Abstract
Hairy cell leukemia (HCL) is a chronic lymphoproliferative disorder characterized by somatic BRAFV600E mutations. The malignant cell in HCL has immunophenotypic features of a mature B cell, but no normal counterpart along the continuum of developing B lymphocytes has been delineated as the cell of origin. We find that the BRAFV600E mutation is present in hematopoietic stem cells (HSCs) in HCL patients, and that these patients exhibit marked alterations in hematopoietic stem/progenitor cell (HSPC) frequencies. Quantitative sequencing analysis revealed a mean BRAFV600E-mutant allele frequency of 4.97% in HSCs from HCL patients. Moreover, transplantation of BRAFV600E-mutant HSCs from an HCL patient into immunodeficient mice resulted in stable engraftment of BRAFV600E-mutant human hematopoietic cells, revealing the functional self-renewal capacity of HCL HSCs. Consistent with the human genetic data, expression of BRafV600E in murine HSPCs resulted in a lethal hematopoietic disorder characterized by splenomegaly, anemia, thrombocytopenia, increased circulating soluble CD25, and increased clonogenic capacity of B lineage cells—all classic features of human HCL. In contrast, restricting expression of BRafV600E to the mature B cell compartment did not result in disease. Treatment of HCL patients with vemurafenib, an inhibitor of mutated BRAF, resulted in normalization of HSPC frequencies and increased myeloid and erythroid output from HSPCs. These findings link the pathogenesis of HCL to somatic mutations that arise in HSPCs and further suggest that chronic lymphoid malignancies may be initiated by aberrant HSCs.
INTRODUCTION
Hairy cell leukemia (HCL) is a chronic lymphoproliferative disorder characterized as a mature B cell malignancy based on the fact that the hallmark leukemic cell in HCL expresses CD19, surface immunoglobulin (1), and clonal rearrangements of immunoglobulin heavy and light chain genes (2, 3)—all features of mature B cells (4, 5). At the same time, HCL cells also express cell surface markers not present on normal B cells, including CD103 and CD11c, antigens typically expressed on intraepithelial T cells and monocytes, respectively (6, 7). In addition, HCL patients have long been known to have clinical features disparate from most mature B cell malignancies, including the absence of lymph node involvement and frequent splenomegaly due to extramedullary hematopoiesis (EMH) (4). Gene expression microarray studies have not precisely identified a specific B cell population as the cell of origin of HCL (8). Hence, the hematopoietic population that initiates and propagates HCL has not been definitively delineated, and the cell of origin of HCL has remained under debate (9-12).
Tiacci et al. recently identified somatic BRAFV600E mutations in nearly 100% of classic HCL patients (13), a finding since confirmed in multiple studies (14, 15). This seminal observation provided the first genetic insight into HCL pathogenesis and a clonal marker to track the origin and propagation of HCL. We therefore sought to identify the cell of origin of HCL by identifying the point of hematopoietic development at which the BRAFV600E mutation arises.
Here, we identify the presence of the BRAFV600E mutation in hematopoietic stem cells (HSCs) in patients with HCL, as well as consistent abnormalities in hematopoietic stem and progenitor cell (HSPC) frequencies in HCL patients. We also use murine genetic models to functionally characterize the effects of mutant BRafV600E expression at various stages of development and hematopoietic differentiation, revealing that many of the characteristic clinical features of HCL are seen only with expression of BRafV600E in cells less mature than committed B cells. Finally, we evaluated the effect of mutant BRAF inhibition on hematopoiesis in our murine models, as well as in patients with BRAF-mutant HCL using material from patients in an ongoing clinical trial of vemurafenib for relapsed/refractory HCL.
RESULTS
Altered HSPC composition and the presence of BRAFV600E mutations in HSCs from HCL patients
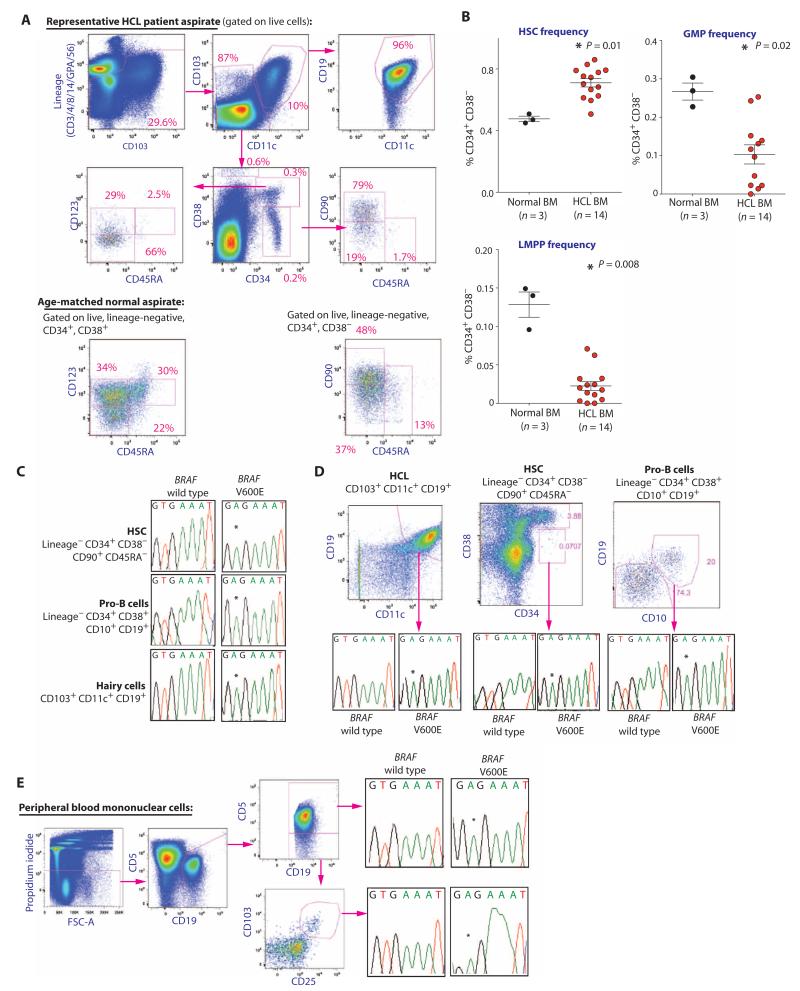
To identify the cell population along the hematopoietic hierarchy in which the BRAFV600E mutation arises, we prospectively purified HSCs [lineage-negative (LN) CD34+ CD38− CD90+ CD45RA−], pro-B cells (LN CD34+ CD38+ CD10+ CD19+), hematogones (CD34− CD38++ CD10+ CD19+), myeloid progenitor (MP) cells (LN CD10− CD19− CD34+ CD38+ CD45RA+/− CD123+/−), and HCL cells (CD19+ CD103+ CD11c+) from the bone marrow (BM) of 14 HCL patients and age-matched controls (Fig. 1A and table S1). CD10, CD19, CD103, and CD11c were excluded from the lineage cocktail and stained with different fluorochromes to assess B and HCL cell frequencies. HCL patients were characterized by an expansion of HSCs among CD34+ CD38− LN cells and a marked decrease in the frequency of granulocyte-macrophage progenitor (GMP) cells among MPs, consistent with the neutropenia and monocytopenia characteristic of HCL (Fig. 1B). This stage-specific dropout of GMPs was also linked to a decrease in the lymphoid-primed multipotent progenitor population (LMPP; LN CD34+ CD38− CD90− CD45RA+) (Fig. 1B), which has been shown to have granulocyte/macrophage-restricted myeloid, as well as normal lymphoid differentiation potential (16, 17). We then used an allele-specific oligonucleotide assay to assess for the BRAFV600E mutation (14) in different hematopoietic subsets after double FACS (fluorescence-activated cell sorting) to ensure >97% purity (fig. S1). We identified the BRAFV600E mutation in the HSC, pro-B cell, and HCL cell populations of HCL patients (Fig. 1, C and D). Analysis of sorted chronic lymphocytic leukemia (CLL) cells in addition to HCL cells from one individual harboring both diseases identified the presence of the BRAFV600E mutation in both cell populations, suggesting a potential shared origin of these two leukemias (Fig. 1E). These data suggest that HCL initiates within the HSPC compartment and not in committed lymphoid cells as previously suggested (9-12).
