Abstract
Androgen receptor genes (AR) have been found to have associations with reproductive development, behavioral traits, and disorders in humans. However, the influence of similar genetic effects on the behavior of other animals is scarce. We examined the loci AR glutamine repeat (ARQ) in 44 Grevy's zebras, 23 plains zebras, and three mountain zebras, and compared them with those of domesticated horses. We observed polymorphism among zebra species and between zebra and horse. As androgens such as testosterone influence aggressiveness, AR polymorphism among equid species may be associated with differences in levels of aggression and tameness. Our findings indicate that it would be useful to conduct further studies focusing on the potential association between AR and personality traits, and to understand domestication of equid species.
Keywords: Zebra, Equus, Androgen receptor, VNTR
Introduction
Androgen receptors (AR) mediate the effects of androgens that are responsible for development and maintenance of the male reproductive system. Deleterious mutations in AR can result in syndromes ranging from mild abnormalities to total failure of normal male phenotypic development (McPhaul, 2002a, McPhaul, 2002b). AR also has associations with disease states (such as prostate cancer) and behavior in humans (Collaer and Hines, 1995, Wyce et al., 2010, Zitzmann and Nieschlag, 2003). Androgen receptors (AR) are DNA-binding transcription factors, the main regulators of androgen signaling in the cell, activated mostly by testosterone and 5α-dihydrotestosterone. In humans the AR gene is located on the X chromosome and comprised of three important structural domains: the N-terminal transactivation domain, the central DNA binding domain, and the C-terminal ligand binding domain (Mangelsdorf et al., 1995). The N-terminal domain in AR contains two polymorphic trinucleotide repeat regions, CAG encoding glutamine and GGN encoding glycine (ARQ and ARG, respectively) in humans (Chang et al., 1988). In vitro studies have shown that a relatively short CAG repeat sequence enhances the transcriptional activity of the AR by promoting interactions between the receptor and coactivators (Chamberlain et al., 1994). The CAG repeat polymorphisms have been shown to affect reproductive capability, various disease risks and personality not only in men but also in women (Choong and Wilson, 1998, Giovannucci et al., 1997, Mouritsen et al., 2013, Olsen et al., 2014, Robeva et al., 2013, Tut et al., 1997). Association between polymorphism of the AR gene and various traits (infertility, disease, personality) has also been documented in a number of animal species (Konno et al., 2011, Lai et al., 2008, Lyons et al., 2014, Revay et al., 2012, Yasui et al., 2013).
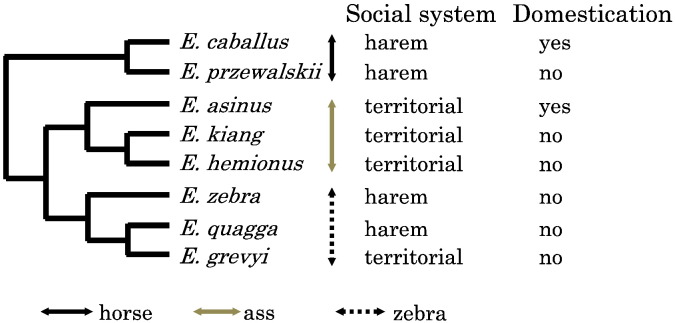
Equidae are comprised of eight species, which all belong to a single genus, Equus. Equus contain four sub-genera, Equus, Asinus, Dolichohippus, and Hippotigris. Sub-genus Equus includes domestic horse and wild horse. Sub-genus Asinus includes donkey, African wild ass, Asian wild ass and kiang. Dolichohippus includes only Grevy's zebra. Hippotigris includes plains zebra and mountain zebra. Together with other factors such as ecology and phylogeny, social structure affects or reflects species difference in personality, such as levels of aggression, affiliation, and pair-bonding. Similarly, social structures could also be reflected in genetic differences. Members of the genus Equus show two patterns of social organization (Rubenstein, 2011) (Fig. 1). In the sub-genus Equus and Hippotigris, a single breeding male is found in constant association with a fixed group of 1–6 unrelated females and their offspring; this is the forming of a harem. On the other hand, in the sub-genus Asinus and Dolichohippus, breeding males have territories, and females do not form stable groups. The diversity of these social systems is not consistent with genetic phylogeny. In addition, humans succeeded only in the domestication of the horse in sub-genus Equus and donkey in sub-genus Asinus, which display different social systems. The other species, including wild asses and zebras, have been considered not suitable for domestication because of their untamable nature.
