Abstract
Osteoporosis is a disease characterized by low bone mass, leading to an increased risk of fragility fractures. GATA4 is a zinc-finger transcription factor that is important in several tissues, such as the heart and intestines, and has recently been shown to be a pioneer factor for estrogen receptor alpha (ERα) in osteoblast-like cells. Herein, we demonstrate that GATA4 is necessary for estrogen-mediated transcription and estrogen-independent mineralization in vitro. In vivo deletion of GATA4, driven by Cre-recombinase in osteoblasts, results in perinatal lethality, decreased trabecular bone properties, and abnormal bone development. Microarray analysis revealed GATA4 suppression of TGFβ signaling, necessary for osteoblast progenitor maintenance, and concomitant activation of BMP signaling, necessary for mineralization. Indeed, pSMAD1/5/8 signaling, downstream of BMP signaling, is decreased in the trabecular region of conditional knockout femurs, and pSMAD2/3, downstream of TGFβ signaling, is increased in the same region. Together, these experiments demonstrate the necessity of GATA4 in osteoblasts. Understanding the role of GATA4 to regulate the tissue specificity of estrogen-mediated osteoblast gene regulation and estrogen-independent bone differentiation may help to develop therapies for postmenopausal osteoporosis.
Keywords: GATA4, ESTROGEN, TGF BETA, BMP, OSTEOBLAST
Introduction
Fifty percent of women over the age of 50 yr will experience an osteoporotic fracture in their lifetime.(1) The highest risk factor for osteoporosis is being postmenopausal. Thus, understanding the mechanism of action of estrogens in bone is key to preventing and treating osteoporosis. 17β-estradiol (E2) induces apoptosis in bone resorbing osteoclasts and is antiapoptotic in osteoblasts, leading to an overall building of bone.(2) Toward this end, we showed that E2, via estrogen receptor alpha (ERα), induces transcription of Fas ligand (FasL) in osteoblasts, resulting in a paracrine signal to induce osteoclast apoptosis.(3) E2 also induces transcription of alkaline phosphatase and Bmp2 in osteoblasts, thereby regulating osteoblast differentiation.(4)
The GATA family of transcription factors is a conserved set of proteins that bind to the DNA sequence (A/T)GATA(A/G). Gata4 is expressed early in embryogenesis and is a key regulator of mesodermal and endodermal development.(5) Gata4 was recently described to be expressed in osteoblasts, and to be regulated by ERα.(6) However, a role for GATA4 in the commitment of bone progenitors, differentiation of preosteoblasts, or mineralization of bone has not been previously described.
TGFβ and BMP family members play important roles in skeletal development.(7) TGFβ-1, TGFβ-2, and TGFβ-3 are important in the maintenance and expansion of osteoblast progenitors, and BMP-2, BMP-4, BMP-5, BMP-6, and BMP-7 induce osteoblast differentiation.(8) TGFβ signaling leads to the phosphorylation of SMAD2/3, which then stimulates proliferation and early osteoblast differentiation, while inhibiting terminal differentiation.(9) BMP signaling leads to the phosphorylation of SMAD1/5/8 and activation of the expression and activity of Runx2 and other genes(7) necessary for osteoblast differentiation. Although the actions of both TGFβ and BMP signaling in bone have been fairly well characterized, the mechanisms regulating their transcriptional regulation are poorly understood.
GATA4 and SMAD signaling pathways regulate transcription in the heart, gut, and ovaries. In heart development, BMP4 signaling regulates Gata4 expression(10) and conversely, GATA4 regulates Bmp4 expression.(11) Furthermore, GATA4 and SMADs co-activate transcription of heart-specific genes such as NKX2-5.(12) GATA4 and TGFβ signaling crosstalk in the gut, where they synergize to regulate epithelial gene expression(13); similarly, in granulosa cells of the ovary, TGFβ upregulates Gata4 and then GATA4 and SMAD3 cooperate to regulate inhibin-alpha.(14)
Global knockout of Gata4 in the mouse reveals heart and gut defects and lethality between embryonic day 7.5 (E7.5) and E10.5.(15,16) Owing to the early embryonic lethality of these models, the importance of GATA4 for bone development in vivo was never studied. In order to investigate osteoblast-specific effects of GATA4, we analyzed the estrogen-dependent and estrogen-independent effects of GATA4 in vitro, and then selectively ablated GATA4 in osteoblasts in vivo. These results show that GATA4 in osteoblasts is necessary for survival and proper bone development. Furthermore, we demonstrate that GATA4 regulates TGFβ and BMP pathways in osteoblasts. Together, these studies identify GATA4 as a regulator of osteoblast commitment during early development via E2-dependent and E2-independent pathways.
Materials and Methods
Ethics statement
All animal work was approved by the Animal Research Committee at UCLA.
Reagents
E2 was purchased from Sigma-Aldrich Co (St. Louis, MO, USA). All E2 experiments were performed in media without phenol red and with 5% charcoal dextran-treated fetal bovine serum (CDT-FBS) (Omega Scientific, Dallas, TX, USA). The following antibodies were used: ERα (Thermo Fisher Scientific; clone Ab-16), GATA4 (Santa Cruz; Clone G4), RUNX2 (R&D Systems), and β-actin (Sigma-Aldrich Co.). SMAD antibodies were obtained from Cell Signaling (pSmad1/5/8 #9511; pSmad2: #3108; Smad2: #3122; Smad3: #9523; and Smad5: #9517).
Mice
Gata4 flox/flox mice were purchased from Jackson Laboratory (Gata4tm1.1Sad/J) and backcrossed for 10 generations to the FVB background. Type I collagen A1 (Col1A1) (2.3 kb)-Cre mice (FVB-Tg(Col1a1-cre)1Kry/Mmucd) were purchased from the Mutant Mouse Regional Resource Centers (MMRRC).
Primary calvarial osteoblasts
Neonatal CD1 calvaria were obtained 2 d after birth and incubated for 40 min in αMEM-1.0 mg/mL collagenase P-1.25% trypsin at 37°C. These were washed in αMEM, and transferred to αMEM-1.0 mg/mL collagenase P-1.25% trypsin for 1 hour at 37°C.(17) Digestion was stopped by addition of αMEM/10% FBS. The cells from the second digest were allowed to attach for 48 hours and then differentiated in mineralization medium with media replacement every 3 d. Differentiation was confirmed by quantitation of Col1A1, bone sialoprotein (BSP), and osteocalcin mRNA, alkaline phosphatase positivity, and Alizarin Red staining for mineralization.
Pericytes
Perivascular cells (pericytes) were obtained from human abdominal subcutaneous fat or lipoaspirate, fetal lung, fetal muscle, or fetal bone marrow as described.(18) Because specimens were obtained as anonymous and unidentifiable, the activities of the present research did not involve human subjects and therefore did not require IRB review according to UCLA IRB medical committee standards. For in vitro mineralization, cells were differentiated with the HyClone AdvanceSTEM Osteogenic Differentiation Kit (Fisher Scientific).
