Proper regulation of microbial-induced cytokines is critical to intestinal immune homeostasis. Acute stimulation of NOD2, the Crohn's disease-associated sensor of bacterial peptidoglycan, induces cytokines. However, chronic NOD2 stimulation in macrophages decreases cytokines upon pattern recognition receptor (PRR) re-stimulation; cytokine attenuation to PRR stimulation is similarly observed in intestinal macrophages. The role for the transcriptional repressors Twist1 and Twist2 in regulating PRR-induced cytokine outcomes is poorly understood, and has not been reported for NOD2. We found that Twist1 and Twist2 were required for optimal cytokine downregulation during acute and, particularly, chronic NOD2 stimulation of human macrophages. Consistently, Twist1 and Twist2 expression was increased after chronic NOD2 stimulation; this increased expression was IL-10- and TGFβ-dependent. Although Twist1 and Twist2 did not co-regulate each other's expression, they cooperated to enhance binding to cytokine promoters after chronic NOD2 stimulation. Moreover, Twist1 and Twist2 contributed to enhance expression and promoter binding of the pro-inflammatory inhibitor c-Maf and the transcriptional repressor Bmi1. Restoring c-Maf and Bmi1 expression in Twist-deficient macrophages restored NOD2-induced cytokine downregulation. Furthermore, with chronic NOD2 stimulation, Twist1 and Twist2 contributed to the decreased expression and cytokine promoter binding of the transcriptional activators ATF4, C/EBPα, Runx1 and Runx2. Knockdown of these transcriptional activators in Twist-deficient macrophages restored cytokine downregulation after chronic NOD2 stimulation. Finally, NOD2 synergized with additional PRRs to increase Twist1 and Twist2 expression and Twist-dependent pathways. Therefore, after chronic NOD2 stimulation Twist1 and Twist2 coordinate the regulation of both transcriptional activators and repressors, thereby mediating optimal cytokine downregulation.
Introduction
Human nucleotide-binding oligomerization domain 2 (NOD2), an intracellular sensor of bacteria-derived muramyl dipeptide (MDP; a component of peptidoglycan), confers the greatest genetic risk of developing Crohn's disease (CD), a disease of chronic intestinal inflammation (1). When peripheral monocytes enter mucosal sites, such as the intestinal lamina propria, they are continuously exposed to bacterial products, including the NOD2 ligand, peptidoglycan/MDP (2, 3). Initial NOD2 stimulation results in cytokine secretion (3-5). However, ongoing NOD2 stimulation significantly downregulates cytokine secretion upon restimulation through pattern recognition receptors (PRRs) (3-7). This downregulation is impaired in individuals with CD-associated NOD2 polymorphisms (4, 5). Cytokine secretion in intestinal macrophages is similarly attenuated upon PRR stimulation (8), which is important for intestinal immune homeostasis. Moreover, chronic MDP administration to mice in vivo attenuates subsequent experimental colitis (6), thereby demonstrating the beneficial effects of chronic NOD2 stimulation in intestinal immune regulation.
Mechanisms contributing to cytokine downregulation after chronic NOD2 stimulation in human myeloid-derived cells include the upregulation of the intracellular inhibitors IRAK-M (4), Tollip (3), IRF4 (6, 9), and the Tyro3, Axl and Mer tyrosine kinase receptors (10); the NFκB1-dependent upregulation of the transcriptional repressor ATF3 (3); and the secretion of the inhibitory mediators IL-10 and TGFβ (5). Each of these mechanisms contributes only partially to cytokine downregulation and is operational to varying degrees in different individuals (e.g. IRAK-M) (4, 11). Given the dramatic alterations in macrophage functions and importance of downregulating PRR-initiated pathways upon chronic microbial stimulation, we hypothesized that additional critical mechanisms mediating these changes have yet to be identified, including those involving transcriptional regulators.
Twist1 and Twist2 are basic helix-loop-helix transcriptional repressors that bind to E boxes in gene promoters, acting as master regulators in a variety of biological processes, including organogenesis, osteogenesis, cancer progression and hematopoietic cell development (12-14). As such, Twist1−/− mice are embryonic lethal due to a variety of developmental defects (15). Mutations in Twist1 are associated with Saethre-Chotzen syndrome (16, 17), an autosomal dominant disorder characterized by craniofacial and limb anomalies. Although the major focus on Twist function has been in the context of the critical cellular processes described above, there is evidence for a role for Twists in regulating inflammation (14, 18-22). Twist2−/− mice demonstrate increased myeloid lineage development (14). Moreover, Twist2−/− mice or Twist1+/−/Twist2+/− mice demonstrate increased proinflammatory cytokines associated with increased NFκB pathway signaling and perinatal death, and defects in the type I IFN-mediated suppression of pro-inflammatory cytokines in macrophages (18, 22). In T cells, Twist1 limits Th1, Th17 and T follicular helper cell development (20, 21). However, the role and mechanisms through which Twist1 and Twist2 regulate PRR-induced cytokine outcomes is poorly understood; Twist regulation of NOD2-induced outcomes has not been reported. Given the repressive functions of the Twist proteins and evidence for their ability to limit inflammation in select situations, we hypothesized that Twist1 and/or Twist2 would contribute to downregulating cytokines during both acute and chronic NOD2 stimulation in primary human macrophages.
We found that Twist1 and Twist2 were required for the downregulation of cytokines during both acute and, in particular, chronic NOD2 stimulation of human monocyte-derived macrophages (MDMs). Consistently, Twist1 and Twist2 were each significantly upregulated after prolonged NOD2 stimulation; this upregulation required autocrine IL-10 and TGFβ and was defective in MDMs from Crohn's disease-associated NOD2 risk carriers. Binding of the upregulated Twist1 and Twist2 transcriptional repressors to cytokine promoters was increased in a cooperative manner after chronic NOD2 stimulation. Moreover, Twist1 and Twist2 upregulated the expression and promoter binding of additional inhibitory transcriptional regulators, including c-Maf and Bmi1, while decreasing the expression and promoter binding of essential activating transcription factors, including ATF4, C/EBPα, Runx1 and Runx2. Complementation of the reduced c-Maf and Bmi1 expression, or reduction in the upregulated ATF4, C/EBPα, Runx1 or Runx2 under Twist-deficient conditions were each sufficient to restore cytokine downregulation under chronic NOD2 stimulation conditions. Therefore, Twist1 and Twist2 contribute to cytokine downregulation during chronic NOD2 stimulation through the coordination of the reciprocal expression and cytokine promoter binding of additional transcriptional repressors and activators.
Materials and Methods
Primary MDM culture and genotyping
Informed consent was obtained as approved by the Yale University Institutional Review Board. Monocytes were purified from healthy individuals or Crohn's disease patients (Supplemental Fig 1E) and differentiated to MDMs for seven days with 10 ng/ml M-CSF (Shenandoah Biotechnology, Warwick, PA) as in(4). We performed NOD2 genotyping by TaqMan (Applied Biosystems, Foster City, CA) or Sequenom platform (Sequenom, San Diego, CA).
mRNA expression
Total RNA was isolated, reverse transcribed and quantitative PCR was performed as described previously(4) with normalization to GAPDH. Primers are available upon request.
Myeloid cell stimulation
For tolerance induction in vitro, human MDMs (0.5×106) were pretreated with 100 μg/ml MDP (Bachem, King of Prussia, PA) for 48h prior to extensive wash and retreated for 24h with 100 μg/ml MDP. In some cases anti-IL-10 or anti-TGFβ neutralizing antibodies (R&D Systems, Minneapolis, MN) or lipid A (Peptide International, Louisville, KY) was used. Supernatants were assayed for cytokine secretion per manufacturer instructions using the following antibodies: TNF, IL-6, IL-8, IL-10 (BD Biosciences, San Jose, CA) or IL-12 (eBioscience, San Diego, CA).
Protein expression analysis
Western blot analysis was performed as in(4) using antibodies to Twist1/Twist2 (ab50887; Abcam, Cambridge, MA), c-Maf (sc7866; Santa Cruz Biotechnology, Santa Cruz, CA), Bmi1 (05-1322; EMD Millipore, Billerica, MA), or anti-GAPDH (EMD Millipore).
