Abstract
Background
Clinical evaluation of CNVs identified via techniques such as array comparative genome hybridisation (aCGH) involves the inspection of lists of known and unknown duplications and deletions with the goal of distinguishing pathogenic from benign CNVs. A key step in this process is the comparison of the individual's phenotypic abnormalities with those associated with Mendelian disorders of the genes affected by the CNV. However, because often there is not much known about these human genes, an additional source of data that could be used is model organism phenotype data. Currently, almost 6000 genes in mouse and zebrafish are, when knocked out, associated with a phenotype in the model organism, but no disease is known to be caused by mutations in the human ortholog. Yet, searching model organism databases and comparing model organism phenotypes with patient phenotypes for identifying novel disease genes and medical evaluation of CNVs is hindered by the difficulty in integrating phenotype information across species and the lack of appropriate software tools.
Methods
Here, we present an integrated ranking scheme based on phenotypic matching, degree of overlap with known benign or pathogenic CNVs and the haploinsufficiency score for the prioritisation of CNVs responsible for a patient's clinical findings.
Results
We show that this scheme leads to significant improvements compared with rankings that do not exploit phenotypic information. We provide a software tool called PhenogramViz, which supports phenotype-driven interpretation of aCGH findings based on multiple data sources, including the integrated cross-species phenotype ontology Uberpheno, in order to visualise gene-to-phenotype relations.
Conclusions
Integrating and visualising cross-species phenotype information on the affected genes may help in routine diagnostics of CNVs.
Introduction
High-throughput technologies are increasingly being applied to the detection of copy number variation in patients with developmental delay or unexplained congenital malformations. Methods such as array comparative genomic hybridisation, SNP genotyping array and genome sequencing enable the genome-wide detection of structural variants. The typical landscape of CNVs in a patient shows from 20 to over 100 duplications and deletions,1 ,2 most of which are relatively small in size, while a few CNVs cover longer regions of the genome. Each CNV can either represent neutral polymorphic variation or convey clinical phenotypes by inducing gene dosage effects or dysregulation of genes.3 In particular, the interpretation and classification of rare CNVs remains difficult.4 Therefore, it is desirable to evaluate each of the CNVs with respect to which of the affected genes might be associated with individual phenotypic features observed in the patient. However, aligning the genes affected by a CNV to clinical and functional phenotypic features is still challenging.5–7
Currently, a typical approach is to prioritise larger CNVs for further investigation,6 under the assumption that size correlates with the number of affected genes (NAG) and that larger CNVs are found less frequently among the general population.8 The more genes affected, the stronger the predicted impact on an organism's molecular machinery. In the diagnostic setting, the focus is often set exclusively on CNVs larger than 400 000 bases (400 kb) as candidates for pathogenic variations.9 ,10 However, this strategy is not reliable, since small CNVs can also have serious phenotypic consequences.6 For example, the minimal critical region of deletions in Phelan–McDermid syndrome covers around 140 000 bp and affects only four genes (see https://decipher.sanger.ac.uk/syndrome/20). Furthermore, around 10% (118 of 1230) of the pathogenic CNVs in the data from the International Standards For Cytogenomic Arrays Consortium (ISCA)11 are smaller than 400 kb (data not shown).
Having additional information on the affected genes is often helpful to interpret the identified CNVs. A common approach is to use phenotype information about the human gene or about its ortholog in a model organism such as mouse or zebrafish.12 There are several databases that store the phenotypic consequences of systematic gene knock-outs in these model organisms, but aligning data from these distributed databases to a given set of patient phenotypes can still be a time-consuming and laborious task. To give an example, consider a patient with an abnormality of the morphology of the nose and a deletion encompassing the gene IGF2BP1. This gene is not currently associated with any human diseases (see OMIM:608288), but its ortholog has been knocked out in mouse. To use model organism information, the physician would have to collect all the phenotype information for the mouse knock-out and manually compare it with the human phenotype data in order to find that the deletion of IGF2BP1 might explain the abnormal nose morphology, because the knock-out mouse is annotated to ‘short snout’. Here, we present a software tool to automatically obtain and visualise such phenotypic alignments using a cross-species phenotype ontology.
Ontologies are knowledge representations that make use of controlled vocabularies for representing knowledge about a domain, thereby enabling automated computer reasoning. We have recently described cross-species ontological methods that use computer reasoning over phenotype ontologies.13 These methods can be used to identify similarities between human disease manifestations and observations made in genetically modified model organisms.14 For example, the mouse phenotype ‘short snout’ would then be inferred to be closely related to the human phenotype ‘abnormal nose morphology’. Integration and alignment of the phenotype ontologies and a diversity of model organism databases is being performed within the context of the Monarch Initiative.15 Notably, we have previously shown the usefulness of the Uberpheno resource for disease and CNV interpretation.13 ,16–18 In this manuscript, we show that integrating phenotype information improves the prioritisation of pathogenic CNVs compared with an evaluation based solely on the overlap with known pathogenic and benign CNVs, the haploinsufficiency (HI) score or the size of the CNVs. Here, we present PhenogramViz, implemented as a Cytoscape app. Cytoscape is an easy-to-use open source software platform for complex network analysis and visualisation. Cytoscape's core features can be enhanced by implementing so-called apps.19 ,20 PhenogramViz facilitates visualisation of a large set of integrated phenotypic data and aids in phenotype-guided interpretation and prioritisation of CNVs.
