture of the hippocampus have been observed in a variety of neurological disorders including schizophrenia (Lawrie and Abukmeil, 1998), epilepsy (Cendes et al., 1993) and Alzheimer’s disease (Jack et al., 1992). Manual outline of the hippocampus in standard T1-weighted (T1W) MR images is difficult and varies among publications (Geuze et al., 2005). Two major factors are responsible for this variation:
Manual segmentation protocol
The hippocampal border is not comprehensively defined in human brain atlases. The manual protocol for drawing the outline on MR images varies as a consequence. Reviews of current protocols provide an excellent overview of methodologies (Geuze et al., 2005; Konrad et al., 2009). Based upon such a comprehensive review (Geuze et al., 2005), a protocol for segmentation of the whole hippocampus was adopted for this study that provided clear guidelines for outlining the structure.
Image quality
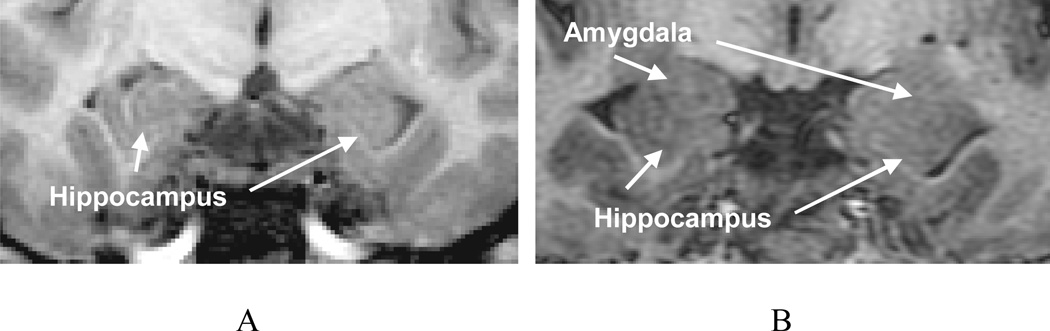
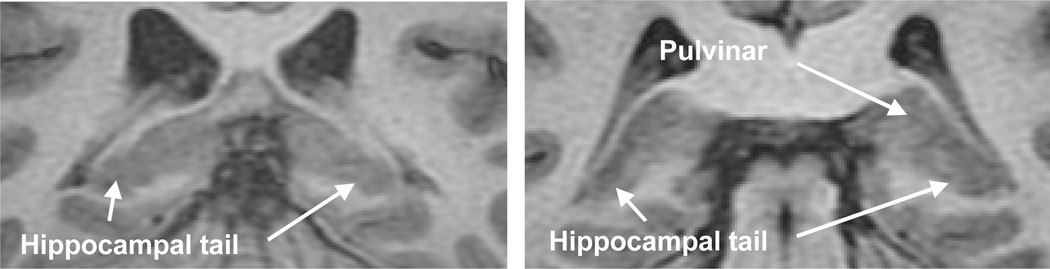
Lack of sufficient quality of the MR image will impede accurate definition of the hippocampal border and introduce subjectivity into the process once the protocol is fixed. Image quality depends on resolution (i.e., pixel size and slice thickness), contrast, SNR and is subject to motion artifact. When definition is not sufficiently clear, external landmarks are used although these may vary in location depending upon the angle between the hippocampal main axis and the imaging plane (Konrad et al., 2009). A partial volume effect (PVE) adds further to the difficulty in low resolution images. This problem is pronounced in the hippocampal tail where the hippocampus curves medially with respect to the coronal plane and, in the hippocampal head where the hippocampus is convoluted and blends with the amygdala. Representative coronal slices of the hippocampal head with different resolutions and acquisition by two MR imaging scanners with field strengths of 1.5 T and 3.0 T illustrate the difficulties encountered in this location because of PVE (Fig. 1). A similar problem can be illustrated in the same fashion with the hippocampal tail (Fig. 2). The border between the hippocampal tail and pulvinar becomes indistinguishable rendering its definition highly subjective. The fimbria, the output tract of the hippocampus, is not very clear in low resolution images either. The temporal horn of the lateral ventricle and the gray matter may occasionally have similar intensities due to the PVE also.
Fig. 1.
Coronal T1W MR image slices of the hippocampal head with two different resolutions. (A) Voxel size of 0.78×0.78×2 mm3 with 1.5 T MR image system. (B) Voxel size of 0.39×0.39×2 mm3 with 3 T MR image system. As shown, there is no clear distinction between the hippocampus and amygdala.
Fig. 2.
Coronal T1W MR image slices of the hippocampal tail in two patients with a voxel size of 0.39×0.39×2 mm3 in both images. A clear distinction between the hippocampal tail and pulvinar is often lacking and the structures appear confluent.
To train an automated segmentation algorithm, a set of MR images with valid hippocampal outlines is required. Such a dataset must be publicly available and two brain MR image datasets with hippocampal outlines are currently provided: (i) the Internet Brain Segmentation Repository (IBSR, http://www.cma.mgh.harvard.edu/ibsr/) and (ii) the LONI Probabilistic Brain Atlas (LPBA40, http://www.loni.ucla.edu/Atlases/LPBA40/) (Shattuck et al., 2008). The IBSR contains T1W MR images of 18 neurologically intact nonepileptic subjects with expert segmentation of 43 bilaterally defined structures including the hippocampi. The voxel size of these images is 0.84×0.84×1.5 mm3. The LPBA40 contains T1W MR images of 40 neurologically intact nonepileptic subjects with expert segmentation of 56 structures including the hippocampi. The voxel size of these images is 0.86×0.86×1.5 mm3. The Alzheimer's Disease Neuroimaging Initiative (ADNI, http://www.adni-info.org/) is another source of hippocampal outlines, but the outlines have been generated semi-automatically using a software tool called SNT (Medtronic Surgical Navigation Technologies, Louisville, CO) and edited manually.
Both of the above datasets contain images of unaffected subjects. In practice, hippocampal segmentation may be problematic due to the shape changes caused by neurological disorders. For instance, in mesial temporal lobe epilepsy (mTLE), atrophy is often observed in the epileptogenic hippocampus. Automatic segmentation of an atrophic structure is a challenging task as its dimensions and shape will differ from those in the training images of nonepileptic subjects. Comparable images in a segmentation algorithm would provide useful indicators of the extent of change encountered both in regards to shape in different planes and overall volume. Other practical problems such as motion artifact and low SNR are issues that might be dealt with by more rigorous application of the actual imaging protocol. The IBSR dataset does include a few samples with motion artifact.
