Abstract
Mutations in ASXL1 are frequent in patients with myelodysplastic syndrome (MDS) and associated with adverse survival yet the molecular pathogenesis of ASXL1 mutations are not fully understood. Recently it has been found that deletion of Asxl1 or expression of C-terminal-truncating ASXL1 mutations (ASXL1-MT) inhibit myeloid differentiation and induce MDS-like disease in mice. Here, we find that SETBP1 mutations (SETBP1-MT) are enriched among patients with ASXL1-mutated MDS patients and associated with increased incidence of leukemic transformation as well as shorter survival, suggesting SETBP1-MT play a critical role in leukemic transformation of MDS. We identify that SETBP1-MT inhibit ubiquitination and subsequent degradation of SETBP1, resulting in increased expression. Expression of SETBP1-MT, in turn, inhibited Pp2a activity, leading to Akt activation and enhanced expression of posterior Hoxa genes in ASXL1 mutant cells. Biologically, SETBP1-MT augmented ASXL1-MT-induced differentiation block, inhibited apoptosis, and enhanced myeloid colony output. SETBP1-MT collaborated with ASXL1-MT in inducing AML in vivo. The combination of ASXL1-MT and SETBP1-MT activated a stem cell signature and repressed the TGF-β signaling pathway, in contrast to the ASXL1-MT-induced MDS model. These data reveal that SETBP1-MT are critical drivers of ASXL1-mutated MDS and identify several deregulated pathways as potential therapeutic targets in high-risk MDS.
INTRODUCTION
Myelodysplastic syndromes (MDS) are heterogeneous clonal diseases of hematopoietic stem cells (HSCs) characterized by dysplastic myeloid cells, ineffective hematopoiesis associated with increased apoptosis, and a predisposition to acute myeloid leukemia (AML).1–3 Transformation of MDS to AML is one of the largest contributors of mortality in MDS patients and a number of studies have suggested genetic alterations which contribute to leukemic transformation of MDS.1, 3, 4 However, the underlying mechanisms for transformation of MDS to AML remain largely elusive and genetically accurate preclinical models for leukemic transformation of MDS are lacking.
With the continuous innovation in the field of next-generation sequencing, a variety of mutations have recently been identified that shed light on the molecular pathogenesis of MDS.5 Several genetic alterations related to epigenetic regulation (including ASXL1, TET2, and EZH2), splicing machinery (including SF3B1, SRSF2, and U2AF1), and cohesion complex (STAG2, RAD21, and SMC3) have been identified in MDS patients in addition to those of transcription factors and oncogenes.6–13 Recent data suggests that even a single alteration of one epigenetic regulator may induce myeloid malignancies such as MDS or MPN (myeloproliferative neoplasms) in vivo.14–20 Using a mouse bone marrow transplantation (BMT) model, we have recently found that expression of C-terminal-truncating ASXL1 mutants (ASXL1-MT), which are found in 15–20% of MDS patients, inhibit myeloid differentiation and induce an MDS-like disease after a long latency by abrogating polycomb repressive complex 2 (PRC2)-mediated methylation of histone H3K27.14 These mice mimicked human MDS in terms of multilineage myelodysplasia, pancytopenia, and hypercellular bone marrow, as well as occasional leukemic transformation. However, given that ASXL1 mutations have been linked to high-risk MDS and leukemic transformation of MDS, further studies to identify the mechanistic basis for transformation of ASXL1-mutated low-risk MDS to high-risk MDS/AML are needed.21–23
In multiple clinical studies with large cohorts of MDS samples, ASXL1 mutations have been reported to be statistically associated with mutations in RUNX1, EZH2, SRSF2, STAG2, NRAS, and SET binding protein 1 (SETBP1).4, 22, 24 In particular, mutations in NRAS and SETBP1 appear to occur after initial establishment of MDS, suggesting that these mutations contribute to disease progression or evolution.4, 25–27 Consistent with this, we previously revealed that ASXL1-MT or ASXL1-knockdown collaborated with NRAS mutations to induce AML.14, 16 The biologic contribution of SETBP1 mutations, in contrast, have not been fully investigated, although several groups have reported that SETBP1 mutations seem to cause gain of function and play oncogenic roles in myeloid malignancies via protection of the protein from degradation by ubiquitination including MDS, chronic myelomonocytic leukemia (CMML) and atypical chronic myeloid leukemia (aCML) and that its mutation is a poor prognostic factor.25, 26, 28, 29 Moreover, in vivo models reflecting oncogenic function of SETBP1 mutation have not been presented. SETBP1 encodes a predominantly nuclear-localized protein that binds the SET oncoprotein and the resulting heterodimer interacts with PP2A, a major serine/threonine phosphatase and tumor suppressor regulating cell proliferation. Formation of a SETBP1-SET-PP2A complex results in PP2A inhibition and AKT activation.25, 30, 31 In addition, Oakley et al. demonstrated that overexpression of Setbp1 promotes the self-renewal of murine progenitors via binding to Hoxa9 and Hoxa10 promoters.32
In this study, we identify genetic and functional evidence for a collaborative association between ASXL1 and SETBP1 mutations among patients with advanced MDS. These data present a novel murine model for leukemic transformation of MDS, provide evidence to support a new role for SETBP1 mutation in leukemic transformation, and identify a number of pathways necessary for leukemic progression of ASXL1/SETBP1-mutated myeloid malignancies.
MATERIALS AND METHODS
Patients
Adult patients (n = 386) diagnosed with de novo MDS according to the 2008 WHO classification at the National Taiwan University Hospital (NTUH) and had cryopreserved bone marrow cells for study were recruited for gene mutation analyses.22, 26 All patients signed informed consents for sample collection in accordance with the Declaration of Helsinki. The clinical portion of this study was approved by the Institutional Review Board of the NTUH (approval 201207075RIB).
Mutation analysis
The coding region of ASXL1 from exon 12 until the stop codon was amplified by three pairs of primers and sequenced by another set of six internal primers. The primer sequences and PCR conditions were described previously.26, 33 Mutation analyses of SETBP1 were performed as described previously.26
Cell culture and differentiation assay
HEK293T and HL60 cells were cultured in DMEM supplemented with 10% FBS and in RPMI-1640 supplemented with 10% FBS, respectively. The murine myeloid cell line 32Dcl3 was grown in RPMI-1640 medium supplemented with 10% FBS and 1 ng/mL IL-3. Before assays for apoptosis and differentiation, transduced 32Dcl3 or HL60 cells were GFP-sorted or subjected to drug selection with 1 µg/mL puromycin and/or 10 µg/mL blasticidin, if necessary.
Analyses of cell growth and apoptosis
The relative rate of viable cells was estimated using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA). Cells were stained with PE-conjugated annexin V (R&D systems, Minneapolis, MN, USA) and apoptosis was determined using flow cytometry. All data with error bars represent the mean ± SEM.
