Abstract
O-6-methylguanine-DNA methyltransferase (MGMT) is an abundantly expressed nuclear protein dealkylating O6-methylguanine (O6-MG) DNA residue, thus correcting the mismatches of O6-MG with a thymine residue during DNA replication. The dealkylating effect of MGMT is relevant not only in repairing DNA mismatches produced by environmental alkylating agents promoting tumor pathogenesis, but also when alkylating molecules are applied in the chemotherapy of different cancers, including glioma, the most common primary tumor of the central nervous system. Elevated MGMT gene expression is known to confer resistance to the treatment with the alkylating drug temozolomide in patients affected by gliomas and, on the contrary, methylation of MGMT gene promoter, which causes reduction of MGMT protein expression, is known to predict a favourable response to temozolomide. Thus, detecting expression levels of MGMT gene is crucial to indicate the option of alkylating agents or to select patients directly for a second line targeted therapy. Further study is required to gain insights into MGMT expression regulation, that has attracted growing interest recently in MGMT promoter methylation, histone acetylation and microRNAs expression. The review will focus on the epigenetic regulation of MGMT gene, with translational applications to the identification of biomarkers predicting response to therapy and prognosis.
Keywords: MGMT, glioblastoma, glioma, temozolomide, microRNA, peptide nucleic acids, locked nucleic acids, methylation, pyrosequencing
1. Role of MGMT in cancer
O6-methylguanine (O6-MG) is one of the major mutagenic and carcinogenic lesions in DNA induced by alkylating mutagens, because of its preference for pairing with thymine instead of cytosine during DNA replication. O6-methylguanine-DNA methyltransferase (MGMT) is a ubiquitously expressed nuclear enzyme which removes alkyl groups from the O6-position of O6-MG (1). Each single alkyl group removed from O6-MG is transferred to a cysteine residue within the active site of MGMT in a stoichiometric second-order reaction, implying the inactivation of one molecule of MGMT enzyme for each alkyl group removed from methylguanine, a process termed suicide inhibition (2). Consequently, the efficiency of O6-MG repair is limited by the number of molecules of MGMT enzyme available, also considering that the dealkylating function of MGMT does not possess redundant or alternative pathways. MGMT-mediated removal of alkyl groups from O6-MG is also relevant in alkylating chemotherapy of glioma, such as with temozolomide and nitrosourea derivatives. Massive DNA alkylation produced by temozolomide causes base mispairing (3,4). If O6-MG is not repaired because of low MGMT expression, O6-MG forms a base pair with thymine. The mismatched O6-MG to thymine base pair is recognized by the pathway involving the repair proteins MLH1, MSH2, MSH6 and PMS2, resulting in futile cycles of repair that lead to cell cycle arrest and cell death (5). On the contrary, the methylation damage produced by temozolomide can be reversed by MGMT, as its DNA repairing activity provides resistance against the cytotoxic effects of guanine methylation. As already shown in clinical trials in patients affected by glioma that have been treated with the alkylating drug temozolomide, whose response to therapy is significantly ameliorated when MGMT expression is reduced because of promoter methylation (6).
2. Nuclear transcription factors regulating the expression of MGMT gene
MGMT gene (ID: 4255, NCBI Ref Seq NM_002412.3) has been cloned (1) and mapped to chromosome 10 in the cytogenetic location 10q26.3, where it spans 15 Kb in length (7). So far 142 single nucleotide variants of MGMT gene have been reported. Transcript of 1265 nucleotides is organized in 5 exons encoding a protein of 238 amino acid residues. A CpG island of 762 base pairs that includes 97 CpG dinucleotides spans the proximal promoter region and the first exon. Different transcription factors have been found to activate the transcription of MGMT gene, including Sp1 (8), NF-κB (9), CEBP (10) and AP-1 (10,11). The cellular tumor antigen p53 has been associated with repression of MGMT transcription (12), possibly via sequestering the Sp1 nuclear transcription factor (13).
