Abstract
In response to DNA damage tissue homoeostasis is ensured by protein networks promoting DNA repair, cell cycle arrest or apoptosis. DNA damage response signaling pathways coordinate these processes, partly by propagating gene expression-modulating signals. DNA damage influences not only abundance of mRNAs, but also their coding information through alternative splicing. Here we show that transcription-blocking DNA lesions promote chromatin displacement of late-stage spliceosomes and initiate a positive feedback loop centered on the signaling kinase ATM. We propose that initial spliceosome displacement and subsequent R-loop formation is triggered by pausing of RNA polymerase at DNA lesions. In turn, R-loops activate ATM which signals to further impede spliceosome organization and augment UV-triggered alternative splicing at genome-wide level. Our findings define the R-loop-dependent ATM activation by transcription-blocking lesions as an important event in the DNA damage response of non-replicating cells and highlight a key role for spliceosome displacement in this process.
INTRODUCTION
The DNA damage response (DDR), an intricate protein network that promotes DNA repair, translesion synthesis, cell cycle arrest or apoptosis, has evolved to counteract the detrimental effects of DNA lesions1-3. In the core of DDR, the ATM and ATR signaling pathways coordinate these processes in response to distinct types of DNA damage; ATR to those processed to single-stranded DNA, and ATM to double-strand DNA breaks (DSBs) and chromatin modifications1,4,5. These signaling networks utilize posttranslational modifications and protein-protein interactions to elicit initial stages of the cellular response. Later DDR stages, involve changes in gene expression. Emerging evidence supports that DNA damage influences not only expression levels of its target genes, by altering transcription rates and mRNA half-life, but also exon selection and ultimately their coding potential6.
Production of mature, protein-coding transcripts depends on the selective intron removal catalyzed by the spliceosome, a dynamic ribonucleoprotein complex consisting of 5 snRNPs (U1, U2, U4, U5 and U6), and a large number of accessory proteins7,8. Exon/intron definition by U1 and U2 snRNPs stimulates the recruitment of pre-assembled U4/U6.U5 snRNP tri-particle and numerous non-snRNP proteins. Following U1/U4 displacement and extensive conformational rearrangements, the two-step splicing reaction is catalyzed by the mature, catalytically active spliceosome composed of U2, U5 and U6 snRNPs8.
The vast majority of mammalian genes are alternatively spliced to produce multiple mRNA variants from a single gene9, expanding thus protein diversity. Numerous mechanisms have evolved to provide the spliceosome the plasticity required for selective exon inclusion, without compromising splicing fidelity9. These range from the presence of cis-acting elements on the transcript itself to post-translational modifications of spliceosomal proteins, which are subject to intracellular and environmental cues. Additionally, since most introns are spliced co-transcriptionally within the chromatin environment, splicing decisions are subject to spatiotemporal control imposed by transcribing polymerases and interaction with chromatin remodelers and histone marks10-12. Exon selection is also influenced by DNA damage6,13. There is evidence for a broad range of damage-induced alternative splicing (AS) events, including alternative exon inclusion and exon skipping, and production of proteins with altered (often pro-apoptotic) function13-16. DNA damage-induced AS has been attributed to changes in the processivity rate of RNA polymerase16 (kinetic coupling) or changes in interaction between the polymerase and splicing regulators14,15 (recruitment coupling), under the assumption that the core spliceosome is largely unaffected. Here we present evidence that DNA damage triggers specific profound changes in spliceosome organization affecting preferentially late-stage spliceosomes. Additionally, we identify a reciprocal regulation between ATM-controlled DDR signaling and the core spliceosome. In response to transcription-blocking DNA lesions, outside of its canonical pathway, ATM contributes to selection of genetic information ultimately included in mature transcripts.
RESULTS
DNA damage targets core spliceosomes
To gain mechanistic insight on the influence of DNA damage to chromatin-associated DDR processes, we used SILAC-based quantitative proteomic17 to characterize UV-irradiation-triggered chromatin composition changes (E.D.fig1a-c). Indirect effects of replication stress were avoided by use of quiescent, human dermal fibroblasts (HDFs). UV-induced photolesions inhibit transcription by impeding RNAPII progression and as anticipated we observed a UV-dependent chromatin-depletion of core splicing factors (SFs). Surprisingly though, this depletion was selective; chromatin abundance of all detected U2 and U5 snRNP-SFs was substantially decreased in irradiated cells while abundance of U1 and U4 snRNP-SFs was not significantly affected (E.D.fig1d; S.I. table1). Considering that spliceosomes containing exclusively U2/U5/U6 snRNPs are formed at later stages of the splicing cycle, following eviction of U1 and U4 from the assembled spliceosome8, we concluded that DNA damage targets preferentially, late maturation-stage spliceosomes unlike chemical transcription inhibition that affects also early-stage spliceosome assembly18.
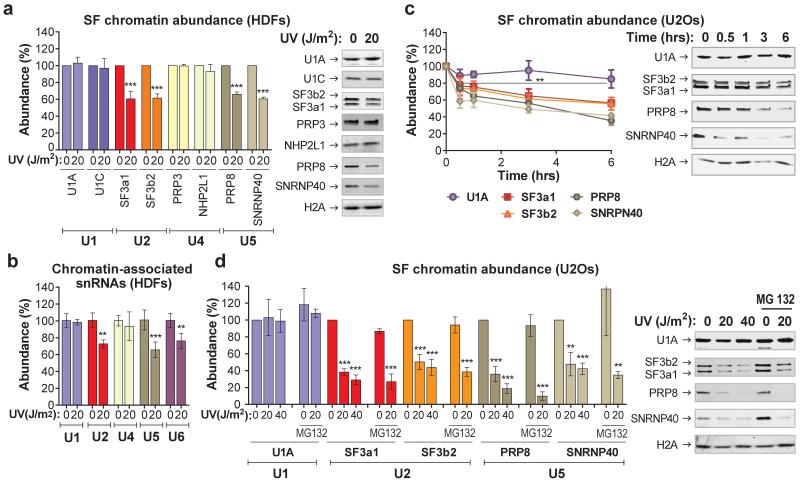
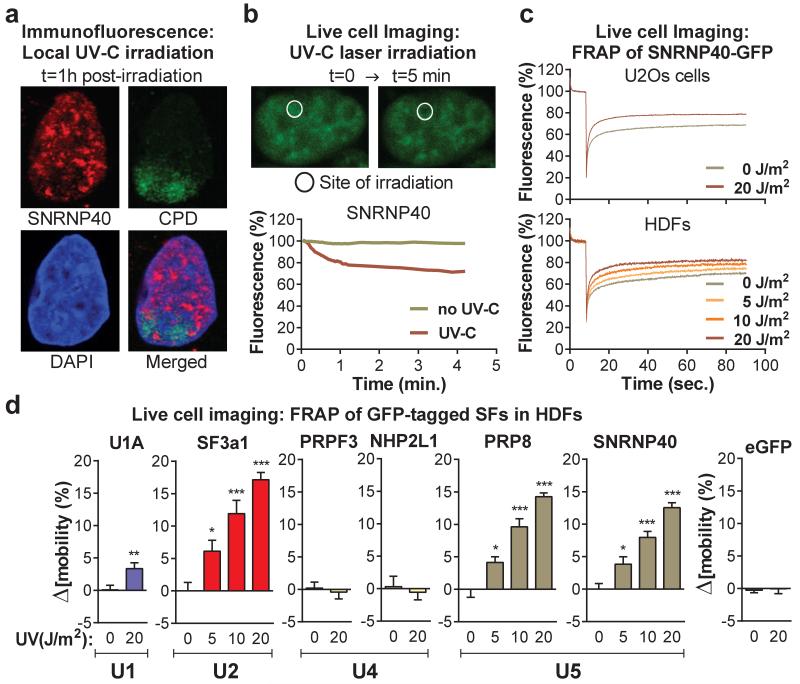
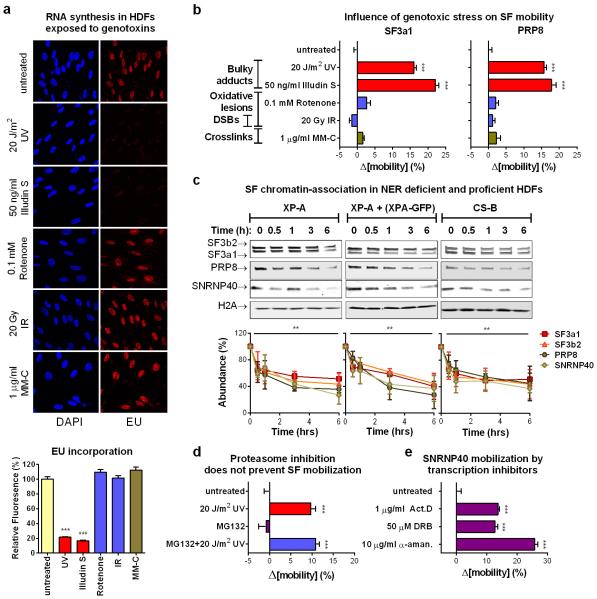
The proteomic results were validated by chromatin fractionation and immunoblotting, for U1 (U1A, U1C), U2 (SF3a1, SF3b2), U4 (PRP3, NHP2L1) and U5 (SNRNP40, PRP8) snRNP-specific proteins8 (fig.1a). We also assayed by qPCR the chromatin-association of all spliceosomal snRNAs. UV-irradiation resulted in preferential chromatin-depletion of U2, U5 and U6 snRNAs, while U1 and U4 were essentially unaffected (fig.1b). Depletion of U2 and U5 snRNP-proteins was time- (fig.1c) and dose-dependent (fig.1d), but independent of proliferation status and cell type (fig.1c, fig.1d). Chromatin-depletion of U2 and U5 snRNP-SFs was independent of proteasome activity (fig.1d), suggesting that depletion was not caused by SF degradation but rather by relocalization. In agreement, total cellular levels of all tested SFs were unaffected by DNA damage (E.D.fig.1e). SF-relocalization was verified by immunofluorescence microscopy after UV-DNA-damage infliction in small subnuclear areas (identified by cyclobutane pyrimidine dimer (CPD)-immunodetection)19. A representative example in fig.2a depicts depletion of SNRNP40 from DNA-damage sites. Relocalization was monitored in real-time, using HDFs stably expressing GFP-tagged members of U2 (SF3a1) and U5 (SNRNP40, PRP8) snRNPs (E.D.fig.2a-d). Subnuclear damage infliction by UV-C microbeam irradiation19 resulted in rapid depletion from irradiated sites of GFP-tagged U2 and U5 snRNP-SFs but not of U1 and U4 (fig.2b; E.D.fig3a-c). Inhibition of transcription-initiation prevented this depletion indicating that the displaced proteins were actively involved in splicing (E.D.fig.3d). Irradiation of the entire cell resulted in prominent changes in SF localization as evidenced by speckle reorganization and enlargement (E.D.fig.4a,b). To further elaborate on the relocalization kinetics of GFP-tagged SFs, we measured their mobility by Fluorescence Recovery after Photobleaching (FRAP). We observed substantial and UV-dose-dependent increases in the mobilities of U2 and U5 snRNP-factors but not of U1 and U4, at one hour post-irradiation (fig.2c,d). In agreement with the chromatin-fractionation assays (fig.1d), mobilization was independent of proteasome activity, confirming that the UV-triggered mobilization is not caused by proteasome-dependent degradation (E.D.fig.5d).