Fig. 1. HSPC abnormalities and the presence of the BRAFV600E mutation in HSCs of HCL patients.
(A) Stem and progenitor flow cytometric analysis of the BM of a representative HCL patient and an age-matched control. The sort schema shown was used for isolation of HCL cells (CD103+ CD19+ CD11c+ cells), HSCs (LN CD34+ CD38− CD90+ CD45RA− cells), hematogones (CD34− CD38++ CD10+ CD19+), and MP cells (LN CD34+ CD38+ CD45RA+/− CD123+/− cells). (B) Frequencies of HSCs, LMPPs (LN CD34+ CD38− CD90− CD45RA+), and GMPs (LN CD34+ CD38+ CD123+ CD45RA+) in 14 patients with HCL and 3 age-matched normal control BM aspirate samples. (C) Prospective cell separation including double sorting to ensure purity and lack of HCL cell contamination followed by allele-specific polymerase chain reaction (PCR) analysis for the presence of the BRAFV600E mutation reveals the mutation in HSCs, pro-B cells, and HCL cells. (D) Similar data from a second HCL patient revealing the mutation in HSC, pro-B, and HCL double-sorted cell populations. (E) Prospective isolation of CLL (CD19+ CD5+ CD103− CD11c− cells) and HCL cell populations (CD19+ CD103+ CD11c+ CD5− cells) from the PB of one individual with both disorders reveals the BRAFV600E mutation in both cell populations. Error bars represent means ± SD. *P < 0.05 (Mann-Whitney U test).
Genetic and functional evaluation of HSPCs in HCL
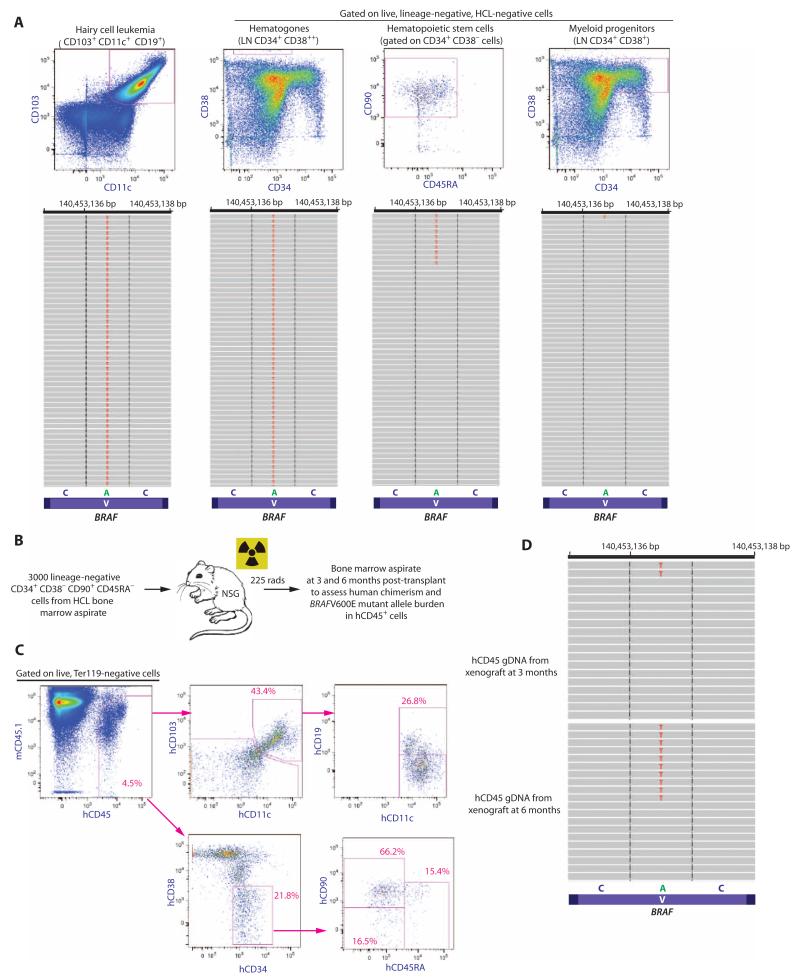
Next, to more quantitatively define the proportion of HSCs bearing mutant BRAF in HCL patients, we performed quantitative sequencing of the region of BRAFV600E in complementary DNA (cDNA) from prospectively purified HCL cells, hematogones, and HSCs (LN CD34+ CD38− CD90+ CD45RA−) from five different HCL patients. At a mean sequencing depth of >1000×, we identified a mean variant allele frequency (VAF) of 63.1% (+12.1%), 23.0% (+22.9%), and 4.97% (±1.9%) in HCL cells, hematogones, and HSCs, respectively (Fig. 2A). The variability of the BRAFV600E VAF in HCL cells was due to the fact that two of the five patients had a homozygous BRAFV600E mutation, whereas the others had a heterozygous mutation. In contrast to the mean VAF of >1% in HCL cells, hematogones, and HSCs, the BRAFV600E mutation was detected at a VAF of only 0.09% (±0.03%) and 0.07% (±0.009%) in MP and CD14+ cells, respectively (table S1).
Fig. 2. Quantitative analysis of the BRAFV600E mutation in HSCs from HCL patients and functional self-renewal capacity of BRAFV600E-mutant HSCs.
(A) Representative FACS analysis and quantitative sequencing analysis revealing the VAF of the BRAF c.T1860A p.V600E mutation in HCL cells, hematogones, HSCs, and MP cells from an HCL patient (for clarity, only 52 reads are displayed). cDNA from double FACS-sorted cell populations were used for MiSeq targeted sequencing. (B) Schema of xenograft experiment where 3000 HSCs from a BRAFV600E-mutant HCL patient were injected into sublethally irradiated NSG mice followed by flow cytometric analysis of human engraftment and HCL cells, as well as quantification of the BRAFV600E mutation by sequencing analysis. (C) At 6 months after transplant, overall human chimerism was 4.5% with the presence of human HSCs (hCD45+ hCD34+ hCD38− hCD90+ hCD45RA−) and a cell population with the immunophenotype of HCL cells (hCD45+ hCD103+ hCD19+ hCD11c+). (D) MiSeq sequencing analysis at 100× coverage reveals BRAFV600E mutation in 4 and 9% of hCD45+ cell genomic DNA at 3 and 6 months, respectively.
Given the high frequency of BRAFV600E mutations in HCL and the fact that the mutation was identified in HSCs, we sought to identify additional co-occurring genetic lesions along the course of hematopoietic differentiation that might cooperate with the BRAFV600E mutation to promote hematopoietic transformation. To this end, we performed targeted mutational analysis of a well-validated panel of 300 known cancer genes (18) (table S2) in genomic DNA from HCL leukemic cells and paired granulocyte samples from three HCL patients. We focused on mutations present in the HCL clone and not in paired granulocytes. This analysis revealed only the BRAFV600E mutation in one patient, BRAFV600E plus an ARID1A p.V1427fs mutation in another patient, and a BRAFV600E plus a MLL3 p.C394Y mutation in a third patient. We then performed sequencing analysis by MiSeq for the BRAFV600E mutation and these additional alterations in HSCs (LN CD34+ CD38− CD90+ CD45RA−), MPs (LN CD19− CD10− CD34+ CD38+), and hematogones (LN CD34+ CD38++) from the respective patients where these somatic mutations were identified in HCL cells. This analysis failed to reveal any mutation in the HSC or hematogone population other than the BRAFV600E mutation.
The identification of the BRAFV600E mutation in purified HSCs and progenitor B cells in HCL patients is highly suggestive of an immature cell of origin in HCL. To define the functional potential of BRAFV600E-mutant HSCs to give rise to HCL, we transplanted 3000 HSCs (LN CD34+ CD38− CD90+ CD45RA−) from the BM aspirate of an untreated HCL patient into sublethally irradiated nonobese diabetic/severe combined immunodeficient interleukin-2 (IL-2) receptor γ chain knockout (NSG) mice (Fig. 2B). Serial flow cytometric and genetic analysis of human CD45+ cells in the BM of engrafted mice at 6 months after transplantation revealed a population of hCD45+ cells with an HCL immunophenotype (hCD103+ hCD11c+ hCD19+) as well as immunophenotypic HSCs (LN CD34+ CD38− CD90+ CD45RA−) (Fig. 2C). Moreover, genetic analysis of unfractionated hCD45+ cells from the BM of engrafted mice revealed the presence of the BRAFV600E-mutant allele at a VAF of 4 and 9% in genomic DNA at 3 and 6 months after transplant, respectively (Fig. 2D). These data indicate that HSCs from HCL patients bearing the BRAFV600E mutation have functional self-renewal capacity, and strongly suggest that these cells give rise to HCL. Moreover, the self-renewal potential of BRAF-mutant HSCs and the fact that the BRAFV600E mutation was detected in HSCs, hematogones, and HCL cells indicate that BRAF-mutant HSCs in HCL patients do not merely represent a subset of HCL cells.
