Abstract
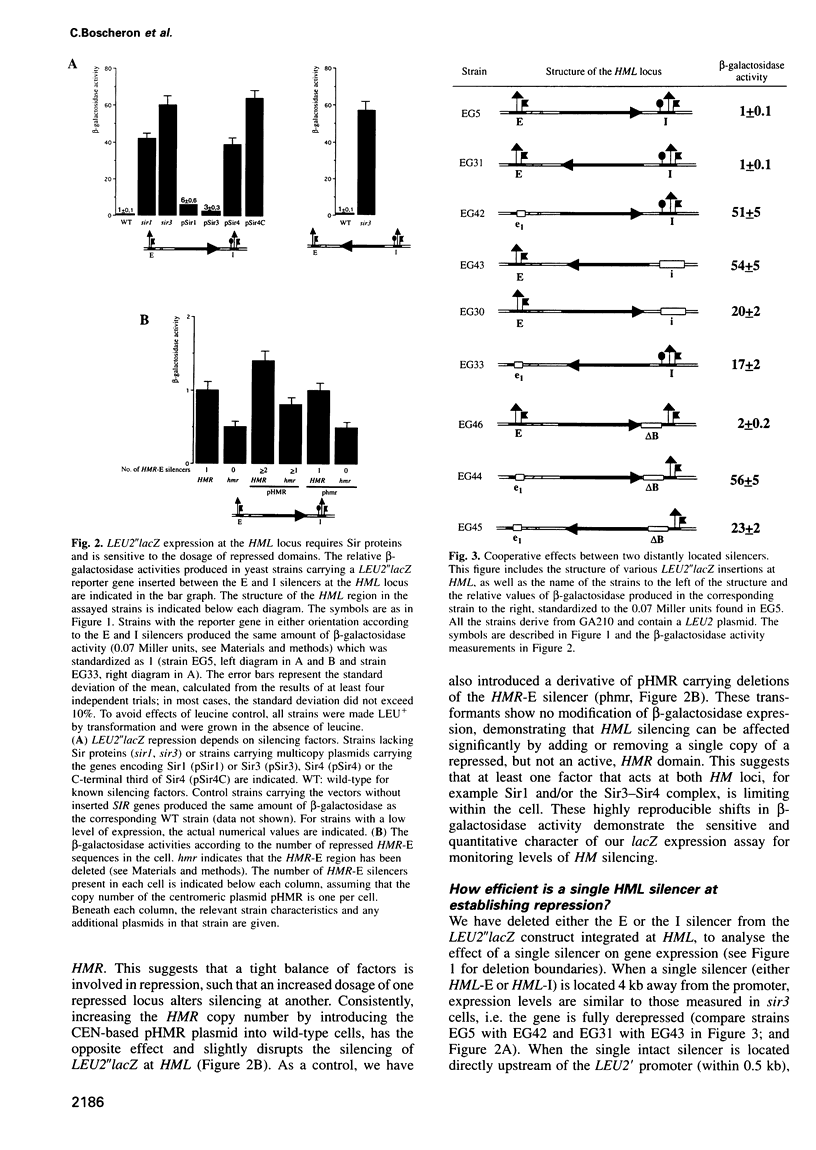
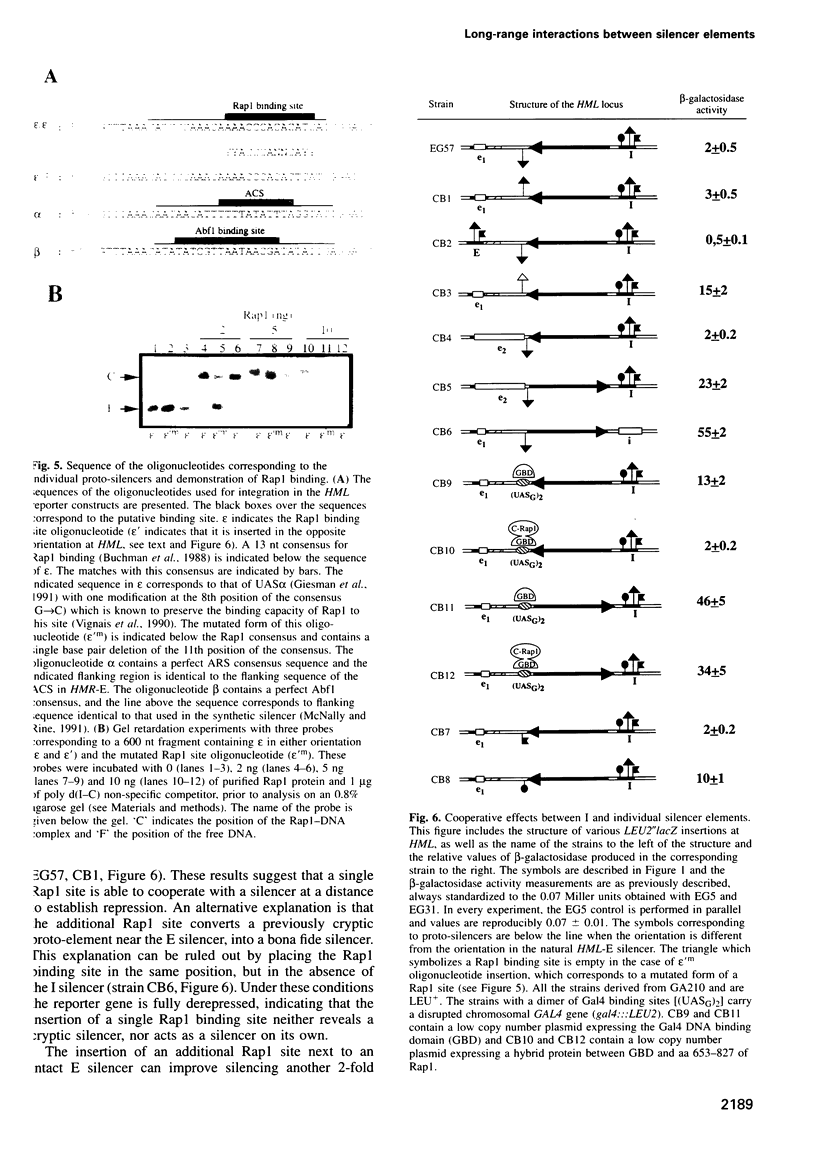
Transcriptional repression at the silent yeast mating type loci is achieved through the formation of a particular nucleoprotein complex at specific cis-acting elements called silencers. This complex in turn appears to initiate the spreading of a histone binding protein complex into the surrounding chromatin, which restricts accessibility of the region to the transcription machinery. We have investigated long-range, cooperative effects between silencers by studying the repression of a reporter gene integrated at the HML locus flanked by various combinations of wild-type and mutated silencer sequences. Two silencers can cooperate over >4000 bp to repress transcription efficiently. More importantly, a single binding site for either the repressor activator protein 1 (Rap1), the autonomous replicating sequence (ARS) binding factor 1 (Abf1) or the origin recognition complex (ORC) can enhance the action of a distant silencer without acting as a silencer on its own. Functional cooperativity is demonstrated using a quantitative assay for repression, and varies with the affinity of the binding sites used. Since the repression mechanism is Sir dependent, the Rap1, ORC and/or Abf1 proteins bound to distant DNA elements may interact to create an interface of sufficiently high affinity such that Sir-containing complexes bind, nucleating the silent chromatin state.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham J., Nasmyth K. A., Strathern J. N., Klar A. J., Hicks J. B. Regulation of mating-type information in yeast. Negative control requiring sequences both 5' and 3' to the regulated region. J Mol Biol. 1984 Jul 5;176(3):307–331. doi: 10.1016/0022-2836(84)90492-3. [DOI] [PubMed] [Google Scholar]
- Amouyal M., Mortensen L., Buc H., Hammer K. Single and double loop formation when deoR repressor binds to its natural operator sites. Cell. 1989 Aug 11;58(3):545–551. doi: 10.1016/0092-8674(89)90435-2. [DOI] [PubMed] [Google Scholar]
- Aparicio O. M., Billington B. L., Gottschling D. E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991 Sep 20;66(6):1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Kobayashi R., Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993 Dec 17;262(5141):1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Micklem G., Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987 Dec 4;51(5):709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- Breeden L., Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harb Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Kimmerly W. J., Rine J., Kornberg R. D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jan;8(1):210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck S. W., Shore D. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 1995 Feb 1;9(3):370–384. doi: 10.1101/gad.9.3.370. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Chambers A., Stanway C., Tsang J. S., Henry Y., Kingsman A. J., Kingsman S. M. ARS binding factor 1 binds adjacent to RAP1 at the UASs of the yeast glycolytic genes PGK and PYK1. Nucleic Acids Res. 1990 Sep 25;18(18):5393–5399. doi: 10.1093/nar/18.18.