Transfection of siRNAs and plasmids
Primary human MDMs were transfected with 100 nM scrambled or ON-TARGETplus SMARTpool siRNA against Twist1, Twist2, c-Maf, Bmi1, ATF4, C/EBPα, Runx1, Runx2, (Dharmacon, Lafayette, CO) (4 pooled siRNAs for each gene), or with 2 μg pT3-EF1a-Bmi1 (Addgene plasmid 31783 kindly deposited by X. Chen (23)), c-Maf (GeneCopoeia, Rockville, MD) or empty vector using Amaxa nucleofector technology (Amaxa, San Diego, CA).
Chromatin immunoprecipitation (ChIP)
ChIP analysis of MDMs was performed according to a modified protocol(3). Primers were designed to amplify genomic sequences at the cytokine gene promoter region (available upon request). Antibodies used included: Twist1 (ab50887), Twist2 (ab66031), Runx1 (ab23980), ATF4 (ab85049), Bmi1 (ab14389) (Abcam), Runx2 (sc10785x), C/EBPα (sc-61x) or c-Maf (sc-7866x) (Santa Cruz Biotechnology).
Statistical analysis
Significance was assessed using a two-tailed Student's t-test. p<0.05 was considered significant. A Bonferroni correction was applied for multiple comparisons. To maintain a consistent pg/ml scale for cytokine concentration measures, the higher levels observed for IL-8 and IL-6 (ng/ml levels) are shown with a multiplication factor.
Results
Twist1 and Twist2 are required for optimal downregulation of cytokines upon acute and chronic NOD2 stimulation
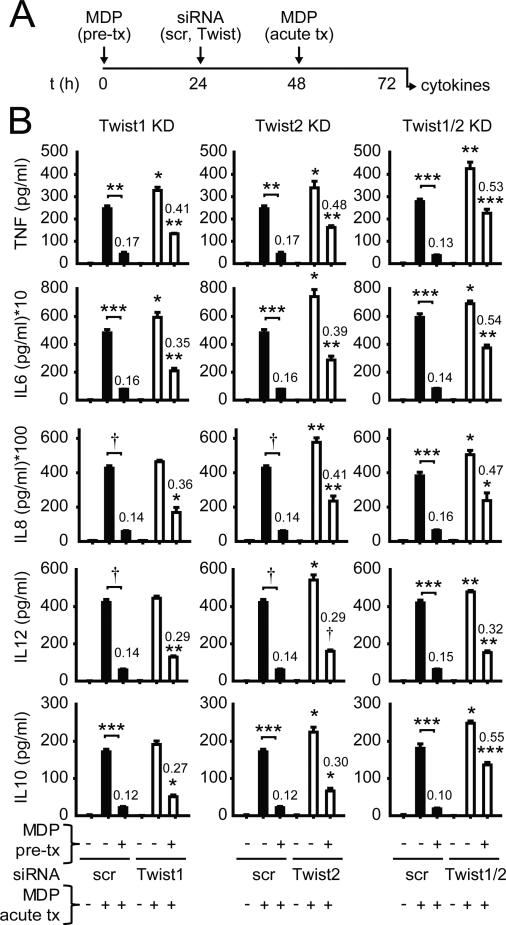
To elucidate the role of Twist1 and Twist2 in cytokine downregulation with acute NOD2 stimulation in human MDMs, we utilized siRNA to reduce the expression of Twist1 and Twist2, alone and in combination. We assessed expression of each Twist with each knockdown condition and found selective downregulation of the targeted Twist (Supplemental Fig 1A), such that Twist1 and Twist2 did not co-regulate the expression of the other. We further confirmed knockdown of Twist1 and Twist2 proteins (Supplemental Fig 1B). Knockdown of Twist1 and Twist2, alone and in combination, led to increased secretion of the pro-inflammatory cytokines TNF, IL6, IL8, IL12p40 (Fig 1A-B) and IL-1β (Supplemental Fig 1C), as well as the anti-inflammatory mediators IL-10 (Fig 1A-B) and IL-1Ra (Supplemental Fig 1C), upon acute NOD2 stimulation of human MDMs. We next evaluated the role of Twist1 and Twist2 in the cytokine downregulation observed after chronic NOD2 stimulation. We previously found that stimulation of NOD2 for 48h is optimal for downregulating cytokine secretion upon re-stimulation through NOD2 or other PRRs(4). We used 100μg/ml MDP as this concentration approximates muramic acid stool levels(24), has been utilized by us and others, and results in optimal cytokine downregulation after chronic NOD2 stimulation(3-5). Upon knockdown of Twist1 or Twist2 after initiation of chronic NOD2 stimulation, and then re-stimulating through NOD2 (Fig 1A), there was significant reversal in the downregulation of both pro-inflammatory cytokines and anti-inflammatory cytokines (Fig 1B). Reversal was further enhanced when Twist1 and Twist2 were knocked-down in combination (Fig 1B). Taken together, Twist1 and Twist2 cooperate to downregulate cytokines upon both acute and chronic NOD2 stimulation of human MDMs.
Figure 1. Twist1 and Twist2 are required for optimal cytokine downregulation upon chronic NOD2 stimulation.
(A) Timeline schematic for MDP pre-treatment of MDMs, Twist1/2 knockdown and subsequent acute MDP treatment. (B) Human MDMs were left untreated (for acute) or pre-treated with 100μg/ml MDP for 24h, then transfected with scrambled, Twist1 or Twist2 siRNA alone or in combination, and 24h later (total 48h MDP pre-treatment), MDMs were treated with 100μg/ml MDP for an additional 24h (acute). Supernatants were examined for cytokines. Mean + SEM for n=4. Similar results were observed for combined Twist1 and Twist2 siRNA for an additional n=8. Numbers on the bars are the ratios of cytokine secretion upon MDP treatment of pre-treated versus non-pretreated MDMs. Statistical significance above the knockdown sample bars is compared to its corresponding scrambled siRNA sample. *, p<0.05; **, p<0.01; ***, p<0.001; †, p<1×10−4. Tx, treatment; scr, scrambled.
Expression of Twist1 and Twist2 increases with chronic NOD2 stimulation in human MDMs in an IL-10- and TGFβ-dependent manner and fails to increase in Crohn's disease NOD2 risk carriers
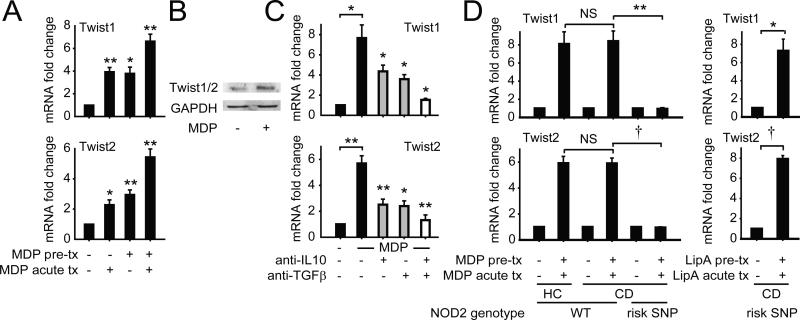
Given the role for Twist1 and Twist2 in downregulating cytokines upon chronic NOD2 stimulation, we questioned if Twist expression is upregulated with chronic NOD2 stimulation of human MDMs. Twist1 and Twist2 mRNA expression was enhanced in 48h MDP pre-treated MDMs and further increased 4h after re-stimulation (Fig 2A). Twist1/2 protein expression was similarly increased after re-stimulation of 48h MDP pre-treated MDMs (Fig 2B). We next sought to define the mechanisms upregulating Twist1 and Twist2 expression upon chronic NOD2 stimulation in human MDMs. We have previously shown that the early secretion of IL-10 and TGFβ is required for subsequent pro- and anti-inflammatory cytokine downregulation during chronic NOD2 stimulation (5). Furthermore, recent studies have demonstrated a necessity for IL-10R on intestinal macrophages for the generation of anti-inflammatory macrophages (25, 26), thereby highlighting the importance of IL-10 in conditioning the inhibitory mechanisms generated in intestinal macrophages. We found that autocrine IL-10 and TGFβ were each required, and moreover cooperated with each other, for optimal upregulation in Twist1 and Twist2 expression upon chronic NOD2 stimulation in human MDMs (Fig 2C). Finally, we questioned if MDMs from Crohn's disease patients similarly upregulate Twist1 and Twist2 upon chronic NOD2 stimulation (see Supplemental Fig 1E for patient characteristics). Twist1 and Twist2 expression in MDMs from wild-type NOD2 Crohn's disease patients was induced to a similar degree as in MDMs from healthy controls (Fig 2D). Furthermore, the range of Twist1 and Twist2 expression upon chronic NOD2 stimulation in these individuals was not significantly different (Supplemental Fig 1D). In contrast, Twist1 and Twist2 expression failed to increase after chronic NOD2 stimulation of MDMs from Crohn's disease loss-of-function NOD2 risk carriers (Fig 2D). MDMs from these NOD2 risk carriers were responsive to stimulation through alternative PRRs, such that Twist1 and Twist2 expression increased in these MDMs upon chronic lipid A treatment (Fig 2D). Therefore, Twist1 and Twist2 increase in an IL-10- and TGF-β-dependent manner upon chronic NOD2 stimulation.