Lentiviral silencing
For shRNA-mediated knockdown of Gata4 expression, cells were plated in six-well plates (1 × 105 cells per well) and infected 24 hours later with lentivirus. Cells were treated with virus in 2 mL MEM per well with a final concentration of 8 μg/mL polybrene. Plates were centrifuged at 1400g at 30°C for 45 minutes. The following day the media was replaced with mineralization medium with media replacement every 3 d. Knockdown of Gata4 was confirmed by qPCR and immunoblotting. Two different shRNA from The RNAi Consortium (TRC) in pLKO vector were used to knockdown mouse Gata4 (TRCN0000095215: CCGGCCCAATCTCGATATGTTTGATCTCGAGATCAAACATATCGAGATTGGGTTTTTG, and TRCN0000095217: CCGGCATCTCCTGTCACTCAGACATCTCGAGATGTCTGAGTGACAGGAGATGTTTTTG) and human GATA4 (TRCN0000020424: CCGGCCAGAGATTCTGCAACACGAACTCGAGTTCGTGTTGCAGAATCTCTGGTTTTT, and TRCN0000020428: CCGGCCCGGCTTACATGGCCGACGTCTCGAGACGTCGGCCATGTAAGCCGGGTTTTT). pLKO-shGFP was used as a negative control for mouse experiments and pLKO-C (shC, with no mammalian target) was used for human experiments.
RNA and qPCR
Cells were hormone-deprived by culture for 3 d in phenol red-free medium (Life Technologies, Grand Island, NY, USA) supplemented with 5% CDT-FBS. Cells were treated with 10 nM E2 or ethanol as a vehicle control for 3 or 24 hours. Total RNA was converted to cDNA with Superscript III First Strand Synthesis Kit according to the manufacturer’s instructions (Life Technologies). Primers were selected using Primer3(19); the sequences are listed in Supporting Fig. 10. cDNA was subjected to quantitative PCR using SYBR Green Mastermix with roxithromycin (ROX). Each RNA sample was collected in triplicate and each PCR reaction was amplified in triplicate.
Microarray
Total RNA was analyzed using the MouseRef-8 v2.0 Expression BeadChips from Illumina (San Diego, CA, USA). Data was analyzed using GenomeStudio (Illumina). Gene ontology was performed by The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7.(20) Upon acceptance, microarray data will be submitted to NCBI Geo Database.
Immunohistochemistry
Formalin-fixed paraffin-embedded samples were processed through standard deparaffinization protocols. The tissue was then incubated in blocking buffer (5% normal goat serum, 2.5% bovine serum albumin [BSA] in PBS at pH 7.5) for 30 min. Primary antibodies were incubated overnight at 4°C in a humidified chamber followed by either the DAKO Envision+ visualization system and counterstaining with hematoxylin or by immunofluorescence with 4,6-diamidino-2-phenylindole (DAPI) counterstaining.
Protein and immunoblotting
Cells were hormone-deprived by culture for 3 d in phenol red–free medium (Invitrogen Corporation) supplemented with 5% CDT-FBS. Cells were treated with 10 nM E2 or ethanol as a vehicle control for 24 hours and then lysed in EBC buffer (50 mM Tris [pH 8], 120 mM NaCl, 0.5% Nonidet P-40) supplemented with a protease inhibitor mixture (Complete; Roche Applied Science, Indianapolis, IN, USA) for 30 min on ice. Proteins were subjected to SDS-PAGE and immunoblotting with antisera to the indicated proteins.
Alkaline phosphatase assay
Osteoblasts were differentiated for the indicated amount of time and then fixed with 3.7% formaldehyde, and stained for alkaline phosphatase activity with SigmaFast 5-bromo-4-chloro-3-indolyl phosphate/4-nitro blue tetrazolium (BCIP/NBT) (Sigma).
Mineralization assay
Osteoblasts were fixed in 50% ethanol for 15 min at 4°C, then 1% Alizarin Red S (wt/vol with 0.1% ammonium hydroxide) was added for 30 min. The stain was washed with water and photographed. The Alizarin Red was eluted with 10% cetylpyridinium chloride and the optical density (OD) was measured at 570 nm.
RNA in situ
Gata4 mRNA was detected by RNAscope technology (2.0 High Definition; Advanced Cell Diagnostics, Hayward, CA, USA) in wild-type FVB embryos according to the manufacturer’s recommendations.
Recombination efficiency
The calvariae of postnatal day 0 (P0) mice were placed on a stainless steel grid in a 12-well tissue culture dish in differentiation media for 7 days. RNA from calvariae was obtained and cDNA was synthesized. Primers were designed to anneal to WT cDNA in exon 5 (which is excised by Cre recombinase) and exon 6 of Gata4 (GCGGAAGGAGGGGATTCAAA and TGAATGTCTGGGACATGGAGC).
Skeletal preparations
Skeletal preparations were performed as described.(21) Briefly, embryos were eviscerated and fixed in 95% EtOH overnight at 4°C, followed by Alcian Blue staining (0.01% Alcian Blue 8GX [Sigma-Aldrich] [wt/vol] in 95% EtOH) overnight at room temperature. Samples were then stained for Alizarin Red (0.05% Alizarin Red S [Sigma-Aldrich] [wt/vol] in 1% KOH) for 3 to 4 hours and cleared in a series of graded KOH in glycerol.
Von Kossa
Deparaffinized sections were incubated with 1% silver nitrate solution under ultraviolet light for 30 minutes. Unreacted silver was removed with 5% sodium thiosulfate. The sections were counterstained with nuclear fast red.
Bone μCT scanning and analysis
Femurs, tibia/fibula, L5 vertebrae, and skulls from a total of 9 P0 WT and Gata4 conditional knockout (cKO) mice were dissected, cleaned of soft tissue, and stored in 70% ethanol before μCT scanning. μCT scanning was performed with a Skyscan 1172 scanner (Skyscan, Kontich, Belgium) with the X-ray energy equaling 30 KVp and 175 μA and a voxel isotropic resolution of 6 μm. Prior to scanning, bones were positioned with gauze in the sample holder. To compare femur measurements of trabecular bone microarchitecture and cortical bone morphology, 65 transaxial slices were reconstructed and measured for each sample covering 370 μm of the distal metaphysis starting 0.290 mm above the distal epiphysis and moving along the shaft, and 66 slices covering 380 μm (33 slices each above and below the mid-diaphysis), respectively. For trabecular bone measurements, contours were drawn in the medullar cavity at a fixed distance from the endosteum to define tissue volume. For L5 vertebrae, 60 slices, covering ~345 μm, were analyzed. Bone volume fraction (BV/TV), trabecular number (Tb.N), trabecular thickness (Tb.Th), and trabecular separation (Tb.Sp) were measured for L5 vertebrae and trabecular bone of the femur, using CTAn software (Skyscan, Kontich, Belgium). For cortical bone measurements, contours were drawn immediately adjacent to the periosteum bone perimeter to define the periosteal envelope. Total cross sectional area (T.Ar), cortical bone area (B. Ar), cortical area fraction (% B.Ar), and mean cortical thickness (Ct. Th) were measured using CTAn software.
Statistical analysis
All experiments represent both biological and experimental triplicates. Error bars represent mean ± SD; **p < 0.01, *p < 0.05 using a Student’s t test.