Vector construction
Human SETBP1-WT and SETBP1-D868N cDNA with TY1 or FLAG tags were subcloned into the pMYs-IRES-GFP (pMYs-IG) retrovirus vector. We constructed and used the retrovirus vectors pMYs-SETBP1-WT-3×TY1-IG, pMYs-SETBP1-D868N-3×TY1-IG, pMYs-SETBP1-D868N-3×FLAG-IG, and pMYs-ASXL1-MT-IRES–nerve growth factor receptor (pMYs-ASXL1-MT-INGFR), in which SETBP1-WT or SETBP1-D868N tagged with a TY1 or FLAG epitope at the C terminus and ASXL1-MT (1900–1922del;E635RfsX15) tagged with a FLAG epitope at the N terminus was inserted upstream of the IRES-EGFP/NGFR cassette of pMYs-IG/pMYs-INGFR.
Western blot analysis
Cell lysates were subjected to immunoblotting using the following antibodies: TY1 (clone MAb-054-050; Diagenode, Denville, NJ, USA), tubulin (clone B-5-1–2; Santa Cruz Biotechnology, Dallas, TX, USA), HA (clone 3F10; Roche, Penzberg, Germany), Akt1 (clone C-20; Santa Cruz Biotechnology), Phospho-Akt (Thr308) (clone D25E6; Cell Signaling Technology, Danvers, MA, USA), Phospho-Akt (Ser473) (clone D9E; Cell Signaling Technology), PP2A (clone 1D6; Millipore, Billerica, MA, USA), Phospho-PP2A alpha (clone E155; Abcam, Cambridge, UK), FLAG (M2 FLAG; Sigma-Aldrich, St. Louis, MO, USA), ERK1/2 (clone C-16/C-14; Santa Cruz Biotechnology), and I2PP2A (SET) (clone E-15; Santa Cruz Biotechnology). Immunoprecipitation was performed in an immunoprecipitation buffer (150 mM NaCl, 50 mM Tris pH 7.5, 1 mM EDTA, 1% Triton, 2 mM sodium orthovanadate, 2 mM PMSF, 50 mM sodium fluoride) as previously reported.14
Flow cytometric analysis
Briefly, cells were stained with the indicated phycoerythrin (PE)-conjugated antibodies (eBioscience, San Diego, CA, USA). Flow cytometric analysis of the stained cells was performed with FACSCalibur Flow (BD Biosciences, San Jose, CA, USA) equipped with FlowJo Version 7.2.4 software (Tree Star, Ashland, OR, USA). All data with error bars represent the mean ± SEM.
Colony-forming assay
Lineage-negative Sca1+ c-Kit+ (LSK) cells were isolated from bone marrow (BM) of C57BL/6 mice according to published methods.34, 35 Retrovirus-infected LSK cells were sorted at 60 hours from the initiation of infection with a FACSAria cell sorter (Becton Dickinson, Franklin Lakes, NJ, USA) for use in the Methocult 3231 colony-forming assay (StemCell Technologies, Vancouver, Canada) supplemented with 100 ng/mL mouse SCF and 20 ng/mL mouse IL-3. A total of 1,000 cells were cultured in duplicate in 2.5-cm dishes. The colony-forming cells were harvested and replated every 7 days and scored for colony formation.
Mice
C57BL/6 (Ly5.1) mice (Sankyo Labo Service Corporation, Tokyo, Japan) and C57BL/6 (Ly5.2) mice (Charles River Laboratories Japan, Yokohama, Japan) were used for BMT experiments. Mouse BMT was performed as described previously.36 Briefly, BM cells derived from 5-fluorouracil-treated Ly5.2 mice were transduced with the retrovirus vectors described above, and the transduced cells were transplanted into sublethally-irradiated Ly5.1 mice.
Transfection and retrovirus production
Retroviral production was done as described previously.36, 37 Briefly, retroviruses were generated by transient transfection of Plat-E packaging cells using the calcium-phosphate co-precipitation method. An alternative method was used for the retroviral production of SETBP1-WT or SETBP1-D868N to obtain an adequate viral titer. The retrovirus vectors and pcDNA3/VSV-G were transfected into a 293gp packaging cell line using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA) and the subsequent viral supernatants were collected. These supernatants were used to infect 293gpg cells, which were then sorted by FACS AriaII (Becton Dickinson) twice to enrich for cells that expressed EGFP at high levels. VSV-G pseudotyped retroviruses were produced by removing tetracycline from the culture media and the viruses were concentrated by centrifugation at 6,000 × g for 16 h. Cell lines 32Dcl3 and HL60 cells were infected with retroviruses as previously described.37
qRT-PCR
As previously described, total RNAs were treated with deoxyribonuclease I (Invitrogen) and reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA).38 qRT-PCR was performed using SYBR Premix EX Taq (Takara Bio, Otsu, Japan) and Rotor-Gene Q (Qiagen, Venlo, Netherlands). Primer sets are described in Supplementary Table 1.
ChIP assay
Chromatin immunoprecipitation (ChIP) assays were performed using anti-TY1 antibodies as described previously.39 Quantitative PCR was performed with a Rotor-Gene Q (Qiagen) using SYBR Premix EX Taq (Takara). Primer sets are described in Supplementary Table 1. All data with error bars indicate the mean ± SEM. Data are representative of three independent experiments.
PP2A phosphatase assay
A phosphatase assay from whole cell lysates was carried out using the PP2Ac Immunoprecipitation Phosphatase Assay kit (Millipore) according to the manufacturer’s protocol as previously described.40
BM serial transplantation and drug model of FTY720
GFP/NGFR-positive BM cells (1 × 105 cells) from leukemic mice were transplanted into sublethally irradiated C57BL/6 (Ly5.2) mice. After confirmation of engraftment (2 weeks after transplant), mice were treated with FTY720 (Selleck, Houston, TX, USA, 10 mg/kg/d, i.p.) or vehicle for 5 days.
Transcriptome analysis
Library preparation for whole transcriptome analysis was performed according to the manufacturer’s instructions (SureSelect Strand Specific RNA Preparation Kit, Agilent Technologies, Santa Clara, CA, USA). Briefly, poly(A) RNA purified from 2 µg of total RNA was chemically fragmented to appropriate sizes. First-strand cDNA was synthesized from the fragmented poly(A)-selected mRNA, followed by the synthesis of second-strand cDNA. Then, adaptor oligo-DNA was ligated to both ends of the double-stranded cDNA. Libraries were prepared with 12 cycles of PCR amplification of adaptor-ligated cDNA using primers that are complimentary to the adaptors. The libraries were sequenced by GAIIx next generation sequencer (Illumina, San Diego, CA, USA) using the single-end 36 bp sequencing protocol. The generated sequence tags were mapped onto the murine genomic sequence (mm9, UCSC Genome Browser) and mRNA expression levels were normalized as reads per kilobase per million (RPKM).