Figure 2. Twist1 and Twist2 expression is increased in MDMs after chronic NOD2 stimulation in an IL-10- and TGFβ-dependent manner whereas this increase is impaired in MDMs from Crohn's disease NOD2 risk carriers.
(A) MDMs (n=4) were left untreated or pre-treated with 100μg/ml MDP for 48h, then stimulated with 100μg/ml MDP for 4h (acute). Fold mRNA expression of Twist1 and Twist2 normalized to untreated cells + SEM. (B) MDMs were left untreated or pre-treated with 100 μg/ml MDP for 48h, then treated with 100μg/ml MDP for 8h and assessed for Twist1/2 expression by Western blot. Representative Western blot from 1 of 3 individuals. GAPDH was used as a loading control. (C) MDMs were cultured with isotype control, neutralizing TGFβ (25μg/ml) or neutralizing IL-10 (5μg/ml) antibodies, alone or in combination, for 1h, then left untreated (for acute) or pre-treated with 100μg/ml MDP for 48h, and then treated with 100μg/ml MDP for an additional 4h (acute). Fold mRNA induction of Twist1 and Twist2 normalized to untreated cells (n=4) + SEM. Statistical significance above the neutralizing antibody sample bars is compared to its corresponding isotype treated sample. (D) MDMs from WT NOD2 healthy controls (WT HC, n=8), WT NOD2 Crohn's disease patients (WT CD, n=8) or Crohn's disease NOD2 risk carriers (Risk SNP, n=3; as per Supplemental Fig 1E) were left untreated or pre-treated with 100μg/ml MDP for 48h, then stimulated with 100μg/ml MDP for 4h (acute). Included is pre-treatment and acute stimulation with 0.1μg/ml lipid A to ensure that MDMs from NOD2 risk patients are responsive to other stimuli. Fold Twist1 and Twist2 mRNA expression is represented by normalizing treated samples to untreated samples + SEM. *, p<0.05; **, p<0.01; †, p<1×10−4. Tx, treatment.
Twist1 and Twist2 cooperate with each other for the enhanced binding to cytokine promoters observed after chronic NOD2 stimulation
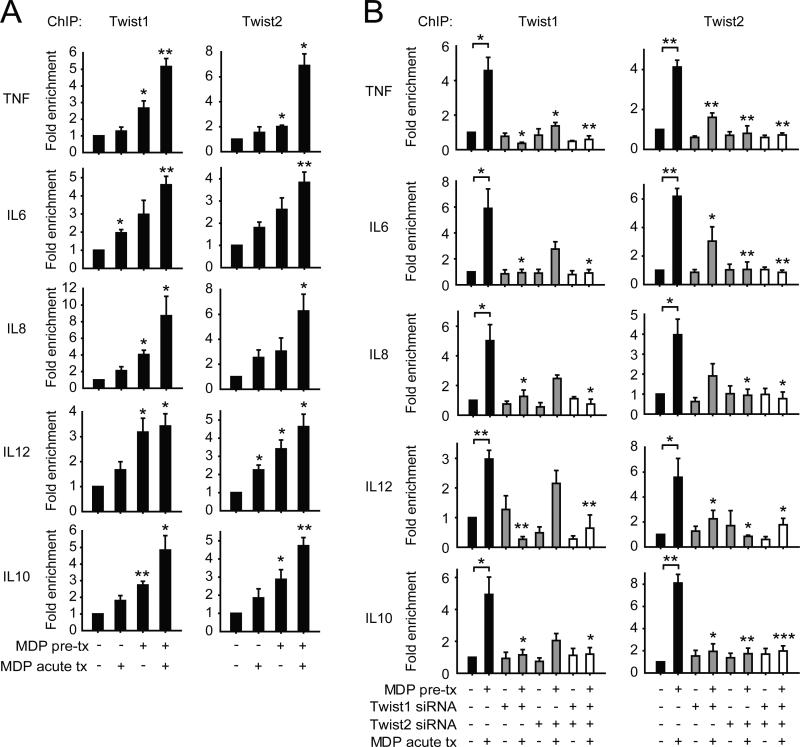
Twist1 and Twist2 transcriptional repressors were upregulated with chronic NOD2 stimulation (Fig 2) and were required for optimal cytokine downregulation after chronic NOD2 stimulation (Fig 1). We therefore questioned if the binding of these transcriptional repressors to cytokine promoters was enhanced after chronic NOD2 stimulation, and if so, if Twist1 and Twist2 cooperated with each other for this enhanced binding. Twist1 and Twist2 binding to both pro-inflammatory and anti-inflammatory cytokine promoters increased particularly 48h after NOD2 stimulation and even more so upon restimulation of MDP-pretreated MDMs (Fig 3A). Upon examining restimulated, MDP-pretreated MDMs where Twist1 and Twist2 binding to cytokine promoters was most enriched, we observed that knockdown of each Twist affected binding not only of itself, but also of the other Twist protein to cytokine promoters (Fig 3B). Taken together, chronic NOD2 stimulation results in significantly enriched and cooperative binding of the transcriptional repressors Twist1 and Twist2 to pro-inflammatory and anti-inflammatory cytokine promoters.
Figure 3. Twist1 and Twist2 cooperate with each other for enhanced binding to cytokine promoters after chronic NOD2 stimulation.
(A) MDMs (n=4) were left untreated or pre-treated with 100μg/ml MDP for 48h, and then treated with 100μg/ml MDP for an additional 4h (acute). (B) MDMs (n=4) were left untreated (for acute) or pre-treated with 100μg/ml MDP for 24h, then transfected with scrambled, Twist1 or Twist2 siRNA alone or in combination, and 24h later (total 48h MDP pre-treatment) MDMs were treated with 100μg/ml MDP for an additional 4h (acute). (A-B) Recruitment of Twist1 and Twist2 to cytokine gene promoters was assessed by ChIP. Fold enrichment normalized to untreated, scrambled siRNA-transfected cells + SEM. (B) Statistical significance above the knockdown sample bars is compared to its corresponding scrambled siRNA sample. *, p<0.05; **, p<0.01; ***, p<0.001. Tx, treatment.
Twist1- and Twist2-dependent upregulation of c-Maf and Bmi1 is critical for cytokine downregulation after chronic NOD2 stimulation
In addition to their direct suppressor activity, transcriptional repressors, including the Twist proteins, can both increase the expression of additional inhibitory molecules and suppress the expression of essential activating molecules; in combination these enable optimal suppressive outcomes (12). We therefore first investigated if Twist1 and Twist2 might be contributing to cytokine downregulation after chronic NOD2 stimulation through upregulating inhibitory molecules previously identified to play a role in NOD2-induced cytokine downregulation. However, Twist1 and Twist2 knockdown, either alone or in combination, did not impair the upregulation of NFκB1, ATF3, IRAK-M, or Tollip after chronic NOD2 stimulation (Supplemental Fig 2A). We further examined proximal signaling pathways that are induced upon acute NOD2 stimulation, and found that combined Twist1 and Twist2 knockdown did not regulate the acute NOD2-induced activation of the ERK, p38 or JNK pathways (Supplemental Fig 2B), or the early secretion of IL-1β (Supplemental Fig 2C) which we have found to be a measure of NOD2-induced caspase-1 activation (27, 28).