Fig. 1.
Phylogeny and sociality in equids.
Phylogenetic tree follows Steiner and Ryder (2011), based on mtDNA and nuclear DNA sequences.
As genus Equus displays different social systems, it is a suitable system to investigate the function of personality- and sociality-related genes in different sub-genera. Because AR variation may influence aggressiveness along with other traits, they are among the ideal candidate genes to study the basis for social system and behavioral differences in equids, which in turn can help us to understand the functions and evolution of these genes. As a first step toward this approach, we analyzed the locus ARQ in three zebra species for comparison with those of horses.
Material and methods
Samples
We obtained blood, muscles, hair and feces from Grevy's zebra (Equus grevyi) (n = 44), plains zebra (Equus quagga) (n = 23) and Hartmann's mountain zebra (Equus zebra hartmannae) (n = 3). DNA was extracted from whole blood, muscles and hair using QIAGEN DNeasy Blood and Tissue Kit (QIAGEN), from feces using QIAGEN DNeasy Stool Kit (QIAGEN). All individuals were kept in captivity at zoos in Japan or the UK. Captive populations of Grevy's and Hartmann's zebra are closely managed under endangered species breeding programs therefore it is highly unlikely that the samples we obtained were from hybridized individuals.
PCR and sequence
We amplified a 220 base pair region of exon1 in equine AR by polymerase chain reaction (PCR). We designed the primer pairs based on horse AR sequence information (GenBank: JN187443, (Revay et al., 2012)) using Primer3 software (Forward primer: GAACAGCAGCCTTCACAACA, Reverse primer: CTGCCTCCCTCGCTCTCC). A 10 μl PCR reaction contained 20 ng DNA, 0.5 μM of each of the primers, 0.5 U LA Taq polymerase, GC buffer I (TaKaRa), and 400 μM of each dNTP. For fecal samples, instead of 20 ng DNA, 2 μl of extracted DNA solution and 0.1 μg of T4 Gene 32 Protein (Nippon Gene) were added. The PCR conditions consisted of an initial denaturation at 95 °C for 2 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 62 °C for 30 s, extension at 72 °C for 1 min, and final extension at 72 °C for 10 min. In fecal samples, the PCR conditions consisted of an initial denaturation at 95 °C for 2 min, followed by 40–45 cycles of denaturation at 95 °C for 30 s, annealing at 62 °C for 30 s, extension at 72 °C for 1 min, and final extension at 72 °C for 10 min. PCR products were purified using a High Pure PCR Product Purification Kit (Roche), and their nucleotide sequences were determined by cycle sequencing using a Big Dye Terminator v3. 1 Cycle Sequencing Kit (Roche) and an Applied Biosystems 3130xl Genetic Analyzer (Applied Biosystems). All determined sequences were checked visually using BioEdit software, version 7. 0. 9. 0. Polymorphisms in the sequences (SNPs and VNTR) were detected by the alignment using MEGA software, version 6.0 (Tamura et al., 2013).
Results
The nucleotide and amino acid sequence alignments are shown in Fig. 2a and b, respectively. In three zebra species, we found 5 lengths in CAG repeat regions, 8, 9, 10, 12, and 15 (Fig. 2). Allele frequencies in three zebra species are presented in Table 1. The data for these sequences were deposited in the DDBJ Genbank database under accession numbers Genbank LC030245-LC030253.
Fig. 2.
a) Sequences of the androgen receptor exon1 CAG repeat region in equid. Locations of VNTR are marked by squares. b) Sequences of amino acid of the androgen receptor exon1 CAG repeat region in equid. Locations of VNTR are marked by squares.
Table 1.
Allele frequencies of androgen receptor in three zebra species.
| Grevy's zebra |
Plains zebra |
Mountain zebra |
Horse |
||||||
|---|---|---|---|---|---|---|---|---|---|
| N | frequency | N | frequency | N | frequency | N | frequency | ||
| VNTR | 5 | 0 | 0.000 | 0 | 0.000 | 0 | 0.000 | 25 | 1.000 |
| (CAG)n | 8 | 16 | 0.198 | 0 | 0.000 | 0 | 0.000 | 0 | 0.000 |
| 9 | 0 | 0.000 | 16 | 0.372 | 1 | 0.250 | 0 | 0.000 | |
| 10 | 65 | 0.802 | 14 | 0.326 | 1 | 0.250 | 0 | 0.000 | |
| 12 | 0 | 0.000 | 13 | 0.302 | 0 | 0.000 | 0 | 0.000 | |
| 15 | 0 | 0.000 | 0 | 0.000 | 2 | 0.500 | 0 | 0.000 | |
| SNP | G | 68 | 0.840 | 43 | 1.000 | 3 | 1.000 | 25 | 1.000 |
| G325C | C | 13 | 0.160 | 0 | 0.000 | 0 | 0.000 | 0 | 0.000 |
N: the number of X chromosomes.