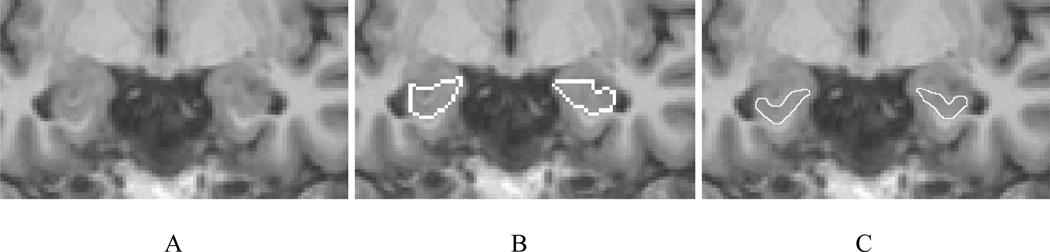
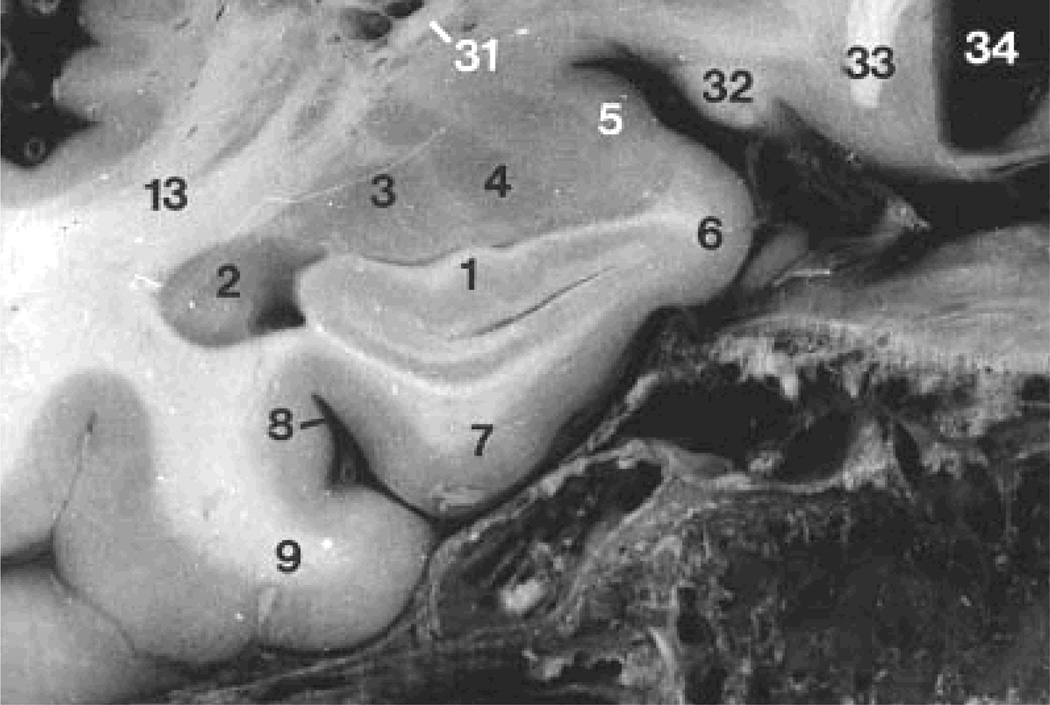
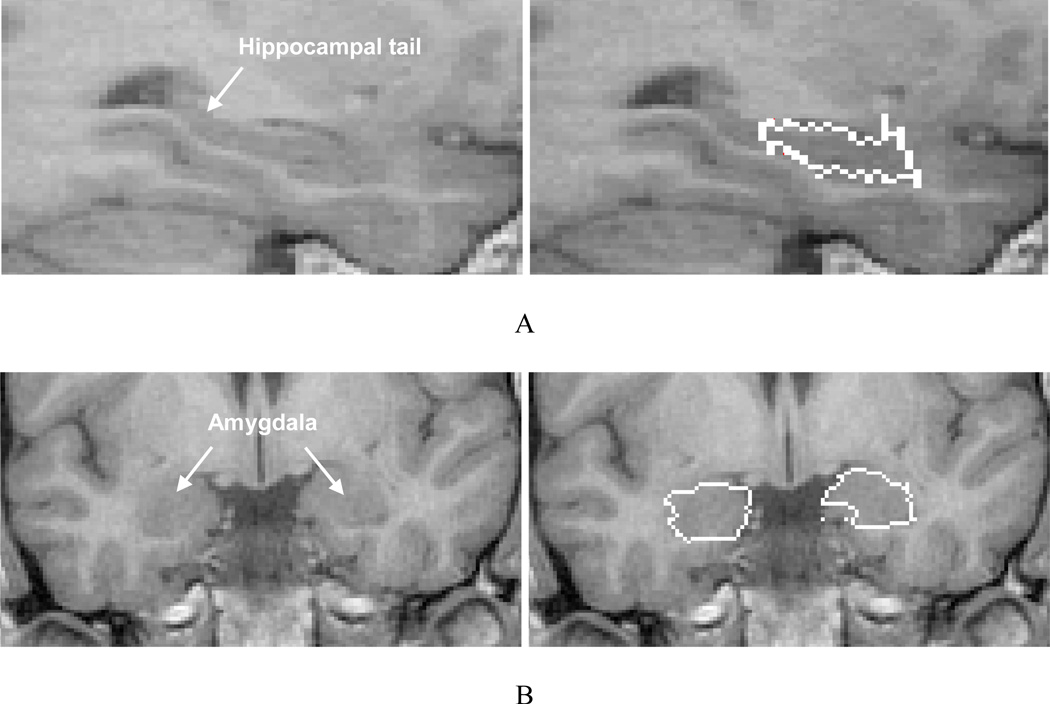
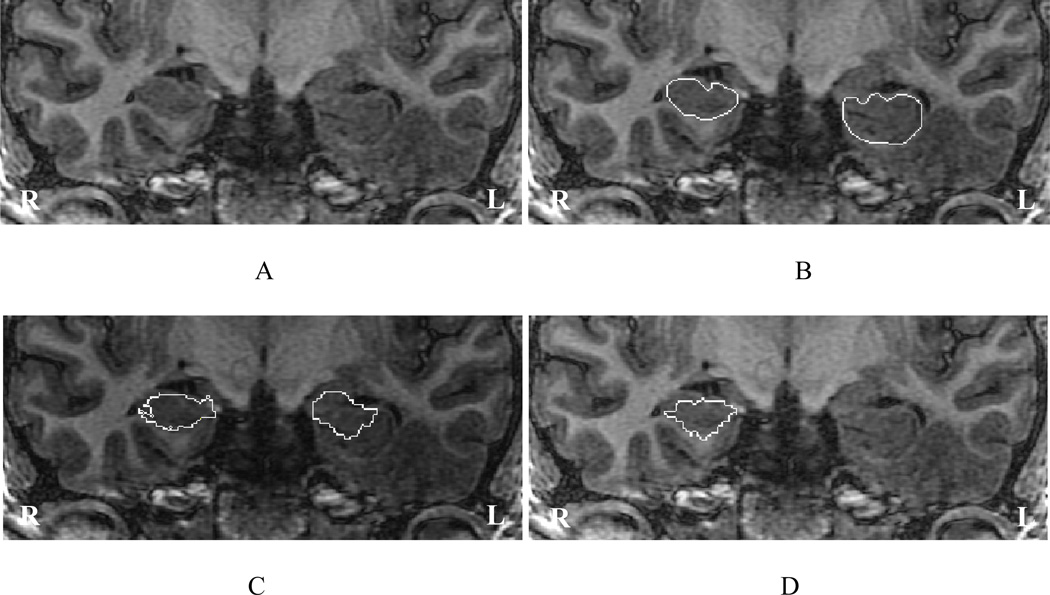
The average hippocampal volume calculated from the IBSR label maps is significantly higher (left side 3637 mm3, right side 3616 mm3) in comparison with our previously reported values for a set of 25 healthy subjects (left side 2507 mm3, right side 2510 mm3) (Jafari-Khouzani et al., 2010). This appears attributable largely to the inclusion of white matter (i.e., alveus and fimbria) and neighboring nonhippocampal tissues in the IBSR segmentations. The alveus is a white matter tract containing axons from hippocampal and subicular neurons that enter the fimbria (Duvernoy, 2005). Fig. 3A–B shows the coronal section of the hippocampal head in slice Number 65 of IBSR_07_ana.img in the IBSR dataset with the manual hippocampal segmentation provided by the IBSR. The alveus and part of the amygdala are included in the hippocampal segmentation. Fig. 3C shows the outline that is used in the present scheme drawn with a thickness smaller than the pixel size to better identify the real boundary. The sectional anatomy of the hippocampal head in a similar slice provided by another source (Duvernoy, 2005) is presented in Fig. 4 and illustrates a distinctive border between the hippocampus and amygdala. Unfortunately, because of a PVE, this distinction is not usually observed in standard T1W MR images.
Fig. 3.
Coronal section of slice Number 65 of image IBSR_07_ana.img from the IBSR data set. (A) The original image; (B) Manual outlines of the hippocampi provided by the IBSR; (C) Our proposed manual outlines.
Fig. 4.
Coronal section of the hippocampal head from an MR image atlas of the human hippocampus (Duvernoy, 2005). Labels 1, 3 and 4 show the hippocampal head and the lateral and basal nuclei of the amygdala, respectively.
The LPBA40 hippocampal segmentation protocol is significantly different from other protocols in the literature. Fig. 5 shows the hippocampal outlines in representative slices of subject S01 from the LPBA40 dataset. The majority of the hippocampal tail is not included in the segmentation (Fig. 5A), whereas part of the amygdala is included (Fig. 5B). The average hippocampal volume calculated from the LPBA40 label maps are 4120 mm3 and 3907 mm3 for the left and right sides, respectively.