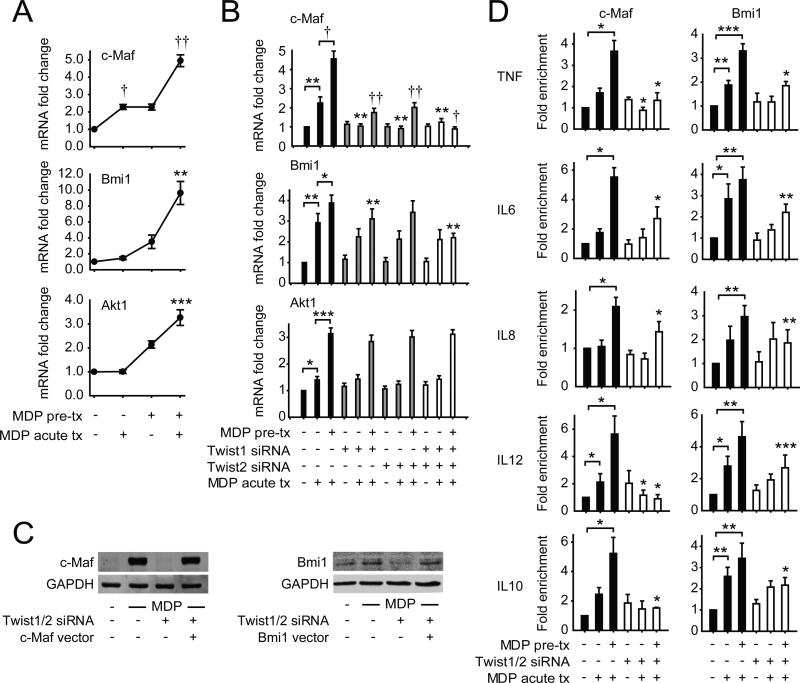
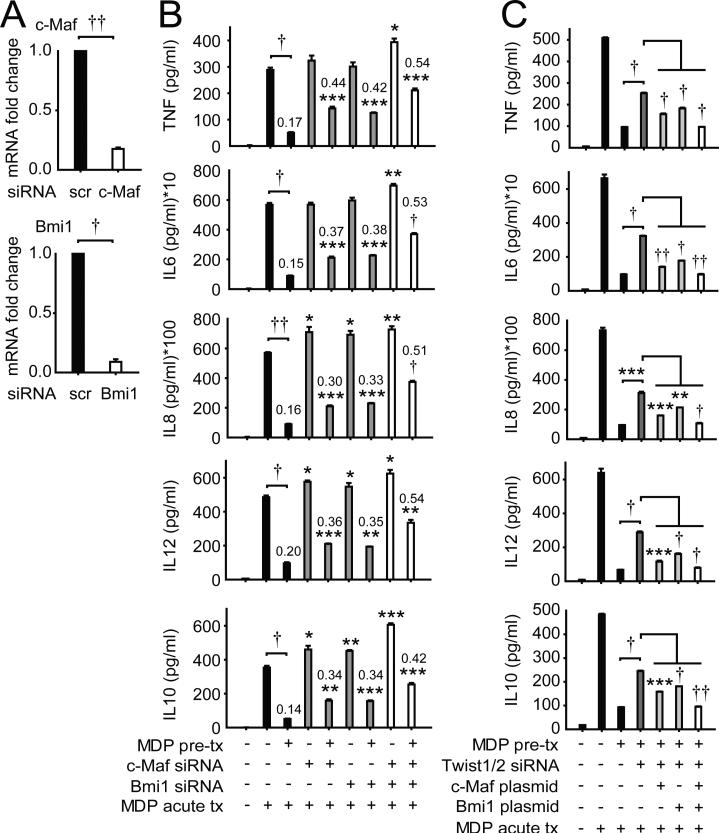
Twist1 and Twist2 have not been well-studied in PRR-induced cytokine regulation. However, they have been well-investigated in carcinogenesis, osteogenesis, and hematopoietic cell development. We therefore questioned if inhibitory molecules upregulated by Twist1 and/or Twist2 identified through these other processes might be similarly Twist-dependent upon chronic NOD2 stimulation in human MDMs, and thereby contribute to the cytokine downregulation observed. During myeloid lineage development, Twist2 can upregulate the pro-inflammatory transcriptional inhibitor c-Maf(14); Twist2 can also bind to the c-Maf promoter with LPS treatment in mouse macrophages (29). However, c-Maf regulation of PRR-initiated pathways has not been well-defined. Twist1 regulates Bmi1 expression and cooperates with Bmi1 in repressor functions during epithelial-mesenchymal transition relevant to cancer metastasis (30); a role for Bmi1 in PRR signaling and outcomes is poorly understood. Further, Twist regulates PI3K pathways (31), and Akt1 can contribute to an ‘anti-inflammatory’ macrophage phenotype (32). We found that c-Maf, Bmi1 and Akt1 were upregulated with chronic NOD2 stimulation (Fig 4A). However, only c-Maf and Bmi1 transcript upregulation was Twist1- and Twist2-dependent (Fig 4B); c-Maf and Bmi1 protein upregulation was similarly Twist-dependent (Fig 4C). Furthermore, the binding of c-Maf and Bmi1 to both pro-inflammatory and anti-inflammatory cytokine promoters was enriched with chronic NOD2 stimulation (Fig 4D); this enhanced promoter binding was Twist1- and Twist2-dependent (Fig 4D). Importantly, through c-Maf and Bmi1 knockdown (Fig 5A), we established that c-Maf and Bmi1 each contributed to the cytokine downregulation observed after chronic NOD2 stimulation (Fig 5B). Moreover, they cooperated with each other for optimal cytokine downregulation (Fig 5B). Finally, to clearly establish the role of c-Maf and Bmi1 in Twist-dependent contributions to chronic NOD2-induced cytokine downregulation in MDMs, we restored both c-Maf and Bmi1 expression in Twist-deficient MDMs to levels similar to those observed after chronic NOD2 stimulation (Fig 4C). Despite the absence of Twist1 and Twist2, reconstitution of each c-Maf and Bmi1 could partially restore the cytokine downregulation observed after chronic NOD2 stimulation (Fig 5C). Moreover, the restoration was further improved with combined expression of both c-Maf and Bmi1 (Fig 5C). Therefore, one mechanism through which Twist1 and Twist2 downregulate cytokines after chronic NOD2 stimulation is through the upregulation in expression and cytokine promoter binding of the transcriptional repressors c-Maf and Bmi1.
Figure 4. Upon chronic NOD2 stimulation, expression and cytokine promoter binding of the anti-inflammatory transcription factors c-Maf and Bmi1 are upregulated in a Twist-dependent manner.