Figure 1. DNA damage-triggered chromatin-displacement of activated spliceosomes.
a,b, UV-induced changes in chromatin-association of spliceosome components in quiescent HDFs; a, Immunoblots (right) and quantification (left) of SF chromatin-association; b, chromatin-associated snRNAs quantified by Q-PCR and normalized to HotAir ncRNA (n=4, mean ± s.d., T-test). d,e, immunoblots (right) and quantification (left) of SF chromatin-association in U2Os cells; d, time post UV-irradiation, e, UV dose-response and lack of influence of proteasome inhibition. b, d, e, Graphs: Signal intensities normalized to H2A. (n=3, mean ± s.d., T-test and one-way ANOVA.
Figure 2. Mobilization and displacement of mature spliceosomes from UV-C induced DNA damage sites.
a, Immunofluorescence detection of SNRNP40 and CPDs in U2Os cells UV-irradiated through porous membranes. b, SNRNP40-GFP depletion from UV-C laser microirradiation sites in U2Os cells; typical image (top) and fluoresence quantification of 20 cells (bottom). c, FRAP of UV-triggered SNRNP40-GFP mobilization in U2Os and quiescent HDFs (n=25). d, FRAP of free eGFP or GFP-tagged SFs in UV-irradiated quiescent HDFs. Δ[mobility] = (Fluorescence irradiated – fluorescence non-irradiated cells) at 1 min post-bleaching (n=25, mean ± s.e.m., T-test and one-way ANOVA).
The UV-dependent chromatin-depletion of snRNAs and proteins participating in late-stage spliceosomes, loss of association with elongating RNAPII (E.D.fig.1f), rapid displacement from DNA damage sites and mobilization of U2 and U5 snRNP-factors, indicate that UV-irradiation influences preferentially, late-stage RNAPII-associated spliceosomes.
NER-independent SF-mobilization
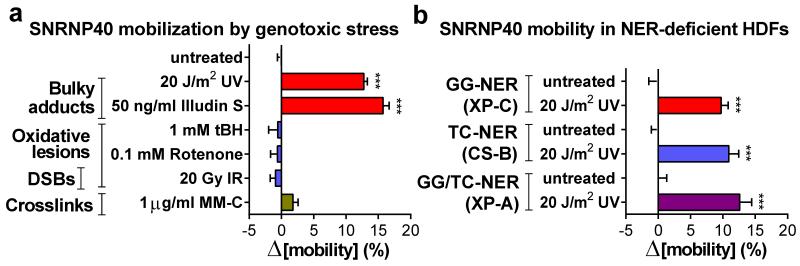
Next we addressed by FRAP, whether spliceosome mobilization is caused by specific DNA lesions or is a general response to macromolecular damage. Significant SF-mobilization was caused by genotoxins inflicting transcription-blocking DNA lesions (UV-irradiation, Illudin S), but not oxidative damage (tert-Butyl-hydroxide, rotenone, ionizing radiation), DSBs (ionizing radiation) or DNA interstrand crosslinks (mitomycin C). This specificity argues that the observed mobilization does not result from non-specific RNA/DNA damage but only from DNA lesions that interrupt transcription (fig.3a; E.D.fig.5a,b) and are substrates of the transcription-coupled nucleotide excision repair (TC-NER) pathway20,21. Interestingly, HDFs deficient in either the Transcription Coupled (TC)-NER, or Global Genome (GG)-NER (lacking CSB and XPC activities respectively), or in both (lacking XPA), show no impairments either in damage-triggered spliceosome mobilization (fig.3b) or in chromatin-displacement of endogenous U2 and U5 snRNP-SFs (E.D.fig.5c). Thus, the influence of transcription blocking lesions in SF localization is independent from NER complex assembly indicating that pausing of elongating RNAPII is necessary and sufficient, to trigger chromatin displacement of late-stage spliceosomes.
Figure 3. Chromatin displacement of mature spliceosomes is caused by RNAPII-blocking lesions and is NER-independent.
a, FRAP of SNRNP40-GFP in quiescent HDFs exposed to genotoxins (n=30, mean ± s.e.m., one-way ANOVA). b, UV-triggered mobilization of SF3a1-GFP and SNRNP40-GFP in HDFs deficient in GG-NER (XP-C), TC-NER (CS-B) or both (XP-A) (n=30, mean ± s.e.m., T-test).
Spliceosome mobilization by DDR signals
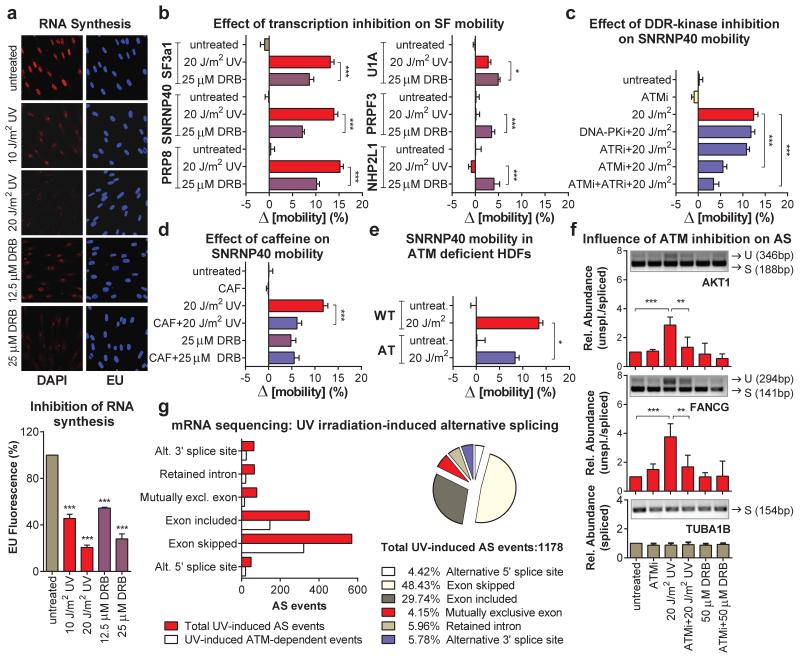
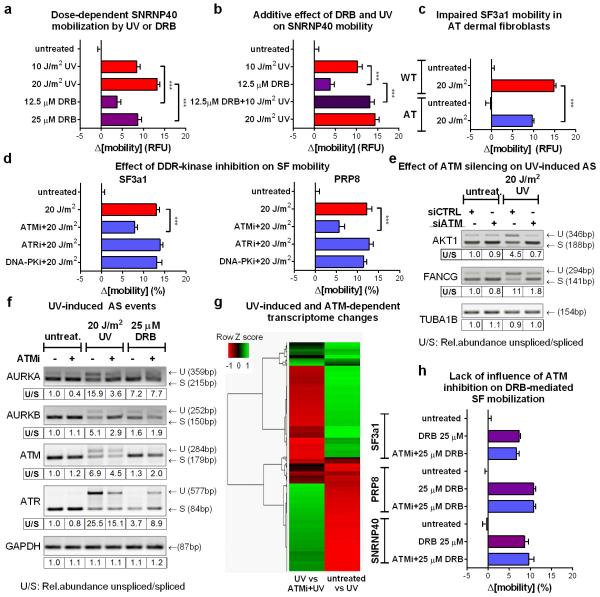
Transcription inhibition by chemicals that target RNAPII, mobilize SFs of all snRNPs unlike UV-irradiation that preferentially targets those participating in late-stage complexes (fig.4b; E.D.fig5e). This preferential mobilization implies distinct mechanisms of action between UV-irradiation-dependent and chemically-induced transcription inhibition. However, to formally exclude the possibility that transcription-blocking DNA lesions mobilize spliceosomes exclusively through RNAPII arrest, we used 5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole (DRB) to inhibit transcription to the same extent as UV-irradiation. Transcription arrest was evaluated by reduced 5-ethynyl-uridine (EU) incorporation in newly synthesized RNA (fig.4a). Both treatments increased spliceosome mobility (fig.4b; E.D.fig.6a) and their combination had an additive effect (ext.fig.6b). Remarkably, UV-irradiation had a more profound SF-mobilization effect than DRB (at equal transcription-inhibiting doses), indicating that transcription inhibition alone is not sufficient to attain the extensive mobilization triggered by UV-irradiation (fig.4b; E.D.fig.6a).
Figure 4. ATM modulates spliceosome mobilization and influences splicing decisions upon DNA damage.
a, RNA synthesis measured by EU pulse-labeling. (n=150, mean ± s.e.m., T-test). b, c, d, e, FRAP of SFs in quiescent HDFs (n=25, mean ± s.e.m., one-way ANOVA); (b) response to UV- or DRB-treatment, (c) UV-irradiation +/− ATM, ATR, or DNA-PK inhibitors, (d), UV- or DRB-treatment +/− Caffeine, (e) HDFs from an AT patient or a healthy donor. f, DRB- or UV-triggered and ATM-dependent intron-inclusion assayed by RT-PCR in quiescent cells. Graphs: signal intensity expressed as unspliced/spliced ratio. (n=4, mean ± s.d., one-way ANOVA). g, Genome-wide identification by RNA-Seq, of UV-induced AS events. Left: Types of AS events. Right: number of total and ATM-dependent events.
Pausing of RNAPII at DNA lesions not only halts transcription, but also activates DDR signaling pathways that modulate the cellular response via posttranslational modifications1,22. Considering that many core SFs have been identified as DDR kinase-substrates22,23 we used the broad range DDR kinase inhibitor caffeine, to evaluate if DDR signaling influences spliceosome organization. Caffeine partially suppressed the UV-dependent spliceosome mobilization but had no influence on the DRB-dependent mobilization confirming that the two processes are, in part, mechanistically distinct (fig.4d).
To dissect which DDR signaling system augments the UV-triggered spliceosome mobilization, cells were treated with specific inhibitors of the major caffeine-sensitive DDR kinases: ATM, ATR and DNA-PK. Neither ATR nor DNA-PK inhibition had a significant effect (fig.4c; E.D.fig.6d). Remarkably, ATM inhibition in non-replicating cells suppressed SF mobilization to levels similar to caffeine (fig.4c, d; E.D.fig.6d), while it had no influence on DRB-mediated mobilization (E.D.fig.6h). The dependency of UV-triggered spliceosome mobilization in ATM signaling was confirmed by the impaired SF mobilization in HDFs derived from an Ataxia Telangiectasia (AT) patient compared to those of a healthy donor (fig.4e; E.D.fig.6c). Thus DNA damage-triggered spliceosome mobilization results from the combined contribution of transcription inhibition and ATM signaling.
To evaluate the impact of ATM-dependent spliceosome mobilization on pre-mRNA processing, we assayed splicing efficiency in a select panel of DDR and cell-cycle related genes24. Quiescent RPE cells were UV-irradiated in the absence or presence of the ATM inhibitor and intron-retention was assayed by RT-PCR24. UV-irradiation resulted in increased ATM-dependent intron-retention (fig.4f; E.D.fig.6f) while transcription inhibition by DRB had minor, and ATM-independent, effects. Specificity of the ATM inhibitor was confirmed by siRNA-mediated ATM silencing which gave identical results (E.D.fig.6e).