Fig. 5.
Hippocampal outlines in representative sagittal (A) and coronal (B) slices of subject S01 from the LPBA40 database. As shown, the hippocampal tail is not included (A) whereas some parts of the amygdala are included (B).
Some researchers use their own segmented set of images to avoid these discrepancies but are unable to evaluate their algorithms against a separate set of images. Most are forced then to use different combinations of the available images as training and testing sets; hence, the reliability of their approach remains uncertain.
In this paper, we introduce a publicly available dataset of T1W MR images (http://www.radiologyresearch.org/) with hippocampal outlines for the validation of automated hippocampal segmentation algorithms. Advantages of this dataset include the following:
Training and testing sets are separate. The researchers are encouraged to submit their segmentation results on the testing set to evaluate their algorithms.
Images are acquired using two different MR imaging systems with different field strengths and thus have different resolutions and contrasts. Twenty of the images have a higher resolution and are acquired with a 3.0 T MR imaging system.
Special care has been taken to minimize the PVE in the manual drawings. The sagittal view and neighboring coronal slices are used to reduce the segmentation inaccuracy due to the PVE.
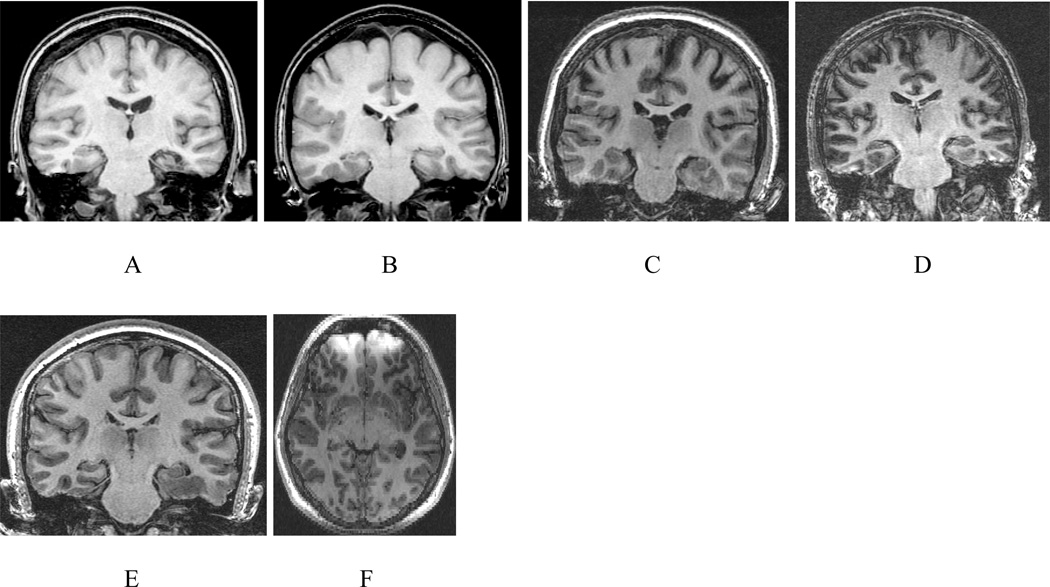
Forty of the images belong to patients with medically intractable temporal lobe epilepsy. The hippocampal outline commonly differs in these circumstances making the segmentation challenging. This provision allows an evaluation of the proposed segmentation algorithm under practical considerations. Fig. 6 shows the slices of a few samples from this dataset under various practical adversities such as atrophy, motion artifact, poor SNR, the presence of a lesion and field inhomogeneity. The latter confounds automated skull-stripping and co-registration.