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C. T., Bartel P. L., Sternglanz R., Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C. T., Buck S., Sternglanz R., Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993 Nov 5;75(3):531–541. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- Cockell M., Palladino F., Laroche T., Kyrion G., Liu C., Lustig A. J., Gasser S. M. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J Cell Biol. 1995 May;129(4):909–924. doi: 10.1083/jcb.129.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman J. B., Hicks J. B., Broach J. R. Identification of sites required for repression of a silent mating type locus in yeast. J Mol Biol. 1984 Oct 5;178(4):815–834. doi: 10.1016/0022-2836(84)90313-9. [DOI] [PubMed] [Google Scholar]
- Foss M., McNally F. J., Laurenson P., Rine J. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science. 1993 Dec 17;262(5141):1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- Fox C. A., Loo S., Dillin A., Rine J. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev. 1995 Apr 15;9(8):911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- Fromental C., Kanno M., Nomiyama H., Chambon P. Cooperativity and hierarchical levels of functional organization in the SV40 enhancer. Cell. 1988 Sep 23;54(7):943–953. doi: 10.1016/0092-8674(88)90109-2. [DOI] [PubMed] [Google Scholar]
- Giesman D., Best L., Tatchell K. The role of RAP1 in the regulation of the MAT alpha locus. Mol Cell Biol. 1991 Feb;11(2):1069–1079. doi: 10.1128/mcb.11.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Laroche T., Gasser S. M. Telomeres and the functional architecture of the nucleus. Trends Cell Biol. 1993 Apr;3(4):128–134. doi: 10.1016/0962-8924(93)90175-z. [DOI] [PubMed] [Google Scholar]
- Gilson E., Roberge M., Giraldo R., Rhodes D., Gasser S. M. Distortion of the DNA double helix by RAP1 at silencers and multiple telomeric binding sites. J Mol Biol. 1993 May 20;231(2):293–310. doi: 10.1006/jmbi.1993.1283. [DOI] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Hardy C. F., Sussel L., Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992 May;6(5):801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- Hecht A., Laroche T., Strahl-Bolsinger S., Gasser S. M., Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995 Feb 24;80(4):583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hill J., Donald K. A., Griffiths D. E., Donald G. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991 Oct 25;19(20):5791–5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J. F., Laroche T., Brand A. H., Gasser S. M. RAP-1 factor is necessary for DNA loop formation in vitro at the silent mating type locus HML. Cell. 1989 Jun 2;57(5):725–737. doi: 10.1016/0092-8674(89)90788-5. [DOI] [PubMed] [Google Scholar]
- Ivy J. M., Klar A. J., Hicks J. B. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Feb;6(2):688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G. H. Position-effect variegation and the new biology of heterochromatin. Curr Opin Genet Dev. 1994 Apr;4(2):281–291. doi: 10.1016/s0959-437x(05)80055-3. [DOI] [PubMed] [Google Scholar]