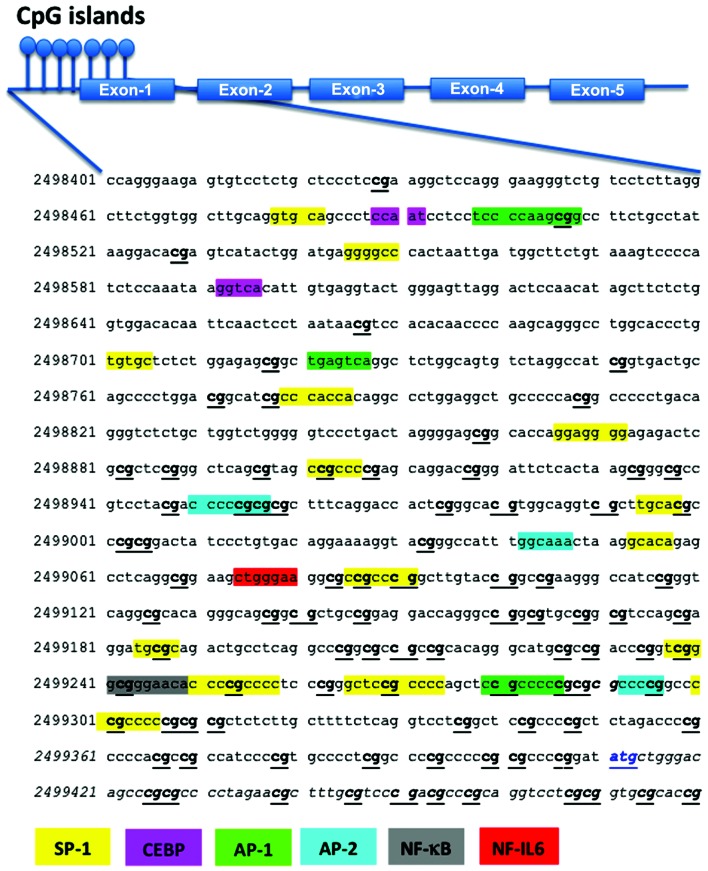
In silico analysis on the putative consensus sequences for the binding of nuclear transcription factors in MGMT promoter sequence reveals further nuclear factors potentially involved in activating MGMT transcription, for instance AP-2, NF-IL6, and ER-α, besides Sp1, AP-1 and c/EBP, as shown in Fig. 1. As some of these consensus sequences include CpG dinucleotides, question has arisen on whether methylation of CpG dinucleotides in key consensus sequences could hinder the binding of the corresponding transcription factors, thus reducing the transcriptional activation of the MGMT gene. For instance, this inhibiting mechanism has been evidenced for Sp1-dependent, but not NF-κB-dependent activation of MGMT transcription (8,9), thus keeping the hypothesis open for other transcription factors. Noteworthy, MGMT expression has been found heterogeneous within histological sections of gliomas, being higher in the inner core of the tumor than in the periphery (14).
Figure 1.
MGMT promoter: schematic of transcription factors and methylation islands. Schematic representation of the CpG dinucleotides (bold underlined) and the putative consensus sequences of the major nuclear transcription factors involved in MGMT transcriptional activation as identified in silico by Transcription Elements Search System (TESS) analysis (http://www.cbil.upenn.edu/tess). Color boxes indicate the position of the nuclear transcription factors SP-1, CEBP, AP-1, AP-2, NF-κB and NF-IL6.
Further analyses correlated the high expression of MGMT with the presence of a hypovascular central core of the tumor, where activation of the hypoxia inducible factor (HIF)-1α will in turn promote expression of MGMT, particularly in the hypoxic glioma stem cells niches (14,15). The MGMT/HIF-1α regulatory axis has been further confirmed, since the expression of the bone morphogenetic protein 2 (BMP2) has been shown to downregulate MGMT expression through HIF-1α-dependent downregulation (16). Moreover, hypoxia intervenes further on MGMT function as HIF-1α is known to induce also the expression of the hypoxia-inducible and steroid-inducible N-myc downstream regulated gene 1 (NDRG1) protein, which binds to and stabilizes MGMT protein (17). In summary, although Sp1, AP-1, CEBP, NF-κB and HIF-1α as single transcription factors have been proved to activate MGMT gene regulation (8–11), the understanding of the synergy within these transcription factors in activating MGMT expression and the composition of the MGMT promoter enhanceosome need further investigation.
3. Effect of histone modifications
Epigenetic modifications of MGMT gene have been found to play a relevant role in MGMT expression in the context of cancer. Histone acetylation and methylation has been extensively investigated in relation to MGMT expression, in different cancer models including gliomas (18–24). Acetylation of lysine residues on histones H3 and H4 (H3Ac and H4Ac), that are associated with open chromatin and active transcription, has been found elevated in cell lines expressing high level of MGMT, suggesting a role for these histone modifications (18). On the contrary, di-methylation of lysine 9 of histone 3 (H3me2K9) has been found relevant in silencing MGMT expression (20).
