Abstract
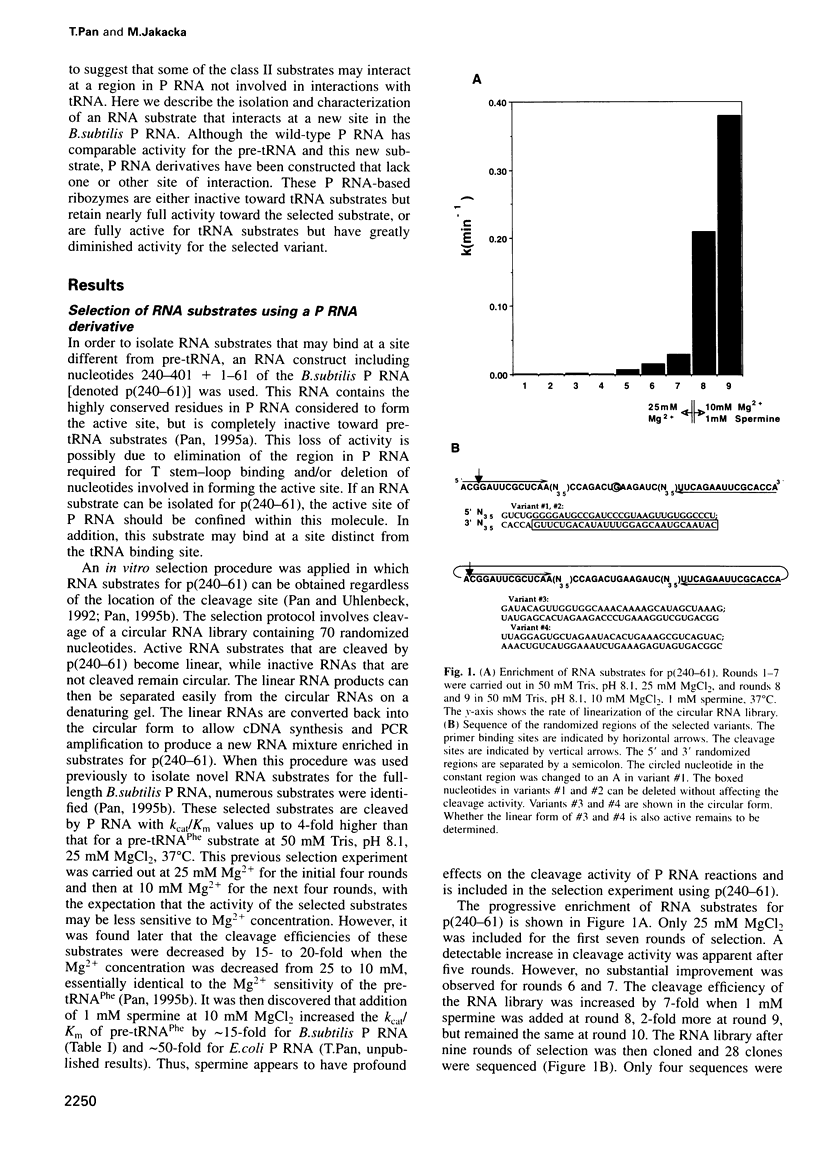
The ribozyme from Bacillus subtilis RNase P (P RNA) recognizes an RNA structure consisting of the acceptor stem and the T stem-loop of tRNA substrates. An in vitro selection experiment was carried out to obtain potential RNA substrates that may interact with the P RNA differently from the tRNA substrate. Using a P RNA-derived ribozyme that contains most, if not all, of the structural elements thought to be involved in active site formation of P RNA, but lacks the putative binding site for the T stem-loop of tRNA, a single RNA substrate was isolated after nine rounds of selection. This RNA is a competent substrate for the ribozyme used in selection as well as for the full-length P RNA. Biochemical characterization shows that this selected substrate interacts at a different site compared with the tRNA substrate. The selection experiment also identified a self-cleaving RNA seemingly different from other known ribozymes. These results indicate that a biological ribozyme can contain different binding sites for different RNA substrates. This alternate binding site model suggests a simple mechanism for evolving existing ribozymes to recognize RNA substrates of diverse structures.