RESULTS
Clinical significance of SETBP1 mutation among ASXL1-mutated MDS patients
We first investigated a possible genetic interaction between SETBP1 and ASXL1 mutations in MDS patients through analysis of ASXL1 and SETBP1 in a cohort of 368 patients with de novo WHO-defined MDS. ASXL1 mutations were detected in 17.39% (64/368) of the cohort. Patients with an ASXL1 mutation had a significantly higher incidence of concurrent SETBP1 mutations than ASXL1-wildtype MDS patients (6/64 (9.38%) vs. 2/304 (0.66%); p=0.0005) (Figure 1a). The mutation patterns of six MDS patients with concurrent ASXL1 and SETBP1 mutations are shown in Supplementary Table 2. Consistent with recent reports, SETBP1D868N (n = 3), E858K (n = 1), G870S (n = 1), and S867R (n = 1) mutations, all of which are within the SKI homologous region, were identified (Figure 1b and Supplementary Table 2). Other than E858K, these mutations were found in the consensus binding region for β-TrCP1, the substrate recognition subunit for E3 ubiquitin ligase.29
Figure 1.
SETBP1 mutations are enriched in ASXL1-mutant de novo MDS patients and inhibit ubiquitination and subsequent degradation of SETBP1. (a) Proportion of SETBP1 mutations among ASXL1-mutated (left, n=64) or ASXL1-wild type (right, n=304) MDS patients are indicated. Red portion shows SETBP1-mutational frequency. (b) Schematic diagram of SETBP1 protein and SETBP1 mutations (asterisk) identified in ASXL1-mutated MDS patients. Three AT hook domains, SKI homologous region, SET binding domain and repeat domain are indicated. (c–d) Cumulative incidence of leukemic transformation (c) and Kaplan-Meier analysis for survival (d) in ASXL1-mutated MDS patients with or without SETBP1 mutations (n=6, solid line, n=58, dashed line, respectively; p-values calculated using a log-rank test).
Next, we analyzed the impact of SETBP1 mutations on the prognosis of ASXL1-mutated cases. Among ASXL1-mutated patients, those harboring SETBP1 mutations had a higher incidence of leukemic transformation than those without (p=0.042; Figure 1c). Furthermore, MDS patients with both mutations had a significantly shorter overall survival compared to those without SETBP1 mutations (median, 10.5 vs. 22.5 months, p= 0.046; Figure 1d). We also performed multivariate analysis for the overall survival in 64 MDS patients with ASXL1 mutations and found that the SETBP1 mutation was an independent poor prognostic factor (relative risk 3.340, 95% CI 1.150–9.698, p =0.027) irrespective of age, 2008 WHO classification and IPSS (International Prognostic Scoring System) classification (Supplementary Table 3).
Increased stability of SETBP1-MT due to decreased ubiquitination
Next, we evaluated the protein expression levels of SETBP1-WT and SETBP1-D868N by Western blot analysis after transfecting HEK293T with empty vector, 3×TY1-SETBP1-WT, or 3×TY1-SETBP1-D868N (Supplementary Figure 1A and 1B). As Piazza et al. showed using G870S mutant, another mutant SETBP1-D868N also exhibited a dramatically increased stability compared to SETBP1-WT. Treatment with MG-132, a protease inhibitor, increased the expression levels of SETBP1-WT.29 These data suggest that SETBP1-D868N can escape ubiquitination and subsequent degradation by inhibiting its binding to the E3 ligase β-TrCP1, as has been previously identified for SETBP1-G870S mutations.29 These results indicate that D868N acts as a gain-of-function mutation.
Expression of the SETBP1 mutant impairs apoptosis and differentiation
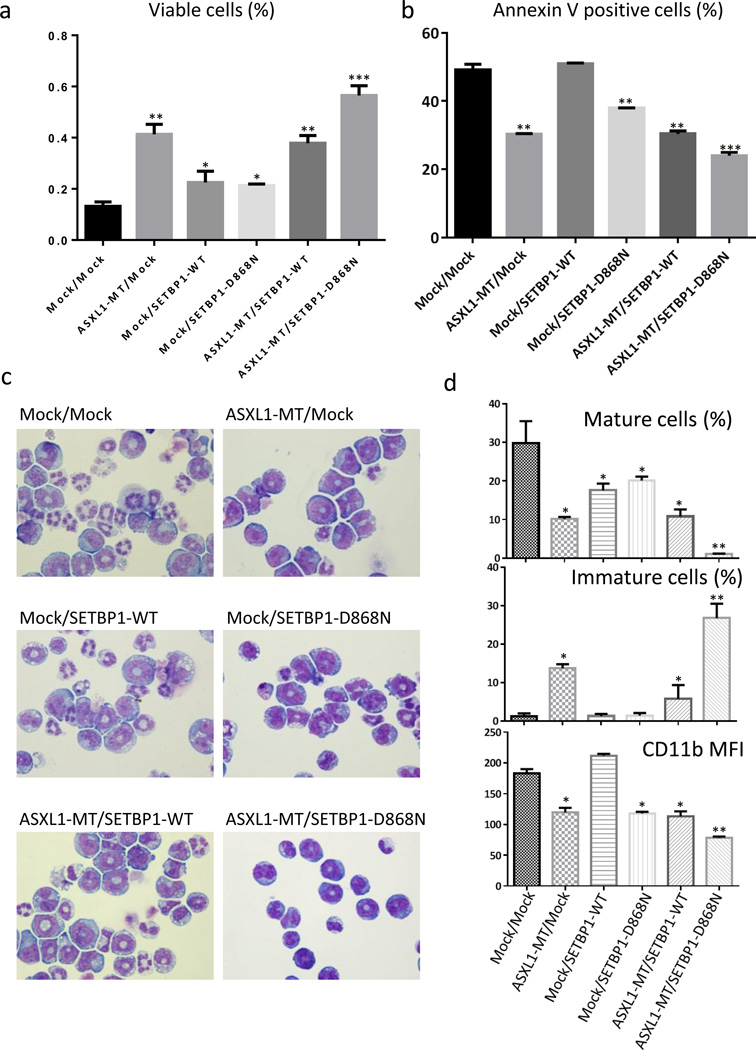
In order to understand the biological contribution of SETBP1 mutations to ASXL1 mutations, we first examined the effect of SETBP1 mutant on apoptosis or differentiation of an ASXL1-mutated clone. We produced IL-3-dependent murine 32Dcl3 myeloid cells overexpressing mock/mock, ASXL1-MT (ASXL1E635RfsX15)/mock, mock/SETBP1-WT, mock/SETBP1-D868N, ASXL1-MT/SETBP1-WT, or ASXL1-MT/SETBP1-D868N using pMYs-IRES-Puro and pMYs-IRES-GFP vectors, whose expressions were confirmed using qRT-PCR (Supplementary Figure 2A and 2B). As shown in Figure 2a and 2b and Supplementary Figure 2C, expression of both ASXL1-MT and SETBP1-D868N attenuated apoptosis of 32Dcl3 cells induced by IL-3 withdrawal, although it did not induce factor-independent growth nor change the growth rate of 32Dcl3 cells in the presence of IL-3 (data not shown). Cell cycle analysis of the transfectants revealed that simultaneous expression of ASXL1-MT and SETBP1-D868N resulted in a slight increase of cells in G2/M and S phases and concomitant reductions of cells in the G1 phase (Supplementary Figure 2D), suggesting that the additional expression of SETBP1-D868N induces cell cycle progression.