Phenotypic analysis of mice with pan-hematopoietic versus B cell-restricted expression of BRafV600E
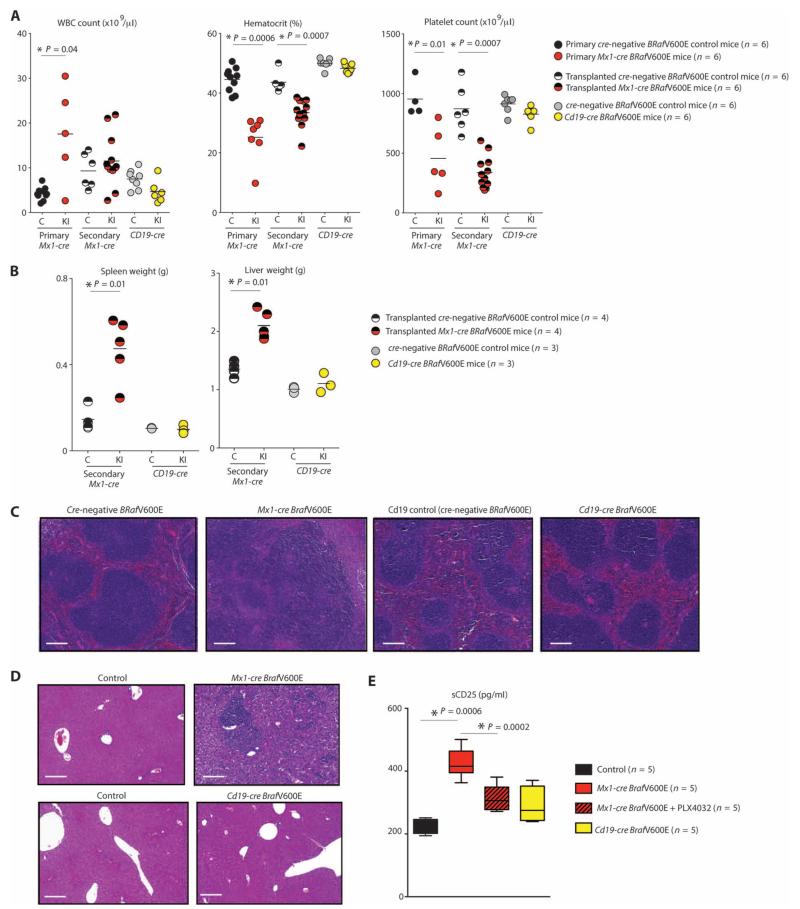
We next sought to assess the effects of expressing BRafV600E in different hematopoietic compartments in vivo. We used a conditional BRafV600E murine model in which the mutant allele is expressed from the endogenous BRaf locus after Cre-mediated deletion of a lox-stop-lox cassette (19). BRafV600E mice were crossed with Mx1-cre, Vav-cre, and Cd19-cre transgenic mice to delineate the effects of mutant BRaf expression in postnatal HSPCs, prenatal hematopoietic cells, and B lineage cells, respectively. Mx1-cre BRafV600E mice developed a lethal hematopoietic disorder characterized by splenomegaly due to EMH [as previously reported (19)] and hepatomegaly (also due to EMH), anemia due to impaired erythroid differentiation, and thrombocytopenia by 3 weeks of age (Fig. 3, A to D, and fig. S2, A and B). The hematopoietic phenotype of Mx1-cre BRafV600E was recapitulated when whole BM was transplanted into lethally irradiated recipient mice, consistent with a cell-autonomous phenotype (Fig. 3, A and B, and fig. S2A) (blood counts, spleen, and liver weights were analyzed 6 weeks after transplantation in 4-week-old lethally irradiated recipient mice to allow time for engraftment; spleen and liver weights were analyzed at 3 weeks of age in Cd19-cre BRafV600E mice). In addition, mice with pan-hematopoietic BRafV600E expression were marked by increased circulating soluble CD25 (sCD25) (Fig. 3E), a well-characterized tumor marker of HCL (6). sCD25 levels were significantly down-regulated (P = 0.002, Mann-Whitney U test) after therapy with the RAF inhibitor PLX4720. Because of a previously described high frequency of spontaneous Cre induction in the Mx1-cre BRafV600E mice (19), we used tamoxifen-inducible mutant BRaf expression in the Cre-ERT BRafV600E model (which has minimal spontaneous excision) to characterize the latency of cell-autonomous hematopoietic abnormalities arising after Cre induction. BM cells from Cre-ERT BRafV600E or BRafV600E Cre-negative control mice were transplanted into lethally irradiated recipients. Blood samples were taken from recipient mice 4 weeks after transplantation (at which time there were no significant differences between Cre-ERT BRafV600E and BRafV600E cre-negative control mice), and then mice were treated with tamoxifen to activate expression of BRafV600E. Two weeks after tamoxifen administration, Cre-ERT BRafV600E mice developed significant anemia (P = 0.002) and thrombocytopenia (P = 0.02) relative to control mice (fig. S2C).
Fig. 3. Phenotypic analysis of mice with pan-hematopoietic versus B lineage–restricted expression of BRafV600E.
(A) White blood cell (WBC) count, hematocrit, and platelet count in 3-week-old primary Mx1-cre BRafV600E mice, lethally irradiated CD45.2 recipient mice 6 weeks after transplantation with Mx1-cre BRafV600E BM, and 3-week-old primary Cd19-cre BRafV600E mice (C, Cre-negative BRafV600E control; KI, Cre+ BRafV600E knock-in; bar represents mean). (B) Weights of spleens and livers of lethally irradiated CD45.2 recipient mice 6 weeks after transplantation with Mx1-cre BRafV600E BM versus 3-week-old Cd19-cre BRafV600E mice (bar represents mean). (C and D) Histological evaluation of spleens (C) and livers (D) from the same mice as in (B). Scale bars, 200 μm. (E) Quantification of serum concentrations of sCD25 in CD45.2 mice 6 weeks after transplantation with cre-negative BRafV600E BM followed by 10 days of vehicle [5% dimethyl sulfoxide (DMSO), 1% methylcellulose] treatment (n = 4), Mx1-cre BRafV600E BM followed by 10 days of vehicle (5% DMSO, 1% methylcellulose; n = 5), or PLX4720 treatment at 50 mg/kg twice daily (n = 5), or 12-week-old Cd19-cre BRafV600E mice treated with vehicle (n = 5). sCD25 levels were significantly (P = 0.001) elevated in Mx1-cre BRafV600E mice treated with vehicle compared with all other groups, and PLX4720 administration resulted in significant (P = 0.002) down-regulation of sCD25 (box and whiskers plot is shown with bottom and top of the box representing the first and third quartiles, and the band inside the box representing the median). *P < 0.05 (Mann-Whitney U test).
Expression of BRafV600E in fetal hematopoietic cells using the Vav-cre transgene resulted in 100% embryonic lethality (fig. S3A). Analysis of embryos generated from crossing Vav-cre transgenic mice to BRafV600E mice revealed complete lethality of Vav-cre BRafV600E embryos beyond day 12.5. At embryonic day 12.5, Vav-cre BRafV600E embryos were observed at Mendelian ratios but were characterized by gross pallor, fetal liver necrosis, and a marked impairment of erythroid differentiation consistent with in utero hematopoietic transformation (fig. S3, B to E).
In contrast to the effects of BRafV600E expression in HSPCs, conditional BRafV600E expression in B lineage cells with Cd19-cre did not result in reduced survival or in an overt hematopoietic phenotype. Mice sacrificed at 1 year of age had no overt phenotype outside of the B lineage, despite clear activation of mitogen-activated protein kinase (MAPK) signaling in B lineage cells (Fig. 3, A to D, and fig. S3, F and G). Cd19-cre BRafV600E mice also had minimal elevation of sCD25 (Fig. 3E). These data demonstrate that restricting BRafV600E expression to committed B lymphoid cells does not result in malignant transformation, suggesting that the phenotype of HCL is driven by alterations in HSPCs.