Methods
Data preparation
We use four different resources providing links between a gene g and phenotype terms: OMIM,21Orphanet,22 Mouse Genome Informatics (MGI)23 and Zebrafish Information Network (ZFIN).24 OMIM and Orphanet contain links between genes (g) and human monogenic syndromes. These syndromes, in turn, are linked to terms of the Human Phenotype Ontology (HPO) representing the abnormalities of patients with that disease.25 MGI and ZFIN provide phenotype annotations of knock-out or knock-down experiments for mouse and zebrafish genes. Aligned and integrated data from these sources are being made available as part of the Monarch Initiative.15 Especially, the Uberpheno ontology allows information from mouse and zebrafish to be integrated in the following way:
We assign an information content value, IC(t), to each phenotype term t of the Uberpheno ontology. This value is defined as the negative logarithm of the frequency of annotations to that term.26 Here, that frequency is the probability of annotations to term t among all annotated genes in human, mouse and zebrafish. A high IC(t) indicates a high specificity of t because only a few genes are linked to t.
We also obtained HI scores27 for each gene, which are used for visualisation and benchmarking.
We downloaded the CNV data provided by Database of Genomic Variants (DGV)28 and the ISCA.11 We excluded CNVs with no affected genes. We claim that larger CNVs are easily identifiable using standard prioritisation methods such as NAG. Thus, we focused on CNVs with no more than 30 affected genes.
From the resulting data, we generated a set of pathogenic CNVs and a set of benign CNVs. Our list of benign CNVs comprises all CNVs listed in DGV entries and CNVs from ISCA that are marked as ‘benign’ or ‘likely benign’. Our list of pathogenic CNVs comprises all CNVs listed in ISCA that are marked as ‘pathogenic’ or ‘likely pathogenic’.
Affected genes
For each CNV, we compile a list of affected genes that comprises any gene whose start position or end position (coding sequence) is located within the CNV interval. Thus, partially affected genes are included in order to assess possible effects caused by disruption of gene sequences at the CNV boundaries.
Construction of phenograms
PhenogramViz supports generation of hypotheses regarding which affected gene g of a deleted or duplicated region might cause which phenotypic feature of the phenotypic spectrum of a patient.
That means, given a patient with a phenotypic spectrum P={t1, t2,...} for each t∈P, we attempt to find a gene g that explains t. To construct a phenogram (Ψ), we iterate over each gene g in the CNV and obtain all phenotype annotations tg from the four resources described above. For each of these annotations, we search for the best (ie, highest IC) common ancestor (tCA) with the patient's phenotypic features (tp) in the Uberpheno ontology. Using this approach, we obtain two types of links between g and tp: direct and indirect links. If tg is a descendant of tp (ie, tCA=tp, meaning that the phenotype associated with the gene is identical to a phenotype seen in the patient or is a more specific subclass of it), we can infer a direct link from g to tp. An example for a direct link is gene DTNA in figure 3, which is annotated to ‘Ventricular septal defect’, which is a subclass of tp ‘Abnormality of cardiac ventricle’. If tg and tp are both descendants of tCA, then there is an indirect link between tg and tp. Note that in this case IC(tCA) must be greater than a specified threshold λ, which is used to exclude unspecific matches. An example for an indirect link is gene TTR in figure 3, which is annotated to ‘Nystagmus’ (tg). Both ‘Nystagmus’ and ‘Strabismus’ (tp) are descendants of ‘Abnormality of eye movement’ (tCA). Adding nodes and directed edges iteratively results in a network of genes and phenotypes, which we call phenogram.13
Figure 3.
Example phenogram. Human genes located in the selected CNV are displayed as green diamonds, phenotypes found in the patient are displayed as blue circles and the common ancestors between a gene's phenotype annotation and the patient's phenotypes are displayed as orange triangles. The size of a gene node is proportional to its haploinsufficiency score.27Edges from genes are labelled by the phenotypes (human, mouse or zebrafish) associated with that gene and that were used for linking the gene directly or indirectly to the patient phenotypes. A right-click on nodes opens a context menu that provides either a link to the corresponding term-information page (here: HPO Term Browser) for a phenotype (1) or a link to the corresponding entries at the EntrezGene website for a gene (2). A right-click on edges opens a context menu that provides external links to the primary annotation resources, here: to MGI (3) and to the OMIM entry (4) associated with the gene. HPO, Human Phenotype Ontology; MGI, Mouse Genome Informatics; MPO, Mammalian Phenotype Ontology; ZFIN, Zebrafish Information Network.
Phenotype-dependent scorings
Aside from allowing for investigation of CNVs using cross-species phenotype information, the app additionally allows individual CNVs to be prioritised using a score that illustrates their relevance to the patient’s phenotypes. This score, called phenogram-score (PHS), is calculated for each of the patient's CNVs. It is defined as the size of the phenogram, i.e. the number of nodes that the phenogram contains. Formally, PHS=|{t∈Ψ}|+|{g∈Ψ}|.