Figure 2.
SETBP1-MT impairs differentiation and apoptosis in vitro. (a–b) IL-3 dependent murine myeloid 32Dcl3 cells transduced with pMYs-IP/pMYs-IG, pMYs-IP-ASXL1-MT/mock, mock/pMYs-IG-SETBP1-WT, mock/pMYs-IG-SETBP1-D868N, pMYs-IP-ASXL1-MT/pMYs-IG-SETBP1-WT, or pMYs-IP-ASXL1-MT/pMYs-IG-SETBP1-D868N were cultured in RPMI-1640 supplemented with 10% FBS. Proportions of viable cells (a) and Annexin V positive cells (b) 24 hours after IL-3 withdrawal are shown. All data with error bars indicate the mean + SEM from duplicated experiments. (c–d) The transduced 32Dcl3 cells were cultured with 50 ng/ml of G-CSF for 6 days. Cytospin preparations of these cells assessed by Wright-Giemsa staining (c) are shown. Original magnification, ×400. Proportions of mature (segmented) cells (%, top) and immature (blastic) cells (%, middle) or mean fluorescent intensity (MFI) of CD11b estimated by FACS analysis (bottom) are shown. All data with error bars indicate the mean + SEM from duplicated experiments. Compared to the control, *p<0.05, ** p<0.01, ***p<0.001, Student’s t test.
We previously reported that ASXL1-MT inhibited G-CSF-induced differentiation of 32Dcl3 toward neutrophils.14 Interestingly, the acquisition of SETBP1-D868N augmented the differentiation block in ASXL1-MT-expressing 32Dcl3 cells (Figure 2c and 2d). In accordance with this, all-trans retinoic acid–induced (ATRA-induced) granulocytic differentiation of HL60 cells, a human AML cell line, was also synergistically attenuated by the expression of ASXL1-MT and SETBP1-D868N (Supplementary Figure 2E and 2F).
SETBP1-MT enhances clonogenic capacity of myeloid progenitor cells
To determine whether expression of SETBP1-D868N has an influence on differentiation or self-renewal in hematopoietic stem cells, we compared colony formation ability between LSK cells transduced with mock/mock, ASXL1-MT/mock, mock/SETBP1-WT, mock/SETBP1-D868N, ASXL1-MT/SETBP1-WT, or ASXL1-MT/SETBP1-D868N using pMYs-IRES-GFP and pMYs-IRES-hNGFR vectors. ASXL1-MT/SETBP1-D868N–transduced LSK cells produced significantly more colonies with a relatively immature morphology, although overexpression of SETBP1-D868N or SETBP1-WT did not immortalize LSK cells (Figure 3a and 3b), contrary to previous reports.25
Figure 3.
Expression of both ASXL1-MT and SETBP1-D868N increased myeloid colony output. Colony-forming potentials of LSKs transduced with indicated plasmids in methylcellulose supplemented with SCF (100 ng/ml) and IL-3 (20ng/ml) are shown. Four columns represent numbers of serially replated colonies from the first (left) to the fourth (right) plating (a). All data with error bars indicate the mean ± SEM from duplicated cultures. Cytospin preparations of cultured cells in the second replating colonies assessed by Wright-Giemsa staining are shown (b). Compared to the control, *p< 0.05, **p<0.01, ***p<0.001, Student’s t test.
SETBP1-D868N collaborates with ASXL1-MT in inducing AML
We next examined the in vivo effect of simultaneous expression of ASXL1-MT and SETBP1-D868N using mouse BMT model. Intriguingly, in 3 independent experiments, all of the mice transplanted with BM cells expressing both ASXL1-MT and SETBP1-D868N died of acute leukemia with remarkable hepatosplenomegaly after a short latency (median survival, 73 days), while all of the mice transplanted with BM cells expressing either ASXL1-MT or SETBP1-D868N survived for 6 months after transplantation (Figure 4a and 4b). We confirmed high expression levels of both mutants in the BM cells of AML mice (Supplementary Figure 3A and 3B). Morphological analysis of BM or spleen cells in leukemic mice displayed frequent immature blasts with slight differentiation toward myeloid lineage. The blasts were positive for GFP and NGFR (Figure 4c), indicating they expressed SETBP1-D868N and ASXL1-MT. The phenotypic markers of the blast were CD11b–positive, and Gr1, c-kit, ScaI, and CD34-weak positive (Figure 4d). These leukemic mice exhibited remarkable leukocytosis, anemia, thrombocytopenia, macrocytosis, hepatosplenomegaly, and hypercellular BM compared to other control mice (Figure 4e-4k). Consistent with our previous report, mice with ASXL1-MT alone developed mild pancytopenia and hepatosplenomegaly with hypercellular BM mimicking early stages of MDS.14 On the other hand, we observed no significant change other than BM cellularity or splenomegaly in mice with SETBP1-D868N alone up to 6 months (Figure 4e-4k) or up to 12 months (data not shown). In addition, leukemic cells from mice with both ASXL1-MT and SETBP1-D868N were serially transplantable in sublethally irradiated recipients (data not shown) and grew rapidly without a change in phenotype in vitro, while those of the mice with ASXL1-MT or mock could not survive under the same conditions (Supplementary Figure 3C-3E).
Figure 4.
SETBP1-MT collaborated with ASXL1-MT in inducing AML. (a) Kaplan-Meier analysis for the survival of mice transplants with BM cells transduced with empty vector (Mock, n=6, blue circle), pMYs-ASXL1-MT-IG (ASXL1-MT, n=6, yellow triangle), pMYs-SETBP1-D868N-IG (SETBP1-D868N, n=6, green triangle), and pMYs-SETBP1-D868N-IG and pMYs-ASXL1-MT-INGFR (ASXL1-MT/SETBP1-D868N, n=9, red square). P values were calculated using a log-rank test. (b) Macroscopic findings of sacrificed mice transplanted with BM cells transduced with ASXL1-MT and SETBP1-D868N. Representative photographs are shown. (c) Cytospin preparations of BM cells derived from mice with ASXL1-MT and SETBP1-D868N were stained with Wright-Giemsa. Representative photographs are shown. Original magnification, ×400. (d) Flow cytometric analyses of BM cells derived from mice with ASXL1-MT and SETBP1-D868N. (e–k) Mice transplanted with both ASXL1-MT and SETBP1-D868N (ASXL1-MT/SETBP1-D868N, n=8) expressing cells displayed progressive leukocytosis (e), anemia (f), macrocytosis (g), thrombocytopenia (h), hepatosplenomegaly (i–j) and hyperplastic BM (k) compared with the control mice transplanted with empty vectors (mock, n=6), ASXL1-MT (n=6), or SETBP1-D868N (n=5). BM cells were isolated from the femurs and tibias of the sacrificed mice. *p< 0.05, **p<0.01, ***p<0.001, Student’s t test.