Grevy's zebra had two alleles; 8-repeat allele and 10-repeat allele. Plains zebra had three alleles; 9-repeat, 10-repeat, and 12-repeat allele. Mountain zebra had three alleles: 9-repeat, 10-repeat, and 15-repeat allele. In each zebra species, VNTRs were observed, where none have been reported in horses (Revay et al., 2012) (Fig. 2). All alleles detected were longer than that of the horse (5 repeat). The longest allele (15 CAG repeats) had a deletion of AAA (Lysin) at 156 bp (Fig. 2).
There was only one SNP (G6C) observed among the three zebra species: this SNP was a non-synonymous substitution (V2L), and was found only in the 10-repeat allele in Grevy's zebra. There was one fixed non-synonymous substitution observed between zebra and horse (nucleotide G36A; amino acid G12S).
Discussion
In this study, we surveyed the allele frequencies and nucleotide sequence differences in CAG repeat regions of the AR gene in three zebra species and found significant differences to the existing horse reference data. In horses, VNTRs at the ARQ locus have not been reported and only a single allele including 5 CAG repeats has been previously described (Revay et al., 2012). Our study provides the first information about AR in other equids.
Association between ARQ variation and aggressiveness has been demonstrated in humans (Vermeersch et al., 2010) and previous reports have shown that individuals with shorter CAG repeat in humans and dogs tend to be more aggressive (Jonsson et al., 2001, Konno et al., 2011). We found that locus ARQ in three zebra species had longer alleles than that in horses. As zebras have not been domesticated it might be expected that they are more aggressive than horses. The increased allele length observed in zebras in this study therefore does not correspond to the pattern of allele length and aggression observed in humans and dogs. Although this unexpected result remains to be explained, and there is a need for analysis of more samples in each species to increase confidence in the observed patterns, in vitro studies have shown that medium-length CAG repeats had high transcriptional activity of AR compared with both shorter and longer CAG repeats (Nenonen et al., 2010). Indeed, in the study that compared between two species (chimpanzee and bonobo), chimpanzees that had more aggressive traits had generally shorter alleles than bonobos (Garai et al., 2014). It would be interesting to investigate the association between AR activity and gene polymorphisms. Furthermore, it will be useful to conduct a standardized assessment of aggressiveness in zebra species and horse to compare personality in equids.
All samples used in this study were derived from captive populations, so might not reflect the number of alleles and allele frequencies of native populations, due to the founder effect and genetic drift. It is interesting to note that polymorphism was maintained in each zebra species, despite these populations being derived from a small number of founders, suggesting that diversity in natural populations could be higher. In contrast the lack of diversity previously observed in horses may relate to its long history of domestication.
This study could not identify if these polymorphisms occurred in the common ancestor of horse and zebra or were only generated in zebra species after they diverged. In the first case, the AR gene might have undergone positive selection in horse domestication. To clarify the derivation of these polymorphisms, we must investigate other Equus species. Recently, the gene groups that are associated with the domestication of the horse were reported (Schubert et al., 2014). One gene group is related to muscular and limb development, articular junctions, and cardiac systems. The second group relates to cognitive functions including social behavior, learning capabilities, fear response, and agreeableness. Genes involved in tameness and social behavior are important in domestication, so it is thought that AR gene contributing to the activity of testosterone might be significant in domestication. Moreover, the analysis of the relationship between the AR gene and reproductive ability may be useful to the conservation of endangered Equus species. Intra species polymorphisms that we found in zebra species will be valuable to investigate the relationship between genetic polymorphism and function (responsiveness and metabolism).
In this study, we found evidence for variation between zebra species and horse at locus ARQ that may relate to differences in aggression and social systems, although the number of samples in this study does limit interpretation of these findings. To understand the evolution of AR in equids, it would be useful to conduct further studies on the association of these traits with the polymorphisms that we found in zebra species and to compare the results with those of other equids.