The relevance of the role of histone acetylation on MGMT expression has been recently confirmed by testing in vitro the effect of the histone deacetylase (HDAC) inhibitor suber-oylalanide hydroxamic acid (SAHA), which increased MGMT expression, thus strengthening resistance to the alkylating agent temozolomide (22). However, considering the multiplicity of the target genes that may be modulated by HDAC inhibitors, these drugs could have opposite effects when used in chemotherapy. For instance, the treatment of U251 glioma cells with the HDAC inhibitor LBH589 increased the sensitivity to temozolomide. This could be explained with an increased expression of the heat shock protein 90 (HSP90), which in turn induced downregulation of expression of the epidermal growth factors receptor (EGFR) and the phosphoprotein p-Akt. This could be responsible for an increase of the pro-apoptotic effect of temozolomide, independently of the levels of expression of MGMT (23).
The major role of histone in the regulation of expression of MGMT is summarized in Fig. 2. Of note, histone acetylation and the expression levels of HDAC are therefore increasing in relation to the regulation of expression of MGMT, consequently improving the response to therapy and the overall prognosis of patients affected by glioma (24).
Figure 2.
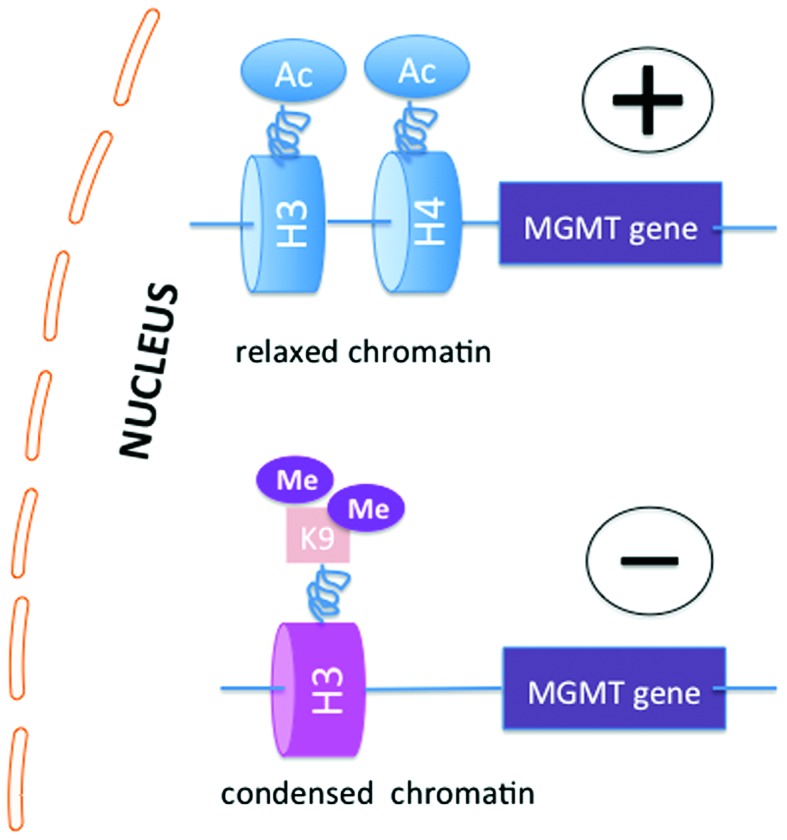
Effect of histone modification on MGMT gene expression. Acetylation of histones H3 and H4 promotes MGMT transcription (18), whereas di-methylation of lysine 9 on histone H3 represses MGMT transcription (20).