Acquisition of SETBP1 mutation results in Pp2a inhibition
To determine whether SETBP1-D868N also interacts with SET and inactivates PP2A as previously reported for other SETBP1 mutants,29 coimmunoprecipitation analysis using the anti-TY1 antibody was carried out in HEK 293T cells transfected with mock, SETBP1-WT, or SETBP1-D868N vectors. Given that none of the SETBP1 mutations occurs within the SET-binding domain (Figure 1b), SETBP1-D868N and SETBP1-WT were shown to associate with SET and PP2A (Figure 5a). To further investigate whether SETBP1-D868N induced PP2A inactivation in BM cells in vivo, we analyzed the phosphorylation levels of Pp2a and Akt in FACS-purified c-kit-positive cells from the BM of mice transplanted with BM cells expressing empty vector (control), ASXL1-MT (MDS model), or both ASXL1-MT and SETBP1-D868N (AML model). As expected, we observed phosphorylation of tyrosine 307 of Pp2a in AML mice. Consistent with this finding, threonine 308 and serine 473 residues of Akt were highly phosphorylated in AML mice, without affecting their expression levels, relative to control or MDS mice, indicating that impairment of Pp2a phosphatase activity, at least in part, resulted in leukemogenesis in our mice model as well as previously reported in vitro experiments (Figure 5b and 5c).29–31 Next, to assess the therapeutic potential of targeting Pp2a against disease progression, we treated the AML mice with FTY720, a Pp2a activator, in vitro and in vivo (Figure 5d and Supplementary Figure 3A). As expected, FTY720 reduced phosphorylation of tyrosine 307 of Pp2a and inhibited, in a dose dependent manner, the proliferation of cell lines transduced with both ASXL1-MT and SETBP1-D868N (Figure 5d and Supplementary Figure 4A). However, the therapeutic effect of FTY720 on AML mice was limited in spite of decreased phoporylation of Pp2a, suggesting that pathways other than Pp2a inhibition also contribute to leukemogenesis in these mice (Supplementary Figure 4A and 4B).
Figure 5.
SETBP1 mutation resulted in Pp2a inhibition and increased Hoxa9 and Hoxa10 expression. (a) HEK293T cells were transiently transfected with empty vector, SETBP1-WT-3×TY1 and SETBP1-D868N-3×TY1, followed by IP of TY1 epitope and Western blotting for SET and PP2A. Cell lysates were also subject to immunoblotting with anti-TY1 Ab, anti-SET Ab, anti-PP2A Ab, or anti-tublin Ab. (b) Cell lysates from FACS-purified c-Kit+ cells from the BM of mice transplanted with BM cells transduced with empty vector (control), ASXL1-MT (MDS model), or both ASXL1-MT and SETBP1-D868N (AML model), were subject to immunoblotting with anti-phospho-Akt (T308/S473) Ab, anti-Akt Ab, anti-phopho-PP2A (Y307)Ab, anti-PP2A Ab, or anti-tublin Ab. (c) PP2A phosphatase assay in c-Kit+ cells from a control mouse (Mock) and a leukemic mice (ASXL1-MT/SETBP1-D868N). Results are representative of 3 independent experiments. *p< 0.05. (d) Relative growth rate of BM cells from leukemic mice transplanted with BM cells transduced with ASXL1-MT and SETBP1-D868N in the presence of 1ng/ml IL-3 and indicated concentration of FTY720. (e) Relative expression levels of Hoxa5/Hoxa7/Hoxa9/Hoxa10 were examined by qRT-PCR in CD3- B220- Ter119-negating BM cells derived from normal controls (Control), MDS mice (ASXL1-MT) and AML mice (ASXL1-MT + SETBP1-D868N). The values were normalized by Gapdh mRNA levels. All data with error bars are presented as mean + SEM of 2 independent experiments. (f–g) Quantitative PCR analyses following anti-TY1 ChIP in HEK293T cells transfected with empty vector, SETBP1-WT-3×TY1, SETBP1-D868N-3×TY1 using anti-TY1 antibody. PCR products specific to various regions and diagrams of tested region of HOXA9 (f) or HOXA10 locus (g) were shown in bottom panels. Results are representative of 3 independent experiments. Transcriptional start sites are indicated as arrows. *p< 0.05, **p<0.01, ***p<0.001, Student’s t test.
SETBP1-D868N activates Hoxa9 and Hoxa10 expression in mice
It has been demonstrated that overexpression of wildtype Setbp1 activates Hoxa9 and Hoxa10 in myeloid progenitors and induces self-renewal of immortalized cells.32 Moreover, we previously reported that ASXL1-MT derepressed the expression of Hoxa9 and Hoxa10 in ASXL1-MT-induced MDS mice.14 Here, we observed further up-regulation of posterior Hoxa genes in ASXL1-MT/SETBP1-D868N-induced AML mice (Figure 5e). In addition, SETBP1-WT and SETBP1-D868N, both of which contain three highly conserved AT-hook motifs (Figure 1b), were shown to directly and specifically bind to these HOXA9 and HOXA10 regions using anti-TY1 ChIP (Figure 5f and 5g and Supplementary Figure 4C and 4D).
Combination of ASXL1-MT and SETBP1-D868N activates a stem cell signature and represses the TGF-β signaling
To elucidate the mechanism for leukemogenesis induced by the combination of ASXL1-MT and SETBP1-D868N, we compared the gene expression profile of CD3–B220–Ter119– BM cells of AML mice induced by both ASXL1-MT and SETBP1-D868N versus those induced by ASXL1-MT alone. Gene set enrichment analysis (GSEA) revealed that addition of SETBP1-D868N significantly altered the expression profile of ASXL1-MT MDS (Figure 6a-6e). GSEA indicated that the gene set induced by simultaneous expression of ASXL1-MT and SETBP1-D868N correlated with that enriched in HSC or early progenitors, and negatively correlated with that of mature cells or lineage markers when compared to the gene set induced by ASXL1-MT alone (Figure 6a-6e and Supplementary Figure 5A-5G). This suggested that the mutation of SETBP1 contributes to acquisition of self-renewal of HSCs or leukemic stem cells and inhibition of differentiation toward neutrophils, consistent with our in vitro results (Figure 6a–c and Supplementary Figure 5A-5C). Interestingly, GSEA also indicated global repression of the Tgf-β signaling pathway and reciprocal up-regulation of Myc-activated genes in leukemic mice (Figure 6d and 6e and Supplementary Figure 5D–F), while the MDS mice exhibited up-regulation of Tgf-β signaling (Supplementary Figure 5G). The repressed genes encoded for the receptors of Tgf-β, Smad proteins, and the major targets of Tgf-β signaling, which were confirmed by qRT-PCR (Figure 6f).