(A) MDMs (n=8) were left untreated or pre-treated with 100μg/ml MDP for 48h, and then treated with 100μg/ml MDP for 4h (acute). Fold increase in c-Maf, Bmi1 and Akt1 mRNA was normalized to untreated MDMs + SEM. (B) MDMs (n=8) were left untreated (for acute) or pre-treated with 100μg/ml MDP for 24h, then transfected with scrambled, Twist1 or Twist2 siRNA, alone or in combination, 24h later (total 48h after MDP pre-treatment) MDMs were treated with 100μg/ml MDP for 4h (acute) and assessed for c-Maf, Bmi1 and Akt1 mRNA expression. Fold mRNA expression normalized to untreated, scrambled siRNA-transfected MDMs + SEM. (C) MDMs were left untreated or pre-treated with 100μg/ml MDP for 24h, then transfected with scrambled, or a combination of Twist1 and Twist2 siRNA ± vectors expressing c-Maf or Bmi1, and 24h later (total 48h MDP pre-treatment), MDMs were treated with 100μg/ml MDP for an additional 8h (acute) and c-Maf or Bmi1 expression was assessed by Western blot. GAPDH was assessed as a loading control. (D) MDMs were left untreated (for acute) or pre-treated with 100μg/ml MDP for 24h, then transfected with scrambled or a combination of Twist1 and Twist2 siRNA, and 24h later (total 48h after MDP pre-treatment) MDMs were treated with 100μg/ml MDP for an additional 4h (acute). Recruitment of c-Maf (n=4) and Bmi1 (n=5) to cytokine gene promoters was assessed by ChIP. Fold enrichment normalized to untreated, scrambled siRNA-transfected MDMs + SEM. (B-C) Statistical significance above the knockdown sample bars is compared to its corresponding scrambled siRNA sample. *, p<0.05; **, p<0.01; ***, p<0.001; †, p<1×10−4; ††, p<1×10−5. Tx, treatment.
Figure 5. Upregulated Bmi1 and c-Maf cooperate to mediate Twist-dependent cytokine downregulation upon chronic NOD2 stimulation of human MDMs.
(A) MDMs (n=4) were transfected with scrambled, Bmi1 or c-Maf siRNA for 24h. Fold mRNA expression normalized to scrambled siRNA-transfected MDMs + SEM. (B) MDMs (n=4) were left untreated (for acute) or pre-treated with 100μg/ml MDP for 24h, then transfected with scrambled, c-Maf or Bmi1 siRNA, alone or in combination, and 24h later (total 48h MDP pre-treatment), MDMs were stimulated with 100μg/ml MDP for 24h (acute) and cytokine secretion was assessed + SEM. Numbers on the bars are the ratios of cytokine secretion upon MDP treatment of the corresponding pre-treated versus non-pretreated MDMs. Statistical significance above the knockdown sample bars is compared to its corresponding scrambled siRNA sample. (C) MDMs were left untreated (for acute) or pre-treated with 100μg/ml MDP for 24h, then transfected with scrambled, or a combination of Twist1 and Twist2 siRNA ± vectors expressing c-Maf or Bmi1 (or empty vector control), and 24h later (total 48h MDP pre-treatment), MDMs were treated with 100μg/ml MDP for an additional 24h (acute) and supernatants were examined for cytokines. Mean concentration + SEM for n=4. Numbers on the bars are the ratios of cytokine secretion upon MDP treatment of pre-treated versus non-pretreated MDMs. *, p<0.01; **, p<0.01; ***, p<0.001; †, p<1×10−4; ††, p<1×10−5. Tx, treatment; scr, scrambled.
Twist1 and Twist2 downregulate the expression and binding to cytokine promoters of the transcriptional activators ATF4, C/EBPα, Runx1, and Runx2
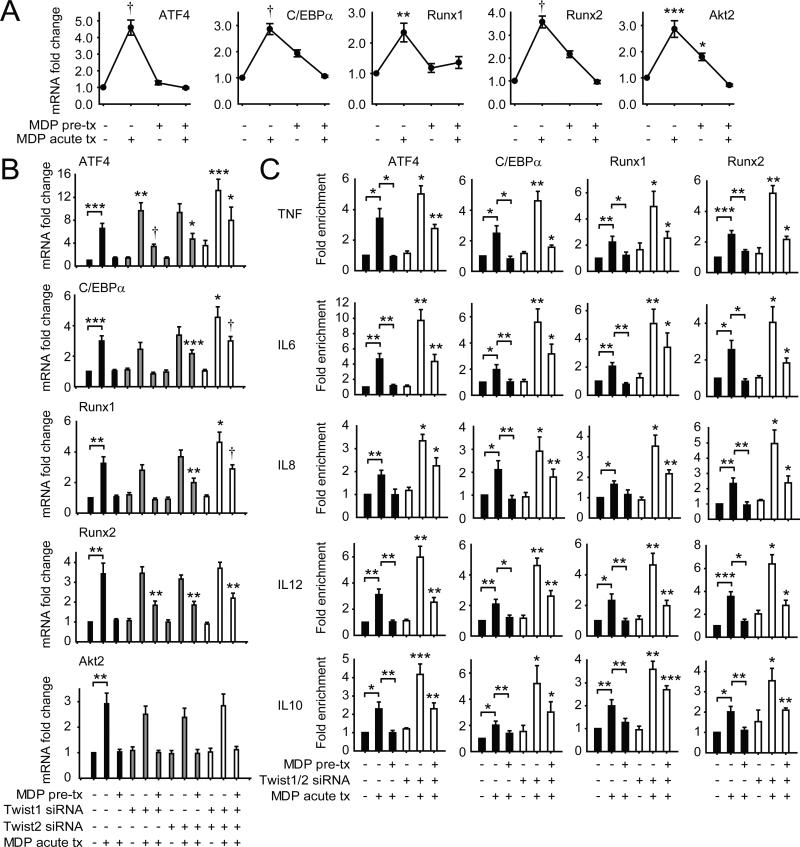
Twist1 and Twist2 can both decrease the expression and compete with promoter binding of activating transcription factors in osteogenesis, carcinogenesis and myeloid differentiation (12). We therefore hypothesized that Twist1 and Twist2 might be similarly downregulating activating transcription factors upon chronic NOD2 stimulation where cytokines are dramatically reduced. How activating transcription factors are regulated with chronic NOD2 stimulation has not been well-defined. We therefore considered the Twist-regulated activating transcription factors defined in other cellular processes. Twist1 can attenuate ATF4 binding to the osteocalcin promoter in osteoclasts (33). Furthermore, Twist1 and Twist2 can inhibit Runx2 transactivation functions during osteoblast differentiation (34). Moreover, Twist2 can negatively regulate myeloid lineage differentiation by interacting with and inhibiting the transcription factors Runx1 and C/EBPα (14). Twist proteins can also regulate Akt2 expression in breast cancer cells (35). To our knowledge, roles for ATF4, C/EBPα, Runx1, and Runx2 in NOD2-induced outcomes have not been reported. We therefore first examined the regulation of each of these transcription factors with acute and chronic NOD2 stimulation of MDMs.
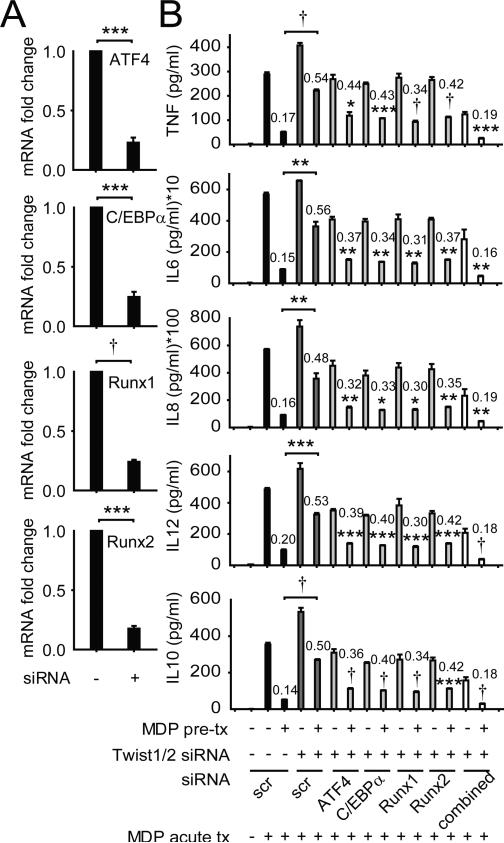
With NOD2 stimulation of MDMs, ATF4, C/EBPα, Runx1, Runx2 and Akt2 were each upregulated acutely, but downregulated by 48h, and they failed to increase on restimulation of NOD2 (Fig 6A), consistent with the attenuated cytokines at this time. We further assessed the Twist-dependency for these downregulated transcription factors, and found that ATF4, C/EBPα, Runx1, and Runx2 downregulation was dependent on the combined expression of Twist1 and Twist2 (Fig 6B). Akt2 downregulation after chronic NOD2 stimulation, in contrast, was Twist-independent (Fig 6B). We therefore pursued the role and regulation of the Twist-dependent transcription factors. ATF4, C/EBPα, Runx1 and Runx2 bound to cytokine promoters with acute NOD2 stimulation, and this binding was dramatically reduced after chronic NOD2 stimulation, consistent with the reduced cytokines observed under these conditions (Fig 6C). Moreover, upon Twist1 and Twist2 knockdown during chronic NOD2 stimulation, the binding of these transcription factors to cytokine promoters failed to undergo reduction relative to scrambled knockdown cells, consistent with the impaired cytokine downregulation (Fig 6C). Finally, to ensure that the relative increase in expression and binding of ATF4, C/EBPα, Runx1 and Runx2 to cytokine promoters in Twist-deficient MDMs was contributing to the failure to downregulate cytokines upon chronic NOD2 stimulation, we utilized siRNA to each of the transcription factors (Fig 7A), alone and in combination. Reduced expression of each ATF4, C/EBPα, Runx1 and Runx2 partially restored cytokine downregulation, and combined knockdown of these Twist-regulated transcription factors fully restored cytokine downregulation in Twist-deficient macrophages after chronic NOD2 stimulation (Fig 7B). Cells were viable under these conditions (data not shown). Therefore, Twist1 and Twist2 contribute to the downregulation in expression and promoter binding of various transcription factors, in particular ATF4, C/EBPα, Runx1 and Runx2, which in turn is necessary for optimal cytokine downregulation after chronic NOD2 stimulation in MDMs.