4. MGMT promoter methylation
Methylation of MGMT promoter is found in 40% of cancer types such as glioma and colorectal cancer and in 25% of non-small cell lung carcinoma, lymphoma and head and neck carcinoma (25). As promoter methylation is one of the major post-transcriptional mechanisms reducing protein expression, the methylation of CpG sites in the promoter and the overall extent of methylation could affect the levels of expression of the protein. Therefore, expression of MGMT protein is significantly reduced in MGMT-methylated cancer cells, as detected by immunohistochemistry (25), and the levels of expression of MGMT protein have been associated with the efficacy of response of cancer cells to alkylating drugs in glioma tumor models in rodents in vivo (26,27). All these pieces of evidence prompted investigations on the relation between MGMT promoter methylation and the response to chemotherapy with alkylating drugs in patients affected by gliomas (6). This original observation was soon extended into a large series of clinical investigations (28–84) in order to assess the potency of MGMT methylation as a predictive marker in relation to alkylating therapy in different conditions of adult and pediatric gliomas, of both high and low grade according to WHO classification (85).
The relevance of MGMT methylation as a biomarker has been strengthened by the widely accepted application of the consensus reached in Phase III clinical trials jointly conducted by European and North-American research networks, that were summarized in the so-termed ‘Stupp protocol’ for the first line treatment of patients affected by gliomas, which includes the post-surgery association of radiotherapy and temozolomide (47). The predictive value of MGMT methylation in response to temozolomide has reached over the years an overall confirmatory consensus, such that analysis of MGMT methylation has been included to stratify patients enrolled in major multicenter international clinical trials (38,47,58,60,71,74,75,86) and leading recommendations have been stated on how to treat patients affected by glioma, where the analysis of MGMT methylation is assumed as one of the key decision points in the therapeutic flow-chart (87). Besides the overall confirmatory consensus, different issues have been considered both to interpret the role and to improve the predictive role of MGMT methylation as clinical biomarker. For instance, the overall survival and the progression-free survival of patients treated with temozolomide is related to the overall level of methylation of the MGMT promoter, assessed by quantitative methylation-specific techniques, as highly methylated samples are significantly associated with the best prognosis (50). However, within the 97 CpG dinucleotides identified in the CpG island of the MGMT proxymal promoter, the extent of methylation can be variable, with different effects on the degree of gene silencing (18).
Moreover, methylation of a specific CpG dinucleotide of the CpG island could have different impact on transcriptional downregulation. For instance, binding of the methyl-CpG binding (MCB) protein 2 (MeCP2) to methylated CpG islands have been found relevant in silencing MGMT expression (20). MGMT methylation could interfere with the activatory role of the transcription factor Sp1 (8), but the identification of the specific CpG dinucleotides involved has not been cleared. The expression of MGMT protein is heterogeneous within the glioma tissue. For instance, MGMT promoter methylation is not different among the concentric layers of glioblastoma specimens (15,88) whereas the core of the glioma often presents higher expression of MGMT protein than the peripheral areas (88), possibly because of the transcriptional activation of expression mediated by hypoxia and activation of HIF-1α pathway (14,15). Thus, MGMT promoter methylation is a very relevant, but not a unique mechanism, to regulate the expression of MGMT and the response to therapy with alkylating agents (89).
5. MicroRNAs in MGMT expression regulation
MicroRNAs (miRs) (www.mirbase.org) belong to a family of small (19 to 25 nucleotides in length) noncoding RNAs that target specific sequences of mRNAs thereby regulating gene expression (90,91), causing translational repression or mRNA degradation, depending on the degree of complementarities between miRs and the target sequences (92,93). Although in silico analysis of the 3′-UTR of the MGMT gene has revealed several potential sequences that could be a site for interaction of miRs (94), miR-dependent regulation of MGMT expression is presently under intensive investigation. Different studies have been recently conducted in order to associate modulation of expression of specific miRs with the response to temozolomide in patients affected by glioblastoma and in experimental cell models in vitro (95–127). However, only some of these investigations can be directly related to the therapeutic axis between temozolomide and MGMT expression.
A genome-wide analysis of expression of 1,146 miRs performed in tissue samples obtained from 82 glioblastoma specimens was correlated with overall survival and further validated in The Cancer Genome Atlas (TCGA) dataset, which includes 424 glioma samples (100). Comparative analysis evidenced the miR-181d expression in glioma tissues as inversely correlated with a favourable prognosis in these patients and that the favourable effect of miR-181d is, at least partially, related to its effect in downmodulating MGMT mRNA expression (100). Furthermore, miR-181d expression inversely correlated with that of MGMT mRNA in glioma tissue specimens; moreover, overexpression of miR-181d in A1207, LN340 and T98G glioblastoma cell lines reduced MGMT mRNA levels and conferred pro-apoptotic sensitivity to temozolomide (100).