To investigate genome-wide the influence of UV-irradiation on AS, as well as the ATM contribution in UV irradiation-dependent gene expression and mRNA processing (AS) changes, we employed RNA Sequencing (RNA-Seq) on cells that were untreated or UV irradiated in the presence or absence of the ATM inhibitor. We observed an impressive anti-correlation in transcript abundance of UV-regulated messages in the presence or absence of ATM activity (E.D.fig.6g) revealing a previously unknown role of ATM signaling in UV-induced gene expression changes. Importantly, UV-irradiation resulted in wide-spread splicing changes, a subset of which (up to 40%) was partly, ATM-dependent demonstrating the genome-wide influence of ATM not only in mRNA abundance but also in UV-induced AS (fig.4g; S.I. table2).
Collectively, these findings demonstrate that UV-irradiation influences gene expression in an ATM-dependent manner and that ATM participates in the selection of the genetic information contained in mature transcripts, thus disclosing a novel non-canonical function of ATM in DDR.
Spliceosome-ATM reciprocal regulation
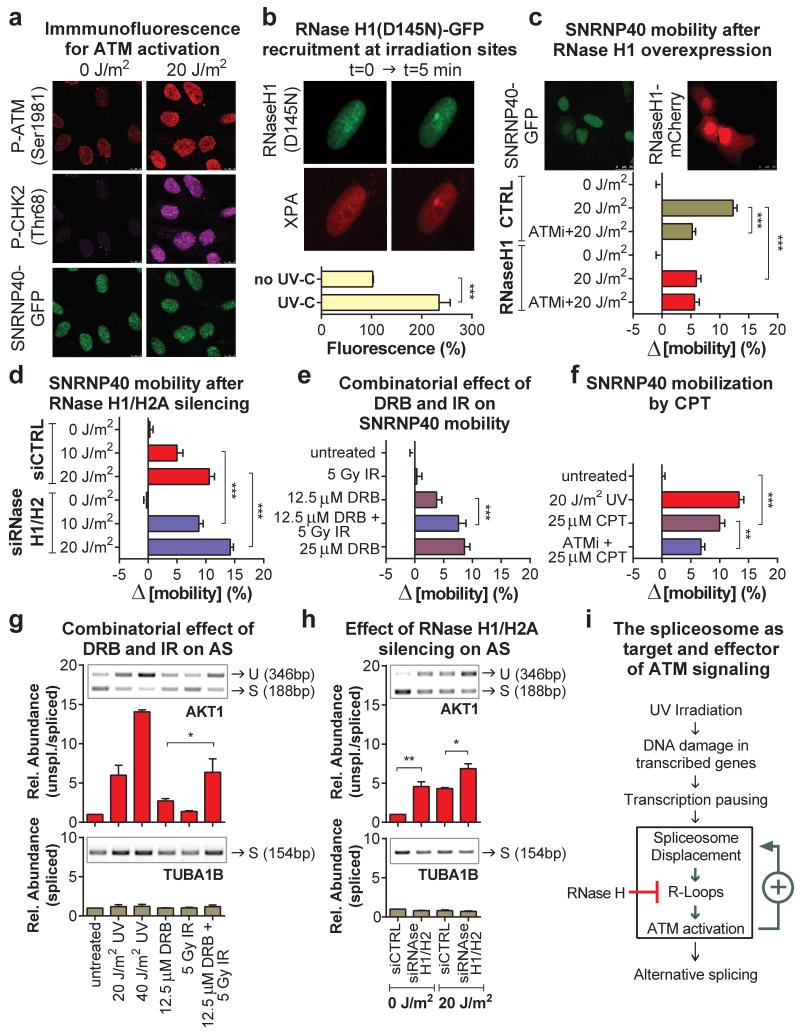
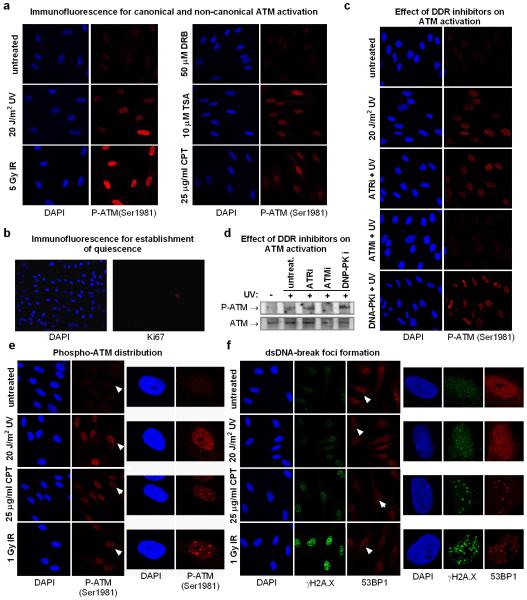
The surprising ATM-dependency of SF-mobilization in quiescent cells indicates that UV-irradiation activates ATM, via a mechanism distinct from its canonical activation by replicative stress and IR-inflicted DSBs4,25. Indeed, UV-irradiation of quiescent HDFs activated ATM, as evidenced by its auto-phosphorylation26 and phosphorylation of CHK227 (fig.5a; E.D.fig.7a-e) to levels similar to the topoisomerase I inhibitor Camptothecin (CPT)28 and the deacetylase inhibitor and non-canonical ATM activator, Trichostatin A (TSA)5 (E.D.fig.7a). Interestingly, in UV-irradiated cells active ATM was dispersed throughout the nucleus, which contrasts to the focal accumulation triggered by DSB-inducing agents such as CPT or IR (E.D.fig.7e). Furthermore, in cells were ATR was also inhibited29, UV-dependent γH2A.X and 53BP1 foci were rare (E.D.fig.7f), suggesting that in non-proliferating cells UV-dependent ATM activation occurs in absence of DSBs.
Figure 5. Reciprocal regulation between spliceosome mobilization and R loop-dependent ATM signaling.
a, Immunofluorescence of ATM activation in quiescent HDFs. b, Recruitment of RNAseH1(D145N)-GFP and mCherry-XPA at UV-C microirradiation sites (n=10, mean ± s.e.m., T-test). c,d,e,f FRAP showing SNRNP40-GFP mobilization in (c) untransfected and mCherry-RNaseH1 expressing U2OS cells, (d) after RNAseH1/H2A silencing, (e) in quiescent HDFs treated with DRB and/or IR and (f) after UV or CPT treatment. (n=30, mean ± s.e.m., one-way ANOVA). g,h, Intron retention assayed by RT-PCR in quiescent cells after (f) silencing of RNaseH1/H2A or (g) combined IR/DRB treatments (n=2, mean ± s.d., one-way ANOVA). (i) Model of UV-triggered and R-loop/ATM-augmented spliceosome mobilization.
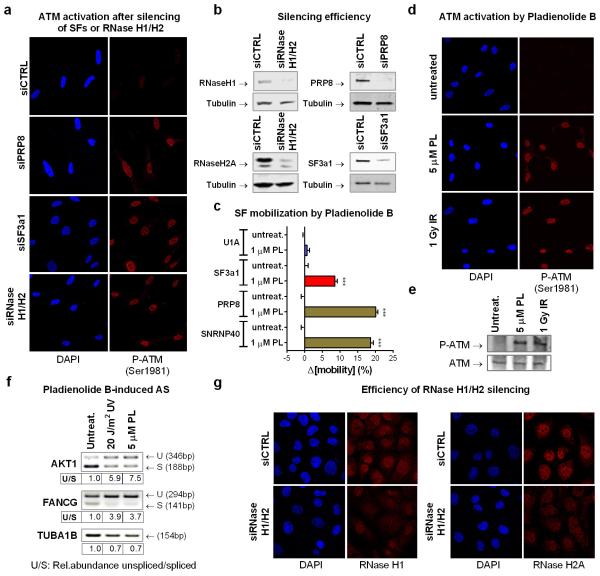
Impairments in co-transcriptional splicing promote hybridization of nascent RNA and single-stranded template DNA at the transcription bubble, resulting in three-nucleic acid-strand structures known as R-loops25. R-loops have been reported to cause genomic instability after SF depletion25,30 and activate ATM in both proliferating and post-mitotic cells28,31. In agreement, siRNA-mediated silencing of U2 or U5 snRNP-SFs, or combined RNase H1/H2A silencing, resulted in ATM activation in absence of other treatments (E.D.fig.8a,b,g). Similarly, treatment of quiescent cells with Pladienolide B32, which arrests late-stage spliceosomes and mobilizes U5, and to a lesser extent U2 snRNP-SFs (E.D.fig.8c), gave a robust ATM activation (E.D.fig.8d,e) and resulted in intron retention levels comparable to UV-irradiation (E.D.fig.8f). To explain our observations we formulated the following hypothesis: RNAPII arrest at DNA lesions displaces a subset of SFs engaged in co-transcriptional splicing. Spliceosome-displacement, in combination with negative supercoiling behind RNAPII, facilitates hybridization of naked pre-mRNA (still containing intronic sequences) to the DNA template-strand. The resulting R-loop activates ATM which then amplifies the mobilization signal and stimulates further spliceosome-displacement either by promoting disassembly or preventing assembly of late-stage spliceosomes. Accordingly, we predicted that: 1) R-loops are formed at sites of UV-DNA damage, and 2) manipulation of R-loop-levels will alter spliceosome mobility.
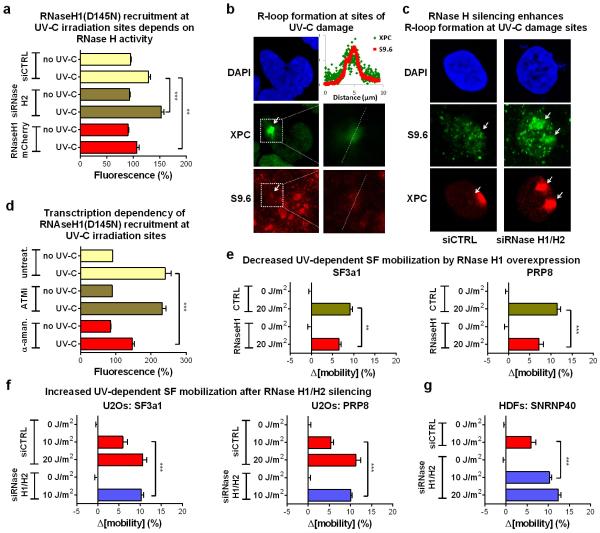
To visualize and resolve R-loops in UV-irradiated cells we exploited the ability of RNaseH to bind and hydrolyze RNA at RNA:DNA duplexes33. For indirect, real-time visualization of R-loop, we used HDFs stably expressing GFP-tagged RNaseH1(D145N), a binding-competent but catalytically inactive RNaseH134. RNaseH1(D145N) was rapidly recruited to UV-C micro-irradiated sites in a transcription-dependent but ATM-independent manner (fig.5B; E.D.fig.9d), suggesting R-loop formation at DNA-damage sites. The ability of RNaseH1(D145N) to detect R-loops, was confirmed by overexpression of active RNaseH1 or by silencing of RNaseH2 which prevented or potentiated respectively, recruitment of RNaseH1(D145N) at UV-C micro-irradiation sites (E.D.fig.9a). Formation of R-loops at these sites was verified using the S9.6 DNA:RNA hybrid-specific antibody35 (E.D.fig.9b). Specificity of immunodetection was confirmed by RNaseH silencing which resulted in enhanced signal at DNA damage sites, at UV-doses when R-loop formation is below detection limits in siRNA control-transfected cells (E.D.fig.9c).