Figure 6. Expression and cytokine promoter binding of the transcriptional activators ATF4, C/EBPα, Runx1 and Runx2 is reduced in a Twist-dependent manner during chronic NOD2 stimulation.
(A) MDMs (n=8) were left untreated or pre-treated with 100μg/ml MDP for 48h, and then MDMs were treated with 100μg/ml MDP for 4h (acute). Fold change in ATF4, C/EBPα, Runx1, Runx2 and Akt2 mRNA normalized to untreated MDMs + SEM. (B) MDMs (n=8) were left untreated (for acute) or pre-treated with 100μg/ml MDP for 24h, then transfected with scrambled, Twist1 or Twist2 siRNA, alone or in combination, and 24h later (total 48h MDP treatment), MDMs were stimulated with 100μg/ml MDP for 4h (acute) and assessed for ATF4, C/EBPα, Runx1, Runx2 and Akt2 mRNA expression. Fold mRNA expression normalized to untreated, scrambled siRNA-transfected MDMs + SEM. (C) MDMs (n=4-6) were left untreated (for acute) or pre-treated with 100μg/ml MDP for 24h, then transfected with scrambled or a combination of Twist1 and Twist2 siRNA, and 24h later (total 48h MDP pre-treatment) MDMs were treated with 100μg/ml MDP for an additional 4h (acute). Recruitment of ATF4, C/EBPα, Runx1 and Runx2 to cytokine gene promoters was assessed by ChIP. Fold enrichment normalized to untreated, scrambled siRNA-transfected cells + SEM. (B-C) Statistical significance above the knockdown sample bars is compared to its corresponding scrambled siRNA sample. *, p<0.01; **, p<0.01; ***, p<0.001; †, p<1×10−4. Tx, treatment.
Figure 7. ATF4, C/EBPα, Runx1, and Runx2 downregulation contributes to Twist-mediated cytokine downregulation after chronic NOD2 stimulation .
(A) MDMs (n=4) were transfected with scrambled, ATF4, C/EBPα, Runx1 or Runx2 siRNA for 24h. Fold mRNA expression normalized to scrambled, siRNA-transfected MDMs + SEM. (B) MDMs (n=4) were left untreated (for acute) or pre-treated with 100μg/ml MDP for 24h, then transfected with scrambled or a combination of Twist1 and Twist2 siRNA + ATF4, C/EBPα, Runx1 or Runx2 siRNA, alone or in combination, and 24h later (total 48h MDP pre-treatment), MDMs were stimulated with 100μg/ml MDP for 24h (acute) and cytokine secretion was assessed. Numbers on the bars are the ratios of cytokine secretion upon MDP treatment of pre-treated versus the corresponding non-pretreated MDMs. Significance comparisons are between MDP treatment of pre-treated, Twist1/2 siRNA-treated cells relative to the same condition in combination with siRNA to each of the transcriptional activators or as indicated. *, p<0.01; **, p<0.01; ***, p<0.001; †, p<1×10−4. Tx, treatment; scr, scrambled.
NOD2 synergizes with TLR2, TLR4 and TLR9 to enhance Twist1 and Twist2 upregulation and induction of Twist-dependent pathways
As intestinal macrophages are exposed to multiple PRRs, we next assessed if chronic stimulation of other PRRs similarly upregulates Twist1 and Twist2 expression in human MDMs, and if these other PRRs cooperate with NOD2 to regulate Twist1- and Twist2-mediated outcomes. Similar to chronic NOD2 stimulation, cytokines are downregulated upon re-stimulation of MDMs after chronic stimulation of TLR2, TLR4 or TLR9 (Supplemental Fig 3A). We found that upon pre-treatment through TLR2, TLR4 or TLR9, and then re-stimulation of each respective TLR, Twist1 and Twist2 expression was increased (Supplemental Fig 3B). The Twist-dependent pathways we had identified were similarly regulated under these conditions, with upregulation of the transcriptional repressors c-Maf and Bmi1 (Supplemental Fig 3C), and downregulation of the transcriptional activators ATF4, C/EBPα, Runx1 and Runx2 (Supplemental Fig 3D). Although low dose MDP treatment synergized with each of the TLR ligands to increase expression of cytokines during acute treatment, cytokines were downregulated upon chronic stimulation of NOD2 with each of the TLRs (Supplemental Fig 3A). Moreover, chronic MDP treatment synergized with each of the TLRs to upregulate Twist1 and Twist2, and the transcriptional repressors c-Maf and Bmi1 (Supplemental Fig 3B-C). Taken together, under the chronic microbial ligand exposure that occurs in the intestinal environment, NOD2 can synergize with other PRRs to upregulate Twist1 and Twist2 expression and Twist-dependent mechanisms that lead to cytokine downregulation.
Discussion
In this study, we demonstrate that Twist1 and Twist2 contribute to the downregulation of pro-inflammatory cytokines upon acute, and particularly chronic NOD2 stimulation of human MDMs. Twist1 and Twist2 expression increased with chronic NOD2 stimulation, consistent with the important role for these transcriptional repressors in the downregulation of cytokines observed after prolonged NOD2 stimulation. Twist upregulation is similarly observed in MDMs from Crohn's disease patients, but is impaired in MDMs from Crohn's disease NOD2 risk carriers. Early secreted IL-10 and TGFβ were critical for Twist1 and Twist2 expression upregulation with chronic NOD2 stimulation. Binding of Twist1 and Twist2 to cytokine promoters was particularly enhanced after chronic NOD2 stimulation and the two proteins cooperated with each other for optimal promoter binding. In addition to the direct binding of the Twist1 and Twist2 transcriptional repressors to cytokine promoters, they also orchestrated a coordinated program in which the expression and cytokine promoter binding of additional transcriptional repressors including c-Maf and Bmi1 was increased, whereas the expression and cytokine promoter binding of various activating transcription factors including ATF4, C/EBPα, Runx1 and Runx2 was reduced. These combined Twist-mediated mechanisms contributed to the ability of Twist1 and Twist2 to downregulate NOD2-induced cytokines (Supplemental Fig 4). Further, NOD2 could synergize with various TLRs to mediate these outcomes, consistent with the cooperative role for NOD2 with other PRRs and cytokines in enhancing downstream immune outcomes (36-42). Therefore, we now elucidate a clear role for Twist1- and Twist2-mediated mechanisms in downregulating cytokines upon acute and chronic NOD2 stimulation of primary human MDMs.