Additional information on miR-dependent regulation of MGMT expression has been provided by an investigation starting from a bioinformatics analysis in the TCGA database related to glioblastoma (128), aimed to search inverse correlation between miRs levels and MGMT mRNA, taking into account also the contribution of the MGMT promoter methylation (106). The bioinformatics analysis confirmed the role of miR-181d and found miR-767-3p and miR-648 as novel potential regulators of MGMT gene expression (106). Validation in glioma tissues ex vivo and in experimental glioblastoma cell models in vitro supported the bioinformatics analyses and indicated that downregulation of MGMT expression by miR-181d and miR-767-3p is due to degradation of the MGMT mRNA whereas miR-648 affects MGMT protein translation (106). Experiments performed in vitro confirmed that the overexpression of these three miRs reduces MGMT protein expression and confers sensitivity to temozolomide (106). A third contribution was focused on the paralogues miR-221 and miR-222 (114), known to be highly expressed in glioblastoma tissues (128,129). They have been found to downregulate MGMT expression and to confer increased sensitivity to temozolomide (114). These results need to be confirmed, since a pro-apoptotic effect of glioblastoma cell lines was obtained using antagomiR molecules against miR-221 and the expression of the 221/222 cluster was associated to chemio- and radio-resistance (130–133).
A fourth major contribution was obtained by testing significant reduction of MGMT protein expression with a genome-wide miR screening performed by transfecting 885 known miRs into the T98G glioblastoma cell line, which is characterized by high levels of the MGMT protein (122). The miRs identified by the first screening were validated by in silico analysis of putative binding sites in the 3′-UTR region of MGMT gene utilizing different algorithms, which restricted the potentially relevant miRs to a limited series, that was verified for inverse correlation of expression with MGMT in the Chinese Cancer Genome Atlas, then tested in LN340 glioblastoma cell line in vitro (122). Some of the previously reported miRs (such as miR-221, -222 and -648) did not consistently downregulate MGMT expression in LN340 cells, whereas miR-181d and miR-767-3p had a positive effect (122).
In addition, the novel regulatory miR-603, which directly interacts with the 3′-UTR region of MGMT gene, produced a 6-fold decrease of both MGMT mRNA and protein upon transfection in glioblastoma cell line in vitro and sensitized glioblastoma cells to temozolomide (122). Therefore, although considering that temozolomide sensitivity of glioblastoma can be modulated by different miRs independently of the regulation of MGMT expression, six miRs have been reported to be involved in the regulation of MGMT protein expression either by degrading MGMT mRNA, namely miR-181d, -767-3p, -221, -222, -603 (100,106,114,122), or affecting the MGMT protein translation, such as miR-648 (106). Considering that this series of miRs have been identified with different approaches and that the inverse correlation expected between MGMT expression and the expression of each miR is not usually characterized by very high and significant correlation coefficient, possibilities are open that further miRs will be revealed as regulators of MGMT expression and that the MGMT downregulation might require the synergy of action of these and/or other miRs, which presently requires further investigation.
6. MGMT expression as predictive biomarker
Inactivation of expression of MGMT gene as an effect of MGMT promoter methylation has been found as a relevant predictive biomarker of the response to the alkylating drug temozolomide in patients affected by glioma (6) and the original observation was basically confirmed by several replication studies (28–84). These studies raised the question on the most reliable method to evaluate MGMT expression, either directly or indirectly, in relation to its clinical predictivity to alkylating therapy of gliomas. MGMT clinical testing assays have been extensively reviewed elsewhere (134) and thus only briefly summarized here. The most direct assay to detect the effect of MGMT in dealkylating O6-MG would be in principle measuring enzyme activity (135). However, the method proposed and originally tested in glioma specimens is cumbersome for routine clinical applications and requires radioactive isotopes and availability of fresh tissue (135). After the initial studies (135,136), it has not been extensively investigated in relation to the response to alkylating agents. Immunohistochemistry of MGMT protein has been first compared with enzyme activity (137) and then correlated with response to temozolomide (2,138–140).