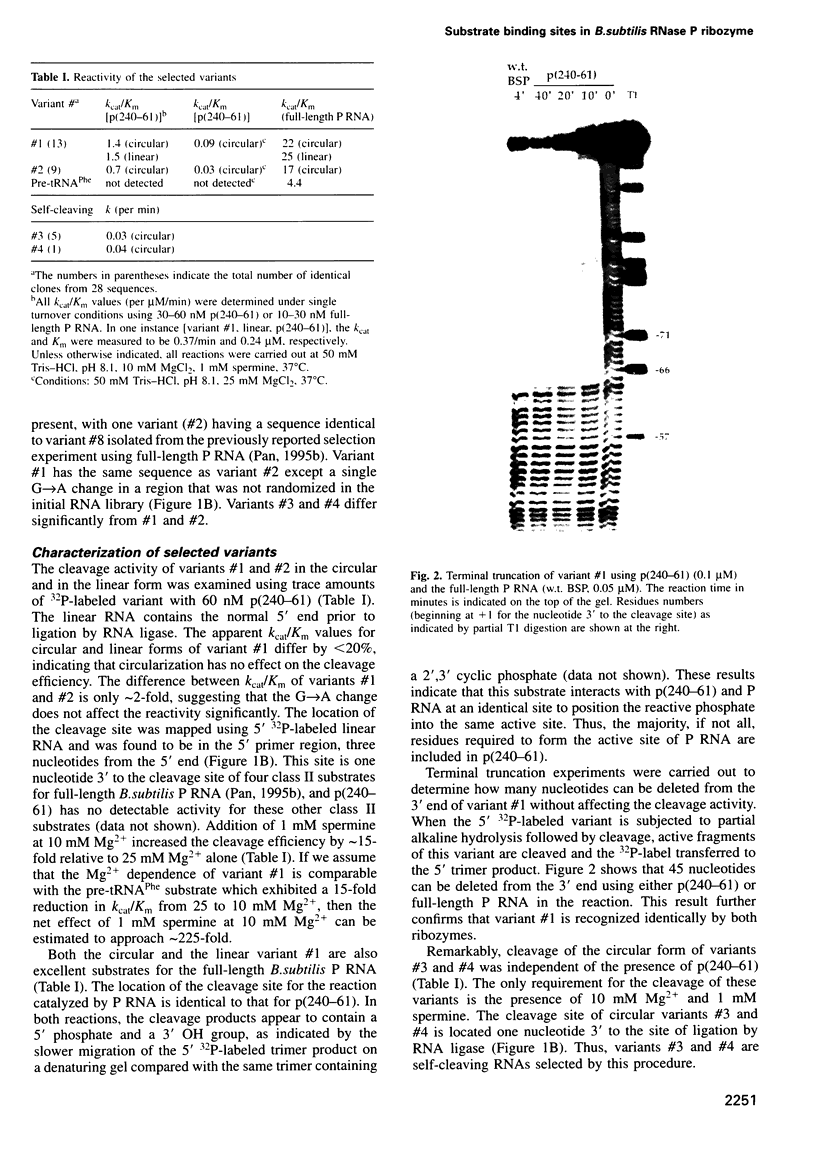
Full text
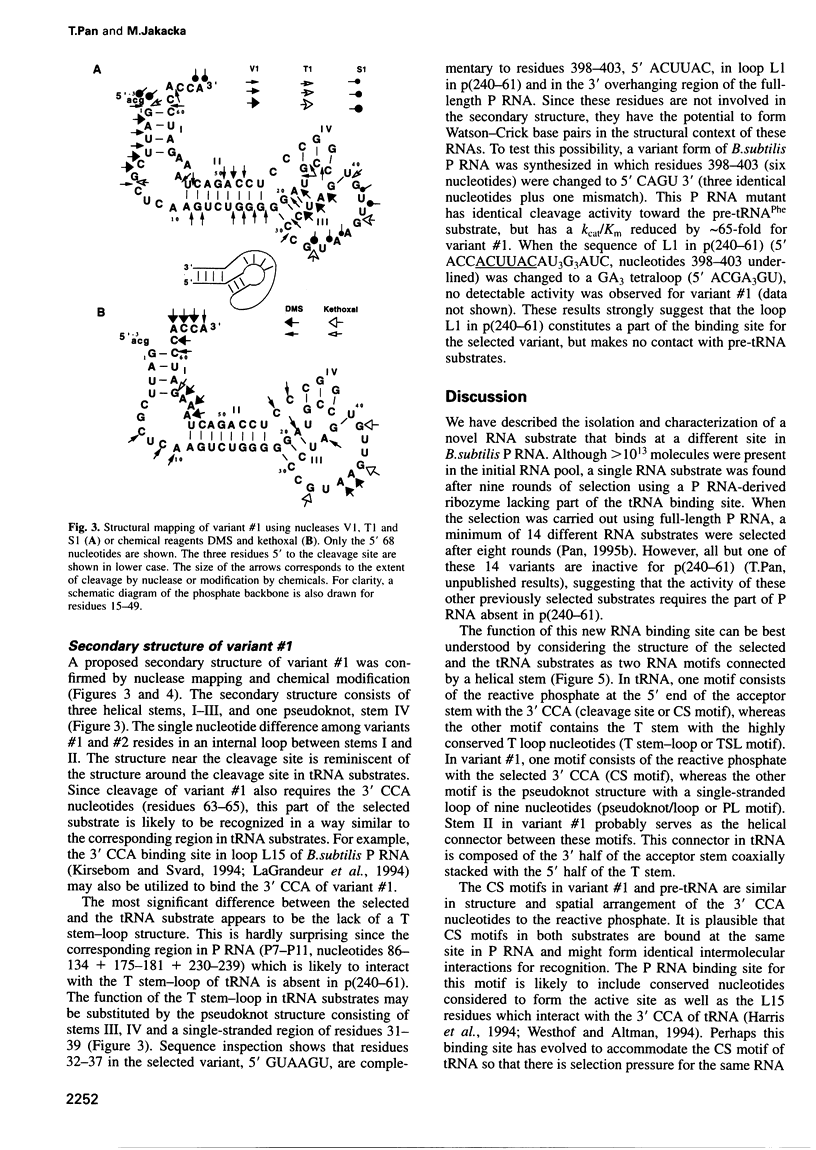
PDF
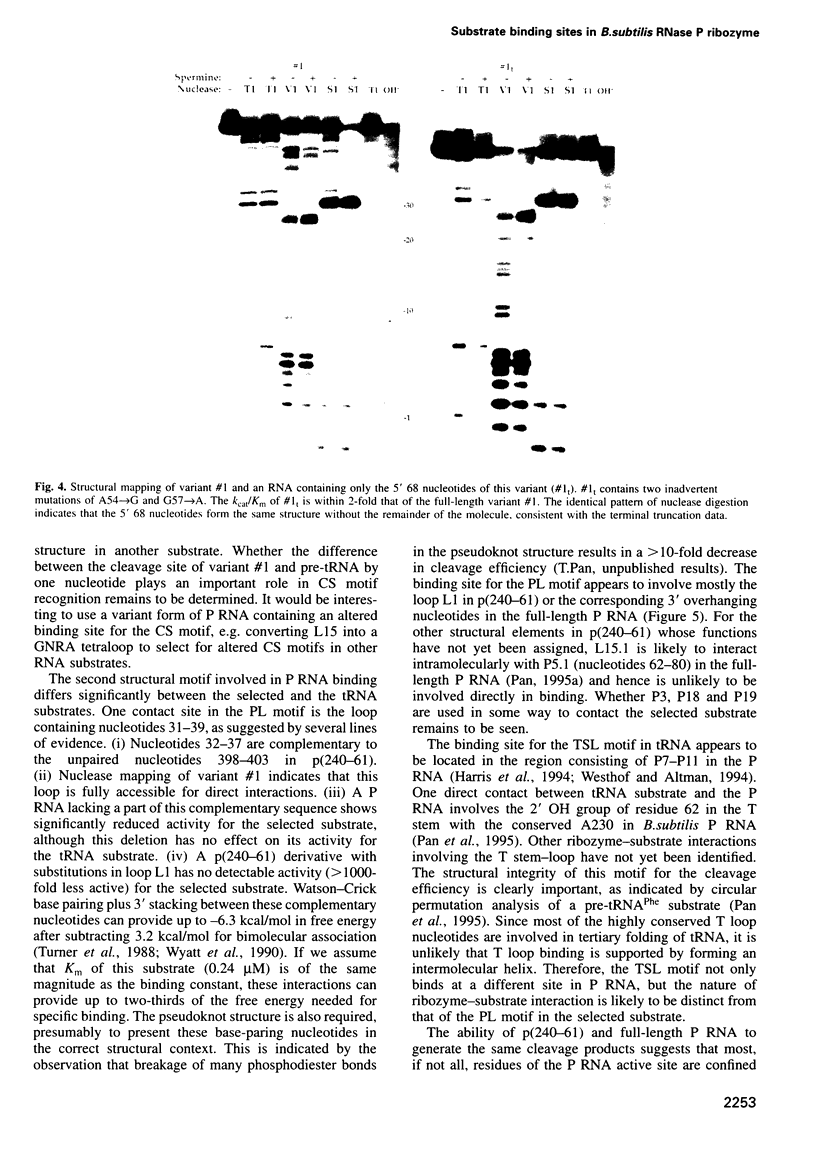

Images in this article
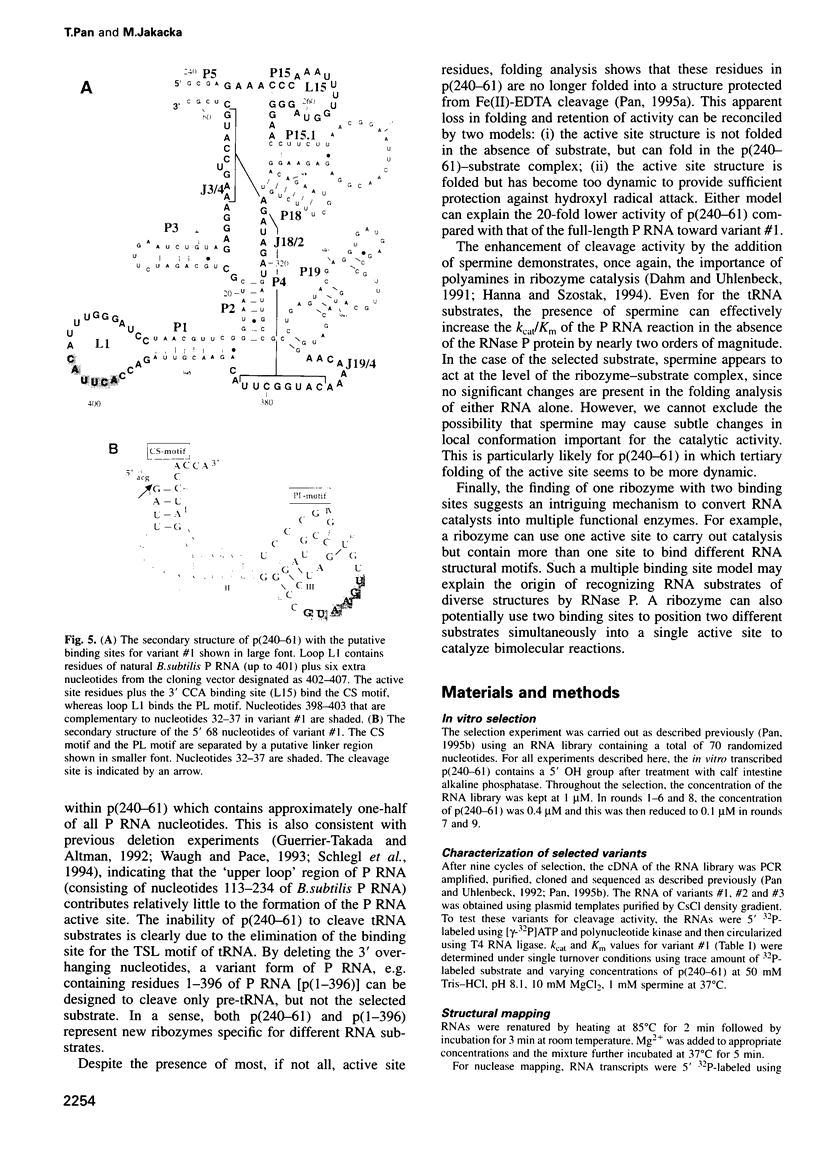
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S., Kirsebom L., Talbot S. Recent studies of ribonuclease P. FASEB J. 1993 Jan;7(1):7–14. doi: 10.1096/fasebj.7.1.7916700. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- Dahm S. C., Uhlenbeck O. C. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry. 1991 Oct 1;30(39):9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- Doudna J. A., Cech T. R. Self-assembly of a group I intron active site from its component tertiary structural domains. RNA. 1995 Mar;1(1):36–45. [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A., Szostak J. W. RNA-catalysed synthesis of complementary-strand RNA. Nature. 1989 Jun 15;339(6225):519–522. doi: 10.1038/339519a0. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Altman S. External guide sequences for an RNA enzyme. Science. 1990 Aug 17;249(4970):783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Altman S. Reconstitution of enzymatic activity from fragments of M1 RNA. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1266–1270. doi: 10.1073/pnas.89.4.