Effect of BRafV600E on HSC function and germinal center response to alloantigen
Given the expansion of HSCs noted in HCL patients, we next assessed the effects of expression of BRafV600E on murine HSPC subsets (fig. S4). FACS analysis of HSPCs from the BM and spleen of secondarily transplanted Mx1-cre BRafV600E mice and Cre-negative control mice revealed an increase in lineage-negative sca-1+ c-kit+ (LSK), LMPP, and common lymphoid progenitor (CLP) populations in the BM of knock-in mice relative to controls and a large increase in all HSPC populations in the spleen of knock-in mice relative to controls (consistent with EMH in BRafV600E knock-in mice) (fig. S4).
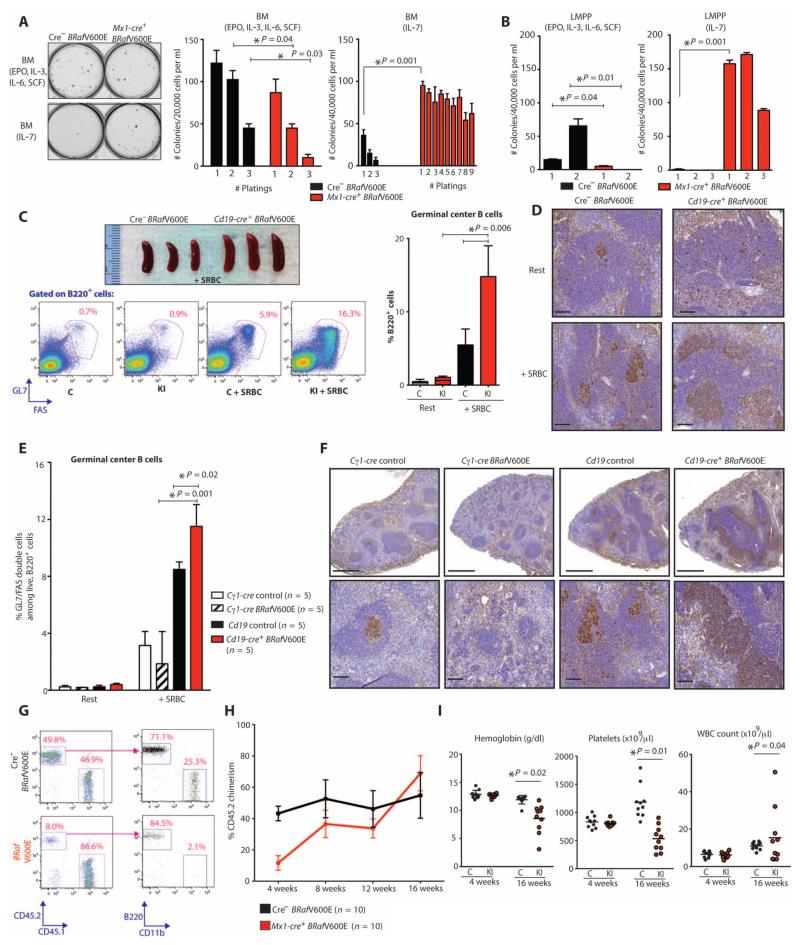
We next sought to understand the effects of BRafV600E expression on HSPC function and lineage specification. We plated unfractionated BM cells from Mx1-cre BRafV600E mice in methylcellulose-containing myeloid and erythroid cytokines [IL-3, IL-6, stem cell factor (SCF), and erythropoietin (EPO)] or the lymphopoietic cytokine IL-7. BRafV600E cells demonstrated impaired colony formation in myeloid and erythroid conditions. However, BRafV600E BM exhibited serial replating capacity (>10 platings) in the presence of IL-7 (Fig. 4A). This increased clonogenic capacity was likewise observed using Cd19-cre BRafV600E BM cells, suggesting increased self-renewal of an early B lineage population (fig. S5A). Accordingly, immunophenotyping of colonies capable of serially replating in IL-7-containing methylcellulose revealed predominantly pre-B cells and their progeny (fig. S5B). Given that LMPPs are poised in hematopoiesis between self-renewing HSCs and lymphoid and granulocyte/macrophage progenitors, we sought to examine the functional effects of BRafV600E expression in this population. Purified LMPP cells (LSK Flk2+) from Mx1-cre BRafV600E mice acquired the ability to serially replate in IL-7-containing methylcellulose but exhibited impaired colony formation relative to control LMPP cells when plated in the presence of myeloid and erythroid cytokines (Fig. 4B). Together, these data suggest that BRafV600E induces aberrant self-renewal in lymphoid precursor cells while impairing myeloid differentiation.
Fig. 4. Effect of BRafV600E mutation on HSPC differentiation, self-renewal, and GC response to alloantigen.
(A and B) Plating of whole BM (A) and sorted LMPP cells (B) in methylcellulose medium containing myeloid and erythroid cytokines (EPO, IL-3, IL-6, and SCF) or IL-7. BRafV600E cells could be replated for >10 platings in the presence of IL-7. Photograph of initial plating shown on left. (C and D) GC response in Cd19-cre BRafV600E (n = 5) and control mice (n = 5) 10 days after SRBC injection by gross photographs of mouse spleens (top), flow cytometric assessment (bottom and bar graph on right) (C), and immunohistochemistry for peanut agglutinin (PNA) (D). Scale bars, 100 μm. C, Cre-negative BRafV600E control; KI, Cd19-cre BRafV600E. (E and F) GC response in Cd19-cre BRafV600E and control mice alongside age-matched mice with GC-restricted BRafV600E expression (Cγ1-cre BRafV600E) by flow cytometry (E) and by PNA stain (F). Scale bars, 500 μm (top) and 100 μm (bottom). (G and H) Competitive transplantation of Mx1-cre BRafV600E (n = 10 recipient mice) compared with Cre-negative BRafV600E whole BM cells (n = 10 recipient mice) 4 weeks (G) and up to 16 weeks (H) after transplantation. (I) Mice transplanted with BRafV600E hematopoietic cells in a competitive manner (n = 10 mice in control and n = 10 mice in knock-in group) developed anemia and thrombocytopenia concomitant with expansion of engrafted BRafV600E HSPCs as shown in (H). Error bars represent means ± SD for (A) to (C), (E), and (H). Bar represents mean value in (I). *P < 0.05 (Mann-Whitney U test).
We next sought to determine the effect of alloantigen perturbation on the B cell phenotype of BRafV600E-mutant mice. Four-week-old mice with B lineage expression of BRafV600E were injected with T cell–dependent antigen sheep red blood cells (SRBCs) to induce germinal center (GC) formation. Analysis performed 10 days later (at which time the GC reaction is at its peak) revealed a significant (P = 0.006) increase in spleen weight, as well as the number and size of GC B cells in BRafV600E mice relative to controls (Fig. 4, C and D). The GC response was more significant (P = 0.02) in Cd19-cre BRafV600E mice compared to mice with GC-restricted expression of BRafV600E (Cγ-cre BRafV600E), suggesting that this enhanced response to antigenic stimulus arises from the effects of BRafV600E in early B lineage cells rather than in mature B cells (Fig. 4, E and F).
Given the human and murine data demonstrating that BRafV600E is acquired in HSCs in HCL, we investigated the effects of mutant BRaf on HSC self-renewal. We assessed the self-renewal of HSCs from CD45.2 Mx1-cre BRafV600E or cre-negative BRaf V600E control mice in competitive repopulation assays. Four weeks after transplantation of equal numbers of Mx1-cre BRafV600E BM cells and cre-negative control cells into lethally irradiated recipient mice, the CD45.2 peripheral blood (PB) chimerism of mice transplanted with CD45.2 Mx1-cre BRafV600E BM was greatly reduced compared to control (Fig. 4, G and H). At the same time point, the proportion of CD45.2 cells that were B220+ B lineage cells was higher in mice transplanted with Mx1-cre BRafV600E CD45.2-positive cells compared to controls (Fig. 4G). Sixteen weeks after transplantation, the proportion of Mx1-cre BRafV600E CD45.2 cells was similar to cre-negative controls, consistent with intact self-renewal of BRafV600E HSCs (Fig. 4H). Mice transplanted with Mx1-cre BRafV600E CD45.2-positive cells developed anemia and thrombocytopenia by 16 weeks after transplantation, a phenotype similar to that which was observed in primary Mx1-cre BRafV600E mice and noncompetitive transplants (Fig. 4I). To exclude an effect of homing or engraftment on the function of BRafV600E BM cells in vivo, we performed competitive transplantation of Cre-ERT BRafV600E BM cells followed by cre-mediated expression of the mutant allele 4 weeks after transplantation, demonstrating a significant (P = 0.006 at 16 weeks after transplantation) competitive advantage of Cre-ERT BRafV600E BM cells (fig. S5C).