Initial analyses were performed by immunofluorescence in order to quantify by digital image analyses the levels of expression, whereas often the immunohistochemistry assay of MGMT protein was restricted to the count of positive cells, thus excluding the information on the real amount of expression in each cells, which is in principle the most relevant piece of information to predict the resistance to temozolomide (2,138–140). Moreover, the quantification of the percentage of positive cells could be discordant between different pathologists (141). Possibly because of these limitations, the reliability of immunohistochemistry of MGMT protein to predict response to alkylating agents in glioma has been strongly criticized (141,142) and not widely utilized in clinical practice. Although MGMT mRNA might not always represent MGMT protein expression, MGMT transcript has been tested in few studies in relation to glioma, mainly by quantitative reverse-transcription polymerase chain reaction (RT-qPCR) and less frequently by in situ hybridization (143–150).
Low MGMT mRNA expression has been found predictive of better response to temozolomide in at least two recent observational studies (89,150), consistently with the elevated methylation pattern of MGMT promoter. However, MGMT mRNA quantitation is not largely utilized as clinical predictive biomarker, possibly because so far these analyses on transcripts were performed in total RNA obtained from fresh tissue samples during neurosurgery (89,150), in order to cope with the instability of RNA. Therefore, more clinical observations on MGMT mRNA and clinical response to temozolomide and the feasibility of performing these analyses either from fresh tissue or from the routine tissue slices of formalin-fixed paraffin-embedded samples will be useful. Thus, for practical reasons mainly related to the stability of DNA, the most widely utilized clinical assay to estimate MGMT expression levels is the analysis of MGMT promoter methylation (151,152). Non-quantitative methylation-specific polymerase chain reaction (MS-PCR) assay after bisulfite conversion of the MGMT promoter has been utilized in the first clinical studies investigating the predictive role of MGMT methylation in response to temozolomide (6,34,38,41,49,52,60,68,149,153–157). MGMT promoter methylation has been also extended from a non-quantitative to a quantitative assay using pyrosequencing technique (158,159).
Noteworthy, the quantitative MGMT methylation, as assessed by pyrosequencing, has been correlated with progression-free survival and overall survival of patients treated with temozolomide. Patients stratified in ranges of percentage of MGMT methylation were significantly correlated with clinical outcome (50), which is consistent with an inverse correlation between promoter methylation and protein expression. Further applications of the quantitative MS-PCR assay in large clinical trials confirmed its reliability (72,74,75). Importantly, different method comparisons concluded that non-quantitative MS-PCR assays are scarcely reproducible within and between laboratories (55,160–163), whereas at the present time quantitative MS-PCR by pyrosequencing technique has become the technique of choice for clinical routine applications (164).
In summary, the most widely utilized assay to analyze the levels of expression of MGMT gene in clinical routine are based on the indirect analysis of MGMT promoter methylation. Evidence are growing on the need of stratifying patients in terms of quantitative methylation, utilizing reliable and reproducible techniques such as pyrosequencing. Further observations on quantitative assays for MGMT mRNA and quantitative MGMT methylation levels will accomplish the role of MGMT expression as biomarker predicting the response to alkylating chemotherapy, in order to provide more rational bases for decisions on the therapeutic options available for patients affected by glioma.
7. Manipulating MGMT expression to improve first line therapy of glioblastoma
Glioblastoma is the most common primary tumor of the central nervous system, accounting for 12–15% of all intracranial tumors and 50–60% of gliomas (165). Patients die within a few months if untreated and surgery followed by radiotherapy in addition to temozolomide prolongs median survival to 12–15 months, although disease progresses within 6–9 months, with 2-years survival <25% (165). The exponential increase of available anti-cancer targeted therapies provides new hope in improving the prognosis of these patients, since repositioning different drugs tested in other cancers to glioblastoma patients are under intensive investigation (166). For instance, anti-angiogenic targeted therapy with bevacizumab was found to increase the median progression-free survival of 4.4 months (167,168). Considering that reduced expression or silencing of MGMT gene due to promoter methylation in glioma specimens increased median overall survival after temozolomide of 6.4 months (34) and of 15.1 months comparing the group of non-methylated versus that of highly methylated tumor samples (50), reduction of MGMT expression in patients presenting non-methylated MGMT promoter could be of relevant benefit in relation to the pace of the advancements in the therapy of this specific malignant tumor.