Fig. 6.
Coronal (A – E) and axial (F) MR image sections of a few samples of the dataset demonstrating various practical problems: (A) Atrophy of the left hippocampus and adjacent parahippocampal gyrus. (B) Atrophy of the right hippocampus. (C) Motion artifact disturbs the definition of the gray-white matter interface. (D) Low SNR reduces contrast and enhances the PVE. (E) A lesion of the left parahippocampal and fusiform gyri may distort the local anatomy and cause difficulty with boundary definitions. (F) Field inhomogeneity will interfere with comparative study of bilateral structures and enhance PVE.
We have evaluated two segmentation algorithms, namely Brain Parser (Tu et al., 2008) and CFL (Aljabar et al., 2007), using the above dataset.
Materials and Methods
Subjects
Fifty subjects comprising 40 patients (13 males, 27 females) with mesial (m)TLE, age range 15–64, (39±12, mean±SD) and 10 nonepileptic subjects (5 males, 5 females), age range 19–54 (34±13) were included. A number of patients had hippocampal atrophy. The nonepileptic subjects were drawn from an archive of control subjects in a sleep research project. The 50 subjects were randomly selected and also reflect practical problems such as motion artifact, low SNR and head tilt.
MR imaging
Thirty of the subjects, including 20 epilepsy patients and 10 nonepileptic subjects, had their MR images acquired with a General Electric 1.5 T Signa system (GE Medical Systems, Milwaukee WI). These subjects underwent coronal T1W MR study using a spoiled gradient-echo (SPGR) sequence with TR/TE/TI = 7.6/1.7/500 ms, flip angle = 20°, field of view (FOV) = 200×200 mm2, matrix size = 256×256, pixel size = 0.78×0.78 mm2, slice thickness = 2.00 mm (voxel size = 0.78×0.78×2.00 mm3), bandwidth = 25 KHz and scanning time of 5 minutes and 45 seconds. The remaining twenty subjects, all epilepsy patients, had their MR images acquired with a General Electric 3.0 T system (GE Medical Systems, Milwaukee WI). These subjects underwent coronal T1W MR study using a SPGR sequence with TR/TE/TI = 10.4/4.5/300 ms, flip angle = 15°, FOV = 200×200 mm2, matrix size = 512×512, pixel size = 0.39×0.39 mm2, slice thickness = 2.00 mm (voxel size = 0.39×0.39×2.00 mm3) and scanning time of 6 minutes.
The images were converted to ANALYZE (Mayo Clinic Analyze 7.5) format. For privacy protection, all images were de-identified before conversion. Furthermore, the subject’s face was manually removed from each image.
Segmentation protocol
A single investigator (KJ) outlined all coronal hippocampal contours (usually 20 slices/case) using Eigentool, an in-house software (http://www.radiologyresearch.org/eigentool.htm). These were then verified by two other investigators (KE and SP). All images were segmented in the coronal plane although the other two planes were used to help find landmarks. All outlines were converted to label maps in ANALYZE format.
Manual hippocampal segmentation protocols vary notably among investigators (Geuze et al., 2005; Konrad et al., 2009). Some protocols exclude the hippocampal tail or head. In others, the extent of inclusion of the hippocampal tail and head vary. The amygdalohippocampal complex may be measured as a single entity. The anatomical boundaries of the hippocampus vary among protocols. An inability to adequately distinguish the gray matter of the hippocampus from the neighboring cerebrospinal fluid of the temporal horn compromises even the delineation of its lateral border. Konrad et al. (Konrad et al., 2009) identify major areas causing differences among protocols as:
Inclusion/exclusion of hippocampal white matter (alveus and fimbria)
Definition of the border between anterior hippocampus and amygdala
Definition of the posterior border and the extent to which the hippocampal tail is included