Effect of BRAF inhibition on BRAF-mutant HSPCs and hematopoietic differentiation
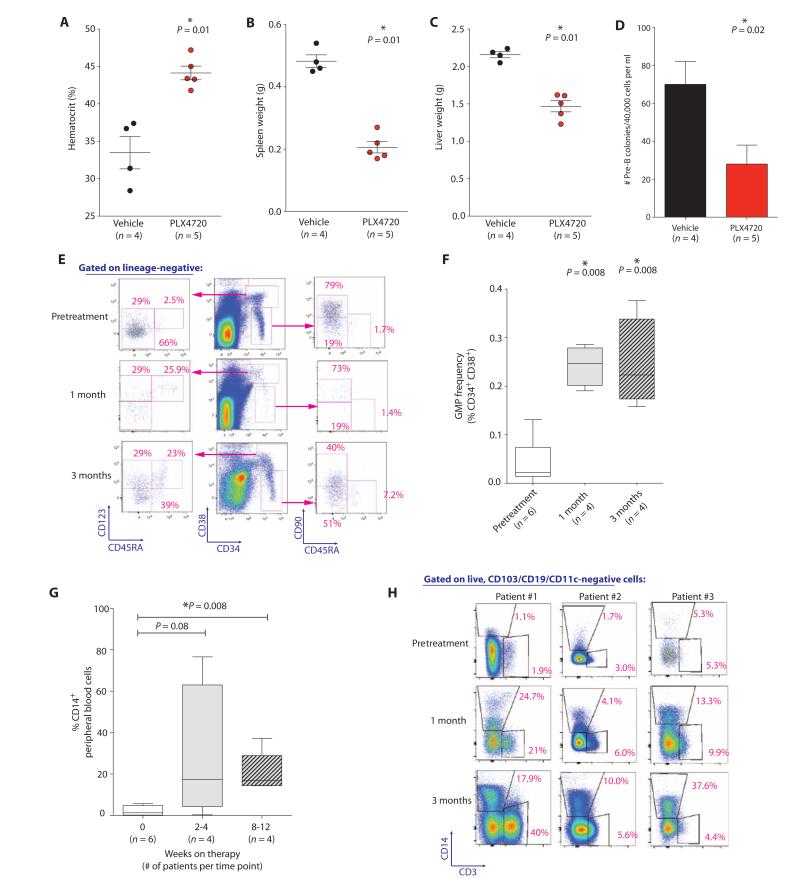
The above genetic and functional data suggest that BRafV600E mutations alter the differentiation and self-renewal potential of HSPCs and that the ensuing lymphoid transformation and block in myeloid and erythroid differentiation result in phenotypic HCL (although cells with classic hairy cell–like morphology were not seen in the murine models). Recent case reports have noted that HCL patients respond to inhibition of mutant BRAF with vemurafenib, as assessed by a reduction in morphologic hairy cells and a reduction in splenomegaly (20, 21). We therefore investigated the effect of vemurafenib on BRAFV600E-mutant HSPCs using murine models and relapsed/refractory HCL patients treated on a phase 2 study of vemurafenib. PLX4720 treatment of wild-type mice transplanted with Mx1-Cre+ BRafV600E-mutant BM cells improved anemia, reduced hepatosplenomegaly, and attenuated B cell clonogenic capacity (Fig. 5, A to D, and fig. S6, A to F). In the clinical context, vemurafenib treatment normalized HSPC frequencies within 3 months of starting therapy (Fig. 5, E and F). Patients treated with vemurafenib had restoration of normal myelopoiesis with BRAF inhibition, demonstrating that the impaired myeloid differentiation in HCL is dependent on mutant BRAFV600E signaling (Fig. 5, G and H). Vemurafenib treatment restored the myeloid colony-forming ability of sorted HSCs (LN CD34+ CD38− CD90+ CD45RA−) and MPs (LN CD34+ CD38+) isolated from HCL patients compared to pretreatment marrows (fig. S6G), suggesting that the improvement in myeloid and erythroid progenitor output in vivo is cell-autonomous.
Fig. 5. Normalization of HSPC compartment and increased myeloid/erythroid output after BRAF inhibition.
(A to D) Effect of 10 days of PLX4720 treatment (50 mg/kg orally twice daily) on hematocrit (A), spleen (B) and liver (C) weights, and ex vivo B cell colony formation (D) relative to vehicle (5% DMSO, 1% methylcellulose) treatment in Mx1-cre BRafV600E mice. (E) Flow cytometric characterization of long-term HSC (LN CD34+ CD38− CD90+ CD45RA−) and GMP frequencies (LN CD19− CD10− CD34+ CD38+ CD123+ CD45RA+) in serial BM aspirates from patients throughout vemurafenib therapy. (F) GMP frequencies throughout vemurafenib therapy in BRAFV600E-mutant HCL patients (four to six patients per time point). BM aspirates were performed before treatment and at 1 and 3 months after vemurafenib as part of an ongoing phase 2 clinical trial of vemurafenib in HCL. (G) Percentage of CD14+ cells among PB mononuclear cells in HCL patients throughout treatment (four to six patients per time point). (H) Analysis of CD14+ and CD3+ cells in PB of three patients throughout therapy. Error bars represent means ± SD in (A) to (D). (F and G) Box plots with band inside box representing median and ends of whiskers representing minimum and maximum values. *P < 0.05 (Mann-Whitney U test).
DISCUSSION
The hallmark leukemic cell in HCL has frequently been considered to be derived from a postgerminal B cell, given that these cells express switched immunoglobulin isotypes (1), with immunoglobulin variable genes that have undergone somatic hypermutation in most patients (3, 22). At the same time, many features of HCL are not consistent with origin from a postgerminal B cell, such as their unique immunophenotype and morphology, as well as decreased hematopoietic output that is often out of proportion to HCL disease burden in the BM. By tracing the origin of a specific somatic aberration characteristic of HCL, we have identified a clear link in the pathogenesis of HCL to an oncogenic disease allele acquired in the HSC compartment. Functional studies with human and murine BRAFV600E-mutant HSCs further demonstrated that the BRAF mutation affects the differentiation and function of different committed hematopoietic progenitors, which may drive the disease phenotype.
Although HCL is a relatively rare malignancy, the present data further demonstrate that mature B cell malignancies can initiate in the HSC compartment. Although the stem cell origin for myeloid malignancies such as myeloproliferative neoplasms, myelodysplastic syndromes, and acute myeloid leukemia (AML) is well established, a link between aberrations in HSPCs and development of mature lymphoid malignancies has been less thoroughly investigated. One reason for this is that, unlike mature myeloid cells, subsets of normal mature B cells are characterized by the capacity to self-renew and differentiate as part of their normal function. For example, the function of memory B cells is to self-renew and generate differentiated progeny in response to antigenic stimuli. Thus, the paradigm of linking B cell malignancies to counterparts in normal B cell development has been a predominant model to describe the cell of origin for these disorders and may have obscured the identification of a more primitive cell of origin. Emerging evidence suggests that HSPCs may play important roles in other neoplasms of mature B cells. For example, multiple myeloma, a disorder considered to be a malignancy of late-stage immunoglobulin-secreting plasma cells, was recently found to contain subpopulations of pre-plasmablasts and CD20+ B cell progenitors, which propagate the disorder and mediate treatment resistance (23). Similarly, Kikushige et al. recently demonstrated that the propensity to generate clonal B cells in patients with the mature B cell malignancy CLL is acquired in the HSC compartment (24). Recent genomic analyses of leukemias of another lymphoid lineage, T cell acute lymphoblastic leukemia (T-ALL), revealed that a specific subset of T-ALL is highly similar to normal and myeloid leukemic HSCs in gene expression and mutational profile (25). Collectively, these findings suggest that genomic and functional analyses of lymphoid malignancies may reveal unexpected alterations in less differentiated HSPC populations.