Figure 6.
Combined expression of SETBP1 and ASXL1 mutations induces a stem cell signature and inhibits expression of genes involved in hematopoietic differentiation and TGF-β signaling. (a–e) Gene set enrichment analysis (GSEA) determining specific gene sets or pathways that are positively or negatively regulated by additional expression of SETBP1-D868N. Compared with MDS mice expressing ASXL1-MT alone (right side), AML mice expressing ASXL1-MT and SETBP1-D868N (left side) negatively correlated with gene sets downregulated in hematopoietic stem cells (a), those of mature hematopoietic cells (b), and those of cell lineage (c). The expression profile of the AML mice also inversely correlated with gene sets of TGF-β signaling pathway (d) and associated with those of Myc pathway (e). The normalized enrichment scores (NES) and P-values are given. (f) qRT-PCR validation of genes related to TGF-β signaling pathway in the BM of control, MDS (ASXL1-MT) or AML (ASXL1-MT + SETBP1-D868N) mice. *p< 0.05, **p<0.01, ***p<0.001, Student’s t test. All data with error bars indicate the mean + SEM from duplicated experiments.
DISCUSSION
New insights into the genetic basis of MDS were obtained in the last decade using next-generation sequencing technology, shedding light on the founding or driver mutations of MDS and the mutations acquired at the later phases of the disease.3–5
Of importance, MDS patients at early stages present with the paradox of a variable cytopenia in spite of hypercellular BM, associating with ineffective hematopoiesis in which MDS cells tend to be apoptotic.41–44 In fact, the incidence of apoptosis in the BM is higher in patients with low-risk MDS compared to a normal counterpart. On the contrary, in more advanced stages, MDS clones acquire the potential of resistance to apoptosis, leading to transformations and therapeutic tolerance.43, 45 As for the underlying mechanism, involvement of mutations in TP53 and NRAS genes, both associated with poor prognosis, has been well investigated.8, 46 For example, NRAS mutations, present in 10% of MDS patients, exhibit an anti-apoptotic effect by increasing sensitivities to cytokines or growth factors leading to the progression of MDS.1 To further investigate how ASXL1-mutated MDS clones transform into leukemia, we examined co-existing mutations of 64 ASXL1-mutated MDS patients. Interestingly, mutations of ASXL1 and SETBP1 not only co-occurred significantly more often than by chance, but also resulted in shorter overall survival and a higher incidence of leukemic transformation when compared with SETBP1 non-mutated cases, indicating that combined mutation of these genes provides a selective advantage and that an additional SETBP1 mutation plays a pivotal role in disease progression.
In vitro expression of SETBP1-D868N enhanced myeloid colony formation of ASXL1-MT-transduced LSK cells and augmented the ASXL1-MT-induced differentiation block of 32Dcl3 cells and primary BM cells. Of note, SETBP1-D868N collaborated with ASXL1-MT to induce AML with a short latency in a mouse BMT model. To the best of our knowledge, this is the first report concerning an in vivo model of the SETBP1 mutation and the first report revealing disease progression of an ASXL1-mutant MDS model. The escape of SETBP1-D868N from ubiquitination and subsequent degradation is most likely a gain-of-function mutation, combined with the observation that overexpression of SETBP1-WT exhibited milder effects than that of SETBP1-D868N. SETBP1 mutations were found in late phase MDS patients, indicating that the acquisition of the SETBP1 mutation is a critical event for ASXL1-mutated MDS during disease progression.26
The formation of the SETBP1-SET-PP2A complex has been shown to result in PP2A inhibition.31 PP2A is a tumor suppressor that inhibits cellular transformation by regulating several signaling activities critical for malignant transformation, including AKT and ERK1/2 pathways.30, 47, 48 Indeed, we showed that both SETBP1-WT and SETBP1-D868N could interact with SET or PP2A. Moreover, BM cells from leukemic mice displayed activation of Akt, and phosphorylation of Pp2A, leading to decreased activity. However, while administration of FTY720, a Pp2a activator, did efficiently repress the growth rate in vitro, the in vivo effect of FTY720 against the ASXL1-MT/SETBP1-D868N MDS/AML model was marginal. This suggests that signaling pathways other than SETBP1-SET-PP2A play a predominant role in the ASXL1-MT/SETBP1-D868N-induced MDS/AML model. In this paper, we have identified potentially dysregulated pathways that could contribute to leukemic transformation.
First, both SETBP1-WT and SETBP1-D868N can bind to the promoter regions of Hoxa9 and Hoxa10, leading to remarkable up-regulation of these genes. This is consistent with previous reports that overexpression of Hoxa9 and Hoxa10 induced immortalizaion of myeloid progenitors and disease progression.49, 50 Further upregulation of Hoxa9 and Hoxa10 by the combination of ASXL1-MT and SETBP1-D868N may play important roles in the leukemic transformation of MDS.
Second, using RNA-seq and GSEA with the in vivo mouse model,51 we identified several deregulated pathways. Among them, we focused on the TGF-β signaling pathway, which is a potent negative regulator of proliferation,52 since there is evidence showing that resistance to the growth inhibitory and apoptotic effects of TGF-β promotes clonal expansion and down-regulation of TGF-β signaling plays a pivotal role in the pathogenesis of AML.53–55 For example, it has been demonstrated that AML1/ETO and PML/RARα prevent DNA binding of Smad3 and Smad2/3 phosphorylation, respectively.53, 54 In accordance with the down-regulation of the TGF-β pathway, we observed up-regulation of the Myc pathway that would also contribute to leukemogenesis and the global down-regulation of Smad2/3 targets as well as TGF-β receptors shown in Figure 6F.56–58 Intriguingly, Zhou et al. reported that SMAD2, a downstream mediator of TGF-β receptors, is constitutively activated and overexpressed in MDS BM precursors and that shRNA-mediated down-regulation or pharmacologic inhibition of TGF-β receptor 1 leads to enhanced hematopoiesis in a variety of MDS subtypes in vitro.59 Thus, it is tempting to consider that the TGF-β signaling pathway inhibits leukemic transformation of MDS and that SETBP1 mutations contribute to the down-regulation of this important pathway.