Acknowledgments
This work was supported by a Grant-in-aid for Scientific Research from JSPS (grant number 21310150, 25290082, 25118005 and 15H00441). We thank the following for providing samples; Kyoto City Zoo, Ishikawa Zoo, Himeji Central Park, Toyama Family Park, Hamamatsu City Zoo, Morioka Zoological Park, Noichi Zoo, Wanpa-ku Zoo, Nogeyama Zoo, Kushiro Zoo, Hirakawa Zoo, Izu Animal Kingdom, Kawasaki Zoo, Toyohashi Zoo, Tama Zoological Park, Tobe Zoo in Japan, Edingburg Zoo, Marwell Wildlife, Whipsnade Zoo, Port Lympne Zoo, and Woburn Safari Park in United Kingdom.
References
- Chamberlain N.L., Driver E.D., Miesfeld R.L. The length and location of CAG trinucleotide repeats in the androgen receptor N-terminal domain affect transactivation function. Nucleic Acids Res. 1994;22(15):3181–3186. doi: 10.1093/nar/22.15.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.S., Kokontis J., Liao S.T. Structural analysis of complementary DNA and amino acid sequences of human and rat androgen receptors. Proc. Natl. Acad. Sci. U. S. A. 1988;85(19):7211–7215. doi: 10.1073/pnas.85.19.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choong C.S., Wilson E.M. Trinucleotide repeats in the human androgen receptor: a molecular basis for disease. J. Mol. Endocrinol. 1998;21(3):235–257. doi: 10.1677/jme.0.0210235. [DOI] [PubMed] [Google Scholar]
- Collaer M.L., Hines M. Human behavioral sex differences: a role for gonadal hormones during early development? Psychol. Bull. 1995;118(1):55–107. doi: 10.1037/0033-2909.118.1.55. [DOI] [PubMed] [Google Scholar]
- Garai C., Furuichi T., Kawamoto Y., Ryu H., Inoue-Murayama M. Androgen receptor and monoamine oxidase polymorphism in wild bonobos. Meta Gene. 2014;2:831–843. doi: 10.1016/j.mgene.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci E., Stampfer M.J., Krithivas K., Brown M., Brufsky A., Talcott J., Hennekens C.H., Kantoff P.W. The CAG repeat within the androgen receptor gene and its relationship to prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 1997;94(7):3320–3323. doi: 10.1073/pnas.94.7.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson E.G., von Gertten C., Gustavsson J.P., Yuan Q.P., Lindblad-Toh K., Forslund K., Rylander G., Mattila-Evenden M., Asberg M., Schalling M. Androgen receptor trinucleotide repeat polymorphism and personality traits. Psychiatr. Genet. 2001;11(1):19–23. doi: 10.1097/00041444-200103000-00004. [DOI] [PubMed] [Google Scholar]
- Konno A., Inoue-Murayama M., Hasegawa T. Androgen receptor gene polymorphisms are associated with aggression in Japanese Akita Inu. Biol. Lett. 2011;7(5):658–660. doi: 10.1098/rsbl.2011.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.L., L'Eplattenier H., van den Ham R., Verseijden F., Jagtenberg A., Mol J.A., Teske E. Androgen receptor CAG repeat polymorphisms in canine prostate cancer. J. Vet. Intern. Med. 2008;22(6):1380–1384. doi: 10.1111/j.1939-1676.2008.0181.x. [DOI] [PubMed] [Google Scholar]
- Lyons R.E., Loan N.T., Dierens L., Fortes M.R., Kelly M., McWilliam S.S., Li Y., Bunch R.J., Harrison B.E., Barendse W., Lehnert S.A., Moore S.S. Evidence for positive selection of taurine genes within a QTL region on chromosome X associated with testicular size in Australian Brahman cattle. BMC Genet. 2014;15:6. doi: 10.1186/1471-2156-15-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf D.J., Thummel C., Beato M., Herrlich P., Schutz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R.M. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhaul M.J. Androgen receptor mutations and androgen insensitivity. Mol. Cell. Endocrinol. 2002;198(1–2):61–67. doi: 10.1016/s0303-7207(02)00369-6. [DOI] [PubMed] [Google Scholar]
- McPhaul M.J. Molecular defects of the androgen receptor. Recent Prog. Horm. Res. 2002;57:181–194. doi: 10.1210/rp.57.1.181. [DOI] [PubMed] [Google Scholar]