Results
GATA4 regulates ERα and ERα target genes in osteoblasts
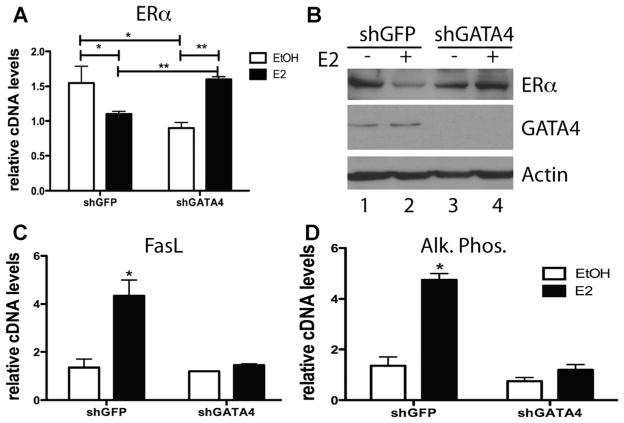
Maximal binding of GATA4 to osteoblast-specific target gene enhancers precedes ERα binding, and GATA4 is necessary for open chromatin (marked by histone 3 lysine 4 dimethylation) at ERα binding sites. In accordance, knockdown of GATA4 led to a reduction of ERα binding to DNA at these sites. Together, these results suggest that GATA4 is a pioneer factor (pioneer factors are a special class of transcription factor that can associate with compacted chromatin to facilitate the binding of additional transcription factors(22)) for ERα in osteoblast-like cells.(6) Therefore, we performed knockdown of Gata4 in primary murine calvarial osteoblasts to determine the effects of GATA4 on ERα and ERα target gene regulation. Gata4 can be successfully knocked down with lentivirally expressed short hairpins directed at Gata4 mRNA. Two different short hairpins (designated #2 and #4) reduced the level of Gata4 mRNA by more than two-thirds (Supporting Fig. 1A) compared to shGFP-infected cells. For consistency, all figures show shGATA4 #2 and supporting figures show shGATA4 #4. shGATA4 lentivirus induced apoptosis in 5.8% of the cells (Supporting Fig. 2) compared to 0% of the shGFP-infected cells. However, after 2 wk of differentiation, both the shGATA4 and shGFP-infected cells were at confiuence (data not shown). In vehicle (ethanol, EtOH)-treated cells, shGATA4 reduced the basal levels of ERα mRNA and protein (Fig. 1A, B). Additionally, shGATA4 affected the E2 response of ERα in these cells. ERα protein and mRNA levels are negatively regulated following E2 treatment in E2-treated shGFP calvarial osteoblasts, as observed in other cell types.(23,24) Loss of GATA4 in primary calvarial osteoblasts treated with E2 led to an increase in ERα mRNA and protein compared to vehicle-treated cells (Fig. 1A, B). Together, these data demonstrate a misregulation of ERα signaling in GATA4-ablated cells.
Fig. 1.
GATA4 regulates ERα and E2 target genes. (A) Calvarial osteoblasts were infected with lentivirus expressing either shGFP or shGATA4. Cells were then differentiated for 2 wk and RNA was obtained. qPCR was performed for ERα and normalized to actin mRNA. (B) Calvarial osteoblasts were infected with lentivirus expressing either shGFP or shGATA4. Cells were then differentiated for 2 wk and then treated with 10 nM E2 for 24 hours. Protein was obtained and immunoblots for ERα, GATA4, and actin were performed. (C) RNA was obtained as in A, and qPCR was performed with primers to FasL and normalized to actin mRNA. (D) RNA was obtained as in A, and qPCR was performed with primers to alkaline phosphatase and normalized to actin mRNA. *p < 0.05; **p < 0.001. Alk. Phos. = alkaline phosphatase.
FasL and alkaline phosphatase are two important targets of ERα in osteoblasts(3,25) (Fig. 1C, D). When GATA4 is knocked down, E2 can no longer induce FasL and alkaline phosphatase mRNA (Fig. 1C, D), suggesting that GATA4 protein is necessary for expression of E2-mediated gene targets in primary calvarial osteoblasts. E2 also increases Gata4 mRNA, and after shGATA4, Gata4 mRNA is not increased (Supporting Fig. 1B). Therefore, GATA4 is necessary to regulate E2 targets in primary osteoblasts, regulating both osteoblast differentiation (via alkaline phosphatase) and osteoclast apoptosis (via FasL).
ERα-independent roles for GATA4 in bone mineralization
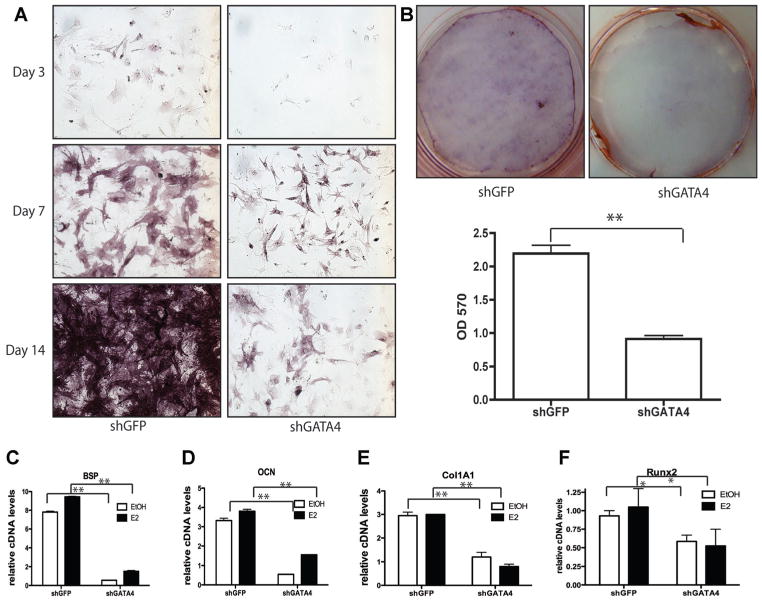
Because GATA4 regulates osteoblast differentiation genes such as alkaline phosphatase (Fig. 1D), we hypothesized that GATA4 may be necessary for proper differentiation and mineralization. To test this, primary calvarial osteoblasts were differentiated for 14 d following Gata4 knockdown, and alkaline phosphatase activity and mineralization were quantitated (Fig. 2A, B). Knockdown of Gata4 reduced the amount of alkaline phosphatase activity as early as 3 d after differentiation and throughout the time course of differentiation (Fig. 2A). Furthermore, knockdown of Gata4 led to a greater than twofold reduction in mineralization, as compared to control shGFP (p < 0.001; Fig. 2B). An independent shRNA targeting of Gata4 (shGATA4 #4) confirmed a significant reduction in mineralization (Supporting Fig. 3). The data suggest an essential role for GATA4 in differentiation and/or mineralization in primary osteoblasts. Interestingly, E2 does not increase mineralization in calvarial osteoblast cultures in vitro (data not shown and Almeida and colleagues(26)). In addition, E2 treatment does not increase expression of bone differentiation markers such as bone sialoprotein (BSP (Spp1)), osteocalcin (OCN (Bglap)), Col1A1, or RUNX2 (Fig. 2C–F). However, knockdown of Gata4 significantly reduced the levels of BSP, OCN, Col1A1, and RUNX2 mRNA, corresponding to the differentiation and mineralization assays, indicating E2-independent effects for GATA4 in osteoblast differentiation.
Fig. 2.
GATA4 regulates differentiation and mineralization in vitro. (A) Primary calvarial osteoblasts were infected with lentivirus expressing an shRNA directed to GFP (shGFP) or to Gata4 (shGATA4). The following day the cells were placed in mineralization media and allowed to differentiate for 3, 7, or 14 d. Cells were then fixed and assayed for alkaline phosphatase. (B) Primary calvarial osteoblasts were infected with lentivirus expressing an shRNA directed to GFP (shGFP) or to Gata4 (shGATA4). The following day the cells were placed in mineralization media and allowed to differentiate for 2 wk. Cells were then fixed and stained using Alizarin Red. Alizarin Red was eluted and the mineral content was measured at an OD of 570 nm. Silencing was performed in three wells and the average OD is displayed. (C–F) Calvarial osteoblasts were infected with lentivirus expressing either shGFP or shGATA4. Cells were then differentiated for 2 wk and RNA was obtained. qPCR was performed for the indicated genes and normalized to actin mRNA. *p < 0.05; **p < 0.001. BSP = bone sialoprotein; OD = optical density; OCN = osteocalcin; Col1A1 = type I collagen A1; Runx, Runt-related transcription factor.