In the studies by Kikushige et al., xenotransplantation of HSCs from CLL patients gave rise to engraftment of mono- and oligoclonal cell populations with a CLL-like phenotype, but the resulting clones were genetically disparate from the original CLL clones seen in the leukemic cells originally isolated from patients. One potential explanation for this finding is that additional genetic abnormalities acquired along the course of hematopoiesis might be essential in giving rise to the clonal CLL disorder (24, 26). Similarly, here, although BRafV600E expression in HSPCs resulted in increased clonogenic capacity of early B cell populations, as well as anemia, thrombocytopenia, and EMH, no cells with a morphologic phenotype of hairy cells were seen in any murine model studied. This suggests that the development of additional co-occurring genetic alterations along the course of hematopoiesis may be necessary to give rise to mature HCL cells. Alternatively, the level of expression and/or functional role of BRAF in hematopoiesis may be incompletely conserved between mouse and human.
There are several limitations of this study to note. First, we attempted to address the question of whether there is a hierarchy of additional genetic alterations co-occurring with the BRAFV600E mutation in HCL patients. Targeted sequencing analysis of 300 cancer-associated genes in three HCL samples revealed only two additional somatic mutations co-occurring with the BRAFV600E mutation in HCL cells, neither of which were present in HSCs from the same individual. These data are consistent with the BRAFV600E mutation representing an early or inciting event in HCL pathogenesis. This is analogous to recently described preleukemic HSCs from AML patients who harbor somatic mutations in frequently mutated genes such as DNMT3A (27, 28). In such a scenario, a preleukemic HSC clone in HCL might acquire subsequent additional genetic alterations in HSCs, B cell progenitors, or mature B cells, resulting in the appearance of a mature B cell clone that undergoes characteristic immunoglobulin rearrangement and eventually proliferates to manifest as clinically apparent HCL. However, a more extensive mutational analysis of HCL cells and paired HSPCs will be needed to more definitively address this question. Moreover, our use of granulocyte DNA as matched somatic tissue may have obscured additional mutations acquired early in the hematopoietic compartment and present at similar frequencies in granulocyte and HCL DNA. Second, although our analyses of the VAF of the BRAFV600E mutation demonstrated the presence of the BRAF mutation in HSPC subsets from HCL patients, these analyses used cDNA, where the level of wild-type and mutant BRAF expression may differ from the VAF at the level of genomic DNA in these cell subsets.
Data from the murine models studied here and flow cytometric characterization of the BM of HCL patients suggest that the cytopenias seen in HCL patients are due in part to HSPC-intrinsic effects of the BRAFV600E mutation on erythropoiesis, megakaryopoiesis, and myelopoiesis. Moreover, these data suggest that the use of therapies targeting MAPK signaling in HCL may lead to durable remissions not only through effects on mature leukemic cells but also through targeted inhibition of signaling and survival in mutant HSPCs. Indeed, treatment of both murine models and HCL patients with RAF inhibition resulted in improvements in myelopoiesis and erythropoiesis, as well as restoration of aberrant HSPC frequencies. Our results predict that therapeutic mutant BRAF inhibition will have the capability to induce durable remissions and restoration of normal hematopoiesis in HCL.
MATERIALS AND METHODS
Study design and patients
The goal of this study was to understand the cell of origin of HCL through the use of (i) genetic analysis of specific hematopoietic subsets from the BM and blood of HCL patients, (ii) comparison of the effects on hematopoiesis of activating the BRafV600E mutation in various hematopoietic cell subsets in mice, and (iii) analysis of the effects of targeting vemurafenib on hematopoiesis in patients with HCL treated longitudinally with vemurafenib and in BRafV600E mice treated in vivo with vemurafenib.
Patient samples were obtained from patients with newly diagnosed or relapsed HCL seen at Memorial Sloan Kettering Cancer Center. The effect of vemurafenib on stem cell number, function, and hematopoietic reconstitution in vivo was studied using patient samples from an ongoing phase 2 clinical trial of vemurafenib in relapsed or refractory HCL ("A phase II study of the BRAF inhibitor, vemurafenib, in patients with relapsed or refractory hairy cell leukemia," ClinicalTrials.gov identifier NCT01711632). The study is ongoing and being conducted according to the Declaration of Helsinki and with approval from the Institutional Review Board of Memorial Sloan Kettering Cancer Center. All patients had HCL refractory to standard therapy and met all inclusion and exclusion criteria. All provided written informed consent before participating in this study. This study involves exposure to vemurafenib at a dose of 960 mg orally twice a day in back-to-back cycles of 4 weeks (28 days) for at least three cycles. A BM aspirate and/or biopsy is performed before treatment and after the first cycle. After the completion of the third cycle, a repeat BM aspirate and/or biopsy is performed for assessment of response and evaluation of minimal residual disease. If either morphologic or minimal residual disease is evident, an additional three cycles of vemurafenib are given for a maximum of six cycles total.
Experiments analyzing human hematopoietic cell subsets pretreated with vemurafenib (Fig. 1) involved 14 individual HCL patients and 3 normal age-matched controls (table S1). Analysis of the effects of vemurafenib therapy on hematopoiesis longitudinally in clinical trial specimens involved four to six patient samples per group (Fig. 5, F and G) or one to three representative patient samples (Fig. 5, E and H).
All murine experiments were done using 3 to 10 animals per group in each experiment, and all experiments were performed at least three times except where noted. For murine drug experiments, animals were randomly allocated to drug treatment or vehicle treatment, and treatment was administered in an unblinded fashion.
Antibodies and FACS analysis of human and murine cells
Details of all antibodies (for both human and mouse studies) used for FACS and Western blot analyses are located in the Supplementary Materials.
Animals, in vivo studies, and in vivo drug treatment of mice
All animals were housed at Memorial Sloan Kettering Cancer Center. All animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees at Memorial Sloan Kettering Cancer Center.
Mice with conditional expression of BRafV600E (LSL-BRafV600E) from its endogenous locus were originally created and described by Mercer et al. (19). LSL-BRafV600E mice on a pure C57BL/6 background were crossed to interferon-α–inducible Mx1-cre mice (The Jackson Laboratory), hematopoietic-specific Vav-cre mice, B lineage Cd19-cre mice, GC-restricted Cγ-cre mice, and tamoxifen-inducible pCMVCre-ERT mice (described as Cre-ERT in the article) (29-33). All Cre strains listed above were on a pure C57BL/6 background. Further details of transplantation analyses and in vivo drug studies in mice are listed in the Supplementary Materials.
NSG xenotransplantation assay
FACS-purified cells were transplanted by retro-orbital injection into 6-week-old NSG mice (The Jackson Laboratory) conditioned with 225 rads of irradiation. BM aspirates were performed at 3 and 6 months after transplantation, and cells were then analyzed for human engraftment (hCD45+) that was further characterized for human HSC markers (hCD34, hCD38, hCD90, hCD45RA) and HCL markers (hCD19, hCD103, hCD11c).
BRAFV600E allele-specific PCR analysis and quantitative sequencing
Allele-specific oligonucleotide PCR analysis for detection of the BRAFV600E mutation was performed as described previously (14), using cDNA generated from RNA of sorted cell populations. After amplification of PCR products, Sanger sequencing of PCR products generated from amplification using wild-type forward (5′-TAGGTGATTTTGGTCTAGCTACCGT-3′) and common reverse primer (5′-GTAACTCAGCAGCATCTCAGGG-3′), as well as using mutant-specific forward (5′-TAGGTGATTTTGGTCTAGCTACCGA-3′) and common reverse primer, was performed. The presence of amplified PCR products using the mutant-selective forward primer and common reverse primer with clear evidence of the BRAFV600E mutation in Sanger sequencing electropherograms indicated detection of the BRAFV600E mutation.