Temozolomide itself has been shown to partially deplete the extent of expression of MGMT protein in tumor specimens of one patient affected by metastatic melanoma (2), possibly as a result of the inactivation and degradation of one molecule of MGMT protein each cycle of dealkylation of O6-MG, but the large series of clinical trials performed in patients affected by glioblastoma considering MGMT methylation indicated this mechanism is not sufficient in routinary clinical applications aimed to overcome the resistance to temozolomide in MGMT unmethylated glioma cells (38,47,58,60,71,74,75,86). Different alternative small organic molecules to bypass the limitations of temozolomide in MGMT unmethylated patients have been devised, as reviewed extensively elsewhere (169), such as 1,3-bis(2-chloroethyl)1-nitrosourea (BCNU), also known as carmustine, or 3-[(4-amino-2-methyl-5-pyrimidinyl) methyl]-1-(2-chloroethyl)-1-nitrosourea hydrochloride (ACNU), also known as nimustine. However, the frequent severe hematological side effects of these drugs, such as thrombocytopaenia and neutropenia in 18–23% of patients, are presently limiting the application of these drugs in patients with glioblastoma (169).
A further alternative was to sensitive tumor cells to temozolomide by concomitant use of the pseudosubstrate O6-benzylguanine (O6-BG) which also depletes MGMT by activating its ‘suicidal’ dealkylation mechanism (170). However, a Phase II clinical trial with temozolomide and concomitant O6-BG produced grade 4 hematological adverse events in 48% of the patients, halting further attempts to use this concomitant therapy (171). Thus, alkylating drugs alternative to temozolomide or MGMT pseudosubstrates to bypass or potentiate temozolomide are presently under scrutiny in terms of advantages for patients with unmethylated MGMT gene and suggest the need of exploring other approaches.
The present knowledge on the mechanisms of regulation of MGMT gene expression at different transcriptional and post-transcriptional levels could be utilized to design innovative tools to manipulate its expression. For instance, RNA-based molecules and analogues, locked nucleic acids (LNAs) and peptide nucleic acids (PNAs), are novel tools to upmodulate or downmodulate miRs expression (172–176), by molecular mimicry or competitive sequestration, respectively, with potential experimental therapeutic applications on post-transcriptional gene expression modulation, in principle feasible also in glioblastoma models (130).
8. Conclusions
MGMT expression in patients affected by glioblastoma is a double-edged sword as low levels favour cancer pathogenesis by affecting repair of DNA from environmental alkylating agents whereas high levels are responsible for the resistance to the most effective drug presently utilized, the alkylating molecule temozolomide.
Some of the molecular pathways of regulation of expression of MGMT gene have been now cleared, as summarized in Fig. 3, being the post-transcriptional regulation by miRs still an open field of investigation. MGMT expression is widely utilized in clinical practice as a predictive marker for the response to alkylating chemotherapy, and quantitation of methylation of MGMT promoter by pyrosequencing is the method of choice to identify non responders to temozolomide that can take advantage of alternative second line targeted therapies.
Figure 3.
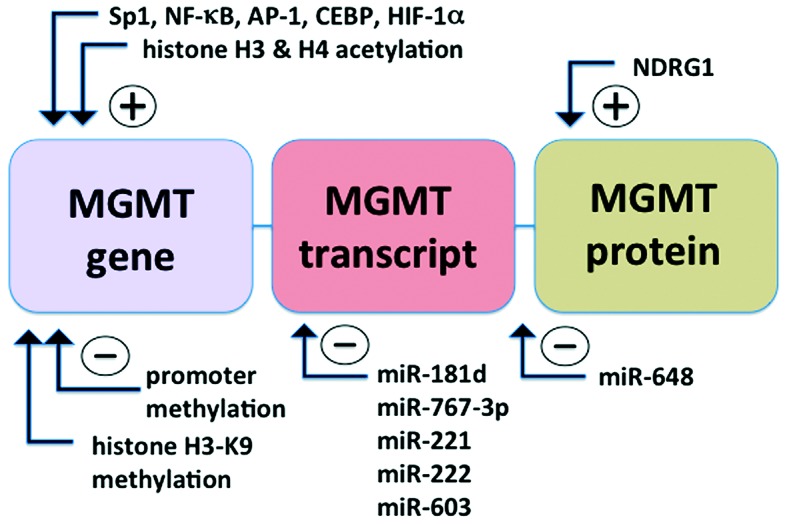
Summary of different known modulators of MGMT gene expression. MGMT expression is increased by different nuclear transcription factors (Sp1, NF-κB, AP-1, CEBP and HIF-1α) (8–13), together with the acetylation of histones H3 and H4 (18) and the stabilization by binding of N-myc downstream regulated gene 1 (NDRG1) protein (17). On the contrary, MGMT expression is downregulated by different mechanisms, namely methylation of the CpG islands in the promoter (25), di-methylation of histone H3K9 (20), degradation of mRNA by miR-181d, -767-3p, -221, -222, -603 (100,106,114,122), interference with protein translation by miR-648 (106).