Definition of the inferomedial border of the hippocampus
Use of varying arbitrary lines whenever the border is not clear
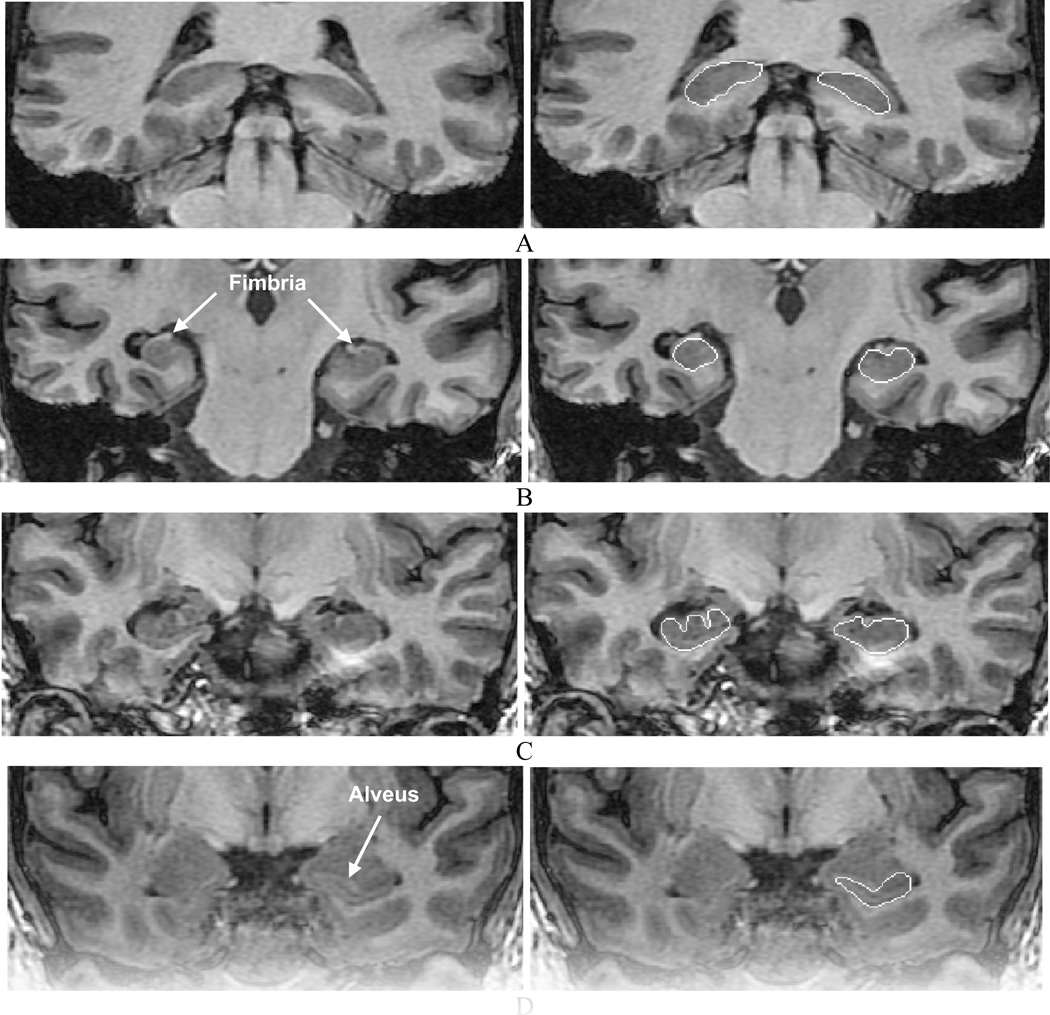
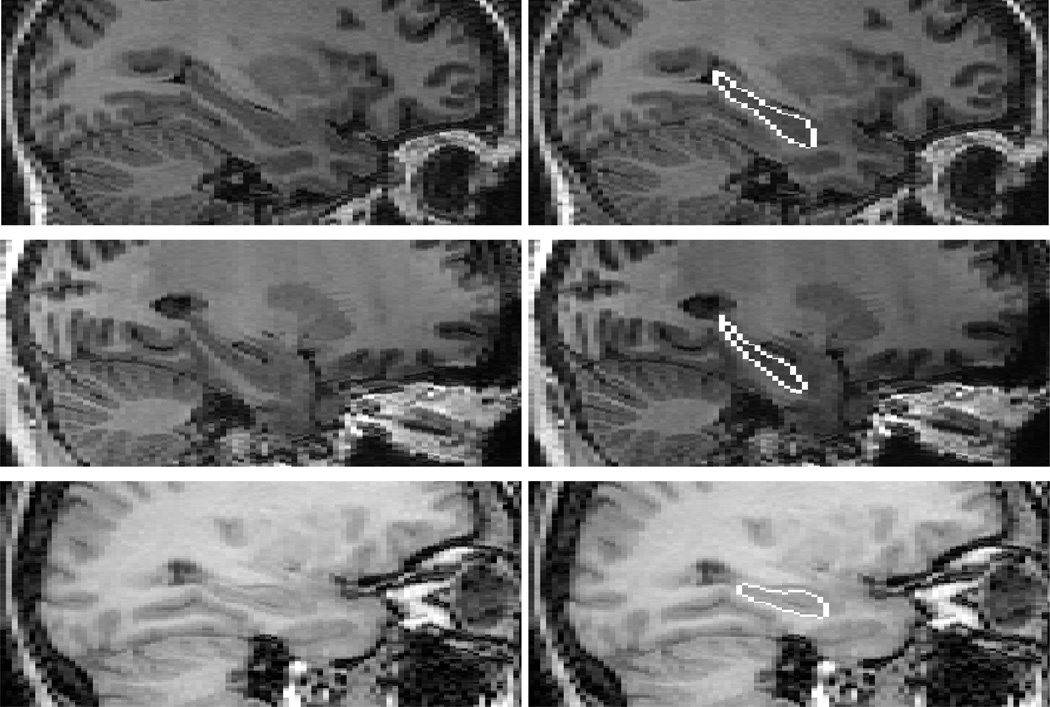
In our protocol, the whole hippocampus, including head, body and tail, is outlined. White matter tracts, such as the alveus and fimbria, are not included. Examples of the hippocampal outlines are provided in Fig. 7. The alveus is used as a landmark separating the amygdala and hippocampus (Fig. 7D). The amygdala and temporal horn of the lateral ventricle are not included in the segmentation whereas the subiculum is included. The most anterior coronal slice is taken where, in high quality images, the alveus is still detectable as the head of the hippocampus tapers below the amygdala. Otherwise, any remaining anterior boundary is estimated based on experience and with the help of the sagittal view. The hippocampal tail is included to the point where it narrows and curves medially towards the crus. The gray-white matter interface is used as the inferior and lateral border. This interface may become smooth in some slices due to the PVE. The sagittal view and neighboring coronal slices are used to reduce the segmentation inaccuracy attributable to the PVE. We also limited the use of arbitrary lines and, instead, estimated the hippocampal border using intensity information as well as neighboring slices in cases where the border was not clear. As outlines were drawn in the coronal plane, care was taken to avoid jagged boundaries in other views (Fig. 8).
Fig. 7.
Manual segmentation of the hippocampi in representative coronal T1-weighted MR images of the hippocampal (A) tail, (B) and (C) body and (D) head.
Fig. 8.
Sagittal view of hippocampal outlines of three patients. As shown, jagged boundaries have been avoided. Grainy boundaries are attributed to a slice thickness that is larger than the pixel dimension.
Data dissemination
Twenty-five sets of images consisting of 20 from epilepsy patients, 10 taken with the 1.5 T scanner and 10 with the 3 T scanner, and 5 from nonepileptic subjects were randomly selected as training samples and were provided with hippocampal label files. The remaining 25 sets of images, similar in distribution as that of the training set, were provided as testing samples but without label files. The users may use the training samples to train their algorithms, submit the outcome of their segmentation algorithms on the testing images and receive the results based on a number of metrics listed below.
Segmentation metrics
Using a manually segmented image as our gold standard, an automatic segmentation may be evaluated by the calculation of segmentation similarity metrics. If X and Y represent the manual and automatic segmentation label sets, respectively, we define true positive as TP = X ∩ Y, true negative as TN = X̅ ∩ Y̅, false positive as FP = X̅ ∩ Y and false negative as FN = X ∩ Y̅. The following 11 metrics then are calculated to evaluate the automatic segmentation outcome (Dice, 1945; Munkres, 2000):
Relative Absolute Value Difference
Hausdorff distance dH (X, Y) = max {D1, D2} where , d(x, y) is the Euclidian distance, and ∂ is the boundary operator.
Hausdorff 95 distance dH95 (X, Y) is similar to dH (X, Y) with the difference that 5% of the outliers are removed before calculation.
Mean distance
Average Symmetric Surface Distance:
Root Mean Square Distance:
where |X| is the size of set X, and . Metrics 1–5 are between zero and unity and increase with increasing quality of segmentation. For a perfect segmentation, metrics 1–5 equate to unity and metrics 6–11 equate to zero. Although there is redundancy among these outcome measures, the intent here is to provide a comprehensive listing of metrics from which authors may select those they feel to be most suitable for publication.