Quantitative sequencing analysis was performed by first preparing libraries with cDNA or genomic DNA from sorted hematopoietic cell subsets from HCL patients using the Kapa Biosystems Library Preparation Kit. In some instances, cDNA was amplified before library preparation using Kapa HiFi PCR kit followed by Agencourt AMPure bead purification (Beckman Coulter). Libraries were pooled equimolarly and sequenced on an Illumina MiSeq. Sequencing was performed at a median depth of >1000× to identify mutations with VAF of >1% (variants present at VAF<1% were not considered informative).
Sequencing analysis of mutations co-occurring with BRAFV600E mutation in HCL We used standard techniques to extract genomic DNA from flow-sorted HCL and granulocyte cells. Barcoded, massively parallel sequencing libraries were prepared (New England Biolabs, Kapa Biosystems), and exon capture was performed on barcoded pools (NimbleGen Seq-Cap) according to the manufacturer’s directions. Briefly, we designed and synthesized synthetic DNA probes complementary to the coding sequence of 300 genes known to undergo somatic genomic alterations in cancer (table S2). Genomic DNA libraries were subjected to solution-phase hybrid capture using the DNA probes, followed by massively parallel sequencing on the Illumina HiSeq 2500. We sequenced 100 bases from both ends of library DNA fragments, achieving about 15 million purity filtered reads per sample. Paired reads were aligned to the reference human genome (hg19) using the Burrows-Wheeler Alignment tool (34) and postprocessed using the Genome Analysis Toolkit according to best practices (35). Single-nucleotide variants were called using MuTect (36), and indels were called using SomaticIndelDetector (35). All alterations were manually reviewed using the Integrative Genomics Viewer (37).
In vitro colony-forming assays
Details of all methylcellulose colony assays using human or mouse hematopoietic cells are described in the Supplementary Materials.
Histological analyses
Mice were sacrificed and dissected to harvest sternum, femurs, tibiae, spleen, and liver. Tissue samples were fixed for 24 hours in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Paraffin blocks were sectioned at 4 μm and stained with hematoxylin and eosin. Cytospins were performed by resuspending cell pellets in warm phosphate-buffered saline. Cells were then spun onto 3 × 1-inch frosted microscope slides (Fisher Scientific) at 350g for 5 min. Slides were air-dried and stained using the Giemsa-Wright method (38). Images were acquired using a Zeiss Axio Observer A1 microscope.
sCD25 receptor measurement
sCD25 concentration was measured in serum from PB using a previously validated Luminex assay (39). Luminex assays were then carried out using the FLEXMAP 3D multiplexing platform (Luminex xMAP system). Serum samples were prepared according to the manufacturer’s instructions.
Statistical analysis
Values reported represent means ± SD (or ±SEM, where noted). P values were calculated with GraphPad Prism, with P < 0.05 considered significant. Fisher’s exact test (two-tailed) or the Mann-Whitney U test (two-tailed, unpaired) was used to compare blood cell counts, organ weights, and cell frequencies determined by FACS. Experiments were done three to five times except where noted, and the particular statistical analyses used in the experiments are noted in the figure captions. Statistics were performed to illustrate significance between groups where n ≥ 3.
Supplementary Material
Acknowledgments
We thank C. Pritchard for permission to use the BRafV600E conditional lox-stop-lox model.
Funding: This work was supported by a grant from the Geoffrey Beene Cancer Foundation to J.H.P. and O.A.-W. O.A.-W. is supported by an NIH K08 Clinical Investigator Award (1K08CA160647-01), a U.S. Department of Defense Postdoctoral Fellow Award in Bone Marrow Failure Research (W81XWH-12-1-0041), the Josie Roberston Clinical Investigator Program, and a Damon Runyon Clinical Investigator Award with Support from the Evans Foundation. S.S.C. is supported by a Young Investigator Award from the American Society of Clinical Oncology and a U.S. Department of Defense Postdoctoral Fellow Award in Bone Marrow Failure Research (BM120096). J.H.P. is supported by a Special Fellow Award in Clinical Research from the Leukemia and Lymphoma Society.
Footnotes
www.sciencetranslationalmedicine.org/cgi/content/full/6/238/238ra71/DC1
Materials and Methods
Fig. S1. HSPC abnormalities in HCL and the presence of BRAFV600E in HSCs.
Fig. S2. Effect of BRafV600E expression at different time points and stages of hematopoiesis.
Fig. S3. Effect of BRafV600E expression on fetal hematopoiesis and in mice with B cell–restricted expression of the mutant allele.
Fig. S4. Effect of BRafV600E expression on HSPC numbers and frequencies.
Fig. S5. Effect of BRafV600E expression on B cell development and self-renewal.
Fig. S6. Normalization of HSPC compartment and increased myeloid/erythroid output from BRAFV600E-mutant HSPCs after BRAF inhibition.
Table S1. Genotyping results of cell populations sorted from HCL patient BM aspirates.
Table S2. Three hundred genes sequenced in three HCL leukemic cell and granulocyte genomic DNA samples.
Author contributions: S.S.C., E.K., J.H.P., M.S.T., C.Y.P., and O.A.-W. designed the study. S.S.C., C.Y.P., W.H., and O.A.-W. performed sorting of human samples. E.K., L.T., Y.R.C., and M.K.K. helped with animal generation, genotyping, and maintenance, as well as experiments involving animals. P.L. and N.R. provided PLX4720 and assistance and advice with drug studies. W.B., C.D., and A.M.M. helped with GC studies and immunohistochemistry. J.T.-F., S.M., and C.Y.P. performed phenotypic and histologic analysis of tissues. J.H.P., R.R., M.P., and M.S.T. helped with collection of patient materials for study. K.H., N.B., and M.F.B. performed genetic analyses on sorted cell populations. S.S.C., E.K., Y.R.C., and O.A.-W. analyzed the data. S.S.C., E.K., and O.A.-W. prepared the manuscript with input from the other authors.
Competing interests: A.M.M. serves as a consultant for Celgene, is part of the speaker bureau for Genentech Basic Science Education Program, and is on the scientific advisory board for Bio-Reference Laboratories. The other authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Golomb H, Davis S, Wilson C, Vardiman J. Surface immunoglobulins on hairy cells of 55 patients with hairy cell leukemia. Am. J. Hematol. 1982;12:397–401. doi: 10.1002/ajh.2830120411. [DOI] [PubMed] [Google Scholar]
- 2.Forconi F, Sahota S, Raspadori D, Mockridge C, Lauria F, Stevenson F. Tumor cells of hairy cell leukemia express multiple clonally related immunoglobulin isotypes via RNA splicing. Blood. 2001;98:1174–1181. doi: 10.1182/blood.v98.4.1174. [DOI] [PubMed] [Google Scholar]
- 3.Maloum K, Magnac C, Azgui Z, Cau C, Charlotte F, Binet J, Merle-Béral H, Dighiero G. VH gene expression in hairy cell leukaemia. Br. J. Haematol. 1998;101:171–178. doi: 10.1046/j.1365-2141.1998.00653.x. [DOI] [PubMed] [Google Scholar]
- 4.Polliack A. Hairy cell leukemia: Biology, clinical diagnosis, unusual manifestations and associated disorders. Rev. Clin. Exp. Hematol. 2002;6:366–388. doi: 10.1046/j.1468-0734.2002.00304.x. [DOI] [PubMed] [Google Scholar]
- 5.Tiacci E, Liso A, Piris M, Falini B. Evolving concepts in the pathogenesis of hairy-cell leukaemia. Nat. Rev. Cancer. 2006;6:437–448. doi: 10.1038/nrc1888. [DOI] [PubMed] [Google Scholar]
- 6.Janik J. Tumor markers in hairy cell leukemia. Leuk. Lymphoma. 2011;52(Suppl. 2):69–71. doi: 10.3109/10428194.2011.568651. [DOI] [PubMed] [Google Scholar]