Phenotype-independent scorings
Here, four different scorings were considered: NAG, number of overlapping benign CNVs (OBE), number of overlapping pathogenic CNVs (OPA) and HI score. NAG counts the number of genes located in the CNV region. HI takes the maximum haploinsufficiency score (see above) for the affected genes as an indicator for their pathogenicity. Although the HI score was designed for deletions of genes, we assume that for a given gene, the HI score is generally a good indicator for dosage sensitivity caused by either threshold effects or altered stoichiometry.27
OBE reflects the amount of overlap with benign CNVs found in DGV and ISCA for each CNV in a patient. Here, an overlap means that at least 80% of the patient's CNV is covered by the benign CNV. Based on the number of overlapping CNVs, the OBE score is calculated (table 1A).
Table 1.
Mapping overlaps of pathogenic and benign CNVs to scores and colours
| (A) Incorporation of the overlap with benign CNVs into the score OBE and its visual feedback in the app. | ||
|---|---|---|
| CNV overlaps with... | OBE score | Colour |
| no benign CNVs | 0 | Red |
| one benign CNV | 1 | Orange |
| two or three benign CNVs | 2 | Yellow |
| more than 3 benign CNVs | 3 | Green |
| (B) Incorporation of the overlap with pathogenic CNVs into the score OPA and its visual feedback in the app. | ||
|---|---|---|
| CNV overlaps with... | OPA score | Colour |
| no pathogenic CNVs | 0 | Green |
| one pathogenic CNV | 1 | Yellow |
| two or three pathogenic CNVs | 2 | Orange |
| more than 3 pathogenic CNVs | 3 | Red |
A CNV of a patient can be tested for overlap with known benign and known pathogenic CNVs seen in other individuals.
OBE, number of overlapping benign CNVs; OPA, number of overlapping pathogenic CNVs.
OPA reflects the amount overlapping pathogenic CNVs found in ISCA for each CNV in a patient (see table 1B). An overlap is considered if the ratio of their intersecting region to their joined region is greater than 0.1. This ensures that both an overlap exists and the lengths of the two CNVs are similar.
Ranking of CNVs
The scores described above (NAG, OBE, OPA, HI and PHS) can be used to rank CNVs found in a patient, whereby for NAG, OPA, HI and PHS the CNVs are ranked in descending order, for OBE the CNVs are ranked in ascending order. Aside from individual rankings, a combined ranking is computed, where several individual ranks of a CNV are averaged. These averaged ranks are taken to rank the CNVs again in descending order. Here, we used PHS together with OPA, OBE and HI (PHS+OPA+OBE+HI). Note that in case of ties, we determined the average rank, i.e. if five CNVs obtain the highest score, they all get the rank 3.
Benchmark test for CNV prioritisation
The number of CNVs per individual is estimated between 20 and 100.1 ,2 We performed a benchmark test, where we chose a pathogenic CNV together with the associated HPO terms according to the criteria described below. For each pathogenic CNV, we simulated 100 test cases. In each test case, we added 49 random benign CNVs from the set of benign CNVs described above. We ranked the CNVs in each of the sets according to the values calculated by the five methods described above. The analysis was regarded as successful whenever the pathogenic CNV was ranked first. The pathogenic CNVs came from the list of pathogenic CNVs described earlier. We removed all pathogenic CNVs, which were annotated with less than three phenotype terms from the HPO. Thus, in the benchmark test we used 278 pathogenic CNVs, 71 being duplications and 207 being deletions. As mentioned above, we generated 100 tests per CNV, corresponding to a total of 27 800 test cases. We used Uberpheno build #171 (April 2014). An IC-cut-off λ of 1 was used to calculate the phenograms. Files containing the data used in this manuscript are accessible through the PhenogramViz website (http://compbio.charite.de/contao/index.php/phenoviz.html).
Implementation of PhenogramViz as a Cytoscape app
Cytoscape is an open source software platform originally designed for visualising molecular interaction networks and biological pathways and for integrating these networks with annotations, gene expression profiles and other sources of information. It has become a general platform for complex network analysis and visualisation. The Cytoscape core distribution provides basic functionality to layout and query a network with the central organising principle being a network graph, represented as nodes and as edges between nodes. Additional features can be made available as apps (also referred to as plug-ins).19 ,20
PhenogramViz features can be accessed through a Cytoscape Control Panel (see figure 2).
Figure 2.
PhenogramViz Control Panel and workflow. The control panel is used to enter the phenotypic abnormalities of the patient coded as Human Phenotype Ontology (HPO) terms by loading a file (1) or by entering terms via a search window (2). Users should then enter a list of CNVs as a file (3) or as a list of genes that correspond to one or more CNVs (4). The CNVs are then ranked according to the combined score, with the CNV predicted most likely to be pathogenic being placed at top of the list (5). See table 1 for information on colour codes and see Methods on how to access underlying data. Users can double-click individual CNVs to display the phenogram. HPO terms and genes that could not be incorporated into the phenogram are displayed in red font. Users can adjust the specificity filter (λ) to hide or show unspecific matches. To protect privacy, we have ‘greyed out’ the exact positions of the CNVs found in the patient.