Overexpression of active RNaseH1 attenuated the UV-induced spliceosome mobilization to levels identical to ATM inhibition (fig.5c; E.D.fig.9e). No additional effect was observed when the two manipulations were combined (fig.5C) arguing that RNaseH1 mitigates the UV-triggered spliceosome mobilization by preventing ATM activation. Conversely, silencing of RNaseH1 and H2A, which resolve the majority of RNA:DNA duplexes within the cell30,36, results in ATM activation (E.D.fig.8a,b) and augments the UV-triggered R-loop formation (E.D.fig.9c), spliceosome mobilization (fig.5d; E.D.fig.9f,g) and intron retention (fig.5h).
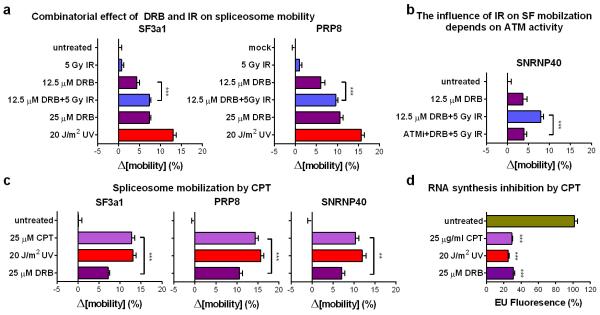
ATM is required for a substantial fraction of the UV-triggered spliceosome mobilization. Regardless, ATM activation alone, by e.g. IR, does not influence spliceosome mobility (fig.3a; fig5e; E.D.fig.5b), indicating that ATM controls a positive feedback mechanism that enhances, but cannot trigger spliceosome displacement (fig.5i). We hypothesized that UV-dependent transcription inhibition acts as the initiating mechanism for spliceosome mobilization which is then enhanced by a secondary ATM-dependent signal. To test this, we used treatments (DRB and IR) that each can specifically influence one process; DRB inhibits transcription (fig.4a) but does not activate ATM (E.D.fig.7a) while IR activates ATM (E.D.fig.7a,e; E.D.fig.8d,e) but does not interfere with global transcription (ext.fig.5a). Combination of DRB and IR had additive effects in both spliceosome mobilization and intron retention (fig.5e,g; E.D.fig10a,b) indicating that ATM amplifies (but does not initiate) a mobilization signal imposed by transcriptional arrest. In agreement, treatment of quiescent HDFs with CPT, which promotes formation of transcription-blocking lesions (E.D.fig.10d) and R-loop-dependent ATM activation28 (E.D.fig.7e), can also efficiently mobilize spliceosomes to levels higher than expected by transcription inhibition alone (fig.5f; E.D.fig.10c).
DISCUSSION
Here we present evidence that the core spliceosome is a target and an effector of the cellular response to transcription-blocking DNA damage and we define a previously uncharacterized ATM-dependent branch of genome surveillance. Transcription-blocking DNA lesions cause selective chromatin-displacement of late-stage spliceosomes by a two-step mechanism involving a stochastic (cis-) and an ATM signaling-mediated (trans-) stage. Our hypothesis is that displacement of assembled co-transcriptional spliceosomes is required to remove steric inhibition that would otherwise prevent back-tracking (or removal) of RNAPII from DNA lesions, which is critical for subsequent DNA repair22. The initial spliceosome displacement likely results in naked (intron-retaining) pre-mRNA readily available for hybridization with template ssDNA at the transcription bubble. This culminates in R-loop formation at damaged DNA sites, which in turn activate ATM. Previously, R-loop mediated ATM activation has been linked to replication-induced DSBs because of collision of arrested transcription-complexes with the replication machinery30,37. Here, we demonstrate that neither DSBs nor replication are required for R-loop-dependent ATM activation. While the exact mode of UV-triggered ATM activation remains to be determined, it does have significant biological consequences. It influences gene expression and plays a fundamental role in augmenting spliceosome-displacement and alternative pre-mRNA splicing at genome-wide level.
ATM activation and spliceosome displacement are subject to a reciprocal regulation, which has two unanticipated implications. First, in response to transcription-blocking lesions, changes in spliceosome organization activate ATM signaling irrespective of replication. Second, ATM modulates DDR, not only by controlling expression levels of its target genes, but also pre-mRNA processing. These observations provide new insights on the mechanisms and consequences of ATM activation in post-mitotic tissues, which is critical for proper cellular function, as evidenced by the severe neurodegeneration in Ataxia-Telangiectasia patients38.
METHODS
Materials
MNase and all chemicals were purchased from Sigma-Aldrich unless otherwise specified. DNA modifying enzymes were from Roche Applied Sciences. Pladienolide B was from Santa Cruz Biotechnology, the ATR inhibitor VE821 from TINIB-Tools, and the ATM inhibitor KU55933 and DNA-PK inhibitor NU7441 from R&D Systems. Antibodies used were against: PRP8 (H300), XPA/p62 (FL-273), p89/XPB (S-19) and β-Tubulin from Santa Cruz Biotechnology; SNRPC/U1C (NBP1-96048), NHP2L1 (NBP1-32732), SF3a1 (NB100-79847), SF3b2 (NV100-79843), RNaseH1 (NBP2-20171), and RNaseH2A (NBP1-76981) from Novus Biologicals; SNRNP40 (SAB2701506) and SRSF2/SC35 (clone SC-35) from Sigma; SNRPA/U1A (3F9-1F7) from ABGENT; PRPF3 (ab187535), RNPII CTD (phospho-S2) (ab5095), RNAPII (ab5095), PCNA (PC-10), Ki67 (ab833) from abcam®; CPD (TDM-2) from MBL International; GFP (11 814 460 001) from Roche; H2A (07-146) from Millipore Corp.; phosphor-ATM(1981)(05-740) from Upstate Biotechnology, Phospho-CHK2(Thr68) (2661) from Cell Signaling. Anti-XPC (rabbit-polyclonal ab) was in-house developed. Odyssey-compatible IRDye680- and IRDye800–conjugated secondary antibodies were from LI-COR. Secondary antibodies conjugated to Alexa Fluorochromes-488, -568, -594 and-647 were from Invitrogen. GFP-tagged proteins were immunoprecipitated with GFP-Trap® beads (ChromoTek).
Cell Culture, SILAC Labeling and Cell treatments
Cell lines used in this study were: Ataxia Telengectasia patient (AT2) and healthy adult donor (C5Ro) derived Human Dermal Fibroblasts (HDFs); SV-40 transformed XP-A (XP12RO), XP-C (XP4A) and CS-B (CS1AN) patient-derived HDFs; hTERT immortalized HDFs (C5Ro-T), VH10 human foreskin fibroblasts (VH10-T) and human retinal pigmented epithelial cells (RPE1, ATCC), human osteosarcoma cells (U2Os); and the amphotropic retroviral packaging cell line Gryphon A (Allele Biotechnology). Cells were subcultivated under standard culture conditions (37°C, 5% CO2) in a humidified incubator. U2OS, Gryphon A, and SV40-transformed cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM, Lonza), supplemented with 10% v/v fetal bovine serum (FBS, Fisher Scientific) and 1% v/v penicillin-streptomycin (PS, Lonza). Primary and TERT-immortalized HDFs and RPE-1 cells were cultured in Ham’s F10 (Lonza) supplemented with 15% FBS and 1% PS. When applicable, cells were synchronized in quiescence by 72 hr serum-deprivation. For FRAP and immunofluorescence experiments, cells were seeded on 25 mm-diameter glass slides. For UV-C laser/live cell imaging experiments cells were seeded on quartz coverslips (010191T-AB; SPI Supplies). For Stable Isotopic Labeling with Amino-acids in Culture (SILAC), C5Ro-T cells were cultured for >5 population doublings (PD) in lysine-, arginine- and leucine-free DMEM (AthenaES) supplemented with antibiotics, non-essential amino-acids (LONZA), 10% dialyzed FBS (Invitrogen) and 105 μg/ml leucine and either 73 μg/ml light [12C6]-lysine and 42 μg/ml [12C6, 14N4]-arginine or with heavy [13C6]-lysine and [13C6, 15N4]-arginine (Cambridge Isotope Laboratories). In each subcultivation, cell numbers were determined using a Beckman Z2 coulter counter (Beckman Coulter, Inc.), and 0.5 × 104 cells were seeded per cm2 of growth surface area. The increase in population doubling (ΔPD) was calculated using the formula ΔPD = Log10 (number of cells harvested/number of cells seeded]/log102.
Cells were UV-C irradiated (254 nm, TUV Lamp, Philips) at the indicated doses. For local DNA damage infliction, cells were UV-irradiated (60 J/m2) through isopore polycarbonate membranes containing 5 μm-diameter pores (Millipore). Chemicals were added directly in the growth media at the indicated concentrations. In FRAP experiments cells were assayed 1 hr after initiation of treatment with the exception of Illudin S and Rotenone, which were assayed at 6hrs. Pre-/incubation with Caffeine (10mM), DDR-kinase inhibitors (10 μM) and MG132 (50μM), started 1 hr prior to genotoxic treatments and lasted throughout the experiment. α-Amanitin treatments were for >24 hrs. For exon-specific RT-PCR cells were lysed 6 hrs after treatment.
Mass spectrometry and data analysis
Nanoflow liquid chromatography-tandem mass spectrometry (LC-MS/MS) and data analysis were a as described39. Briefly, samples containing MNase-digested chromatin were size-fractionated by SDS-PAGE, gels were cut in 2-mm slices, and subjected to dithiotreitol-reduction, iodoacetamide-alkylation and trypsin-digestion. LC-MS/MS was performed on an 1100 series capillary liquid chromatography system (Agilent Technologies) coupled to an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific) operating in positive mode. Raw MS data were analyzed using the MaxQuant software. A false discovery rate of 0.01 for proteins and peptides and a minimum peptide length of 6 amino acids were set. The Andromeda search engine was used to search MS/MS spectra against the International Protein Index (IPI) human database. Statistical analysis was performed with Perseus (1.5.0.30)17.