- Kayne P. S., Kim U. J., Han M., Mullen J. R., Yoshizaki F., Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988 Oct 7;55(1):27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- Kimmerly W., Buchman A., Kornberg R., Rine J. Roles of two DNA-binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. EMBO J. 1988 Jul;7(7):2241–2253. doi: 10.1002/j.1460-2075.1988.tb03064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion G., Liu K., Liu C., Lustig A. J. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993 Jul;7(7A):1146–1159. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K., Käs E., Poljak L., Adachi Y. Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr Opin Genet Dev. 1992 Apr;2(2):275–285. doi: 10.1016/s0959-437x(05)80285-0. [DOI] [PubMed] [Google Scholar]
- Laurenson P., Rine J. Silencers, silencing, and heritable transcriptional states. Microbiol Rev. 1992 Dec;56(4):543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Mao X., Lustig A. J. Mutational analysis defines a C-terminal tail domain of RAP1 essential for Telomeric silencing in Saccharomyces cerevisiae. Genetics. 1994 Dec;138(4):1025–1040. doi: 10.1093/genetics/138.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J., Kotarski M. A., Tartof K. D. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics. 1988 Sep;120(1):181–198. doi: 10.1093/genetics/120.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo S., Rine J. Silencers and domains of generalized repression. Science. 1994 Jun 17;264(5166):1768–1771. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- Mahoney D. J., Broach J. R. The HML mating-type cassette of Saccharomyces cerevisiae is regulated by two separate but functionally equivalent silencers. Mol Cell Biol. 1989 Nov;9(11):4621–4630. doi: 10.1128/mcb.9.11.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney D. J., Marquardt R., Shei G. J., Rose A. B., Broach J. R. Mutations in the HML E silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes Dev. 1991 Apr;5(4):605–615. doi: 10.1101/gad.5.4.605. [DOI] [PubMed] [Google Scholar]
- McBroom L. D., Sadowski P. D. Contacts of the ABF1 protein of Saccharomyces cerevisiae with a DNA binding site at MATa. J Biol Chem. 1994 Jun 10;269(23):16455–16460. [PubMed] [Google Scholar]
- McNally F. J., Rine J. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Nov;11(11):5648–5659. doi: 10.1128/mcb.11.11.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P., Freeman K., Coodly L., Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994 Oct 1;8(19):2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- Müeller-Storm H. P., Sogo J. M., Schaffner W. An enhancer stimulates transcription in trans when attached to the promoter via a protein bridge. Cell. 1989 Aug 25;58(4):767–777. doi: 10.1016/0092-8674(89)90110-4. [DOI] [PubMed] [Google Scholar]
- Orlando V., Paro R. Chromatin multiprotein complexes involved in the maintenance of transcription patterns. Curr Opin Genet Dev. 1995 Apr;5(2):174–179. doi: 10.1016/0959-437x(95)80005-0. [DOI] [PubMed] [Google Scholar]
- Palladino F., Gasser S. M. Telomere maintenance and gene repression: a common end? Curr Opin Cell Biol. 1994 Jun;6(3):373–379. doi: 10.1016/0955-0674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Palladino F., Laroche T., Gilson E., Axelrod A., Pillus L., Gasser S. M. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993 Nov 5;75(3):543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Chromatin complexes regulating gene expression in Drosophila. Curr Opin Genet Dev. 1995 Aug;5(4):466–472. doi: 10.1016/0959-437x(95)90050-q. [DOI] [PubMed] [Google Scholar]