Cytoscape supports a variety of automated network layout algorithms. We chose its ‘Prefuse Force Directed’ layout to be the default for our phenograms. Whereas the network layout determines the location of the nodes and edges, an attribute-to-visual mapping allows data attributes to control the appearance of their associated nodes and edges. Using Cytoscape's Vizmapper feature, we assigned different shapes and colours for genes affected by the selected CNV, phenotypes found in the patient and common ancestors between a gene's phenotype annotation and the patient's phenotypic feature. Edges emanating from a gene are labelled by the phenotype annotation used for creating the edge. For example, in figure 3 the gene CDH2 is labelled with ‘dilated heart left ventricle (M)’ because a mouse model for this gene is associated with this term and the term is a descendant of the patient phenotype ‘Abnormality of cardiac ventricle’. The user can adjust our ‘PhenogramViz’ visual style through Cytoscape's ‘Style’ Control Panel.
We introduced colour codes for the number of overlaps of a CNV with known pathogenic CNVs from ISCA as well as for the number of overlaps with known benign CNVs from DGV and ISCA (table 1). Thus, the colour green applies to CNVs that are more likely to be benign (no overlap with pathogenic CNVs, and multiple overlaps with benign CNVs), and the colour red to CNVs that are more likely to be pathogenic. PhenogramViz contains extensive links and information on the data sources used in the form of tooltips and links to external websites. The app together with further documentation (eg, video tutorials) is available from the website (http://compbio.charite.de/contao/index.php/phenoviz.html).
Results
PhenogramViz is a Cytoscape app that facilitates a phenotype-guided interpretation of CNVs. One of its most fundamental features is visualising gene-to-phenotype associations as a 2D network of nodes and edges, which we call phenograms. Phenograms visualise which gene or genes affected by a deleted or duplicated region are most likely to contribute to the observed phenotypic abnormalities of the patient. For example, in Williams–Beuren syndrome, the deletion of gene ELN is responsible for supravalvular aortic stenosis and gene BAZ1B is linked to hypercalcaemia.29 Thus, in a phenogram, the BAZ1B node would have an edge to the hypercalcaemia node. Genes and phenotypes are connected by directed edges, whereby the cross-species phenotype ontology Uberpheno was used to align patient phenotypes to phenotypes of the genes affected by the CNV.
Scores based on a phenogram are used to rank the CNVs in a patient according to predicted clinical relevance. To test the performance of our method, we simulated 27 800 cases in which a single pathogenic CNV was spiked into a random set of 49 benign CNVs (see Methods). We have compared six methods for scoring the CNVs. One is phenotype-based (PHS), four are phenotype-independent scores (NAG, OBE, OPA and HI) and one is a combined method (PHS+OPA+OBE+HI).
NAG counts the number of affected genes. PHS computes the number of nodes in the corresponding phenogram, connecting affected genes to patient phenotypes. OPA/OBE scores CNVs by the number of overlapping pathogenic/benign CNVs. HI ranks the CNVs according to the maximal haploinsufficiency score found among the affected genes.
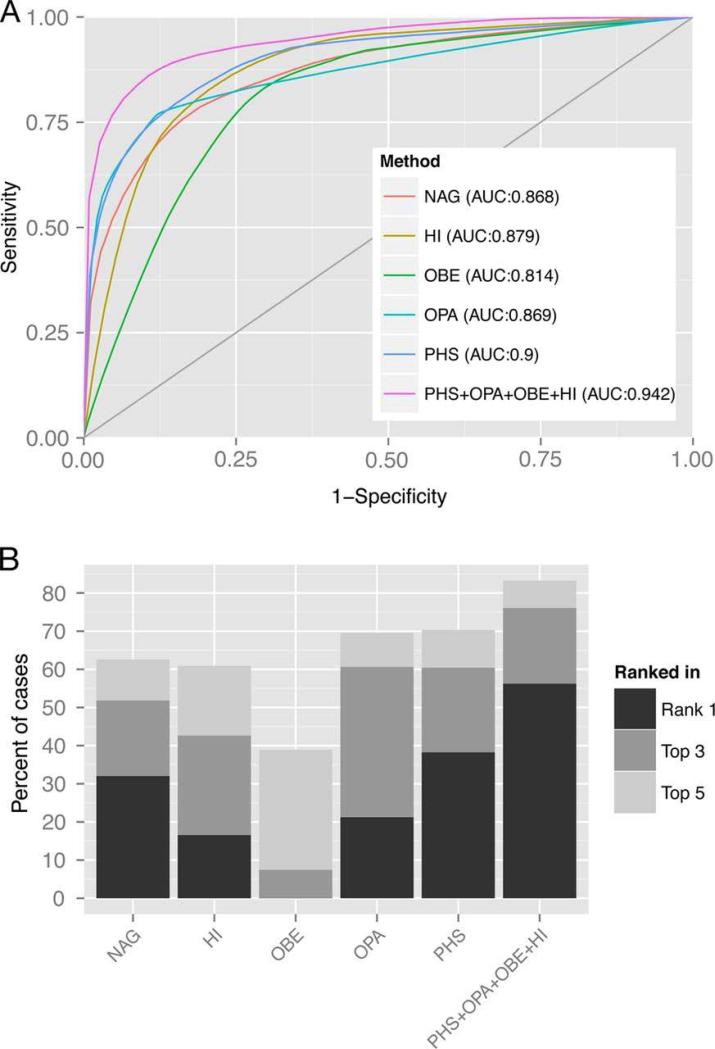
For the 27 800 test cases with 50 CNVs each, we generated the receiver operating characteristic (ROC), which is a plot of the true positive rate against the false positive rate for all different possible cut-offs. The area under the ROC curve (AUC) is an indicator of the performance of a classifier, whereby an AUC of one indicates the best performance (figure 1A) and an AUC of 0.5 indicates a random classification (grey line in figure 1A). Among the individual methods, PHS achieves the best AUC (0.9), followed by OPA (0.869), NAG (0.866), HI (0.879), and OBE (0.814). The combined method PHS+OPA+OBE+HI achieves an AUC of 0.94, outperforming all other methods.