Cloning
Human full-length cDNA clones used for subcloning were; PRP8/PRPF8/DHX16 (CS116070), SF3A1 (SC321295), SNRPN40 (SC112670) and RNASEH1 (SC319446) from Origene and U1A/SNRPA (MHS6278-202826119), NHP2L1 (MHS6278-202839330) and PRP3/PRPF3 (MHS6278-202826220) from Dharmacon. To generate vectors expressing GFP- and mCherry-tagged proteins the open reading frames (minus the STOP codon) of human U1A, SF3a1, PRP3, NHP2L1, PRP8 and SNRNP40 were PCR amplified using oligonucleotides containing restriction enzyme sites. PCR products where subcloned into a pLHCX retroviral expression vector (Clontech Laboratories) modified to contain eGFP lacking the initiation codon. XPA and RNase H1 lacking the mitochondrial localization signal (amino acids 1-28) were subcloned in modified pLHCX vectors containing either eGFP or mCherry lacking their STOP codons. PCR amplifications were performed on a MJ Scientific, Inc., PTC-100 Thermocycler using high-fidelity Phusion polymerase (Bioke). Amplified cDNAs were purified using the Promega Wizard kit. Following restriction digestion of inserts and vectors, shrimp alkaline phosphatase treatment of the vectors, and agarose gel electrophoresis, the gel-excised DNAs were purified using the Promega Wizard kit. DNA inserts were ligated into vectors at a 3:1 molar ratio. Plasmid DNAs were validated by restriction digestion and sequencing.
Infections/Transfections
C5Ro-T, C5Ro, AT-2, U2Os and VH-10T cell lines stably- expressing GFP-tagged proteins were generated by retroviral infection followed by hygromycin selection. For retrovirus production Gryphon-A cells were transfected with the appropriate expression vector using FuGENE 6 (Roche) according to the manufacturer’s instructions. Viral supernatants were harvested 48 hrs post transfection, filtered through 0.45 μm filters (Millipore Corp.) and used immediately to infect subconfluent cell cultures in the presence of 5 μg/ml polybrene. U2Os cells were transiently transfected with RNaseH1-mCherry [pLHCX] using FuGENE 6. For gene silencing the following siRNAs were purchased from Thermo Scientific as SMARTpools: ON-TARGETplus Human RNASEH2A siRNA (L-003535-01-0005) targeting the catalytic subunit A of RNaseH2, ON-TARGETplus RNASEH1 siRNA (L-012595-01-0005), On-TARGETplus PRP8 siRNA (L-012252-01-0005), On-TARGETplus SF3a1 siRNA (L-016051-01-0005), On-TARGETplus ATM siRNA (L-003201-00-00005) and a control/scrambled siRNA duplex (D-001210-05-05). For gene silencing, U2Os cells were transfected with RNAiMAX (Invitrogen), and C5RoT and RPE cells with HiPerfect (Qiagen), as recommended by the manufacturers. To inhibit endogenous RNaseH activity cells were transfected with a (1:1) mixture of siRNAs targeting RNAseH1 and RNaseH2A.
Preparation of whole cell lysates, Chromatin fractionation, and Immunoprecipitations
Whole cell lysates were prepared by lysis of equal cell numbers in 60 mM Tris-Cl pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol and 0.01% bromophenol blue. Crude chromatin was isolated after Triton-X 100 extraction and MNase digestion. All fractionation steps were performed at 4 ° C. Cell pellets were suspended in a non-denaturing isosmotic buffer [10 mM PIPES pH 7.0, 3 mM MgCl2, 100 mM NaCl, 300 mM Sucrose, 0.5 mM Na2VO4, 5mM NaF, 5mM Na4P2O7, 10mM beta-glycerolphosphate, 0.1 mM PMSF, 1mM EGTA, 1× Protease inhibitor cocktail EDTA-free (Roche), 15 μM MG132, 10 mM N-Ethylmaleimide and 20 μM PR-619 (LifeSensors)] and extracted in the same buffer with 0.5 % (v/v) Triton-x 100 for 5 min. Following centrifugation (650g, 5 min), nuclei depleted from soluble nucleoplasm were washed with MNase digestion buffer [50 mM Tris-Cl pH 7.5, 4 mM MgCl2, 50 mM KCl, 300 mM Sucrose, 0.5 mM Na2VO4, 5mM NaF, 5mM Na4P2O7, 10mM beta-glycerolphosphate, 1mM PMSF, 1mM, EGTA and 1× EDTA-free Protease inhibitor cocktail] and subsequently incubated with 0.3 U MNase (Sigma)/1×106 nuclei, and 1 mM CaCl2 (37° C, 10 min). Addition of (NH4)2SO4 to a final concentration of 250 mM was used to facilitate extraction of stably DNA-bound proteins. EGTA and EDTA were added to 5 mM and samples were centrifuged at 16,000g for 20 min. Protein concentrations were determined using a modified Bradford method (Bio-Rad). For GFP-immunoprecipitations, cells were lysed in 20 mM Tris-Cl (pH 7.5), 5 mM MgCl2, 150 mM NaCl, 0.5% Triton X-100, 1× phosphatase inhibitor (Roche) and 1× protease inhibitor cocktail. Chromatin was mechanically sheared by passing through a 27G syringe, 40 times. Particulate matter was removed by centrifugation (20 min at 16,000g) and supernatants containing equal amounts of proteins were used for immunoprecipitation. GFP-tagged proteins were immunoprecipitated directly or after MNase digestion which was used to cleave DNA and RNA and disrupt ternary complexes. Samples were incubated (2 hrs, 4°C) with pre-equilibrated GFP-Trap® coupled to agarose beads (ChromoTek), and after extensive washing (10 mM Tris-Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40), immunocomplexes were dissociated from the beads by heating for 10 min at 95° C, in 120 mM Tris-Cl pH 6.8, 4% SDS, 20% glycerol, 10% β-mercaptoethanol, 0.01% bromophenol blue. For Immunoprecipitation of elongating RNAPII, cells were treated and extracted as for isolation of crude chromatin with the exception that instead of MNase digestion, chromatin was mechanically sheared. Immunoprecipitations were performed by O/N incubation with either the anti-RNAPII CTD phospho-ser2 antibody or rabbit IgG, followed by incubation with Protein A/Protein G Agarose beads (Upstate Biotechnology).
Immunoblotting
Protein samples were size-fractionated on 5-20% gradient SDS-polyacrylamide gels (BioRad) and electro-transferred onto nitrocellulose membranes using a Bio-Rad Mini-Protean electrophoresis system. Abundance of proteins of interest was assayed using antibodies at concentrations recommended by their manufacturers. Membrane were incubated with primary antibodies in Tween 20/Tris-buffered saline (20 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Tween 20) containing 3% w/v nonfat dry milk or, when the p-ATM antibody was used, 3% BSA. Following binding of the appropriate anti-mouse or anti-rabbit Alexa fluorochrome-conjugated secondary antibody and extensive washing, proteins of interest were visualized using the Odyssey CLx Infrared Imaging System (LI-COR Biosciences). Signal intensities were quantified using the ImageQuant TL software (GE Healthcare Life Sciences).
RNA synthesis
Transcription levels were determined following incubation with 2-hr ethynyluridine (EU)40 added directly in the culture (serum-free) media. EU incorporation was visualized using Click-iT conjugation of Alexa Fluor 647 (Invitrogen) according to the manufacturer’s protocol. Images were obtained using a Zeiss Axio Imager Z2 upright laser-scanning confocal microscope equipped with a 63× Plan-Apochromat 1.4 NA oil immersion lens (Carl Zeiss Inc.) Fluorescence-signal intensities were quantified using the ImageJ software (NIH). In each experiment >150 cells per condition were analyzed.
Immunofluorescence and live-cell confocal laser-scanning microscopy
For immunofluoresence experiments, cells were fixed with 3.7% paraformaldehyde (PFA)/PBS and permeabilized in 0.5% Triton-X 100/PBS. For detection of splicing factors and XPC, cells were pre-extracted with 0.5 % Triton-X 100/PBS prior to fixation (0.5 min). For CPD immunodetection, nuclear DNA was denatured with 0.07 N NaOH, 5 min. For SRSF2/SC35 immunodetection cells fixed in 2% PFA/ 0.2% Triton-X 100/PBS were treated with 100% acetone (5 min, −20° C). Non-specific antigens were blocked in 3% bovine serum albumin/PBS. R-loop immunodetection with the S9.6 antibody was as described35. Briefly, PFA-fixed cells were permeabilized by Triton X-100, followed by extraction with 0.5% SDS. Cells were blocked with 3% BSA, 0.1% Tween 20 in 4 × SSC. Hybridization of primary antibodies was overnight at 4°C, and with secondary Alexa Fluorochrome-conjugated antibodies for 1 hr at RT. Coverslips were mounted on glass-slides using 4,6-diamidino-2-phenylindole (DAPI)-containing ProLong Gold antifade reagent (Molecular probes) and imaged on a Zeiss Axio Imager Z2 upright laser-scanning confocal microscope.
Live cell imaging experiments were performed with a Leica TCS SP5 AOBS laser scanning confocal microscope equipped with an environmental chamber (37°C, 5% CO2). Kinetic studies of GFP-tagged proteins were performed using UV-C (266 nm) laser-irradiation for local DNA damage infliction19. Briefly, a 2-mW pulsed (7.8 kHz) diode-pumped solid-state laser emitting at 266 nm (Rapp OptoElectronic) was connected to the confocal microscope with an Axiovert 200M housing adapted for UV by all-quartz optics. By focusing the UV-C laser inside cell nuclei without scanning, only a limited area within the nucleus (diffraction limited spot) was irradiated. Cells were imaged and irradiated through a 100×, 1.2 NA Ultrafluar quartz objective lens. Images obtained prior to and post UV-C laser irradiation were analyzed using the LASAF software (Leica). Fluorescence intensity in the irradiated area or a non-irradiated area in the nucleus was normalized to levels in the same area, prior to irradiation. Data were expressed as percent change in relative fluorescence intensity. In each experiment at least 10 cells were analyzed and all experiments were performed a minimum of three times.
Mobility of GFP-tagged proteins was measured by strip-FRAP as described41. In brief, a narrow (~1 μm) strip spanning the width of the nucleus was photobleached at ≈20% of the initial GFP-signal intensity using a 488 nm-laser at 100% power. Recovery of fluorescence in the strip was monitored at 25-ms intervals. Images obtained were analyzed using the LASAF software (Leica). FRAP data were normalized to the fluorescence levels prior to photobleaching after subtraction of the background signal. In each experiment 8-10 cells/per condition were analyzed and all experiments were performed at least three times. A negative (untreated) and positive (20 J/cm2 UV) control were included in all experiments.
Chromatin-associated RNA isolation and snRNA Q-PCR
Chromatin-associated RNA was isolated by a modification of the method developed by Wuarin, J. and Schibler42 for the isolation of ternary-complex associated nascent RNA. Briefly, cell pellets were re-suspended in 20 mM HEPES (pH 7.5), 10mM KCl, 250 mM Sucrose, 5 mM MgCl2, 1 mM EGTA, 1mM PMSF, 1 μl/ml RNasin (Invitrogen), 1× phosphatase inhibitor (PhosStop, Roche) and 1× protease inhibitor cocktail (Roche), and lysed by the addition of Digitonin to 200 μg/ml final concentration (10 min., 4°C). Nuclei were pelleted by centrifugation (650g, 5 min) and following re-suspension in a buffer containing 20 mM Tris-HCL pH 7.5, 75 mM NaCl, 0.5 mM EGTA, 50% Glycerol, 1mM PMSF, 1 μl/ml RNasin, 1× protease and 1× phosphatase inhibitors, were extracted for 10 min. at 4°C by the addition of 10 volumes of a solution containing 20mM HEPES, pH 7.6, 7.5 mM MgCl2, 0.2 mM EGTA, 300 mM NaCl, 1M Urea and 1% NP40. Pelleted nuclei were re-suspended in Qiazol reagent (Qiagen) and RNA was isolated as recommended by the manufacturer. Following digestion with DNase I (Qiagen) RNA was cleaned-up with the Qiagen RNeasy mini-kit with a second, on-column, DNase I digestion. Equal amounts of RNA from each sample were reverse-transcribed (RT) using random-hexamers and SuperScript® III (Invitrogen). Q-PCR reactions, were performed using primers complementary to human snRNAs (described and validated by Galiveti et al43) or the chromatin-associated HotAir ncRNA (control for data normalization), using the IQ™SYBR® Green Supermix (Bio-Rad) in CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad). Absence of contaminating genomic DNA was verified by the lack of amplified products for all sample/primer-sets by inclusion of mock RT reactions in which no enzyme was added.