Reliability of manual segmentation
To test the reliability of manual segmentation, 10 subjects were randomly selected from the entire group to undergo outlining of the hippocampus. Eight subjects were epileptic, four of whom were imaged with the 1.5 T scanner and four with the 3 T scanner. Two subjects who were not epileptic were imaged with the 1.5 T scanner. In order to avoid reproducibility through familiarity with the initial exercise, six months were allowed to elapse before repetition by the same investigator (KJ). This experiment resulted in the following outcome metrics: Jaccard Index=0.81±0.01, Dice coefficient=0.90±0.01, sensitivity=0.88±0.03, specificity=1.00±0.00, precision =0.91±0.03, RAVD=−0.03±0.05, Hausdorff distance=3.79±1.33, 95% Hausdorff distance=0.22±0.02, mean distance=1.04±0.09, ASSD=0.24±0.03, RMS=0.53±0.06. The majority of discrepancies between the two segmentations were observed in the hippocampal head and tail where the PVE resulted in poor image contrast. The accuracy of automatic segmentation was, in turn, limited by the reliability of manual segmentation.
Testing two segmentation algorithms
We tested two segmentation algorithms, namely Brain Parser (Tu et al., 2008) and CFL (Aljabar et al., 2007) using the dataset. Both algorithms were trained by the training set and tested on the testing set.
Brain Parser
Brain Parser software (Tu et al., 2008) segments regions of interest based on a training set of data and generates label volumes. The software comes pretrained on a provided dataset but can be retrained to work with desired regions of interest. This software was trained using the training images put forward in the current work. One set of training images of good quality (HFH_021) was selected as a template to which all other training images were then coregistered using an affine transformation. Brain Extraction Tool (BET) (Smith, 2002) was used for skull-stripping and FLIRT (Jenkinson and Smith, 2001) for affine coregistration. For subjects with poor BET outcome, manual skull-stripping and correction was applied.
Classifier Fusion and Labeling (CFL)
In this method, multiple atlases were coregistered to the query image and their labels propagated to give a segmentation of the query image. The propagated labels were used as classifiers and combined to yield the label volume (Aljabar et al., 2007). In this approach, all training images (i.e., atlases) were skull-stripped using BET and coregistered to a reference image, the MNI single subject image (Cocosco et al., 1997) with an affine transformation using FLIRT. The query image was also coregistered to the reference image using an affine transformation. Similarity between the coregistered atlases and the query image was calculated using normalized mutual information (Studholme et al., 1999) over the hippocampal region. The atlas images were ranked based on their similarity with the query image. The top-ranked 15 images were selected as classifiers and coregistered nonrigidly to the query image using a free-form deformation model (Rueckert et al., 1999). The labels were then propagated from the selected classifiers to the query image and fused using the vote rule (Rohlfing et al., 2004).
Results
Table 1 shows the average evaluation metrics using both Brain Parser and CFL algorithms on the testing dataset. Using Brain Parser, the average Dice coefficient was 0.64 for the testing set while, for CFL, it increased to 0.75.
Table 1.
Average evaluation metrics for the hippocampus segmentation using the BrainParser and CFL algorithms on the test dataset.
| Jaccard Index | Dice coefficient | Sensitivity | Specificity | Precision | RAVD | Hausdorff distance (mm) |
Hausdorff 95 distance (mm) |
Mean distance (mm) |
ASSD | RMSD | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BrainParser | Mean | 0.47 | 0.64 | 0.92 | 1.00 | 0.50 | 0.94 | 9.23 | 4.58 | 1.58 | 1.33 | 1.88 |
| StD | 0.06 | 0.06 | 0.06 | 0.00 | 0.08 | 0.37 | 4.98 | 1.68 | 0.50 | 0.33 | 0.54 | |
| CFL | Mean | 0.60 | 0.75 | 0.74 | 1.00 | 0.77 | −0.04 | 5.09 | 2.58 | 0.62 | 0.72 | 1.10 |
| StD | 0.07 | 0.07 | 0.10 | 0.00 | 0.07 | 0.15 | 1.62 | 1.08 | 0.17 | 0.27 | 0.39 | |