- Renauld H., Aparicio O. M., Zierath P. D., Billington B. L., Chhablani S. K., Gottschling D. E. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993 Jul;7(7A):1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- Rudolph H., Koenig-Rauseo I., Hinnen A. One-step gene replacement in yeast by cotransformation. Gene. 1985;36(1-2):87–95. doi: 10.1016/0378-1119(85)90072-1. [DOI] [PubMed] [Google Scholar]
- Shei G. J., Broach J. R. Yeast silencers can act as orientation-dependent gene inactivation centers that respond to environmental signals. Mol Cell Biol. 1995 Jul;15(7):3496–3506. doi: 10.1128/mcb.15.7.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M., Roth S. Y., Szent-Gyorgyi C., Simpson R. T. Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J. 1991 Oct;10(10):3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994 Nov;10(11):408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. S., Gottschling D. E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994 Oct 21;266(5184):404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Stavenhagen J. B., Zakian V. A. Internal tracts of telomeric DNA act as silencers in Saccharomyces cerevisiae. Genes Dev. 1994 Jun 15;8(12):1411–1422. doi: 10.1101/gad.8.12.1411. [DOI] [PubMed] [Google Scholar]
- Stone E. M., Swanson M. J., Romeo A. M., Hicks J. B., Sternglanz R. The SIR1 gene of Saccharomyces cerevisiae and its role as an extragenic suppressor of several mating-defective mutants. Mol Cell Biol. 1991 Apr;11(4):2253–2262. doi: 10.1128/mcb.11.4.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartof K. D., Henikoff S. Trans-sensing effects from Drosophila to humans. Cell. 1991 Apr 19;65(2):201–203. doi: 10.1016/0092-8674(91)90153-p. [DOI] [PubMed] [Google Scholar]
- Thierry A., Fairhead C., Dujon B. The complete sequence of the 8.2 kb segment left of MAT on chromosome III reveals five ORFs, including a gene for a yeast ribokinase. Yeast. 1990 Nov-Dec;6(6):521–534. doi: 10.1002/yea.320060609. [DOI] [PubMed] [Google Scholar]
- Thompson J. S., Johnson L. M., Grunstein M. Specific repression of the yeast silent mating locus HMR by an adjacent telomere. Mol Cell Biol. 1994 Jan;14(1):446–455. doi: 10.1128/mcb.14.1.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. S., Ling X., Grunstein M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature. 1994 May 19;369(6477):245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- Tu H., Casadaban M. J. The upstream activating sequence for L-leucine gene regulation in Saccharomyces cerevisiae. Nucleic Acids Res. 1990 Jul 11;18(13):3923–3931. doi: 10.1093/nar/18.13.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais M. L., Huet J., Buhler J. M., Sentenac A. Contacts between the factor TUF and RPG sequences. J Biol Chem. 1990 Aug 25;265(24):14669–14674. [PubMed] [Google Scholar]
- Wright J. H., Gottschling D. E., Zakian V. A. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev. 1992 Feb;6(2):197–210. doi: 10.1101/gad.6.2.197. [DOI] [PubMed] [Google Scholar]