Exon-specific RT-PCR: RNA extraction, Reverse Transcription and PCR
Experiments were performed as described by Ahn et al24. Briefly, RNA was isolated from quiescent RPE-1 cells using the RNeasy kit (QIAGEN). Equal RNA amounts from each sample were reverse transcribed using random hexamers and SuperScript™ III RT and cDNAs were PCR-amplified using the indicated primers and Taq DNA Polymerase (NEB). PCR products were size-fractionated by gel electrophoresis and visualized by ethidium bromide staining. Signal intensities of amplified fragments containing either the unspliced or the spliced intron, were normalized to the levels of the respective fragments in untreated cells and expressed as fold change in relative abundance. All experiment were repeated a minimum of three times. Amplification of constitutive exons from TUBA1B and GAPDH were used as controls for general splicing efficiency. Primers-sets used for amplifications were: FANCG (Exon5-6) 5′-GGATGTCCTCCTGACAGCAT-3′ and 5′-GCTGTGTACACCTGGACCAA-3′; AKT1 (Exon11-12) 5′-ACAAGGACGGGCACATTAAG-3′and 5′-ACCGCACATCATCTCGTACA-3′; AURKA (Exon9-10) 5′-AATGATTGAAGGTCGGATGC-3′ and 5′-TCTGGCTGGGATTATGCTTC-3′; AURKB (Exon6-7) 5′-TGCAGAAGAGCTGCACATTT-3′and 5′-TCTTCAGCTCTCCCTTGAGC-3′; TUBA1B (Exon2-3) 5′-CCGGGCTGTGTTTGTAGACT-3′ and 5′-GATCTCCTTGCCAATGGTGT-3′; ATM (exon19-20) 5′-AAGGAGCTTCCTGGAGAAGAG-3′ and 5′-AACTGTCCTTGAGCATCCCTT-3′; ATR (exon33-34) 5′-AAGGAGCCTATCCTGGCTCTC-3′ and 5′-CTACCCTGGCACTCTGCAGCC-3′.
Paired-end RNA-Seq and data analysis
RPE-1 cells were grown to confluence and serum deprived for 72 hrs to ensure quiescence. Cells were then mock-treated or UV-irradiated with 20 J/m2 UV-C (245nm), in the presence or absence of 10 μM of the ATM inhibitor. Each treatment was performed in duplicate plates that were used as biological replicates. RNA was isolated 6 hrs post-irradiation with the RNeasy kit (QIAGEN) and PolyA RNA was isolated using the Dynabeads mRNA purification kit (Invitrogen). Sample integrity was verified by the Agilent 2100 Bioanalyzer (Agilent Technologies). For all samples the Bioanalyzer RNA Integrity Scores (RIN) were 9.3-10 indicating excellent RNA quality. For each sample a cDNA Library was prepared and validated using the Illumina TruSeq RNA sample preparation kit v2 according to the manufacturer’s instructions. Briefly, equal amounts (200 ng) of polyA-RNA were chemically fragmented, cDNA was generated using random hexamers as primers and adapters were ligated. RNA fragmentation efficiency and similarity between samples was confirmed by the Bioanalyzer after adaptor ligation (average fragment sizes were 317bp-344bp). Following PCR amplification RNA-Seq was performed according to the Illumina TruSeq v3 protocol on the HiSeq2500 platform, generating paired-end, 100 bp reads (9×107 reads/sample). Raw reads were aligned against the human genome assembly (hg19) using TopHat44. Uniquely mapped reads were used for the identification of alternative splicing events using Multivariate Analysis of Transcript Splicing (MATS) as described45. AS events that were significantly increased (p < 0.05, N=2) by UV-irradiation (untreated vs UV-irradiated cells) by a minimum of 10% difference were considered to be UV-induced. Each individual AS event identified in the first analysis to be UV-induced that also decreased by a minimum of 10% in UV irradiated cells in absence of ATM activity (UV irradiated cells vs ATM inhibitor-treated/UV irradiated cells) was considered to be (partly) dependent on ATM activity.
Statistical Analysis
All data presented were reproduced in at least three independent experiments. Statistical analysis was performed using the PRIZM GraphPad software unless otherwise stated. Significance of differences was evaluated with either Student T-test, when only 2 groups were compared, or one-way ANOVA for more than two groups. One-way ANOVA was followed by post-hoc analysis by either the Dunnett’s test (for comparison of experimental conditions to control) or Bonferroni’s test (comparison between groups). Indicated as (*) is p<0.05, (**) p<0.01, and (***) p<0.001. Proteomic-data statistical analysis was performed with Perseus (1.5.0.30)17. Peptides with Significance B value p ≤0.05 in either in the forward or reverse experiments were considered significant and indicated by (+) in the figures. Significant alternative-splicing events were identified by MATS45; only UV-triggered events with p<0.05 were used for further analysis.
Extended Data
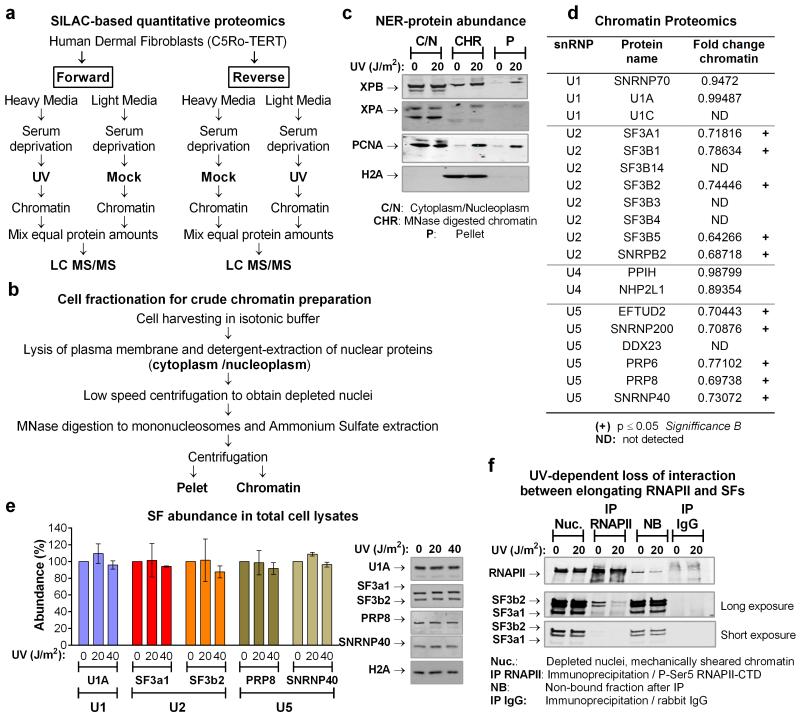
Extended data figure 1.
a. Schematic overview of the proteomic experiments for the identification of proteins that display UV-dependant chromatin association. b. Schematic outline of cell fractionation. c. Validation of chromatin isolation protocol for NER proteins that are reqruited to chromatin in response to DNA damage. Mock-treated or UV irradiated quiescent HDFs (20 J/m2, 1hr post-irradiation) were fractionated as outlined in (b). Equal protein amounts from each fraction were analyzed by immunobloting using antibodies against the indicated NER proteins. Abundance of H2A is shown as a control for chromatin isolation efficiency. d. UV-tiggered changes in chromatin association of core SFs, identifed by quantitative SILAC-proteomics. Proteomic experiments were performed with HDFs as outlined in (a). The table lists representative examples of SFs that participate in distinct snRNP complexes and their chromatin association in response to UV-irradiation (20 J/m2, 1h). U2 ans U5 snRNP-SFs, show signifficantly reduced chromatin association (p≤0.05, Significance B17) and are indicated with a cross. ND=non-detected. e. Abundance of SFs in total cell lysates. Total lysates were prepared from U2Os cells that were mock treated or UV irradiated (20 J/m2, 1hr post-irradiation) and SFs abundance was assayed by immunobloting. Abundance of H2A is shown as a loading control. Right: immunoblots. Left: quantification of SF signal intensities normalized to H2A (n=3, mean ± S.D., one-way ANOVA / Bonferroni). f. UV-dependent interaction of splicing proteins with elongating RNAPII. Quiescent HDFs were prepared as outlined in (b) exept that, instead of MNase digestion, chromatin was mechanically sheared. Elongating RNAPII was immunoprecipitated with an antibody that recognizes specifically the ser2-phosphorylated RNAPII C-Terminal Domain (CTD) and its interaction with the U2 snRNP-SFs SF3a1 and SF3b2 was assayed by immunoblotting.
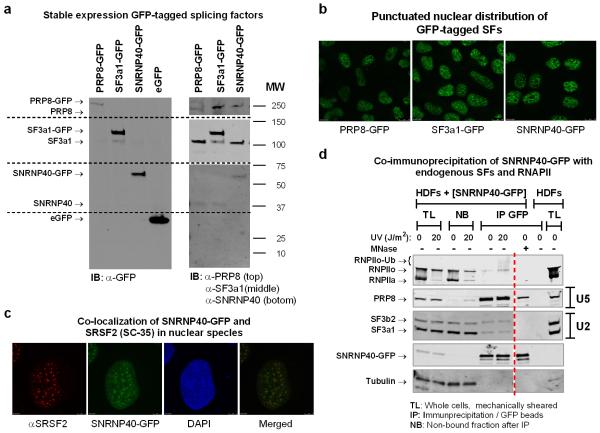
Extended data figure 2. Validation of HDFs stably expressing GFP-tagged splicing factors.
a. Total cell lysates from HDFs stably expressing eGFP tagged PRP8, SF3a1, SNRNP40 or free eGFP, were analyzed by immunoblotting using antibodies against GFP (left) or against PRP8, SF3a1 and SNRNP40 (right). Ectopically produced proteins were expressed at near or below endogenous levels. b. Fluorescent microscopy images of GFP-tagged splicing factors showing the expected punctuated nuclear distribution. c. Localization of SNRNP40-GFP in nuclear speckles which were visualized by immunofluorescence detection of the speckle marker SRSF2/SC35. d. Interaction of SNRNP40-GFP with endogenous splicing factors and elongating RNAPII. Quiescent SNRNP40-GFP expressing HDFs were mock-treated or irradiated with 20 J/m2 UV-C. After a three-hour recovery period, cells were lysed under native conditions and chromatin was sheared by mechanical force. SNRNP40-GFP was immunoprecipitated from total cell lysates using GFP-TRAP® agarose beads and its association with endogenous splicing factors and the large subunit of RNAPII was assayed by immunoblotting. Untransfected cells are shown as negative control. SNRNP40-GFP interacts with U2 and U5 snRNP components, arguing that the GFP-tag does not interfere with complex formation. Interaction of SNRNP40 with its U5 snRNP partner PRP8 is partially maintained even after MNase digestion, consistent with its presence in U4/U6.U5 tri-snRNP complexes. Participation of SNRNP40-GFP in co-transcriptional splicing complexes is confirmed by co-immunoprecipitation of the active (hyperphosphorylated RNAPIIo) large subunit of RNAPII.