GATA4 is necessary in mesenchymal stem cells for osteoblast differentiation
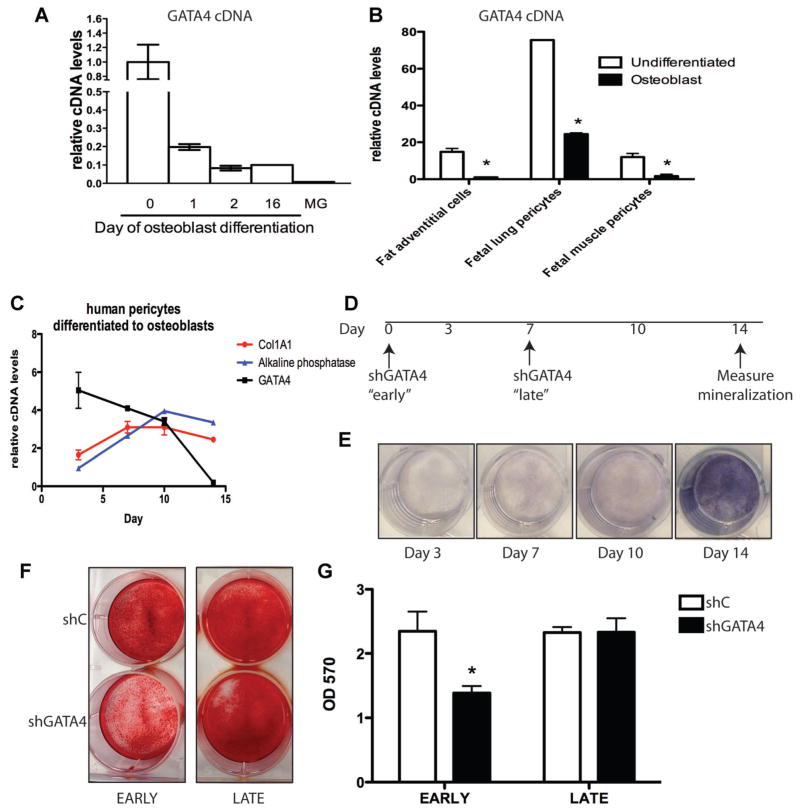
The expression of Gata4 during osteoblast differentiation was further investigated in order to determine if GATA4 has stage-specific effects on osteoblast formation and/or activity. Gata4 mRNA is reduced by fivefold after 1 d of osteoblast differentiation, and by more than 10-fold after 16 d of differentiation (Fig. 3A). However, mature osteoblasts still express significantly more Gata4 mRNA than mammary gland cells used as the control tissue for the absence of Gata4 expression. This suggests that GATA4 might influence early stage “precursor” decisions upstream of mature terminally differentiated osteoblast cells. To assess the translational potential of the model, we next verified that Gata4 is expressed more highly in human mesenchymal stem cells (MSCs) than in differentiated osteoblasts. Human fat adventitial cells, fetal lung pericytes, fetal muscle pericytes, and fetal bone marrow pericytes (the native ancestors of MSCs) were left undifferentiated or differentiated into osteoblasts.(18) Gata4 mRNA was significantly lower in terminally differentiated osteoblasts than in undifferentiated cells (Fig. 3B, C). There is an inverse relationship between osteoblast differentiation markers (Col1A1 and alkaline phosphatase mRNA) and Gata4 mRNA expression (Fig. 3C). Therefore, Gata4 is expressed more highly in mesenchymal stem-like cells than in differentiated, mineralizing osteoblasts.
Fig. 3.
GATA4 regulates bone mineralization early in the differentiation process. (A) Bone marrow stromal cells were differentiated for 0, 1, 2, or 16 d. RNA was obtained and qPCR was performed for Gata4 and normalized to actin mRNA. MG RNA was also obtained for comparison. (B) Fat adventitial cells, fetal lung pericytes, and fetal muscle pericytes were left undifferentiated or differentiated to mineralizing osteoblasts. RNA was obtained and qPCR was performed for Gata4 and normalized to actin mRNA. (C) Human fetal bone marrow CD146+ cells (pericytes) were cultured in osteoblast differentiation media for 3, 7, 10, and 14 d. RNA was obtained and qPCR was performed for Gata4, Col1A1, and alkaline phosphatase cDNA and normalized to actin mRNA. (D) Design of experiment for parts E–G. (E) Human fetal bone marrow CD146+ cells (pericytes) were cultured in osteoblast differentiation media for 3, 7, 10, and 14 d. At the indicated times cells were fixed and assayed for alkaline phosphatase. (F) Pericytes were infected with lentivirus expressing shC or shGATA4 either “early” at day 0 or “late” at day 7. All cells were placed in mineralization media at day 1 and allowed to differentiate for 2 wk. Cells were then fixed and stained using Alizarin Red. (G) Alizarin Red from part F was eluted and the mineral content was measured at an OD of 570 nm. Silencing was performed in three wells and the average OD is displayed. *p < 0.05. OD = optical density; Col1A1 = type I collagen A1; MG = mammary gland.
The observation that Gata4 expression is higher prior to differentiation raises the possibility that GATA4 plays a critical role at the phase of osteoblast commitment. To test this, Gata4 was either knocked down “early” (in undifferentiated pericytes) or “late” in differentiating osteoblasts (1 wk after initiating differentiation; Fig. 3D). The corresponding alkaline phosphatase levels, indicating the amount of differentiation of WT pericytes, is demonstrated in Fig. 3E. The cells were selected with puromycin to ensure efficient lentiviral infection, and then induced to mineralize. Cells infected with control lentivirus showed efficient mineralization, as visualized by Alizarin Red staining, whether infected early or late (Fig. 3F, G). In contrast, cells infected with shGATA4 before differentiation exhibited a significant reduction in the amount of mineralization compared to control infected cells. However, osteoblasts infected with shGATA4 “late” in differentiation showed no loss of mineralization, suggesting that GATA4 plays a role in either osteoblast progenitors or early in differentiation. Taken together, the above results suggest that in vitro in both human and mouse osteoblasts, GATA4 elicits its strongest effects on “early” progenitor and/or precursor cells.
GATA4 expression in vivo
GATA4 has been described in detail in the developing heart, intestines, and several other tissues.(5) Indeed, analysis of Gata4 mRNA in E18.5 embryos reveal highest expression in these tissues (Supporting Fig. 4, and data not shown). However, based on the in vitro data described above, Gata4 expression was analyzed in the developing skull and hind limb. Using highly sensitive in situ RNA hybridization (RNAscope technology), Gata4 mRNA is shown to be abundant in the ventricular zone of the developing brain in E13.5, E14.5, and E16.5 mice, before ossification of the skull (Fig. 4). At E18.5 Gata4 mRNA has decreased significantly in the brain and is present near and/or within the ossifying skull. Gata4 mRNA is also present in proliferating chondrocytes in E13.5, E14.5, and E16.5 hind limbs, which has not been previously reported. At E18.5 Gata4 mRNA is expressed at the ossification front of the growth plate and periosteum. Although the precise identity of the Gata4-positive cells in these locations is as yet unclear, the data are consistent with a direct role for GATA4 at various times and stages in normal embryonic bone development.
Fig. 4.