Figure 1.
Performance evaluation. To evaluate our method, we spiked one pathogenic CNV into a set of 49 benign CNVs and generated a ranked list with each of the methods ‘number of affected genes’ (NAG), ‘overlap benign’ (OBE), ‘overlap pathogenic’ (OPA), ‘haploinsufficiency’ (HI), ‘phenoscore’ (PHS) and a combination of selected methods (PHS+OPA+OBE+HI). A total of 27,800 test cases were generated. (A) Receiver operating characteristic (ROC) curves of the rankings obtained by the different methods (the grey line indicates random ranking). (B) A stacked bar chart showing how often the different methods ranked the pathogenic CNV on first place, among the first three CNVs, and among the first five CNVs. AUC, the area under the ROC curve.
In addition, the combined method ranks the pathogenic CNV in first place in ~56% of the simulations and in the range between rank 1 and rank 5 in over 80% of the simulations (figure 1B). The single methods rank the pathogenic CNV on top in 38% (PHS), 32% (NAG), 21% (OPA) and 17% (HI) of the simulated cases (see figure 1B). We did not find a remarkable difference for duplications vs. deletions, e.g. 55.7% of the pathogenic duplications ranked first place and 56.6% of the pathogenic deletion ranked first place.
Our method is well able to rank smaller pathogenic CNVs better than larger benign CNVs: in repeated simulation runs, we found that ~ 18 000 (of 27 800) patients had at least one benign CNV that was larger (contained more genes) than the sought-after pathogenic CNV. In 98% of the cases (~ 17 700), we ranked a smaller pathogenic CNV better than a larger benign CNV.
In summary, we claim that given sufficient phenotypic information about the patient and the presence of phenotype annotations for the affected genes in human or the orthologous genes in mouse or zebrafish, a score based on the phenotypic alignment (PHS) between the patient's symptoms and the known phenotype annotations of the genes is able to prioritise pathogenic CNVs. PHS in combination with other well-known indicators of CNV pathogenicity (OPA, OBE and HI) is found to reliably identify pathogenic CNVs in a set of benign CNVs.
Usage example and data format
PhenogramViz was implemented as a Cytoscape app, and users first have to install Cytoscape (V.3.0.2 or newer) before installing the app. For the analysis, the user must provide the patient's phenotype and CNV data (figure 2). For each CNV, the PHS is calculated. All CNVs are then ranked by the combined score (PHS+OPA+OBE+HI). If a CNV overlaps with known pathogenic or benign CNVs from ISCA and DGV, this is displayed as a colour code (see figure 2(5)). Double clicking a CNV opens a network view of the corresponding phenogram. Phenograms are constructed with a default λ of 2.5. Figure 3 shows the phenogram resulting from phenotype and CNV data of a patient with a large deletion on chromosome 18 and several smaller deletions and duplications. The patient presented for genetic evaluation because of developmental delay, especially affecting expressive language skills. Clinical examination showed a long philtrum and thin upper lip vermillion, strabismus convergens and hypotelorism, tapering fingers, brachydactyly, oedematous feet and short stature. The parents reported feeding difficulties in infancy. Sonography had revealed an unusual asymmetry in cardiac ventricle size. All of these characteristics were encoded as HPO terms and entered into our app, as well as the list of 28 deletions and duplications. Most of the deletions and duplications were either devoid of genes or annotated as non-pathogenic CNVs. There was one 14.45 Mb deletion containing 55 genes on chromosome 18. Previous reports30 described similar features in patients with overlapping deletions, strongly suggesting a causal role of this deletion in the pathogenesis of the patient's disorder. Analysis of the data with PhenogramViz shows that five genes of the large deletion on chromosome 18 align with central phenotypic characteristics of the patient (figure 3). Some phenotypic features cannot be incorporated into the phenogram, suggesting that they are caused by reduced dosage of gene products whose phenotypic effects in humans or model organisms have not yet been elucidated. As expected, the score of the chromosome 18 deletion ranks substantially better than all other CNVs in this patient (figure 2), emphasising its probable pathogenic role.