A role for Twist repressors in PRR-induced outcomes has been relatively unexplored. However, Twist1- and Twist2-mediated transcriptional mechanisms identified to regulate suppression in biological processes such osteogenesis, hematopoiesis and carcinogenesis provided the opportunity to now dissect if these pathways are Twist-dependent and similarly or differentially regulate outcomes upon PRR stimulation of human macrophages. A number of the downstream molecules contributing to Twist-dependent regulation in these other cell processes have not been previously implicated in NOD2-induced outcomes. One such example is the role we now identify for Twist-dependent c-Maf regulation in downregulating cytokines after chronic NOD2 stimulation. c-Maf has been predominantly studied in the context of T cell differentiation, in particular Tr1 cells (43). However, a few studies have also identified a role for c-Maf as an anti-inflammatory transcription factor in myeloid cells; c-Maf downregulates transcriptional activation of pro-inflammatory cytokines, while upregulating transcriptional activation of IL-10 in myeloid-derived cells (44, 45). In contrast, we found that in primary human macrophages, c-Maf downregulated both pro- and anti-inflammatory cytokines upon NOD2 stimulation (Fig 5). Studies on Bmi1 have focused on its contributions to carcinogenesis. The role of Bmi1 in regulating PRR-induced outcomes has been poorly defined. To our knowledge, there is only one such study; this study identified a role for Bmi1 in suppressing IL-10 in mouse macrophages (46). We now define a broader role for Bmi1 in suppressing a range of both pro- and anti-inflammatory cytokines upon PRR stimulation, with a Twist-dependent role for Bmi1 in the robust downregulation of cytokines after chronic PRR stimulation (Fig 5). Although neither Akt1 nor Akt2 was regulated by Twist proteins, we observed that Akt1 was upregulated (Fig 4A), whereas Akt2 was downregulated (Fig 6A) with chronic NOD2 stimulation. This pattern of expression regulation is consistent with the role of Akt1 in polarization to ‘anti-inflammatory’ macrophages and of Akt2 in polarization to ‘pro-inflammatory’ macrophages (32).
We identified a number of transcription factors, including ATF4, C/EBPα, Runx1 and Runx2 whose expression and binding to cytokine promoters was upregulated with acute NOD2 stimulation, but downregulated in a Twist-dependent manner with chronic NOD2 stimulation. Restoring the downregulation of these transcription factors in Twist-deficient MDMs was able to restore the cytokine downregulation observed with chronic NOD2 stimulation. Runx1 and Runx2 have been well-studied in cell proliferation and differentiation (47), and C/EBPα has been identified as a lineage-specific transcription factor in the hematopoietic system (48). However, the role of these transcription factors in PRR-induced outcomes has been poorly defined. We now identify each of these transcription factors as being essential in the induction of cytokines after acute NOD2 stimulation; their downregulation after chronic NOD2 stimulation contributes to the overall cytokine downregulation under these conditions. Limited studies have examined the role of ATF4 downstream of PRR. ATF4 was found to contribute to cytokine secretion upon acute TLR4 stimulation (49). In contrast, upon pre-stimulation of macrophages through TLR4, TRIF-dependent signaling led to the downregulation of the ATF4-CHOP branch of the unfolded protein response (50). We similarly observed differential regulation of ATF4 with acute and chronic NOD2 stimulation. Taken together, these studies elucidate an important role for Twist1 and Twist2 in regulating NOD2- and PRR-induced cytokine secretion under both the initial acute stimulation as well as under the chronic stimulatory conditions that are present in the intestinal environment, thereby providing insight into the inhibitory mechanisms critical for regulating the Crohn's disease associated NOD2.
Supplementary Material
Footnotes
The authors have no financial conflicts of interest
References
- 1.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng BS, He SH, Zheng PY, Wu L, Yang PC. Mast cells play a crucial role in Staphylococcus aureus peptidoglycan-induced diarrhea. Am J Pathol. 2007;171:537–547. doi: 10.2353/ajpath.2007.061274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng S, Abraham C. NF-kappaB1 inhibits NOD2-induced cytokine secretion through ATF3-dependent mechanisms. Mol Cell Biol. 2013;33:4857–4871. doi: 10.1128/MCB.00797-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hedl M, Li J, Cho JH, Abraham C. Chronic stimulation of Nod2 mediates tolerance to bacterial products. Proc Natl Acad Sci U S A. 2007;104:19440–19445. doi: 10.1073/pnas.0706097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hedl M, Abraham C. Secretory Mediators Regulate Nod2-Mediated Tolerance in Human Macrophages. Gastroenterology. 2011;140:231–241. doi: 10.1053/j.gastro.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe T, Asano N, Murray PJ, Ozato K, Tailor P, Fuss IJ, Kitani A, Strober W. Muramyl dipeptide activation of nucleotide-binding oligomerization domain 2 protects mice from experimental colitis. J Clin Invest. 2008;118:545–559. doi: 10.1172/JCI33145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kullberg BJ, Ferwerda G, de Jong DJ, Drenth JP, Joosten LA, Van der Meer JW, Netea MG. Crohn's disease patients homozygous for the 3020insC NOD2 mutation have a defective NOD2/TLR4 cross-tolerance to intestinal stimuli. Immunology. 2008;123:600–605. doi: 10.1111/j.1365-2567.2007.02735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, Benjamin WH, Orenstein JM, Smith PD. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 2005;115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe T, Asano N, Meng G, Yamashita K, Arai Y, Sakurai T, Kudo M, Fuss IJ, Kitani A, Shimosegawa T, et al. NOD2 downregulates colonic inflammation by IRF4-mediated inhibition of K63-linked polyubiquitination of RICK and TRAF6. Mucosal Immunol. 2014 doi: 10.1038/mi.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng S, Hedl M, Abraham C. TAM Receptor-Dependent Regulation of SOCS3 and MAPKs Contributes to Proinflammatory Cytokine Downregulation following Chronic NOD2 Stimulation of Human Macrophages. J Immunol. 2015;194:1928–1937. doi: 10.4049/jimmunol.1401933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park SH, Park-Min KH, Chen J, Hu X, Ivashkiv LB. Tumor necrosis factor induces GSK3 kinase-mediated cross-tolerance to endotoxin in macrophages. Nat Immunol. 2011;12:607–615. doi: 10.1038/ni.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin Q, Xu Y, He T, Qin C, Xu J. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res. 2012;22:90–106. doi: 10.1038/cr.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miraoui H, Marie PJ. Pivotal role of Twist in skeletal biology and pathology. Gene. 2010;468:1–7. doi: 10.1016/j.gene.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 14.Sharabi AB, Aldrich M, Sosic D, Olson EN, Friedman AD, Lee SH, Chen SY. Twist-2 controls myeloid lineage development and function. PLoS Biol. 2008;6:e316. doi: 10.1371/journal.pbio.0060316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen ZF, Behringer RR. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- 16.el Ghouzzi V, Le Merrer M, Perrin-Schmitt F, Lajeunie E, Benit P, Renier D, Bourgeois P, Bolcato-Bellemin AL, Munnich A, Bonaventure J. Mutations of the TWIST gene in the Saethre-Chotzen syndrome. Nat Genet. 1997;15:42–46. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- 17.Howard TD, Paznekas WA, Green ED, Chiang LC, Ma N, Ortiz de Luna RI, Garcia Delgado C, Gonzalez-Ramos M, Kline AD, Jabs EW. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat Genet. 1997;15:36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- 18.Sosic D, Richardson JA, Yu K, Ornitz DM, Olson EN. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 2003;112:169–180. doi: 10.1016/s0092-8674(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 19.Niesner U, Albrecht I, Janke M, Doebis C, Loddenkemper C, Lexberg MH, Eulenburg K, Kreher S, Koeck J, Baumgrass R, et al. Autoregulation of Th1-mediated inflammation by twist1. J Exp Med. 2008;205:1889–1901. doi: 10.1084/jem.20072468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pham D, Vincentz JW, Firulli AB, Kaplan MH. Twist1 regulates Ifng expression in Th1 cells by interfering with Runx3 function. J Immunol. 2012;189:832–840. doi: 10.4049/jimmunol.1200854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pham D, Walline CC, Hollister K, Dent AL, Blum JS, Firulli AB, Kaplan MH. The transcription factor Twist1 limits T helper 17 and T follicular helper cell development by repressing the gene encoding the interleukin-6 receptor alpha chain. J Biol Chem. 2013;288:27423–27433. doi: 10.1074/jbc.M113.497248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharif MN, Sosic D, Rothlin CV, Kelly E, Lemke G, Olson EN, Ivashkiv LB. Twist mediates suppression of inflammation by type I IFNs and Axl. J Exp Med. 2006;203:1891–1901. doi: 10.1084/jem.20051725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu CR, Lee S, Ho C, Bommi P, Huang SA, Cheung ST, Dimri GP, Chen X. Bmi1 functions as an oncogene independent of Ink4A/Arf repression in hepatic carcinogenesis. Mol Cancer Res. 2009;7:1937–1945. doi: 10.1158/1541-7786.MCR-09-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vavricka SR, Musch MW, Chang JE, Nakagawa Y, Phanvijhitsiri K, Waypa TS, Merlin D, Schneewind O, Chang EB. hPepT1 transports muramyl dipeptide, activating NF-kappaB and stimulating IL-8 secretion in human colonic Caco2/bbe cells. Gastroenterology. 2004;127:1401–1409. doi: 10.1053/j.gastro.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Shouval DS, Biswas A, Goettel JA, McCann K, Conaway E, Redhu NS, Mascanfroni ID, Al Adham Z, Lavoie S, Ibourk M, et al. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity. 2014;40:706–719. doi: 10.1016/j.immuni.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zigmond E, Bernshtein B, Friedlander G, Walker CR, Yona S, Kim KW, Brenner O, Krauthgamer R, Varol C, Muller W, et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity. 2014;40:720–733. doi: 10.1016/j.immuni.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Hedl M, Abraham C. Distinct Roles for Nod2 Protein and Autocrine Interleukin-1{beta} in Muramyl Dipeptide-induced Mitogen-activated Protein Kinase Activation and Cytokine Secretion in Human Macrophages. J Biol Chem. 2011;286:26440–26449. doi: 10.1074/jbc.M111.237495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hedl M, Abraham C. A TNFSF15 disease-risk polymorphism increases pattern-recognition receptor-induced signaling through caspase-8-induced IL-1. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1404178111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Bosch MW, Palsson-Mcdermott E, Johnson DS, O'Neill LA. LPS induces the degradation of PDCD4 to release Twist2, activating c-Maf transcription to promote IL-10 production. J Biol Chem. 2014 doi: 10.1074/jbc.M114.573089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY, Yang WH, Huang CH, Kao SY, Tzeng CH, Tai SK, et al. Bmi1 is essential in Twist1-induced epithelialmesenchymal transition. Nat Cell Biol. 2010;12:982–992. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- 31.Li J, Zhou BP. Activation of β-catenin and Akt pathways by Twist are critical for maintenance of EMT associated cancer stem cell-like characters. BMC Cancer. 2010;11:49. doi: 10.1186/1471-2407-11-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arranz A, Doxaki C, Vergadi E, Martinez de la Torre Y, Vaporidi K, Lagoudaki ED, Ieronymaki E, Androulidaki A, Venihaki M, Margioris AN, et al. Akt1 and Akt2 protein kinases differentially contribute to macrophage polarization. Proc Natl Acad Sci U S A. 2012;109:9517–9522. doi: 10.1073/pnas.1119038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danciu TE, Li Y, Koh A, Xiao G, McCauley LK, Franceschi RT. The basic helix loop helix transcription factor Twist1 is a novel regulator of ATF4 in osteoblasts. J Cell Biochem. 2012;113:70–79. doi: 10.1002/jcb.23329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, et al. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- 35.Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD, Wang LH. Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res. 2007;67:1979–1987. doi: 10.1158/0008-5472.CAN-06-1479. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Moran T, Swanson E, Julian C, Harris J, Bonen DK, Hedl M, Nicolae DL, Abraham C, Cho JH. Regulation of IL-8 and IL-1beta expression in Crohn's disease associated NOD2/CARD15 mutations. Hum Mol Genet. 2004;13:1715–1725. doi: 10.1093/hmg/ddh182. [DOI] [PubMed] [Google Scholar]
- 37.van Heel DA, Ghosh S, Hunt KA, Mathew CG, Forbes A, Jewell DP, Playford RJ. Synergy between TLR9 and NOD2 innate immune responses is lost in genetic Crohn's disease. Gut. 2005;54:1553–1557. doi: 10.1136/gut.2005.065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Netea MG, Azam T, Ferwerda G, Girardin SE, Walsh M, Park JS, Abraham E, Kim JM, Yoon DY, Dinarello CA, et al. IL-32 synergizes with nucleotide oligomerization domain (NOD) 1 and NOD2 ligands for IL-1{beta} and IL-6 production through a caspase 1-dependent mechanism. Proc Natl Acad Sci U S A. 2005;102:16309–16314. doi: 10.1073/pnas.0508237102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fritz JH, Girardin SE, Fitting C, Werts C, Mengin-Lecreulx D, Caroff M, Cavaillon JM, Philpott DJ, Adib-Conquy M. Synergistic stimulation of human monocytes and dendritic cells by Toll-like receptor 4 and NOD1- and NOD2-activating agonists. Eur J Immunol. 2005:2459–2470. doi: 10.1002/eji.200526286. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 41.Wu X, Lahiri A, Haines GK, 3rd, Flavell RA, Abraham C. NOD2 Regulates CXCR3-Dependent CD8+ T Cell Accumulation in Intestinal Tissues with Acute Injury. J Immunol. 2014;192:3409–3418. doi: 10.4049/jimmunol.1302436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lahiri A, Abraham C. Metallothionein-mediated zinc accumulation enhances autophagy and bacterial clearance upon innate receptor stimulation. Gastroenterology. 2014;147:835–846. doi: 10.1053/j.gastro.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pot C, Apetoh L, Kuchroo VK. Type 1 regulatory T cells (Tr1) in autoimmunity. Semin Immunol. 2011;23:202–208. doi: 10.1016/j.smim.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao S, Liu J, Chesi M, Bergsagel PL, Ho IC, Donnelly RP, Ma X. Differential regulation of IL-12 and IL-10 gene expression in macrophages by the basic leucine zipper transcription factor c-Maf fibrosarcoma. J Immunol. 2002;169:5715–5725. doi: 10.4049/jimmunol.169.10.5715. [DOI] [PubMed] [Google Scholar]
- 45.Cao S, Liu J, Song L, Ma X. The protooncogene c-Maf is an essential transcription factor for IL-10 gene expression in macrophages. J Immunol. 2005;174:3484–3492. doi: 10.4049/jimmunol.174.6.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sienerth AR, Scheuermann C, Galmiche A, Rapp UR, Becker M. Polycomb group protein Bmi1 negatively regulates IL-10 expression in activated macrophages. Immunol Cell Biol. 2011;89:812–816. doi: 10.1038/icb.2010.160. [DOI] [PubMed] [Google Scholar]
- 47.Chuang LS, Ito K, Ito Y. RUNX family: Regulation and diversification of roles through interacting proteins. Int J Cancer. 2013;132:1260–1271. doi: 10.1002/ijc.27964. [DOI] [PubMed] [Google Scholar]
- 48.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 49.Zhang C, Bai N, Chang A, Zhang Z, Yin J, Shen W, Tian Y, Xiang R, Liu C. ATF4 is directly recruited by TLR4 signaling and positively regulates TLR4-trigged cytokine production in human monocytes. Cell Mol Immunol. 2013;10:84–94. doi: 10.1038/cmi.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woo CW, Cui D, Arellano J, Dorweiler B, Harding H, Fitzgerald KA, Ron D, Tabas I. Adaptive suppression of the ATF4-CHOP branch of the unfolded protein response by toll-like receptor signalling. Nat Cell Biol. 2009;11:1473–1480. doi: 10.1038/ncb1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.