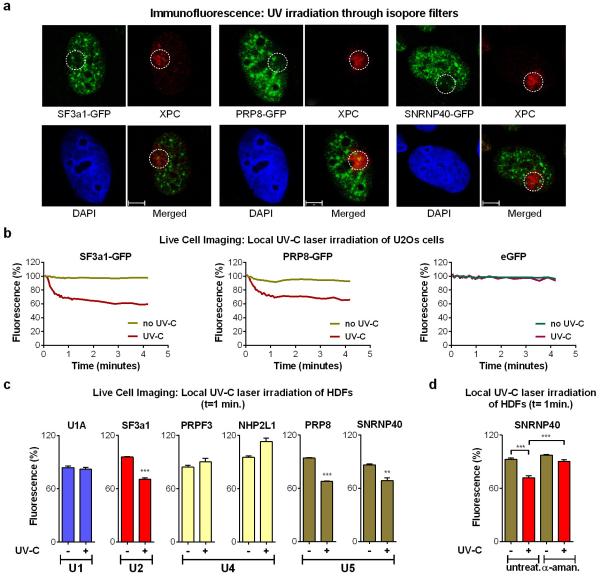
Extended data figure 3. Displacement of mature spliceosomes from subnuclear sites of UV-inflicted DNA damage.
a. U2Os cells stably expressing GFP-tagged SFs were UV irradiated (60 J/m2) through isopore membranes resulting in DNA-lesion formation in small subnuclear areas. DNA damage sites (circled) were visualized by immunofluorescence using an antibody against the NER recognition factor XPC. Scale bar: 5 μm. b. SF3a1-GFP and PRP8-GFP depletion from UV-C laser micro-irradiation sites. Quantification of 20 cells from two independent experiments. eGFP localization at sites of DNA damage is used to demonstrate that depletion of eGFP-tagged SFs is not caused by photobleaching. c. UV-C laser micro-irradiation results in preferential displacement of U2 and U5-associated SFs from DNA damage sites. Quiescent HDFs were irradiated in an ≈1μm diameter nuclear area via a UV-C laser. GFP signal intensity, reflecting the abundance of GFP-tagged U1, U2, U4 and U5 snRNP components at UV-C DNA-damage sites, was quantified in the irradiated and in an unirradiated nuclear area (undamaged control). Plotted is the fluorescence signal intenisty expressed as % of that prior to irradiation, at the 1 min. time point. Cells expressing free eGFP were used as negative control. Representative from three independent experiment (n=12, mean ± s.e.m., paired T-test). d. Depletion of SFs from UV-C laser irradiation sites depends on active transcription. Transcription initiation was inhibited in quiescent HDFs by prolonged α-amanitin treatment (10 μM, ≥24h) prior to subnuclear UV-C laser irradiation. Plotted is the SNRNP40-GFP abundance in irradiated and unirradiated nuclear areas at 1 min post-irradiation. Represenative from three independent experiments (n=12, mean ± s.e.m., one-way ANOVA / Bonferroni).
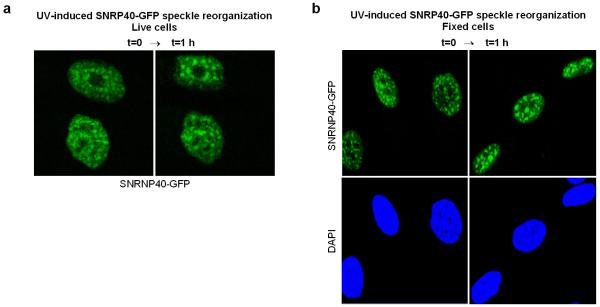
Extended data figure 4. SNRNP40 reorganization and speckle enlargement in response to UV irradiation.
Representative microscopic images showing SNRNP40-GFP distribution in quiescent HDFs prior to, and one hour post UV-C irradiation with 20 J/m2. a. Live cells. b. Fixed cells.
Extended data figure 5.
a. RNA synthesis is inhibited preferentially by genotoxins that inflict bulky DNA lesions. Influence of genotoxins on RNA synthesis of quiescent HDFs was measured by EU-pulse labeling combined with Click chemistry. Top: representative images. Bottom: quantification of fluorescence intensity (n=150, mean ± s.e.m., one-way ANOVA / Bonferroni). b. Mobilization of U2 and U5 snRNPs by genotoxins inflicting transcription blocking-DNA lesions. Mobilization of GFP-tagged SF3a1 (left) and PRP8 (right) assayed by FRAP in quiescent HDFs exposed to different types of genotoxins (n=30, mean ± s.e.m., one-way ANOVA / Dunnett’s). c. Chromatin displacement of mature spliceosomes is not TC-NER dependent. Left: Chromatin abundance of U2 and U5 snRNP-SFs was assayed by immunoblotting in XPA deficient (left), XPA-GFP corrected (middle) and CSB deficient (right) HDFs. Cells were UV-irradiated (20 J/m2) and chromatin was isolated at the indicated times. Top: Immunoblots; bottom: quantification of SF signal intensities normalized to H2A (n=3, mean ± s.d., one-way ANOVA / Bonferroni). d. Proteasome activity is not required for UV-damage induced spliceosome mobilization. Mobilization of SNRNP40-GFP assayed by FRAP in quiescent HDFs exposed to UV radiation in the presence or absence of the proteasome inhibitor MG132 (50μM) (n=30, mean ± s.e.m., T-test). e. SNRNP40-GFP mobilization by transcription inhibition. FRAP of SNRNP40-GFP in quiescent HDFS after inhibition of transcription initiation (10 μg/ml α-amanitin, 24h) or elongation (1 μg/ml Actinomycin D or 50 μM DRB, 1h) (n=30, mean ± s.e.m., one-way ANOVA / Dunnett’s).
Extended data figure 6.
a. UV-irradiation and DRB-dependent mobilization of SNRNP40. Quiescent HDFs expressing SNRNP40-GFP were UV-irradiated or DRB treated with doses that inhibit transcription to similar levels. SF mobility was assayed by FRAP. b. Additive effect of combined UV and DRB treatments. FRAP of SNRNP40-GFP in quiescent HDFs treated with DRB, UV, or a combination of both, each at a dose that inhibits RNA synthesis by ≈50%. c. Impaired UV-dependent SF3a1 mobilization in cells lacking ATM activity. SF3a1-GFP mobilization was measured by FRAP in quiescent HDFs derived from an AT patient or a healthy donor. d. ATM-dependent spliceosome mobilization. Quiescent HDFs were treated with 10 μM ATM (KU55933), ATR (VE821) or DNA-PK (NU7441) inhibitors prior to irradiation. GFP-tagged SF3a1 or PRP8 mobility was assayed by FRAP. ATM, but not ATR or DNA-PK inhibition partially prevented the UV-induced SF-mobilization. (a, b, c, d) n=25, mean ± s.e.m., one-way ANOVA / Bonferroni. e. Reduced UV-induced intron retention in response to ATM silencing. Intron inclusion in RPE cells transfected either with control or ATM silencing siRNAs and subsequently mock-treated or UV irradiated (20 J/m2, 6 hrs) was assayed by RT-PCR. f. ATM-dependent changes in intron retention. Intron inclusion was assayed by RT-PCR in untreated, UV irradiated and DRB treated quiescent cells in the presence or absence of 10 μM ATM inhibitor. g. Heatmap of UV-triggered and ATM-dependent transcriptome changes. Quiescent cells were mock-treated or UV-irradiated in the presence or absence of the ATM inhibitor. Transcriptome profiles were generated by RNA-seq. Differentially expressed genes between untreated and UV-irradiated cells (p<0.05) and UV-irradiated cells in the presence or absence of the ATM inhibitor (p<0.05), were clustered in a Heatmap using Pearson correlation. N=1676 differentially expressed transcripts. The observed anti-correlation indicates that UV-inducible transcriptome changes can be, in part, prevented by ATM inhibition. h. Lack of influence of ATM inhibition on DRB-dependent SF mobility. SF mobility was measured by FRAP in untreated or DRB treated HDFs in the presence or absence of 10 μM ATM inhibitor (n=30, mean ± s.e.m., one-way ANOVA / Bonferroni).
Extended data figure 7. Canonical and non-canonical ATM activation.
a. ATM autophosphorylation (Ser 1981) was assayed in quiescent HDFs one hour after the indicated treatments. In non-replicating cells UV and Trichostatin A (TSA) activate ATM via non-canonical pathways. Transcription inhibition by DRB has no influence on ATM activity. b. The quiescent status of serum deprived HDFs was verified by immunodetection of the cell cycle marker Ki67, which is not expressed by quiescent (G0) cells. c. Immunofluorescence detection of active ATM in quiescent HDFs treated with DDR kinase inhibitors. d. Immunoblotting analysis of nuclear extracts derived from quiescent HDFs treated as in (c) using a phospho-specific ATM (S1981) antibody (top) and an antibody recognizing ATM (bottom). e. Differences in autophosphorylated-ATM distribution in quiescent HDFs treated with various ATM activators. Left: multiple cells; right: single cell magnification illustrating pan-nuclear localization of phosphorylated ATM after UV irradiation and focal accumulation after CPT or IR treatments. Magnified cells are indicated by arrows (left panel). f. Differences in amounts of DNA damage-foci formation indicative of DSBs, in response to CPT, UV-and IR. Quiescent HDFs were pre-treated with the ATR inhibitor (10 μM, 1 hr) and subsequently exposed to the indicated genotoxins. DSB-foci were visualized by immunofluorescence using antibodies against γH2A.X and p53BP1. Left: multiple cells; right: single cell magnification. Magnified cells are indicated by arrows in the left panel.
Extended data figure 8. ATM activation by interference with spliceosome assembly or RNaseH1/H2A silencing.
a. ATM autophosphorylation was assayed by immunofluorescence in HDFs after silencing of SF3a1, PRP8 or combined silencing of RNaseH1 and RNaseH2A. b. Immunoblotting analysis of silenced proteins in total cell lysates. Tubulin is shown as a loading control. c. SF mobilization by the spliceosome inhibitor Pladienolide B was assayed by FRAP in quiescent HDFs. Consistent with its function in interfering with spliceosome maturation following pre-spliceosome assembly, cell treatment with pladienolide B resulted in extensive mobilization of U5 snRNP factors (PRP8 and SNRNP40), partial mobilization of the U2 snRNP SF3a1, and had no influence on the U1 snRNP factor U1A (n=30, mean ± s.e.m., one-way ANOVA / Bonferroni). d and e ATM activation by Pladienolide B. Quiescent HDFs were either treated with 5 μM Pladienolide B or exposed to 1 Gy IR and autophosphorylated ATM was detected by: (d) immunofluorescence, (e) immunoblotting. f. Effect of Pladienolide treatment on intron retention. RNA isolated from mock-treated, UV irradiated or Pladienolide B treated RPE cells. Intron retention assayed by RT-PCR on transcripts of the indicated genes, shows that Pladienolide B influences splicing to the same extend as UV-irradiation. U/S: Ratio of relative abundance of unspliced (U) to spliced (S) introns. g. Efficiency of RNaseH1 and H2A silencing at single cell level assayed by immunofluorescence.