Conclusion
The classification of CNVs as benign or pathogenic is not trivial. Often, medical analysis focuses on the set of affected genes and aims to align knowledge about these genes with patient phenotypes. With its easy-to-use graphical interface, PhenogramViz greatly facilitates the integration of phenotypic data from humans and model organisms in the evaluation of CNVs of uncertain pathogenic significance and thereby allows physicians access to a knowledge base whose utilisation is otherwise difficult in routine diagnostics. One of the major features of our app is the easy access to information and data on which predictions and visualisation are based. For example, every link in a phenogram can be explored and every element of the rankings can be explained (see figure 3). The visualisations can be used as part of the provenance for the data explanations themselves and could be considered as justification for further evaluation, treatments, etc., and towards this end, can be included in the medical record.
Several methods exist for the prediction of pathogenic CNVs among a set of benign CNVs, including Genomic Classification of CNVs Objectively (GeCCO),31 NETwork-Based Analysis of Genetic associations (NETBAG)32 and a ranking based on HI scores.27 GeCCO makes use of several genomic features such as the density of long interspersed nuclear elements or the presence of genes known to be associated with nervous system phenotypes in the mouse and is trained for detecting CNVs related to intellectual disability and is not intended to be a tool for arbitrary diseases associated with CNVs. NETBAG is a functional gene network that has been applied to autism CNVs. It uses the set of affected genes of all CNVs in a patient and tries to identify functionally connected clusters. Again, the phenotypes of the patient are not taken into account. In addition, NETBAG's webpage only allows for loading a set of genes, but no CNV intervals. The aforementioned method that ranks CNVs by HI scores uses a predictive model based on multiple genomic features to distinguish a set of known haploinsufficient and haplosufficient genes. This model can be used to predict specific haploinsufficient genes and thus assesses the pathogenicity of deletions. Again, this method is phenotype agnostic and a software tool to perform predictions is not provided.
In general, it may not be trivial for physicians to apply GeCCO, NETBAG or the HI ranking methods to their own data. We claim that our tool has the advantage of being independent of a specific phenotypic category, since it is not trained for any specific clinical phenotype or disease focus. Also, to our knowledge, ours is the first tool that makes extensive use of the available phenotype resources that exist for human, mouse and zebrafish and aims to perform a phenotypic alignment of the affected genes to the patient phenotypes. Finally, the visual exploration of single gene-to-phenotype relationships for all genes in a particular CNV is a feature unique to our software.
We have demonstrated that a score based on the phenogram can be used for prioritisation of pathogenic CNVs. We furthermore showed that a combination of four scores is best suited for prioritising pathogenic CNVs. Thus, in PhenogramViz the patient's CNVs are ranked per default by the combination of overlap with known pathogenic and benign CNVs, together with a score that reflects how well the phenotype knowledge of the affected genes aligns with the recorded phenotypes of the patient, and the HI score.
Currently, there are 8866 human genes with phenotype annotations directly derived from human diseases or transferred from mouse or zebrafish experiments. Therefore, phenotype associations for about 55% of the approximately 20 000 human protein-coding genes are still to be uncovered. Efforts such as those of the International Mouse Phenotype Consortium33 and Zebrafish Mutation Project34 to provide a comprehensive characterisation of the phenotypic effects of mutation in nearly all mouse genes will further strengthen phenotype-based analyses such as the one presented here. We are also participating in the Monarch Initiative, which aims to provide data integration for a large diversity of phenotype data, including additional model and non-model organisms, which, in future, will greatly expand the phenotype coverage available to PhenogramViz and hence better support clinical interpretation of CNVs.
Acknowledgments
For clinical examination and SNP array analysis of the patient whose data were used for demonstration in the figures, we would like to thank Barbara Mikat (Essen) and Hermann-Josef Lüdecke (Essen).
Funding The project was funded by grants of the Bundesministerium fuer Bildung und Forschung (BMBF project number 0313911) and the European Community's Seventh Framework Programme (Grant Agreement 602300; SYBIL). Further support was provided by the National Institutes of Health (NIH Office of the Director Grant #5R24OD011883).
Footnotes
Contributors US, SK, SCD and PNR: conceived the software. US and SK: programmed the software. SK, JCC and US: tested/evaluated the software. SK, CJM, MAH, DS, JI-S and PNR generated the phenotype ontology and processed phenotype annotation data. SK: performed the benchmark tests. SK, US, JCC, PNR, JI-S, JYH-K, MAH, CJM and DS wrote the manuscript. All authors: approved the manuscript for final submission.
Competing interests None.
Provenance and peer review Not commissioned; externally peer reviewed.
Data sharing statement All the data are publicly available.