Taken together, we detected gain-of-function SETBP1 mutations enriched among ASXL1-mutated MDS and characterized an in vivo MDS/AML model expressing SETBP1 mutations for the first time. Our results indicate that the SETBP1 mutations play critical roles in the leukemic transformation of MDS involving anti-apoptosis, differentiation block, and increased self-renewal through several mechanisms including PP2A inactivation, up-regulation of posterior HOXA, and inhibition of the TGF-β signaling pathway. Given that SETBP1 seems to be directly or indirectly involved in transcription, the binding partners among transcriptional factors and epigenetic modifiers should be elucidated to understand the precise mechanism of the transformation. Our data also suggests deregulated pathways induced by SETBP1 mutations could be potential targets for future therapies.
Supplementary Material
Acknowledgements
This work is mainly supported by Grants-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan. O.A-W. is supported by an NIH K08 Clinical Investigator Award (1K08CA160647-01), a US Department of Defense Postdoctoral Fellow Award in Bone Marrow Failure Research (W81XWH-12-1-0041), the Josie Robertson Investigator Program, and a Damon Runyon Clinical Investigator Award with support from the Evans Foundation. Clinical studies are partially sponsored by grant 100-2314-B-002-057-MY3 from the National Science Council (Taiwan).
Footnotes
Conflict of Interest
The authors report no competing conflicts of interest.
Supplementary information is available at Leukemia’s website.
REFERENCES
- 1.Raza A, Galili N. The genetic basis of phenotypic heterogeneity in myelodysplastic syndromes. Nat Rev Cancer. 2012;12:849–859. doi: 10.1038/nrc3321. [DOI] [PubMed] [Google Scholar]
- 2.Nimer SD. Myelodysplastic syndromes. Blood. 2008;111:4841–4851. doi: 10.1182/blood-2007-08-078139. [DOI] [PubMed] [Google Scholar]
- 3.Cazzola M, Della Porta MG, Malcovati L. The genetic basis of myelodysplasia and its clinical relevance. Blood. 2013;122:4021–4034. doi: 10.1182/blood-2013-09-381665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bejar R, Levine R, Ebert BL. Unraveling the molecular pathophysiology of myelodysplastic syndromes. J Clin Oncol. 2011;29:504–515. doi: 10.1200/JCO.2010.31.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Massé A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 7.Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 8.Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bejar R, Stevenson KE, Caughey BA, Abdel-Wahab O, Steensma DP, Galili N, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30:3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 11.Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2012;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kon S, Minegishi N, Tanabe K, Watanabe T, Funaki T, Wong WF, et al. Smap1 deficiency perturbs receptor trafficking and predisposes mice to myelodysplasia. J Clin Invest. 2013;123:1123–1137. doi: 10.1172/JCI63711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamazaki J, Issa JP. Epigenetic aspects of MDS and its molecular targeted therapy. Int J Hematol. 2013;97:175–182. doi: 10.1007/s12185-012-1197-4. [DOI] [PubMed] [Google Scholar]
- 14.Inoue D, Kitaura J, Togami K, Nishimura K, Enomoto Y, Uchida T, et al. Myelodysplastic syndromes are induced by histone methylation-altering ASXL1 mutations. J Clin Invest. 2013;123:4627–40. doi: 10.1172/JCI70739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muto T, Sashida G, Oshima M, Wendt GR, Mochizuki-Kashio M, Nagata Y, et al. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. J Exp Med. 2013;210:2627–2639. doi: 10.1084/jem.20131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22:180–193. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YR, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med. 2013;210:2641–2659. doi: 10.1084/jem.20131141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Li Z, He Y, Pan F, Chen S, Rhodes S, et al. Loss of Asxl1 leads to myelodysplastic syndrome-like disease in mice. Blood. 2014;123:541–553. doi: 10.1182/blood-2013-05-500272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelsi-Boyer V, Brecqueville M, Devillier R, Murati A, Mozziconacci MJ, Birnbaum D. Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J Hematol Oncol. 2012;5:12. doi: 10.1186/1756-8722-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen TC, Hou HA, Chou WC, Tang JL, Kuo YY, Chen CY, et al. Dynamics of ASXL1 mutation and other associated genetic alterations during disease progression in patients with primary myelodysplastic syndrome. Blood Cancer J. 2014;4:e177. doi: 10.1038/bcj.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thol F, Friesen I, Damm F, Yun H, Weissinger EM, Krauter J, et al. Prognostic significance of ASXL1 mutations in patients with myelodysplastic syndromes. J Clin Oncol. 2011;29:2499–2506. doi: 10.1200/JCO.2010.33.4938. [DOI] [PubMed] [Google Scholar]
- 24.Meggendorfer M, Bacher U, Alpermann T, Haferlach C, Kern W, Gambacorti-Passerini C, et al. SETBP1 mutations occur in 9% of MDS/MPN and in 4% of MPN cases and are strongly associated with atypical CML, monosomy 7, isochromosome i(17)(q10), ASXL1 and CBL mutations. Leukemia. 2013;27:1852–1860. doi: 10.1038/leu.2013.133. [DOI] [PubMed] [Google Scholar]
- 25.Makishima H, Yoshida K, Nguyen N, Przychodzen B, Sanada M, Okuno Y, et al. Somatic SETBP1 mutations in myeloid malignancies. Nat Genet. 2013;45:942–946. doi: 10.1038/ng.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou HA, Kuo YY, Tang JL, Chou WC, Yao M, Lai YJ, et al. Clinical implications of the SETBP1 mutation in patients with primary myelodysplastic syndrome and its stability during disease progression. Am J Hematol. 2014;89:181–186. doi: 10.1002/ajh.23611. [DOI] [PubMed] [Google Scholar]
- 27.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thol F, Suchanek KJ, Koenecke C, Stadler M, Platzbecker U, Thiede C, et al. SETBP1 mutation analysis in 944 patients with MDS and AML. Leukemia. 2013;27:2072–2075. doi: 10.1038/leu.2013.145. [DOI] [PubMed] [Google Scholar]
- 29.Piazza R, Valletta S, Winkelmann N, Redaelli S, Spinelli R, Pirola A, et al. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet. 2013;45:18–24. doi: 10.1038/ng.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cristóbal I, Garcia-Orti L, Cirauqui C, Alonso MM, Calasanz MJ, Odero MD. PP2A impaired activity is a common event in acute myeloid leukemia and its activation by forskolin has a potent anti-leukemic effect. Leukemia. 2011;25:606–614. doi: 10.1038/leu.2010.294. [DOI] [PubMed] [Google Scholar]