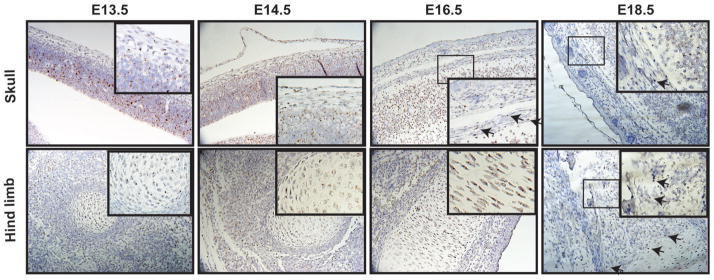
In situ analysis of Gata4 expression. Sagittal sections of E13.5, E14.5, E16.5, and E18.5 wild-type FVB embryos were probed for Gata4 expression (brown) and counterstained with hematoxylin (blue). Arrows indicate Gata4 expression in bone.
GATA4 is necessary for bone development in embryogenesis in vivo
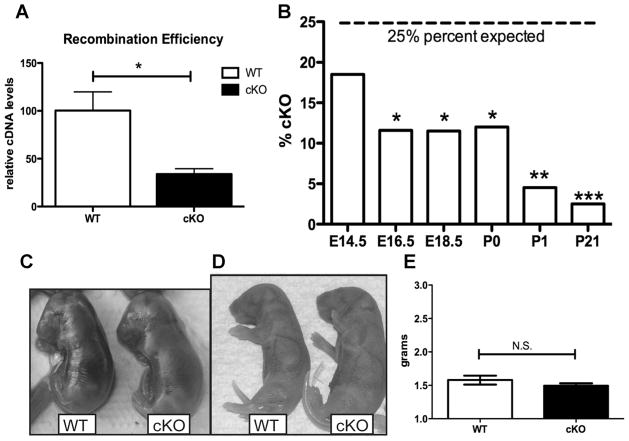
To determine the role of GATA4 in vivo, Gata4 floxed (GATA4Fl/Fl) mice were crossed with mice expressing Cre recombinase under control of a 2.3-kb fragment of the rat Col1a1 gene.(27) Previous studies have shown that Cre-mediated deletion of exons 3, 4, and 5 of the Gata4 gene converts the floxed allele into a recombined allele no longer capable of encoding a functional GATA4 protein.(28) Gata4 recombination efficiency in osteoblasts was determined in vivo by qPCR of mRNA, using primers that cannot detect recombined Gata4 mRNA. Gata4 mRNA was reduced by over 75% in differentiated calvarial organs of GATA4Fl/FlCre+ mice (cKO) (Fig. 5A). The actual recombination of Gata4 in osteoblasts is probably significantly higher, because the calvarial organs consist of other cell types, such as stromal cells.
Fig. 5.
GATA4 cKO mice are not born at expected Mendelian ratios. (A) The recombination efficiency of Cre-mediated excision was measured by qPCR from cDNA from differentiated WT and cKO calvarial organ cultures. (B) Tail DNA was obtained from mice at E14.5, E16.5, E18.5, P0, P1, and P21. Genotyping was performed for the presence of Floxed Gata4 and for Cre recombinase. The percent of mice that were GATA4 Fl/Fl/Cre+ (% cKO) is graphed. *p < 0.05; **p < 0.001; ***p < 0.0001. (C) Photograph of representative WT and cKO mice at E18.5. (D) Photograph of representative WT and cKO mice at P1. (E) Weight, in grams, of newborn WT and cKO mice. cKO = conditional knockout; N.S. = not significant.
Gata4 cKO mice were not present at Mendelian ratios after E16.5 (Fig. 5B). Mutants were morphologically indistinguishable from control littermates’ appearance at any age, including E18.5 (Fig. 5C) or P1 (Fig. 5D), or by weight (Fig. 5E). The mice did not exhibit any obvious heart defects, as might be predicted from leaky Col1a1-Cre expression. Whole hearts and H&E-stained sections of left and right ventricles from P0 animals revealed normal septal structure, and a well-developed myocardium (Supporting Fig. 5, and data not shown). In addition, heart weight–body weight ratios were the same in wild-type and cKO littermate animals (Supporting Fig. 5). Thus, although we cannot at present determine the cause of death of cKO mice, we have no evidence that heart defects are responsible.
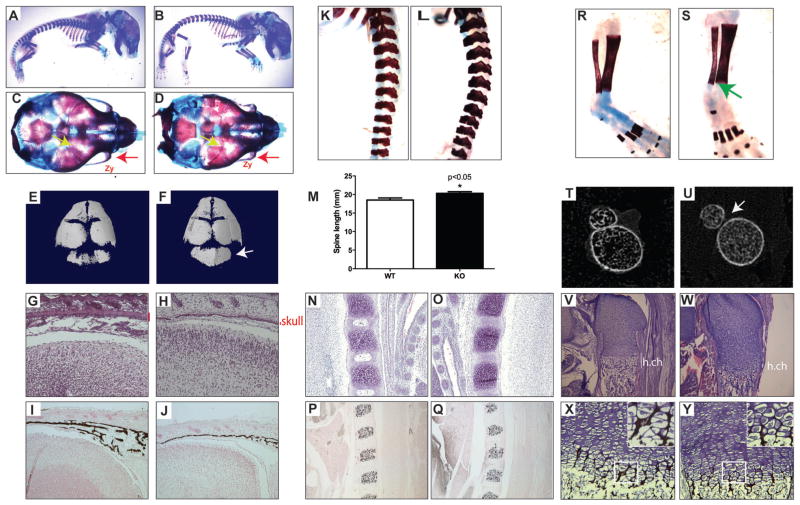
Skeletal preparations of E18.5 mice revealed no gross abnormalities (Fig. 6A, B). However, closer examination revealed multiple skull defects (Fig. 6C–J, Supporting Fig. 5), including smaller zygomatic bones (Fig. 6C, D; red arrows), a more mineralized occipital one (Fig. 6D, F) and decreased thickness of cranial bone (Fig. 6G, H; red lines). Furthermore, Von Kossa staining and μCT analysis demonstrate a reduced mineralization of the skull in the cKO mice (Fig. 6I, J; Supporting Fig. 5F, G).
Fig. 6.
GATA4 cKO mice have skeletal defects. (A–D) Skeletal preparations of E18.5 WT (A, C) and cKO (B, D) mice. (A, B) Whole body. (C, D) Superior view of skull. Red arrow indicates decreased zygomatic bone size in cKO mice. Yellow arrows highlight a suture defect in the cKO skull. (E, F) μCT images of WT (E) and cKO (F) skulls. Arrow points to occipital bone. (G, H) H&E staining of sagittal sections of WT (G) and cKO (H) heads. Red line indicates thickness of skull bone. (I, J) Von Kossa staining of sagittal sections of WT (I) and cKO (J) heads. (K, L) Skeletal preparations of spine from WT (K) and cKO (L). (M) Total spine length of n = 7 mice. *p < 0.05. (N, O) H&E staining of sagittal sections of WT (N) and cKO (O) vertebrae. (P, O) Von Kossa staining of sagittal sections of WT (P) and cKO (Q) vertebrae. (R, S) Skeletal preparations of tibia and fibula from WT (R) and cKO (S). Green arrow indicates lack of fusion of tibia and fibula. (T, U) μCT images of WT (T) and ckO (U) tibia and fibula at their closest distance. (V, W) H&E staining of sagittal sections of WT (V) and cKO (W) femurs. (X, Y) Von Kossa staining of sagittal sections of WT (X) and cKO (Y) femurs. Zy = zygomatic bone; h.ch = hypertrophic chondrocytes. KO = knockout; cKO = conditional knockout.