Relatively high sensitivity with either segmentation method indicates the likelihood that the automated versions enclose the manually segmented version. The actual hippocampal volumes determined by both automatic segmentation algorithms are indeed significantly higher compared to that obtained by manual segmentation (Table 2). High specificity is due to the small size of the hippocampus compared to that of the whole image.
Table 2.
Average hippocampal volume (Mean±StD, mm3) based on manual, Brain Parser, and CFL segmentations for the test dataset.
| Manual | Brain Parser | CFL | ||
|---|---|---|---|---|
| Testing set | Left | 2469±841 | 4559±793 | 3169±880 |
| Right | 2575±645 | 4618±606 | 3400±587 |
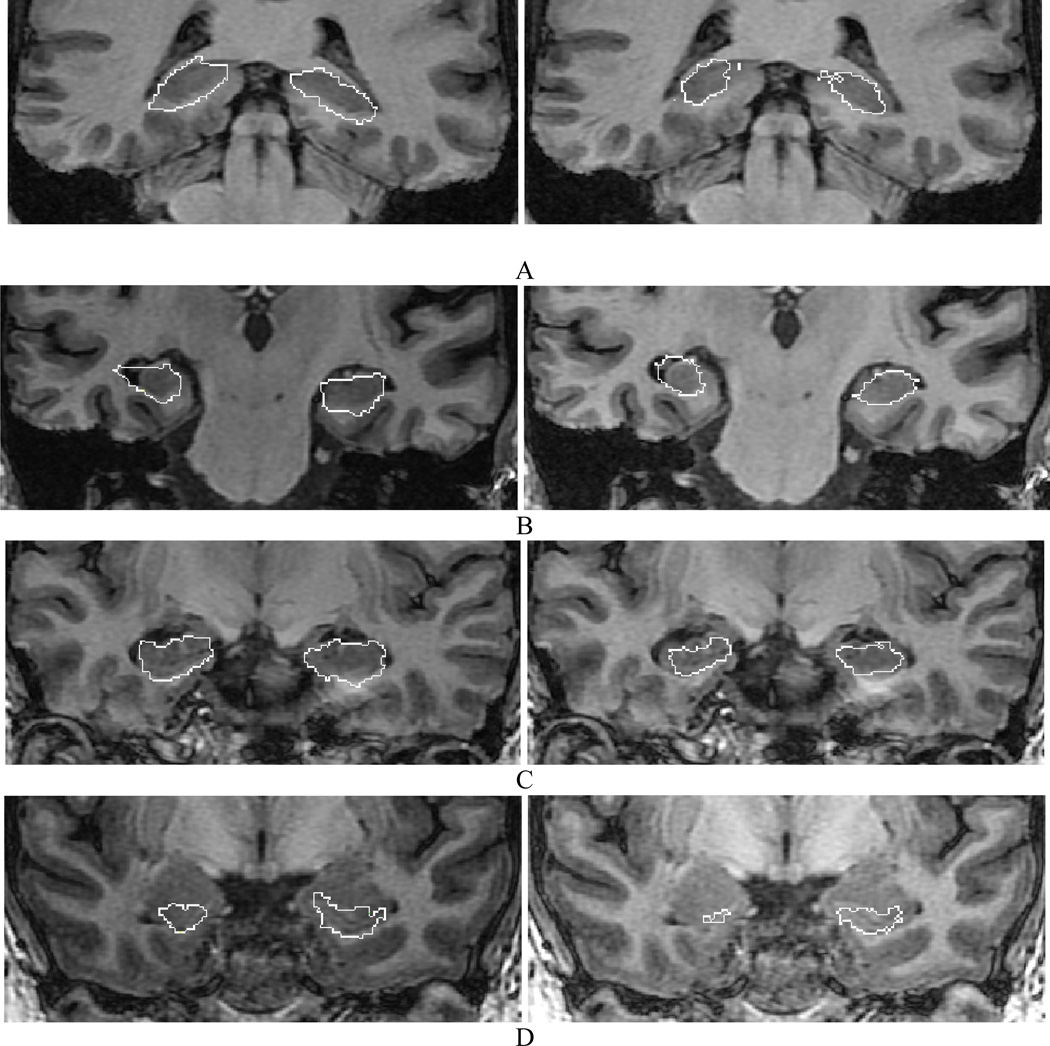
Visual comparisons of hippocampal outlines achieved by automated methods and those manually generated illustrate the distinctions between the two (Fig. 9). The coarse outline in both automated versions is due to reslicing of the images to a voxel size of 1 mm3 before segmentation and mapping the generated labels back to the original images. Whereas distinct white matter borders, such as the alveus, were used in the manually segmented images to demarcate the hippocampal border, this was not the case for either Brain Parser or CFL. Both include the alveus in their outlines (Fig. 9D). Neither of the automated segmentations appreciated the hippocampal folds which characteristically define its anterior appearance (Fig. 9C). An accurate automated segmentation of the hippocampus in such locations is somewhat challenging due to the PVE.
Fig. 9.
Automated segmentation outcomes of the images in Fig. 7 using BrainParser (left) and CFL (right). Although both applications approximate the boundary of the hippocampus reasonably well, the coarseness of the outline, a manifestation of reformatting, reduces accuracy with the inclusion of surrounding borderzones. Absence of definition of borderzone white matter tracts such as the alveus (C, D), in particular, unnecessarily adds to the intended hippocampal volume. The lack of detail in the definition of the interdigitations of the pes hippocampi anteriorly further confounds the ability to accurately measure this site which is often affected by a localization-related epileptogenicity.
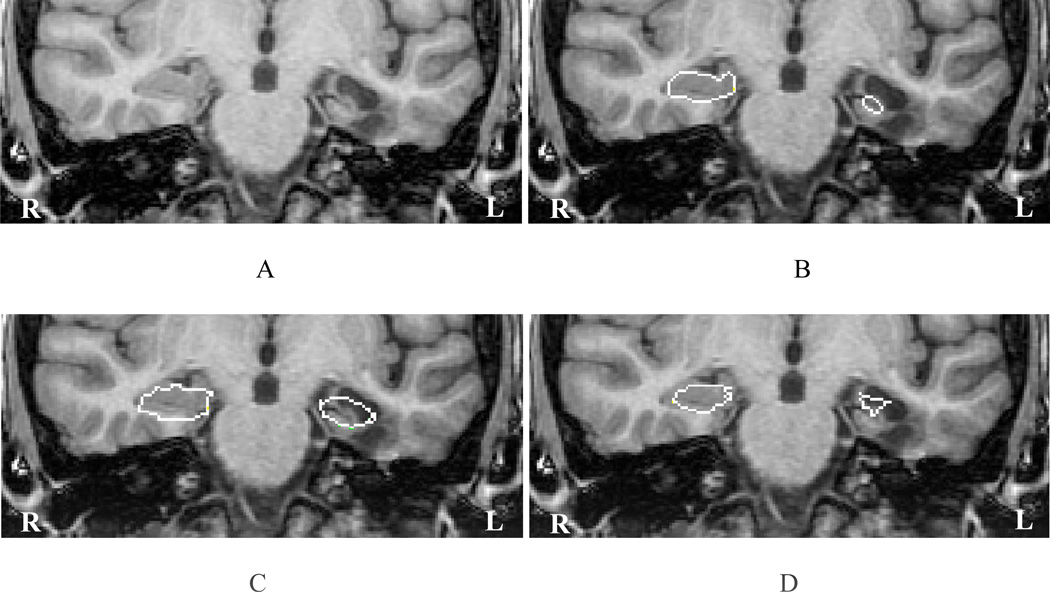
To further study the capabilities of automated segmentation algorithms, several examples of hippocampal deformation were selected (Figs. 10, 11). The presence of an extrinsic lesion, in particular, may be shown to produce sufficient local hippocampal distortion to significantly affect outcome using an automated approach (Fig. 10A). This is made apparent in the case of both Brain Parser (Fig. 10C) and CFL (Fig. 10D) using such an example. Hippocampal deformation by an intrinsic anomaly seems equally likely to affect a poor outcome with either Brain Parser (Fig. 11C) or CFL (Fig. 11D).
Fig. 10.
A representative slice (A) showing hippocampal outlines using manual segmentation (B), Brain Parser (C) and CFL (D). The presence of an extrinsic lesion in the basal left temporal lobe has caused a poor automated segmentation outcome in both applications; even more, in the case of CFL.
Fig. 11.
A representative slice (A) showing hippocampal outlines using manual segmentation (B), Brain Parser (C) and CFL (D) in the case of an intrinsic lesion-induced deformation of the left hippocampus. The deformation has resulted in a poor automated segmentation outcome in both cases.
Discussion
In addition to IBSR and LPBA40, there are other publicly available datasets for the validation of segmentation algorithms, notably for the caudate (Ginneken et al., 2007) and whole brain (Shattuck et al., 2009). The Caudate Segmentation Evaluation (Cause07) (Ginneken et al., 2007) contains separate training and testing sets from healthy controls and patients with schizotypal personality disorder. Labels are provided for the caudate and only for the training samples. No label is provided for the testing set. Advantages of Cause07 include (i) separate training and testing samples, (ii) images acquired from two different centers and (iii) inclusion of pediatric samples. Segmentation Validation Engine (Shattuck et al., 2009) contains 40 T1W MR imaging sets of unaffected subjects used for validation of brain extraction algorithms. Whole brain masks have been generated manually. Furthermore, 56 structures have been segmented semiautomatically. No label is provided; thus, all images are used for testing.