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas E. S., Brown J. W., Pitulle C., Pace N. R. Further perspective on the catalytic core and secondary structure of ribonuclease P RNA. Proc Natl Acad Sci U S A. 1994 Mar 29;91(7):2527–2531. doi: 10.1073/pnas.91.7.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna M., Szostak J. W. Suppression of mutations in the core of the Tetrahymena ribozyme by spermidine, ethanol and by substrate stabilization. Nucleic Acids Res. 1994 Dec 11;22(24):5326–5331. doi: 10.1093/nar/22.24.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M. E., Nolan J. M., Malhotra A., Brown J. W., Harvey S. C., Pace N. R. Use of photoaffinity crosslinking and molecular modeling to analyze the global architecture of ribonuclease P RNA. EMBO J. 1994 Sep 1;13(17):3953–3963. doi: 10.1002/j.1460-2075.1994.tb06711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann R. K., Heinrich J., Schlegl J., Schuster H. Precursor of C4 antisense RNA of bacteriophages P1 and P7 is a substrate for RNase P of Escherichia coli. Proc Natl Acad Sci U S A. 1995 Jun 20;92(13):5822–5826. doi: 10.1073/pnas.92.13.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine Y., Kitabatake M., Yokogawa T., Nishikawa K., Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc Natl Acad Sci U S A. 1994 Sep 27;91(20):9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrandeur T. E., Hüttenhofer A., Noller H. F., Pace N. R. Phylogenetic comparative chemical footprint analysis of the interaction between ribonuclease P RNA and tRNA. EMBO J. 1994 Sep 1;13(17):3945–3952. doi: 10.1002/j.1460-2075.1994.tb06710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh B. K., Pace N. R. Interaction of the 3'-end of tRNA with ribonuclease P RNA. Nucleic Acids Res. 1994 Oct 11;22(20):4087–4094. doi: 10.1093/nar/22.20.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Loria A., Zhong K. Probing of tertiary interactions in RNA: 2'-hydroxyl-base contacts between the RNase P RNA and pre-tRNA. Proc Natl Acad Sci U S A. 1995 Dec 19;92(26):12510–12514. doi: 10.1073/pnas.92.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T. Novel RNA substrates for the ribozyme from Bacillus subtilis ribonuclease P identified by in vitro selection. Biochemistry. 1995 Jul 4;34(26):8458–8464. doi: 10.1021/bi00026a029. [DOI] [PubMed] [Google Scholar]
- Pan T., Uhlenbeck O. C. In vitro selection of RNAs that undergo autolytic cleavage with Pb2+. Biochemistry. 1992 Apr 28;31(16):3887–3895. doi: 10.1021/bi00131a001. [DOI] [PubMed] [Google Scholar]
- Schlegl J., Hardt W. D., Erdmann V. A., Hartmann R. K. Contribution of structural elements to Thermus thermophilus ribonuclease P RNA function. EMBO J. 1994 Oct 17;13(20):4863–4869. doi: 10.1002/j.1460-2075.1994.tb06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Moazed D., Noller H. F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Talbot S. J., Altman S. Gel retardation analysis of the interaction between C5 protein and M1 RNA in the formation of the ribonuclease P holoenzyme from Escherichia coli. Biochemistry. 1994 Feb 15;33(6):1399–1405. doi: 10.1021/bi00172a016. [DOI] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Freier S. M. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Westhof E., Altman S. Three-dimensional working model of M1 RNA, the catalytic RNA subunit of ribonuclease P from Escherichia coli. Proc Natl Acad Sci U S A. 1994 May 24;91(11):5133–5137. doi: 10.1073/pnas.91.11.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt J. R., Puglisi J. D., Tinoco I., Jr RNA pseudoknots. Stability and loop size requirements. J Mol Biol. 1990 Jul 20;214(2):455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]