In addition to cranial defects, we also observed vertebral and appendicular defects. Individual lumbar vertebrae were larger and misshapen (Fig. 6K, L), leading to an overall increase in spine length of cKO mice (Fig. 6M). The vertebral bodies of the cKO mice have a decrease in hypertrophic chondrocytes (Fig. 6N, O) and irregular mineralization (Fig. 6P, Q). There is a lack of fusion of the distal tibia and fibula in cKO mice as observed by both skeletal preparations (Fig. 6R, S) and μCT (Fig. 6T, U). There is also a decrease in hypertrophic chondrocytes and mineralization in the femurs of cKO mice (Fig. 6V–Y).
To quantify the mineralization effects in Fig. 6, μCT analysis was performed on the femurs and vertebrae of WT and cKO newborn mice, which demonstrated that cKO mice had reduced trabecular bone (Fig. 7A–L). The trabecular bone volume (BV/TV), trabecular number (Tb.N), and trabecular thickness (Tb.Th) were significantly reduced in cKO femurs compared to wild-type littermates (Fig. 7C–E). Trabecular spacing was increased in the cKO mice, although it did not reach statistical significance (Tb.Sp; Fig. 7F). Cortical bone parameters (total cross-sectional area, cortical bone area, cortical bone area fraction, or cortical thickness) were not statistically different between wild-type and cKO littermate animals (Supporting Fig. 7). μCT analysis of lumbar vertebrae (L5) confirmed the irregular shape of cKO vertebrae (Fig. 7G, H) and revealed a significant reduction in the BV/TV and Tb.Th (Fig. 7I, K). Tb.N was decreased in the cKO mice, although it did not reach statistical significance (Fig. 7J). Together, the skeletal preparations, histology, and μCT analysis indicate defects in intramembranous and endochondral bone formation.
Fig. 7.
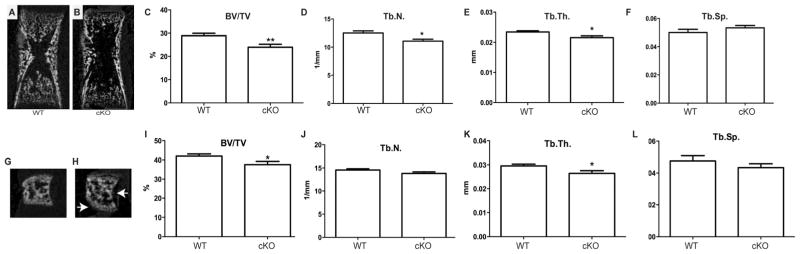
Comparison of trabecular bone structure in P0 WT and cKO mice assessed by μCT. (A, B) Representative μCT images of WT and cKO femurs. (C) BV/TV, (D) Tb.N, and (E) Tb.Th were decreased in femurs of cKO mice. (F) There was no statistical increase in Tb.Sp in cKO mice. (G, H) Representative μCT images of WT and cKO L5 vertebrae. Arrows indicate defects in shape. (I) BV/TV, (J) Tb.N, (K) Tb.Th, and (L) Tb.Sp of L5 vertebrae in WT and cKO L5 vertebrae. n = 9; *p < 0.05. cKO = conditional knockout; BV/TV = bone volume/total volume; Tb.N = trabecular number; Tb.Th = trabecular thickness; Tb.Sp = trabecular spacing.
GATA4 regulates TGFβ and BMP pathways
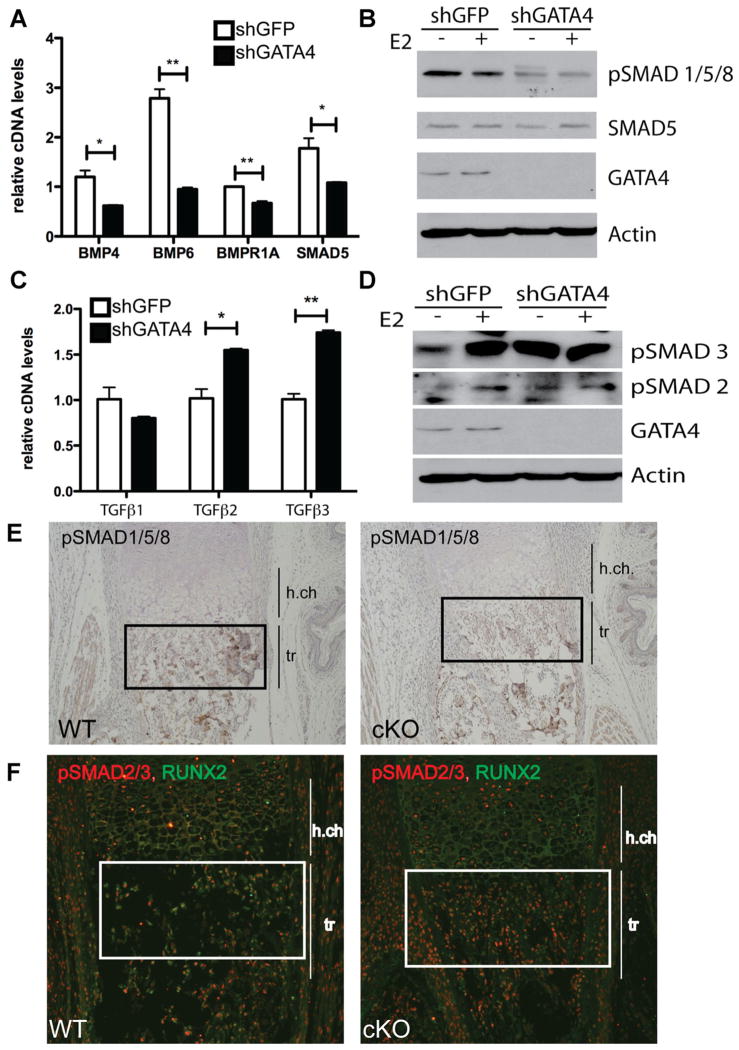
To investigate mechanisms underlying the role of GATA4 in osteoblasts, we examined global gene expression changes following Gata4 knockdown in primary murine calvarial osteoblasts (Supporting Fig. 1A). Osteoblasts were differentiated for 2 wk and RNA was analyzed by genomewide expression profiling (MouseRef-8 v2.0 Expression BeadChips; Illumina). Loss of Gata4 resulted in a decrease in expression of 1610 genes by twofold or greater and an increase in expression of 2399 genes by twofold or greater. Gene ontology analysis by DAVID software revealed several categories of differentially expressed genes.(20) The most downregulated genes in shGATA4 cells encode extracellular matrix proteins (p = 3.2 × 10−16) and drive bone development (p = 3.6 × 10−8), thereby confirming a role for GATA4 in osteoblasts.
Loss of Gata4 resulted in misregulation of a large number of TGFβ superfamily pathway genes (Supporting Figs. 8 and 9). Key upregulated genes in this pathway were TGFβ2 and TGFβ3, whereas both BMP pathway genes were downregulated. Analysis of mRNA expression in shGATA4-infected calvarial osteoblasts confirmed decreased expression of two BMP ligands (Bmp4 and Bmp6), a receptor (Bmpr1A), and a downstream effector SMAD (Smad5) (Fig. 8A). Because of the downregulation of BMP pathway genes, BMP pathway activation (phosphorylated SMAD [pSMAD] 1/5/8 as a downstream target for BMP-mediated signaling(7)) was analyzed with a pSMAD1/5/8 antibody by immunoblotting. Indeed, shGATA4 cells had a reduced amount of pSMAD1/5/8, illustrating that BMP-mediated signal transduction has been severely attenuated. E2 had no effect on the regulation of pSMAD1/5/8 (Fig. 8B). Conversely, TGFβ2 and TGFβ3 were significantly upregulated in shGATA4 cells (Fig. 8C). Consistent with the upregulation of the ligands, pSMAD2/3 were increased in shGATA4 cells (Fig. 8D), demonstrating activation of the TGFβ pathway. E2 induced phosphorylation of SMAD2 and SMAD3 in shGFP cells, but not in shGATA4 cells.