Web resources The URLs for data presented in the manuscript are as follows: UCSC Genome Browser, http://genome.ucsc.edu/; OMIM, http://www.omim.org; Database of Genomic Variants (DGV), http://projects.tcag.ca/variation/; Cytoscape, http://cytoscape.org. The data in this manuscript were obtained from the ISCA Consortium database (http://www.iscaconsortium.org), which generates this information using NCBI's Database of Genomic Structural Variation (dbVar, http://www.ncbi.nlm.nih.gov/dbvar/), study nstd37. Samples and associated phenotype data were provided by ISCA Consortium member laboratories. ISCA files used: http://www.ncbi.nlm.nih.gov/dbvar/studies/nstd37/,http://www.ncbi.nlm.nih.gov/dbvar/studies/nstd45/ and http://www.ncbi.nlm.nih.gov/dbvar/studies/nstd75/. DGV file:http://dgv.tcag.ca/dgv/docs/GRCh37_hg19_variants_2013-07-23.txt. The HI scores were obtained from http://www.plosgenetics.org/article/fetchSingleRepresentation.action?uri=info:doi/10.1371/journal.pgen.1001154.s002.
References
- 1.Kanduri C, Ukkola-Vuoti L, Oikkonen J, Buck G, Blancher C, Raijas P, Karma K, Lahdesmaki H, Jarvela I. The genome-wide landscape of copy number variations in the MUSGEN study provides evidence for a founder effect in the isolated Finnish population. Eur J Hum Genet. 2013;21:1411–16. doi: 10.1038/ejhg.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castellani CA, Melka MG, Wishart AE, Locke ME, Awamleh Z, O'Reilly RL, Singh SM. Biological relevance of CNV calling methods using familial relatedness including monozygotic twins. BMC Bioinformatics. 2014;15:114. doi: 10.1186/1471-2105-15-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–81. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsuchiya KD, Shaffer LG, Aradhya S, Gastier-Foster JM, Patel A, Rudd MK, Biggerstaff JS, Sanger WG, Schwartz S, Tepperberg JH, Thorland EC, Torchia BA, Brothman AR. Variability in interpreting and reporting copy number changes detected by array-based technology in clinical laboratories. Genet Med. 2009;11:866–73. doi: 10.1097/GIM.0b013e3181c0c3b0. [DOI] [PubMed] [Google Scholar]
- 5.Valsesia A, Mace A, Jacquemont S, Beckmann JS, Kutalik Z. The growing importance of CNVs: new insights for detection and clinical interpretation. Front Genet. 2013;4:92. doi: 10.3389/fgene.2013.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poot M, Hochstenbach R. A three-step workflow procedure for the interpretation of array-based comparative genome hybridization results in patients with idiopathic mental retardation and congenital anomalies. Genet Med. 2010;12:478–85. doi: 10.1097/GIM.0b013e3181e3914a. [DOI] [PubMed] [Google Scholar]
- 7.de Leeuw N, Dijkhuizen T, Hehir-Kwa JY, Carter NP, Feuk L, Firth HV, Kuhn RM, Ledbetter DH, Martin CL, van Ravenswaaij-Arts CM, Scherer SW, Shams S, Van Vooren S, Sijmons R, Swertz M, Hastings R. Diagnostic interpretation of array data using public databases and internet sources. Hum Mutat. Published Online First. 2012 doi: 10.1002/humu.22049. doi: 10.1002/humu.22049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korf BR, Rehm HL. New approaches to molecular diagnosis. JAMA. 2013;309:1511–21. doi: 10.1001/jama.2013.3239. [DOI] [PubMed] [Google Scholar]
- 10.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, Faucett WA, Feuk L, Friedman JM, Hamosh A, Jackson L, Kaminsky EB, Kok K, Krantz ID, Kuhn RM, Lee C, Ostell JM, Rosenberg C, Scherer SW, Spinner NB, Stavropoulos DJ, Tepperberg JH, Thorland EC, Vermeesch JR, Waggoner DJ, Watson MS, Martin CL, Ledbetter DH. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riggs ER, Wain KE, Riethmaier D, Savage M, Smith-Packard B, Kaminsky EB, Rehm HL, Martin CL, Ledbetter DH, Faucett WA. Towards a Universal Clinical Genomics Database: the 2012 International Standards for Cytogenomic Arrays Consortium Meeting. Hum Mutat. 2013;34:915–19. doi: 10.1002/humu.22306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webber C, Hehir-Kwa JY, Nguyen DQ, de Vries BB, Veltman JA, Ponting CP. Forging links between human mental retardation-associated CNVs and mouse gene knockout models. PLoS Genet. 2009;5:e1000531. doi: 10.1371/journal.pgen.1000531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doelken SC, Kohler S, Mungall CJ, Gkoutos GV, Ruef BJ, Smith C, Smedley D, Bauer S, Klopocki E, Schofield PN, Westerfield M, Robinson PN, Lewis SE. Phenotypic overlap in the contribution of individual genes to CNV pathogenicity revealed by cross-species computational analysis of single-gene mutations in humans, mice and zebrafish. Dis Model Mech. 2013;6:358–72. doi: 10.1242/dmm.010322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson PN, Webber C. Phenotype ontologies and cross-species analysis for translational research. PLoS Genet. 2014;10:e1004268. doi: 10.1371/journal.pgen.1004268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monarch Initiative. 2012;33:930–40. Secondary 2014. http://monarchinitiative.org/. Hum Mutat. [Google Scholar]