Extended data figure 9.
a. Recruitment of GFP-RNaseH1(D145N) at local DNA-damage sites depends on endogenous levels of RNaseH activity. DNA damage was inflicted via a UV-C laser in ≈1 μm-diameter subnuclear areas of cells after silencing of RNaseH2A or overexpression of RNaseH1-mCherry. Recruitment of RNaseH1(D145N)-GFP at the irradiated sites was monitored by live-cell imaging. Plotted is the fluorescence intensity of RNaseH1(D145N)-GFP at 1 min. post irradiation, at the irradiated and in a non-irradiated nuclear area. Representative from three independent experiments (n=10, mean ± s.e.m., one-way ANOVA / Bonferroni). b and c. R-loop formation at sites of local UV-C laser irradiation. Immunofluorescence detection of R-Loops using the DNA:RNA hybrid-specific S9.6 antibody. Sites of irradiation are visualized by XPC immunodetection. (b) Dashed boxes indicate the magnified areas shown in the right panels. The dashed lines indicate the line-scan track used to quantify fluorescence intensity of S9.6 and anti-XPC (shown in in the graph). (c) Specificity of the antibody was confirmed by its increased sensitivity after RNase H2A silencing and its ability to detect R-loops when suboptimal doses of UV-C irradiation were applied. d. RNaseH1 accumulation at local DNA-damage sites depends on active transcription but not ATM activity. Transcription initiation was inhibited in quiescent HDFs by α-amanitin (10 μg/ml, 24h) prior to local UV-C laser irradiation. Plotted is the fluorescence intensity at 1 min. post irradiation of RNaseH1(D145N)-GFP at the irradiated and in a non-irradiated nuclear area for untreated, ATM-inhibitor and α-amanitin treated cells. Representative from three experiments (n=10, mean ± s.e.m., one-way ANOVA / Bonferroni). e. RNaseH1 overexpression inhibits the UV-dependent spliceosome mobilization. FRAP of U2Os cells stably expressing GFP-tagged SF3a1 and PRP8 and transiently transfected with RNaseH1-mcherry. f. RNaseH1 and H2A silencing potentiates the UV-dependent spliceosome mobilization. RNaseH1 and H2 were silenced in U2Os cells expressing SF3a1-GFP or PRP8-GFP and SF mobility was assayed by FRAP. g. FRAP of SNRNP40-GFP in quiescent HDFs after RNaseH1/H2 silencing. e, f, g, n=30, mean ± s.e.m., one-way ANOVA / Bonferroni.
Extended data figure 10. Combined transcription inhibition and ATM activation, results in extensive mobilization of mature spliceosomes.
a. Combinatorial effect of DRB and IR on spliceosome mobilization. Quiescent HDFs were exposed to IR in the presence or absence of DRB, and SF3a1-GFP and PRP8-GFP mobility was assayed by FRAP. b. The IR-mediated increase of DRB-dependent spliceosome mobilization depends on ATM activity. FRAP of GFP-tagged SNRNP40 in quiescent HDFs treated with DRB and/or IR in the presence or absence of an ATM inibitor. c. Spliceosome mobilization by CPT. Quiescent HDFs were treated with 25 μg/ml CPT, 25 μM DRB and 20 J/m2 UV at doses that inhibit transcription to approx. 30% and their influence on SF3a1, PRP8 and SNRNP40 mobilization was measured by FRAP. Mobilization of GFP-tagged SF3a1, PRP8 and SNRNP40 in quiescent HDFs was measured by FRAP. a, b, c, n=30, mean ± s.e.m., one-way ANOVA / Bonferroni d. Inhibition of RNA synthesis by the treatments shown in (c) was assayed in quiescent HDFs by EU-incorporation and Click-chemistry (n=150, mean ± s.e.m., one-way ANOVA / Dunnett’s).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the Optical Imaging Center of ErasmusMC for technical support, Drs M. Reijns and A. Jackson, U. of Edinburgh for the S9.6 antibody, L. Marufu for technical assistance and Dr N.G.J. Jaspers for intellectual input. This work was funded by the Netherlands Organization for Scientific Research (NWO) ZonMW TOP Grants 912.08.031 and 912.12.132, Horizon Zenith 935.11.042, ALW 854.11.002 and 823.02.013, the Association for International Cancer Research 10-594, European Research Council Advanced Investigator Grants 233424 and 340988, and an ErasmusMC fellowship.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature
RNA Seq data are deposited to SRA with accession number SRP053034.
The authors declare no competing financial interests.
REFERENCES
- 1.Sirbu BM, Cortez D. DNA Damage Response: Three Levels of DNA Repair Regulation. CSH Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciccia A, Elledge SJ. The DNA Damage Response: Making It Safe to Play with Knives. Molecular Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 4.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Bio. 2013;14:197–210. [PubMed] [Google Scholar]
- 5.Kaidi A, Jackson SP. KAT5 tyrosine phosphorylation couples chromatin sensing to ATM signalling. Nature. 2013;498:70–74. doi: 10.1038/nature12201. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Lenzken SC, Loffreda A, Barabino SM. RNA Splicing: A New Player in the DNA Damage Response. Int J Cell Biol. 2013;2013:153634. doi: 10.1155/2013/153634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoskins AA, Moore MJ. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem Sci. 2012;37:179–188. doi: 10.1016/j.tibs.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valadkhan S, Jaladat Y. The spliceosomal proteome: at the heart of the largest cellular ribonucleoprotein machine. Proteomics. 2010;10:4128–4141. doi: 10.1002/pmic.201000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kornblihtt AR, et al. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Bio. 2013;14:153–165. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- 10.Schor IE, Gomez Acuna LI, Kornblihtt AR. Coupling between transcription and alternative splicing. Cancer Treat Res. 2013;158:1–24. doi: 10.1007/978-3-642-31659-3_1. [DOI] [PubMed] [Google Scholar]
- 11.Zhou HL, Luo G, Wise JA, Lou H. Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms. Nucleic Acids Res. 2014;42:701–713. doi: 10.1093/nar/gkt875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexander R, Beggs JD. Cross-talk in transcription, splicing and chromatin: who makes the first call? Biochem Soc Trans. 2010;38:1251–1256. doi: 10.1042/BST0381251. [DOI] [PubMed] [Google Scholar]
- 13.Dutertre M, Sanchez G, Barbier J, Corcos L, Auboeuf D. The emerging role of pre-messenger RNA splicing in stress responses: sending alternative messages and silent messengers. RNA Biol. 2011;8:740–747. doi: 10.4161/rna.8.5.16016. [DOI] [PubMed] [Google Scholar]
- 14.Paronetto MP, Minana B, Valcarcel J. The Ewing sarcoma protein regulates DNA damage-induced alternative splicing. Mol Cell. 2011;43:353–368. doi: 10.1016/j.molcel.2011.05.035. [DOI] [PubMed] [Google Scholar]
- 15.Dutertre M, et al. Cotranscriptional exon skipping in the genotoxic stress response. Nat Struct Mol Biol. 2010;17:1358–1366. doi: 10.1038/nsmb.1912. [DOI] [PubMed] [Google Scholar]
- 16.Munoz MJ, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–720. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 18.Rino J, et al. A stochastic view of spliceosome assembly and recycling in the nucleus. PLoS Comput Biol. 2007;3:2019–2031. doi: 10.1371/journal.pcbi.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinant C, et al. Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J Cell Sci. 2007;120:2731–2740. doi: 10.1242/jcs.004523. [DOI] [PubMed] [Google Scholar]
- 20.Lagerwerf S, Vrouwe MG, Overmeer RM, Fousteri MI, Mullenders LH. DNA damage response and transcription. DNA Repair (Amst) 2011;10:743–750. doi: 10.1016/j.dnarep.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 21.Vermeulen W, Fousteri M. Mammalian Transcription-Coupled Excision Repair. CSH Perspect Biol. 2013:5. doi: 10.1101/cshperspect.a012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 23.Blasius M, et al. A phospho-proteomic screen identifies substrates of the checkpoint kinase Chk1. Genome Biol. 2011;12:R78. doi: 10.1186/gb-2011-12-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahn EY, et al. SON controls cell-cycle progression by coordinated regulation of RNA splicing. Mol Cell. 2011;42:185–198. doi: 10.1016/j.molcel.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 26.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 27.Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 28.Sordet O, et al. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep. 2009;10:887–893. doi: 10.1038/embor.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanasoge S, Ljungman M. H2AX phosphorylation after UV irradiation is triggered by DNA repair intermediates and is mediated by the ATR kinase. Carcinogenesis. 2007;28:2298–2304. doi: 10.1093/carcin/bgm157. [DOI] [PubMed] [Google Scholar]
- 30.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Kotake Y, et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nature chemical biology. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 33.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu H, Lima WF, Crooke ST. Investigating the structure of human RNase H1 by site-directed mutagenesis. J Biol Chem. 2001;276:23547–23553. doi: 10.1074/jbc.M009676200. [DOI] [PubMed] [Google Scholar]
- 35.Bhatia V, et al. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–365. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- 36.Reijns MAM, et al. Enzymatic Removal of Ribonucleotides from DNA Is Essential for Mammalian Genome Integrity and Development. Cell. 2012:149. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McManus CJ, Graveley BR. RNA structure and the mechanisms of alternative splicing. Curr Opin Genet Dev. 2011;21:373–379. doi: 10.1016/j.gde.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biton S, Barzilai A, Shiloh Y. The neurological phenotype of ataxia-telangiectasia: solving a persistent puzzle. DNA Repair (Amst) 2008;7:1028–1038. doi: 10.1016/j.dnarep.2008.03.006. [DOI] [PubMed] [Google Scholar]
METHODS REFERENCES
- 39.Schwertman P, et al. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet. 2012;44:598–+. doi: 10.1038/ng.2230. [DOI] [PubMed] [Google Scholar]
- 40.Nakazawa Y, Yamashita S, Lehmann AR, Ogi T. A semi-automated non-radioactive system for measuring recovery of RNA synthesis and unscheduled DNA synthesis using ethynyluracil derivatives. DNA Repair. 2010;9:506–516. doi: 10.1016/j.dnarep.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 41.Houtsmuller AB, Vermeulen W. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching. Histochem Cell Biol. 2001;115:13–21. doi: 10.1007/s004180000234. [DOI] [PubMed] [Google Scholar]
- 42.Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Molecular and cellular biology. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galiveti CR, Rozhdestvensky TS, Brosius J, Lehrach H, Konthur Z. Application of housekeeping npcRNAs for quantitative expression analysis of human transcriptome by real-time PCR. RNA. 2010;16:450–461. doi: 10.1261/rna.1755810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen S, et al. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res. 2012;40:e61. doi: 10.1093/nar/gkr1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.