- Mouritsen A., Hagen C.P., Sorensen K., Aksglaede L., Mieritz M.G., Main K.M., Almstrup K., Rajpert-De Meyts E., Juul A. Androgen receptor CAG repeat length is associated with body fat and serum SHBG in boys: a prospective cohort study. J. Clin. Endocrinol. Metab. 2013;98(3):E605–E609. doi: 10.1210/jc.2012-3778. [DOI] [PubMed] [Google Scholar]
- Nenonen H., Bjork C., Skjaerpe P.A., Giwercman A., Rylander L., Svartberg J., Giwercman Y.L. CAG repeat number is not inversely associated with androgen receptor activity in vitro. Mol. Hum. Reprod. 2010;16(3):153–157. doi: 10.1093/molehr/gap097. [DOI] [PubMed] [Google Scholar]
- Olsen N.J., Benko A.L., Kovacs W.J. Variation in the androgen receptor gene exon 1 CAG repeat correlates with manifestations of autoimmunity in women with lupus. Endocr. Connect. 2014;3(2):99–109. doi: 10.1530/EC-14-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revay T., Villagomez D.A., Brewer D., Chenier T., King W.A. GTG mutation in the start codon of the androgen receptor gene in a family of horses with 64, XY disorder of sex development. Sex. Dev. 2012;6(1–3):108–116. doi: 10.1159/000334049. [DOI] [PubMed] [Google Scholar]
- Robeva R., Tanev D., Andonova S., Kirilov G., Savov A., Stoycheva M., Tomova A., Kumanov P., Rashkov R., Kolarov Z. Androgen receptor (CAG)n polymorphism and androgen levels in women with systemic lupus erythematosus and healthy controls. Rheumatol. Int. 2013;33(8):2031–2038. doi: 10.1007/s00296-013-2687-2. [DOI] [PubMed] [Google Scholar]
- Rubenstein D.I. Family equidae (Horses and relatives) In: Wilson D.E., Mittermeier R.A., editors. vol. 2. Lynx Edicions; Spain: 2011. pp. 106–143. (Handbook of Mammals of the World). [Google Scholar]
- Schubert M., Jonsson H., Chang D., Der Sarkissian C., Ermini L., Ginolhac A., Albrechtsen A., Dupanloup I., Foucal A., Petersen B., Fumagalli M., Raghavan M., Seguin-Orlando A., Korneliussen T.S., Velazquez A.M., Stenderup J., Hoover C.A., Rubin C.J., Alfarhan A.H., Alquraishi S.A., Al-Rasheid K.A., MacHugh D.E., Kalbfleisch T., MacLeod J.N., Rubin E.M., Sicheritz-Ponten T., Andersson L., Hofreiter M., Marques-Bonet T., Gilbert M.T., Nielsen R., Excoffier L., Willerslev E., Shapiro B., Orlando L. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proc. Natl. Acad. Sci. U. S. A. 2014 doi: 10.1073/pnas.1416991111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner C.C., Ryder O.A. Molecular phylogeny and evolution of the Perissodactyla. Zool. J. Linnean Soc. 2011;163(4):1289–1303. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tut T.G., Ghadessy F.J., Trifiro M.A., Pinsky L., Yong E.L. Long polyglutamine tracts in the androgen receptor are associated with reduced trans-activation, impaired sperm production, and male infertility. J. Clin. Endocrinol. Metab. 1997;82(11):3777–3782. doi: 10.1210/jcem.82.11.4385. [DOI] [PubMed] [Google Scholar]
- Vermeersch H., T'Sjoen G., Kaufman J.M., Vincke J., Van Houtte M. Testosterone, androgen receptor gene CAG repeat length, mood and behaviour in adolescent males. Eur. J. Endocrinol. 2010;163(2):319–328. doi: 10.1530/EJE-10-0090. [DOI] [PubMed] [Google Scholar]
- Wyce A., Bai Y., Nagpal S., Thompson C.C. Research resource: the androgen receptor modulates expression of genes with critical roles in muscle development and function. Mol. Endocrinol. 2010;24(8):1665–1674. doi: 10.1210/me.2010-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui S., Konno A., Tanaka M., Idani G., Ludwig A., Lieckfeldt D., Inoue-Murayama M. Personality assessment and its association with genetic factors in captive Asian and African elephants. Zoo Biol. 2013;32(1):70–78. doi: 10.1002/zoo.21045. [DOI] [PubMed] [Google Scholar]
- Zitzmann M., Nieschlag E. The CAG repeat polymorphism within the androgen receptor gene and maleness. Int. J. Androl. 2003;26(2):76–83. doi: 10.1046/j.1365-2605.2003.00393.x. [DOI] [PubMed] [Google Scholar]