Fig. 8.
GATA4 regulates the TGFβ and BMP pathways in osteoblasts. (A) Calvarial osteoblasts were infected with lentivirus expressing either shGFP or shGATA4. Cells were then differentiated for 2 wk and RNA was obtained. qPCR was performed for the indicated genes and normalized to actin mRNA. (B) Calvarial osteoblasts were infected with lentivirus expressing either shGFP or shGATA4. Cells were then differentiated for 2 wk and then treated with E2 for 24 hours. Protein was obtained and immunoblots for pSMAD1/5/8, SMAD5, GATA4, and actin were performed. (C) RNA was obtained as in A, and qPCR was performed for the indicated genes and normalized to actin mRNA. (D) Protein was obtained as in B, and immunoblots for pSMAD3, pSMAD2, GATA4, and actin were performed. (E) IHC with an antibody to pSMAD1/5/8 was performed on femurs from P0 WT (Fl/Fl) and cKO (Fl/Fl; Cre+) mice. (F) Immunofluorescence with an antibody to pSMAD1/5/8 was performed on femurs from P0 WT (Fl/Fl) and cKO (Fl/Fl; Cre+) mice. *p < 0.05; **p < 0.001. cKO = conditional knockout; h.ch = hypertrophic chondrocytes; tr = trabecular bone; IHC = immunohistochemistry.
To investigate the possibility of an in vivo role for GATA4 regulation of the TGFβ and BMP pathways, the femurs of control (Fl/Fl) and cKO mice were analyzed by immunohistochemistry (IHC) for pSMAD1/5/8 and pSMAD2/3. cKO bones exhibited a decrease in the amount of pSMAD1/5/8 in the trabecular bone (Fig. 8E). Conversely, pSMAD2/3 was increased in the trabecular bone from cKO mice, as compared to WT bone (Fig. 8F). Our findings thus indicate that GATA4 is critical for the maintenance of osteoblast function, and raise the possibility that this is in part through regulating the balance between BMP and TGFβ pathway activity.
Discussion
We previously identified GATA4 as a pioneer factor for estrogen receptor in osteoblasts.(6) Here we demonstrate that GATA4 is important for the regulation of several E2 targets in osteoblasts, but that GATA4 also has E2-independent functions.
Knockout of GATA4 specifically in osteoblasts leads to embryonic and/or perinatal lethality. Because cKO mice were physically indistinguishable from their WT littermates at all stages of development, the cause of death remains under investigation. The Col1A1 2.3-kb promoter is expressed as early as E14.5, when analyzed by crossing the Col1A1-Cre mouse with the ROSA26 reporter mouse (R26R).(29) The Col1A1-Cre;R26R mouse demonstrates a high level of bone-specific expression of Cre recombinase,(29) as does mRNA from many tissues.(27) Furthermore, the Col1a1 (2.3-kb)-GFP transgene was not detected in the heart or intestines,(30) which are two tissues with high Gata4 expression. However, it is possible that expression could occur at a different age, or at a level below the detection of a Northern blot. The Col1a1 2.3-kb promoter might be active in some growth plate chondrocytes,(27) and the extent of this effect needs to be investigated.
Although rare, Col1A1-Cre–mediated deletion has led to embryonic death.(31) Crossing the Gata4 Fl/Fl mouse with a mouse that expresses Cre later in the osteoblast lineage, such as the osteocalcin promoter,(32) could reveal a role for GATA4 in adult mice and potential roles in estrogen-deficient osteoporosis. Similarly, crossing the Gata4 Fl/Fl mouse with a mouse that expresses Cre earlier in development could further define the “early” role for GATA4 suggested by the above data.
Total Gata4 knockout mice exhibit embryonic lethality from E7.5 to E10.5,(15,16) which is much earlier than for the osteoblast-specific knockout described here. Interestingly, specific knockouts of GATA4 in the heart with Cre-recombinase controlled by the β-myosin heavy promoter (β-MHC) or α-MHC promoter were generated at predicted Mendelian ratios and mice were viable up to 16 wk of age.(33) Gata4 deletion from the developing intestinal epithelium,(34) pancreas,(35) testicular somatic cells,(36) or granulosa cells(37) also resulted in mice born at the expected Mendelian ratios, but with tissue-specific effects.
These results demonstrate that GATA4 is essential for bone mineralization both in vitro and in vivo. μCT analysis shows a decrease in trabecular bone measurements in the cKO mice. Furthermore, the skeletal preparations and μCT show defects, including those in the skull, legs, and spine, in these mice.
Mouse models for TGFβ and BMP signaling in bone have been generated (reviewed in Chen and colleagues(8)). TGFβ2 over-expression leads to lower bone mineral density, in agreement with the Gata4 cKO mice.(38) TGFβ2-null mice have a lack of occipital bone formation,(39) corresponding with the increase in occipital bone formation in Gata4 cKO mice and the suppression of TGFβ2 by GATA4. A limb mesenchyme double KO of both BMP2 and BMP4 (driven by Prx1-cre) leads to severe bone malformation. Interestingly, the tibia and fibula in the BMP4 KO are not fused,(40) as is seen in the Gata4 cKO. Furthermore, an osteoblast-specific KO of SMAD1 leads to an osteopenic phenotype.(41) Together, these mouse models are in agreement with a role for GATA4 in regulating TGFβ and BMP signaling. Genetic epistasis experiments will be required to test the physiological relevance of the altered BMP and TGFβ pathway activity we have observed in Gata4 cKO mice.
GATA4 is currently being used for gene therapy in the heart in combination with heart-specific transcription factors.(42) MSCs show potential for the treatment of osteoporosis, and their optimization could be realized by estrogens, selective estrogen receptor modulators (SERMs), and/or GATA4 to better induce osteoblast formation. In conclusion, GATA4 in osteoblasts plays a key role in the regulation of TGFβ and BMP pathways to control bone differentiation and/or mineralization.
Supplementary Material
Acknowledgments
This work was supported by an ASBMR Junior Faculty Osteoporosis Research Award and 1R56DK090231-01 to SAK and AR044528 to KL. The UCLA Vector Core is supported by JCCC/P30 CA016042 and CURE/P30 DK041301. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Disclosures
All authors state that they have no conflicts of interest.
Authors’ roles: Study conduct: MG, AJG, DR, SR, WW, GZ, VHM, EA, GAMC, and SAK. Study design: AJG, ST, BP, KL, GAMC, and SAK. MG, AJG, ST, BP, KL, GAMC, and SAK participated in drafting the manuscript or revising it critically for important intellectual content. MG, AJG, DR, SR, WW, GZ, ST, BP, KL, GAMC, and SAK approved the final version of the submitted manuscript. SAK agrees to be accountable for all aspects of the work.
Additional Supporting Information may be found in the online version of this article.