- 16.Kohler S, Doelken SC, Rath A, Ayme S, Robinson PN. Ontological phenotype standards for neurogenetics. Hum Mutat. 2012;33:1333–9. doi: 10.1002/humu.22112. [DOI] [PubMed] [Google Scholar]
- 17.Kohler S, Bauer S, Mungall CJ, Carletti G, Smith CL, Schofield P, Gkoutos GV, Robinson PN. Improving ontologies by automatic reasoning and evaluation of logical definitions. BMC Bioinformatics. 2011;12:418. doi: 10.1186/1471-2105-12-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohler S, Doelken SC, Ruef BJ, Bauer S, Washington N, Westerfield M, Gkoutos G, Schofield P, Smedley D, Lewis SE, Robinson PN, Mungall CJ. Construction and accessibility of a cross-species phenotype ontology along with gene annotations for biomedical research. F1000Research. 2013;2:30. doi: 10.12688/f1000research.2-30.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat Methods. 2012;9:1069–76. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM(R)). Hum Mutat. 2011;32:564–7. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- 22.Rath A, Olry A, Dhombres F, Brandt MM, Urbero B, Ayme S. Representation of rare diseases in health information systems: the Orphanet approach to serve a wide range of end users. Hum Mutat. 2012;33:803–8. doi: 10.1002/humu.22078. [DOI] [PubMed] [Google Scholar]
- 23.Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE, Mouse Genome Database G. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42(Database issue):D810–17. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howe DG, Bradford YM, Conlin T, Eagle AE, Fashena D, Frazer K, Knight J, Mani P, Martin R, Moxon SA, Paddock H, Pich C, Ramachandran S, Ruef BJ, Ruzicka L, Schaper K, Shao X, Singer A, Sprunger B, Van Slyke CE, Westerfield M. ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics. Nucleic Acids Res. 2013;41(Database issue):D854–60. doi: 10.1093/nar/gks938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, Black GC, Brown DL, Brudno M, Campbell J, FitzPatrick DR, Eppig JT, Jackson AP, Freson K, Girdea M, Helbig I, Hurst JA, Jahn J, Jackson LG, Kelly AM, Ledbetter DH, Mansour S, Martin CL, Moss C, Mumford A, Ouwehand WH, Park SM, Riggs ER, Scott RH, Sisodiya S, Van Vooren S, Wapner RJ, Wilkie AO, Wright CF, Vulto-van Silfhout AT, de Leeuw N, de Vries BB, Washingthon NL, Smith CL, Westerfield M, Schofield P, Ruef BJ, Gkoutos GV, Haendel M, Smedley D, Lewis SE, Robinson PN. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42(Database issue):D966–74. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Resnik P. Using information content to evaluate semantic similarity in a taxonomy. 1995 arXiv preprint cmp-lg/9511007. [Google Scholar]
- 27.Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42(Database issue):D986–92. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pober BR. Williams-Beuren syndrome. N Engl J Med. 2010;362:239–52. doi: 10.1056/NEJMra0903074. [DOI] [PubMed] [Google Scholar]
- 30.Cody JD, Sebold C, Malik A, Heard P, Carter E, Crandall A, Soileau B, Semrud-Clikeman M, Cody CM, Hardies LJ, Li J, Lancaster J, Fox PT, Stratton RF, Perry B, Hale DE. Recurrent interstitial deletions of proximal 18q: a new syndrome involving expressive speech delay. Am J Med Genet A. 2007;143A:1181–90. doi: 10.1002/ajmg.a.31729. [DOI] [PubMed] [Google Scholar]
- 31.Hehir-Kwa JY, Wieskamp N, Webber C, Pfundt R, Brunner HG, Gilissen C, de Vries BB, Ponting CP, Veltman JA. Accurate distinction of pathogenic from benign CNVs in mental retardation. PLoS Comput Biol. 2010;6:e1000752. doi: 10.1371/journal.pcbi.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koscielny G, Yaikhom G, Iyer V, Meehan TF, Morgan H, Atienza-Herrero J, Blake A, Chen CK, Easty R, Di Fenza A, Fiegel T, Grifiths M, Horne A, Karp NA, Kurbatova N, Mason JC, Matthews P, Oakley DJ, Qazi A, Regnart J, Retha A, Santos LA, Sneddon DJ, Warren J, Westerberg H, Wilson RJ, Melvin DG, Smedley D, Brown SD, Flicek P, Skarnes WC, Mallon AM, Parkinson H. The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res. 2014;42(Database issue):D802–9. doi: 10.1093/nar/gkt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kettleborough RN, Busch-Nentwich EM, Harvey SA, Dooley CM, de Bruijn E, van Eeden F, Sealy I, White RJ, Herd C, Nijman IJ, Fenyes F, Mehroke S, Scahill C, Gibbons R, Wali N, Carruthers S, Hall A, Yen J, Cuppen E, Stemple DL. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496:494–7. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]