- 7.Juliusson G, Lenkei R, Liliemark J. Flow cytometry of blood and bone marrow cells from patients with hairy cell leukemia: Phenotype of hairy cells and lymphocyte subsets after treatment with 2-chlorodeoxyadenosine. Blood. 1994;83:3672–3681. [PubMed] [Google Scholar]
- 8.Basso K, Liso A, Tiacci E, Benedetti R, Pulsoni A, Foa R, Di Raimondo F, Ambrosetti A, Califano A, Klein U, Dalla Favera R, Falini B. Gene expression profiling of hairy cell leukemia reveals a phenotype related to memory B cells with altered expression of chemokine and adhesion receptors. J. Exp. Med. 2004;199:59–68. doi: 10.1084/jem.20031175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson K, Boyd A, Fisher D, Leslie D, Schlossman S, Nadler L. Hairy cell leukemia: A tumor of pre-plasma cells. Blood. 1985;65:620–629. [PubMed] [Google Scholar]
- 10.Burke J, Sheibani K. Hairy cells and monocytoid B lymphocytes: Are they related? Leukemia. 1987;1:298–300. [PubMed] [Google Scholar]
- 11.Thorsélius M, Walsh S, Thunberg U, Hagberg H, Sundström C, Rosenquist R. Heterogeneous somatic hypermutation status confounds the cell of origin in hairy cell leukemia. Leuk. Res. 2005;29:153–158. doi: 10.1016/j.leukres.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 12.van den Oord J, de Wolf-Peeters C, Desmet V. Hairy cell leukemia: A B-lymphocytic disorder derived from splenic marginal zone lymphocytes? Blut. 1985;50:191–194. doi: 10.1007/BF00320293. [DOI] [PubMed] [Google Scholar]
- 13.Tiacci E, Trifonov V, Schiavoni G, Holmes A, Kern W, Martelli M, Pucciarini A, Bigerna B, Pacini R, Wells V, Sportoletti P, Pettirossi V, Mannucci R, Elliott O, Liso A, Ambrosetti A, Pulsoni A, Forconi F, Trentin L, Semenzato G, Inghirami G, Capponi M, Di Raimondo F, Patti C, Arcaini L, Musto P, Pileri S, Haferlach C, Schnittger S, Pizzolo G, Foà R, Farinelli L, Haferlach T, Pasqualucci L, Rabadan R, Falini B. BRAF mutations in hairy-cell leukemia. N. Engl. J. Med. 2011;364:2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arcaini L, Zibellini S, Boveri E, Riboni R, Rattotti S, Varettoni M, Guerrera M, Lucioni M, Tenore A, Merli M, Rizzi S, Morello L, Cavalloni C, Da Vià M, Paulli M, Cazzola M. The BRAFV600E mutation in hairy cell leukemia and other mature B-cell neoplasms. Blood. 2012;119:188–191. doi: 10.1182/blood-2011-08-368209. [DOI] [PubMed] [Google Scholar]
- 15.Schnittger S, Bacher U, Haferlach T, Wendland N, Ulke M, Dicker F, Grossmann V, Haferlach C, Kern W. Development and validation of a real-time quantification assay to detect and monitor BRAFV600E mutations in hairy cell leukemia. Blood. 2012;119:3151–3154. doi: 10.1182/blood-2011-10-383323. [DOI] [PubMed] [Google Scholar]
- 16.Goardon N, Marchi E, Atzberger A, Quek L, Schuh A, Soneji S, Woll P, Mead A, Alford KA, Rout R, Chaudhury S, Gilkes A, Knapper S, Beldjord K, Begum S, Rose S, Geddes N, Griffiths M, Standen G, Sternberg A, Cavenagh J, Hunter H, Bowen D, Killick S, Robinson L, Price A, Macintyre E, Virgo P, Burnett A, Craddock C, Enver T, Jacobsen SE, Porcher C, Vyas P. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19:138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 18.Won HH, Scott SN, Brannon AR, Shah RH, Berger MF. Detecting somatic genetic alterations in tumor specimens by exon capture and massively parallel sequencing. J. Vis. Exp. 2013:e50710. doi: 10.3791/50710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercer K, Giblett S, Green S, Lloyd D, DaRocha Dias S, Plumb M, Marais R, Pritchard C. Expression of endogenous oncogenic V600EB-raf induces proliferation and developmental defects in mice and transformation of primary fibroblasts. Cancer Res. 2005;65:11493–11500. doi: 10.1158/0008-5472.CAN-05-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dietrich S, Glimm H, Andrulis M, von Kalle C, Ho A, Zenz T. BRAF inhibition in refractory hairy-cell leukemia. N. Engl. J. Med. 2012;366:2038–2040. doi: 10.1056/NEJMc1202124. [DOI] [PubMed] [Google Scholar]
- 21.Munoz J, Schlette E, Kurzrock R. Rapid response to vemurafenib in a heavily pretreated patient with hairy cell leukemia and a BRAF mutation. J. Clin. Oncol. 2013;31:e351–e352. doi: 10.1200/JCO.2012.45.7739. [DOI] [PubMed] [Google Scholar]
- 22.Vanhentenrijk V, Tierens A, Wlodarska I, Verhoef G, Wolf-Peeters C. VH gene analysis of hairy cell leukemia reveals a homogeneous mutation status and suggests its marginal zone B-cell origin. Leukemia. 2004;18:1729–1732. doi: 10.1038/sj.leu.2403503. [DOI] [PubMed] [Google Scholar]
- 23.Leung-Hagesteijn C, Erdmann N, Cheung G, Keats JJ, Stewart AK, Reece DE, Chung KC, Tiedemann RE. Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell. 2013;24:289–304. doi: 10.1016/j.ccr.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kikushige Y, Ishikawa F, Miyamoto T, Shima T, Urata S, Yoshimoto G, Mori Y, Iino T, Yamauchi T, Eto T, Niiro H, Iwasaki H, Takenaka K, Akashi K. Self-renewing hematopoietic stem cell is the primary target in pathogenesis of human chronic lymphocytic leukemia. Cancer Cell. 2011;20:246–259. doi: 10.1016/j.ccr.2011.06.029. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, Lu C, Chen SC, Wei L, Collins-Underwood JR, Ma J, Roberts KG, Pounds SB, Ulyanov A, Becksfort J, Gupta P, Huether R, Kriwacki RW, Parker M, McGoldrick DJ, Zhao D, Alford D, Espy S, Bobba KC, Song G, Pei D, Cheng C, Roberts S, Barbato MI, Campana D, Coustan-Smith E, Shurtleff SA, Raimondi SC, Kleppe M, Cools J, Shimano KA, Hermiston ML, Doulatov S, Eppert K, Laurenti E, Notta F, Dick JE, Basso G, Hunger SP, Loh ML, Devidas M, Wood B, Winter S, Dunsmore KP, Fulton RS, Fulton LL, Hong X, Harris CC, Dooling DJ, Ochoa K, Johnson KJ, Obenauer JC, Evans WE, Pui CH, Naeve CW, Ley TJ, Mardis ER, Wilson RK, Downing JR, Mullighan CG. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alizadeh A, Majeti R. Surprise! HSC are aberrant in chronic lymphocytic leukemia. Cancer Cell. 2011;20:135–136. doi: 10.1016/j.ccr.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, McLeod JL, Doedens M, Medeiros JJ, Marke R, Kim HJ, Lee K, McPherson JD, Hudson TJ, Brown AM, Trinh QM, Stein LD, Minden MD, Wang JC, Dick JE. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl. Acad. Sci. U.S.A. 2014;111:2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 30.Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stadtfeld M, Graf T. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development. 2005;132:203–213. doi: 10.1242/dev.01558. [DOI] [PubMed] [Google Scholar]
- 32.Casola S, Cattoretti G, Uyttersprot N, Koralov S, Seagal J, Segal J, Hao Z, Waisman A, Egert A, Ghitza D, Rajewsky K. Tracking germinal center B cells expressing germ-line immunoglobulin γ1 transcripts by conditional gene targeting. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7396–7401. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DePristo M, Banks E, Poplin R, Garimella K, Maguire J, Hartl C, Philippakis A, del Angel G, Rivas M, Hanna M, McKenna A, Fennell T, Kernytsky A, Sivachenko A, Cibulskis K, Gabriel S, Altshuler D, Daly M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cibulskis K, Lawrence M, Carter S, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander E, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorvaldsdóttir H, Robinson J, Mesirov J. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strober W. Wright-Giemsa and nonspecific esterase staining of cells. Curr. Protoc. Cytom. 2001 doi: 10.1002/0471142956.cya03ds11. Appendix 3, Appendix 3D. [DOI] [PubMed] [Google Scholar]
- 39.Russell SE, Moore AC, Fallon PG, Walsh PT. Soluble IL-2Rα (sCD25) exacerbates auto-immunity and enhances the development of Th17 responses in mice. PLOS One. 2012;7:e47748. doi: 10.1371/journal.pone.0047748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.