References
- 1.United States Bone and Joint Decade: The Burden of Musculoskeletal Diseases in the United States. Rosemont, IL: American Academy of Orthopaedic Surgeons; 2008. Available from: http://www.bone-andjointburden.org/chapter_downloads/index.htm. [Google Scholar]
- 2.Kousteni S, Bellido T, Plotkin LI, et al. Nongenotropic, sex-nonspecific signaling through the estrogen or androgen receptors: dissociation from transcriptional activity. Cell. 2001;104(5):719–30. [PubMed] [Google Scholar]
- 3.Krum SA, Miranda-Carboni GA, Hauschka PV, et al. Estrogen protects bone by inducing Fas ligand in osteoblasts to regulate osteoclast survival. EMBO J. 2008;27(3):535–45. doi: 10.1038/sj.emboj.7601984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krum SA. Direct transcriptional targets of sex steroid hormones in bone. J Cell Biochem. 2011;112(2):401–8. doi: 10.1002/jcb.22970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275(50):38949–52. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 6.Miranda-Carboni GA, Guemes M, Bailey S, et al. GATA4 regulates estrogen receptor-{alpha}-mediated osteoblast transcription. Mol Endocrinol. 2011;25(7):1126–36. doi: 10.1210/me.2010-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song B, Estrada KD, Lyons KM. Smad signaling in skeletal development and regeneration. Cytokine Growth Factor Rev. 2009;20(5–6):379–88. doi: 10.1016/j.cytogfr.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen G, Deng C, Li YP. TGF-beta and BMP signaling in osteoblast differentiation and bone formation. Int J Biol Sci. 2012;8(2):272–88. doi: 10.7150/ijbs.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alliston T, Choy L, Ducy P, Karsenty G, Derynck R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 2001;20 (9):2254–72. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rojas A, De Val S, Heidt AB, Xu SM, Bristow J, Black BL. Gata4 expression in lateral mesoderm is downstream of BMP4 and is activated directly by Forkhead and GATA transcription factors through a distal enhancer element. Development. 2005;132 (15):3405–17. doi: 10.1242/dev.01913. [DOI] [PubMed] [Google Scholar]
- 11.Nemer G, Nemer M. Transcriptional activation of BMP-4 and regulation of mammalian organogenesis by GATA-4 and -6. Dev Biol. 2003;254(1):131–48. doi: 10.1016/s0012-1606(02)00026-x. [DOI] [PubMed] [Google Scholar]
- 12.Brown CO, 3rd, Chi X, Garcia-Gras E, Shirai M, Feng XH, Schwartz RJ. The cardiac determination factor, Nkx2-5, is activated by mutual cofactors GATA-4 and Smad1/4 via a novel upstream enhancer. J Biol Chem. 2004;279(11):10659–69. doi: 10.1074/jbc.M301648200. [DOI] [PubMed] [Google Scholar]
- 13.Belaguli NS, Zhang M, Rigi M, Aftab M, Berger DH. Cooperation between GATA4 and TGF-beta signaling regulates intestinal epithelial gene expression. Am J Physiol Gastrointest Liver Physiol. 2007;292(6):G1520–33. doi: 10.1152/ajpgi.00236.2006. [DOI] [PubMed] [Google Scholar]
- 14.Anttonen M, Parviainen H, Kyronlahti A, et al. GATA-4 is a granulosa cell factor employed in inhibin-alpha activation by the TGF-beta pathway. J Mol Endocrinol. 2006;36(3):557–68. doi: 10.1677/jme.1.01962. [DOI] [PubMed] [Google Scholar]
- 15.Kuo CT, Morrisey EE, Anandappa R, et al. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11(8):1048–60. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 16.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11(8):1061–72. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 17.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89 (5):747–54. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 18.Crisan M, Yap S, Casteilla L, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3(3):301–13. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 20.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Retting KN, Song B, Yoon BS, Lyons KM. BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development. 2009;136(7):1093–104. doi: 10.1242/dev.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jozwik KM, Carroll JS. Pioneer factors in hormone-dependent cancers. Nat Rev Cancer. 2012;12(6):381–5. doi: 10.1038/nrc3263. [DOI] [PubMed] [Google Scholar]
- 23.Nawaz Z, Lonard DM, Dennis AP, Smith CL, O’Malley BW. Proteasome-dependent degradation of the human estrogen receptor. Proc Natl Acad Sci U S A. 1999;96(5):1858–62. doi: 10.1073/pnas.96.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Read LD, Greene GL, Katzenellenbogen BS. Regulation of estrogen receptor messenger ribonucleic acid and protein levels in human breast cancer cell lines by sex steroid hormones, their antagonists, and growth factors. Mol Endocrinol. 1989;3(2):295–304. doi: 10.1210/mend-3-2-295. [DOI] [PubMed] [Google Scholar]
- 25.Krum SA, Miranda-Carboni GA, Lupien M, Eeckhoute J, Carroll JS, Brown M. Unique ER{alpha} cistromes control cell type-specific gene regulation. Mol Endocrinol. 2008;22(11):2393–406. doi: 10.1210/me.2008-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Almeida M, Martin-Millan M, Ambrogini E, et al. Estrogens attenuate oxidative stress and the differentiation and apoptosis of osteoblasts by DNA-binding-independent actions of the ERalpha. J Bone Miner Res. 2010;25(4):769–81. doi: 10.1359/jbmr.091017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu F, Woitge HW, Braut A, et al. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int J Dev Biol. 2004;48(7):645–53. doi: 10.1387/ijdb.041816fl. [DOI] [PubMed] [Google Scholar]
- 28.Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci U S A. 2004;101(34):12573–8. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dacquin R, Starbuck M, Schinke T, Karsenty G. Mouse alpha1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev Dyn. 2002;224(2):245–51. doi: 10.1002/dvdy.10100. [DOI] [PubMed] [Google Scholar]
- 30.Kalajzic I, Kalajzic Z, Kaliterna M, et al. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17 (1):15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 31.Gaur T, Hussain S, Mudhasani R, et al. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev Biol. 2010;340(1):10–21. doi: 10.1016/j.ydbio.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.VanKoevering KK, Williams BO. Transgenic mouse strains for conditional gene deletion during skeletal development. IBMS Bonekey. 2008;5:151–70. [Google Scholar]
- 33.Oka T, Maillet M, Watt AJ, et al. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ Res. 2006;98(6):837–45. doi: 10.1161/01.RES.0000215985.18538.c4. [DOI] [PubMed] [Google Scholar]
- 34.Battle MA, Bondow BJ, Iverson MA, et al. GATA4 is essential for jejunal function in mice. Gastroenterology. 2008;135(5):1676–86. e1. doi: 10.1053/j.gastro.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xuan S, Borok MJ, Decker KJ, et al. Pancreas-specific deletion of mouse Gata4 and Gata6 causes pancreatic agenesis. J Clin Invest. 2012;122(10):3516–28. doi: 10.1172/JCI63352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kyronlahti A, Euler R, Bielinska M, et al. GATA4 regulates Sertoli cell function and fertility in adult male mice. Mol Cell Endocrinol. 2011;333(1):85–95. doi: 10.1016/j.mce.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kyronlahti A, Vetter M, Euler R, et al. GATA4 deficiency impairs ovarian function in adult mice. Biol Reprod. 2011;84(5):1033–44. doi: 10.1095/biolreprod.110.086850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erlebacher A, Derynck R. Increased expression of TGF-beta 2 in osteoblasts results in an osteoporosis-like phenotype. J Cell Biol. 1996;132(1–2):195–210. doi: 10.1083/jcb.132.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanford LP, Ormsby I, Gittenberger-de Groot AC, et al. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124(13):2659–70. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bandyopadhyay A, Tsuji K, Cox K, Harfe BD, Rosen V, Tabin CJ. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2(12):e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang M, Jin H, Tang D, Huang S, Zuscik MJ, Chen D. Smad1 plays an essential role in bone development and postnatal bone formation. Osteoarthritis Cartilage. 2011;19(6):751–62. doi: 10.1016/j.joca.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartung S, Schwanke K, Haase A, et al. Directing cardiomyogenic differentiation of human pluripotent stem cells by plasmid-based transient overexpression of cardiac transcription factors. Stem Cells Dev. 2013 Apr 1;22(7):1112–25. doi: 10.1089/scd.2012.0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.