The set of images provided in this work could be improved in several ways. Recently published automated segmentation algorithms, such as HAMMER (Shen and Davatzikos, 2002) and FreeSurfer (Fischl et al., 2002), provide labels for multiple brain structures and the same might be provided here. An algorithm’s performance may vary for different structures and the provision of corresponding manually segmented brain structures in the current dataset would be useful for the validation of automated segmentation algorithms of these structures. As the hippocampus has received the highest attention in the literature, we have provided a more comprehensive standard for this structure, in particular.
In the current dataset, the training and testing sets contain five nonepileptic subjects each. The subjects were scanned using a 1.5 T MRI scanner with 0.78×0.78×2.00 mm3 resolution. Populating the dataset with greater numbers of similar cases and with higher resolution studies would provide for increasingly more accurate human brain imaging atlases. Improvements in field strength of scanners with resultant higher image resolution would promote such development over time also. The time spent in the manual segmentation of greater numbers of available images would be rewarded by the improved boundary definition of individual structures. Public availability of a sufficiently large dataset of high resolution MR images will be the substrate upon which automated algorithms may reliably function with the extraction of optimal metrics.
Automated segmentation algorithms are optimized for segmentation of a structure, such as the hippocampus, free of any local distortion by proximate or intrinsic lesions. Furthermore, the images have highly anisotropic voxels which makes segmentation even more challenging. The availability of a dataset that includes such structural anomalies and anisotropies provides a means for identifying such cases and establishing the extent to which any automated method is able to approximate the correct volume.
Although a separate test set provides a means of comparing the applicability of algorithms using a challenging format, repeated uploading of results may bias the algorithms toward the test set. In other words, the test set becomes a means of training. This problem may be alleviated by providing a second set of test images for algorithms that have already been evaluated using the original test images. The users need only to submit their results once.
Conclusion
We have provided a publicly available MR image dataset of nonepileptic subjects and epilepsy patients with hippocampal outlines for validation of segmentation algorithms. The dataset was divided into training and testing sets. Only the hippocampal outlines of the training set are provided. Investigators may use the training images and their outlines to train their algorithms and then apply their algorithms to the testing set. The segmentation outcomes for the testing set may be submitted for evaluation based on 11 metrics. We evaluated Brain Parser and CFL segmentation algorithms using the dataset. The experiments suggest that the segmentation algorithms require modification in order to better accommodate structural deformation of the hippocampus under a variety of circumstances that befall patients with different neurological disorders.
Acknowledgements
This work was supported in part by NIH grant EB002450.
Footnotes
Information Sharing Statement
The dataset is available at http://www.radiologyresearch.org/HippocampusSegmentationDatabase/. The users are required to register in order to log in and download the dataset. Information about how to submit the segmentation outcomes for the testing set and receive the table of metrics is available online.
References
- Aljabar P, Heckemann R, Hammers A, Hajnal JV, Rueckert D. Classifier selection strategies for label fusion using large atlas databases. Med Image Comput Comput Assist Interv. 2007;10:523–531. doi: 10.1007/978-3-540-75757-3_64. [DOI] [PubMed] [Google Scholar]
- Cendes F, Andermann F, Gloor P, Evans A, Jones-Gotman M, Watson C, Melanson D, Olivier A, Peters T, Lopes-Cendes I, et al. MRI volumetric measurement of amygdala and hippocampus in temporal lobe epilepsy. Neurology. 1993;43:719–725. doi: 10.1212/wnl.43.4.719. [DOI] [PubMed] [Google Scholar]
- Cocosco C, Kollokian V, Kwan R, Evans A. Brainweb: Online interface to a 3D MRI simulated brain database. Neuroimage. 1997;5:425. [Google Scholar]
- Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- Duvernoy HM. The human hippocampus: functional anatomy, vascularization, and serial sections with MRI. 3rd ed. New York: Springer, Berlin; 2005. [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Geuze E, Vermetten E, Bremner JD. MR-based in vivo hippocampal volumetrics: 1. Review of methodologies currently employed. Mol Psychiatry. 2005;10:147–159. doi: 10.1038/sj.mp.4001580. [DOI] [PubMed] [Google Scholar]
- Ginneken Bv, Heimann T, Styner M. 3D Segmentation in the Clinic: A Grand Challenge. In: Heimann T, Styner M, van Ginneken B, editors. 3D Segmentation in the Clinic: A Grand Challenge. 2007. pp. 7–15. [Google Scholar]
- Jack CR, Jr, Petersen RC, O'Brien PC, Tangalos EG. MR-based hippocampal volumetry in the diagnosis of Alzheimer's disease. Neurology. 1992;42:183–188. doi: 10.1212/wnl.42.1.183. [DOI] [PubMed] [Google Scholar]
- Jafari-Khouzani K, Elisevich K, Patel S, Smith B, Soltanian-Zadeh H. FLAIR signal and texture analysis for lateralizing mesial temporal lobe epilepsy. Neuroimage. 2010;49:1559–1571. doi: 10.1016/j.neuroimage.2009.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001;5:143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Konrad C, Ukas T, Nebel C, Arolt V, Toga AW, Narr KL. Defining the human hippocampus in cerebral magnetic resonance images--an overview of current segmentation protocols. Neuroimage. 2009;47:1185–1195. doi: 10.1016/j.neuroimage.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie SM, Abukmeil SS. Brain abnormality in schizophrenia. A systematic and quantitative review of volumetric magnetic resonance imaging studies. Br J Psychiatry. 1998;172:110–120. doi: 10.1192/bjp.172.2.110. [DOI] [PubMed] [Google Scholar]
- Munkres JR. Topology. 2nd ed. Upper Saddle River, NJ: Prentice Hall; 2000. [Google Scholar]
- Rohlfing T, Brandt R, Menzel R, Maurer CR., Jr Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. Neuroimage. 2004;21:1428–1442. doi: 10.1016/j.neuroimage.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage. 2008;39:1064–1080. doi: 10.1016/j.neuroimage.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck DW, Prasad G, Mirza M, Narr KL, Toga AW. Online resource for validation of brain segmentation methods. Neuroimage. 2009;45:431–439. doi: 10.1016/j.neuroimage.2008.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen DG, Davatzikos C. HAMMER: Hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imaging. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studholme C, Hill D, Hawkes D. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recognition. 1999;32:71–86. [Google Scholar]
- Tu Z, Narr KL, Dollar P, Dinov I, Thompson PM, Toga AW. Brain anatomical structure segmentation by hybrid discriminative/generative models. IEEE Trans Med Imaging. 2008;27:495–508. doi: 10.1109/TMI.2007.908121. [DOI] [PMC free article] [PubMed] [Google Scholar]