- 31.Cristóbal I, Blanco FJ, Garcia-Orti L, Marcotegui N, Vicente C, Rifon J, et al. SETBP1 overexpression is a novel leukemogenic mechanism that predicts adverse outcome in elderly patients with acute myeloid leukemia. Blood. 2010;115:615–625. doi: 10.1182/blood-2009-06-227363. [DOI] [PubMed] [Google Scholar]
- 32.Oakley K, Han Y, Vishwakarma BA, Chu S, Bhatia R, Gudmundsson KO, et al. Setbp1 promotes the self-renewal of murine myeloid progenitors via activation of Hoxa9 and Hoxa10. Blood. 2012;119:6099–6108. doi: 10.1182/blood-2011-10-388710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gelsi-Boyer V, Trouplin V, Adélaïde J, Bonansea J, Cervera N, Carbuccia N, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145:788–800. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- 34.Nakahara F, Sakata-Yanagimoto M, Komeno Y, Kato N, Uchida T, Haraguchi K, et al. Hes1 immortalizes committed progenitors and plays a role in blast crisis transition in chronic myelogenous leukemia. Blood. 2010;115:2872–2881. doi: 10.1182/blood-2009-05-222836. [DOI] [PubMed] [Google Scholar]
- 35.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe-Okochi N, Kitaura J, Ono R, Harada H, Harada Y, Komeno Y, et al. AML1 mutations induced MDS and MDS/AML in a mouse BMT model. Blood. 2008;111:4297–4308. doi: 10.1182/blood-2007-01-068346. [DOI] [PubMed] [Google Scholar]
- 37.Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, et al. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
- 38.Enomoto Y, Yamanishi Y, Izawa K, Kaitani A, Takahashi M, Maehara A, et al. Characterization of leukocyte mono-immunoglobulin-like receptor 7 (LMIR7)/CLM-3 as an activating receptor: its similarities to and differences from LMIR4/CLM-5. J Biol Chem. 2010;285:35274–35283. doi: 10.1074/jbc.M110.137166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura H, Hayashi-Takanaka Y, Goto Y, Takizawa N, Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct Funct. 2008;33:61–73. doi: 10.1247/csf.07035. [DOI] [PubMed] [Google Scholar]
- 40.Neviani P, Santhanam R, Trotta R, Notari M, Blaser BW, Liu S, et al. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer Cell. 2005;8:355–368. doi: 10.1016/j.ccr.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 41.Raza A, Mundle S, Shetty V, Alvi S, Chopra H, Span L, et al. Novel insights into the biology of myelodysplastic syndromes: excessive apoptosis and the role of cytokines. Int J Hematol. 1996;63:265–278. doi: 10.1016/0925-5710(96)00455-0. [DOI] [PubMed] [Google Scholar]
- 42.Raza A, Gezer S, Mundle S, Gao XZ, Alvi S, Borok R, et al. Apoptosis in bone marrow biopsy samples involving stromal and hematopoietic cells in 50 patients with myelodysplastic syndromes. Blood. 1995;86:268–276. [PubMed] [Google Scholar]
- 43.Parker J, Fishlock K, Mijovic A, Czepulkowski B, Pagliuca A, Mufti G. ‘Low-risk’ myelodysplastic syndrome is associated with excessive apoptosis and an increased ratio of pro- versus anti-apoptotic bcl-2-related proteins. Br J Haematol. 1998;103:1075–1082. doi: 10.1046/j.1365-2141.1998.01114.x. [DOI] [PubMed] [Google Scholar]
- 44.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 45.Parker J, Mufti G, Rasool F, Mijovic A, Devereux S, Pagliuca A. The role of apoptosis, proliferation, and the Bcl-2-related proteins in the myelodysplastic syndromes and acute myeloid leukemia secondary to MDS. Blood. 2000;96:3932–3938. [PubMed] [Google Scholar]
- 46.Kita-Sasai Y, Horiike S, Misawa S, Kaneko H, Kobayashi M, Nakao M, et al. International prognostic scoring system and TP53 mutations are independent prognostic indicators for patients with myelodysplastic syndrome. Br J Haematol. 2001;115:309–312. doi: 10.1046/j.1365-2141.2001.03073.x. [DOI] [PubMed] [Google Scholar]
- 47.Sablina AA, Hahn WC. The role of PP2A A subunits in tumor suppression. Cell Adh Migr. 2007;1:140–141. doi: 10.4161/cam.1.3.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnold HK, Sears RC. A tumor suppressor role for PP2A–B56alpha through negative regulation of c-Myc and other key oncoproteins. Cancer Metastasis Rev. 2008;27:147–158. doi: 10.1007/s10555-008-9128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 50.Calvo KR, Sykes DB, Pasillas M, Kamps MP. Hoxa9 immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced meis expression. Mol Cell Biol. 2000;20:3274–3285. doi: 10.1128/mcb.20.9.3274-3285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong M, Blobe GC. Role of transforming growth factor-beta in hematologic malignancies. Blood. 2006;107:4589–4596. doi: 10.1182/blood-2005-10-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jakubowiak A, Pouponnot C, Berguido F, Frank R, Mao S, Massague J, et al. Inhibition of the transforming growth factor beta 1 signaling pathway by the AML1/ETO leukemia-associated fusion protein. J Biol Chem. 2000;275:40282–40287. doi: 10.1074/jbc.C000485200. [DOI] [PubMed] [Google Scholar]
- 54.Lin HK, Bergmann S, Pandolfi PP. Cytoplasmic PML function in TGF-beta signalling. Nature. 2004;431:205–211. doi: 10.1038/nature02783. [DOI] [PubMed] [Google Scholar]
- 55.Imai Y, Kurokawa M, Izutsu K, Hangaishi A, Maki K, Ogawa S, et al. Mutations of the Smad4 gene in acute myelogeneous leukemia and their functional implications in leukemogenesis. Oncogene. 2001;20:88–96. doi: 10.1038/sj.onc.1204057. [DOI] [PubMed] [Google Scholar]
- 56.Hoffman B, Amanullah A, Shafarenko M, Liebermann DA. The proto-oncogene c-myc in hematopoietic development and leukemogenesis. Oncogene. 2002;21:3414–3421. doi: 10.1038/sj.onc.1205400. [DOI] [PubMed] [Google Scholar]
- 57.Koinuma D, Tsutsumi S, Kamimura N, Taniguchi H, Miyazawa K, Sunamura M, et al. Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor beta signaling. Mol Cell Biol. 2009;29:172–186. doi: 10.1128/MCB.01038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frederick JP, Liberati NT, Waddell DS, Shi Y, Wang XF. Transforming growth factor beta-mediated transcriptional repression of c-myc is dependent on direct binding of Smad3 to a novel repressive Smad binding element. Mol Cell Biol. 2004;24:2546–2559. doi: 10.1128/MCB.24.6.2546-2559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou L, Nguyen AN, Sohal D, Ying Ma J, Pahanish P, Gundabolu K, et al. Inhibition of the TGF-beta receptor I kinase promotes hematopoiesis in MDS. Blood. 2008;112:3434–3443. doi: 10.1182/blood-2008-02-139824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.