Artificial manipulation to silence MGMT expression in patients with unmethylated MGMT promoter to be treated with temozolomide should be explored with both innovative chemical inhibitors of MGMT function and novels molecular biology tools aimed to reduce MGMT expression by targeting MGMT mRNA stability or MGMT mRNA translation.
Acknowledgements
We are grateful to Alessandra Santangelo for helpful discussions and assistance in graphical summaries, Valentino Bezzerri for TESS analysis, Giuseppe Moretto, Alberto Beltramello, Giampietro Pinna, Mario Meglio, Renzo Mazzarotto, Bruno Bonetti, Francesco Sala, Antonio Nicolato, Carlo Cavedon, Claudio Ghimenton, Albino Eccher, Anna D’Amico, Andrea Talacchi, Laura Belli, Elena Bazzoli, Anna Tamanini, Cinzia Cantù, Susanna Khalil, Silvia Munari, Paola Prandini, Lisa Provezza, Simona Granata, Laura Lattanzio, many Colleagues of the Department of Neurosciences, Aldo Scarpa and ARC-NET Research Center, Stefania Montemezzi, Marco Chilosi and the Colleagues of the Department of Pathology and Diagnostics of the University of Verona, Giovanni Gaviraghi and the Scientific Committee of the Verona Brain Research Foundation, Gianmarco Musciano from Diatech Pharmacogenetics for helpful discussions. This work is granted by CIB, by COFIN-2009 and by AIRC (IG 13575: peptide nucleic acids targeting oncomiR and tumor-suppressor miRNAs: cancer diagnosis and therapy) to R.G. and by Verona Brain Research Foundation (VRBF) to G.C. and M.C.D.
Abbreviations
- MGMT
O6-methylguanine-DNAmethyltransferase
- O6-MG
O6-methylguanine
- O6-BG
O6-benzylguanine
- TMZ
temozolomide
- miR
microRNA
- MLH1
MutL homolog 1
- MSH2
MutL homolog 2
- MSH6
MutL homolog 6
- PMS2
postmeiotic segregation increased 2
- HIF-1α
hypoxia inducible factor-1α
- Sp1
specificity protein 1
- NF-κB
nuclear factor for polypeptide gene enhancer in B cells
- c/EBP
CCAAT/enhancer binding protein
- AP-1
activator protein 1
- AP-2
activator protein 2
- NF-IL6
nuclear factor for interleukin-6
- ER-α
estrogen receptor-α
- BMP2
bone morphogenetic protein 2
- NDRG1
N-myc downstream regulated gene 1
- HDAC
histone deacetylase
- HSP90
heat shock protein 90
- MeCP2
methyl-CpG binding (MCB) protein 2
- TCGA
The Cancer Genome Atlas
- EGFR
epidermal growth factors receptor
- MS-PCR
methylation-specific polymerase chain reaction
- LNA
locked-nucleic acids
- PNA
peptide nucleic acids
- BCNU
carmustine, 1,3-bis(2-chloroethyl)1-nitrosourea
- ACNU
nimustine, 3-[(4-amino-2-methyl-5-pyrimidinyl) methyl]-1-(2-chloroethyl)-1-nitrosourea hydrochloride
References
- 1.Tano K, Shiota S, Collier J, Foote RS, Mitra S. Isolation and structural characterization of a cDNA clone encoding the human DNA repair protein for O6-alkylguanine. Proc Natl Acad Sci USA. 1990;87:686–690. doi: 10.1073/pnas.87.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belanich M, Randall T, Pastor MA, Kibitel JT, Alas LG, Dolan ME, Schold SC, Jr, Gander M, Lejeune FJ, Li BF, et al. Intracellular localization and intercellular heterogeneity of the human DNA repair protein O(6)-methylguanine-DNA methyl-transferase. Cancer Chemother Pharmacol. 1996;37:547–555. doi: 10.1007/s002800050427. [DOI] [PubMed] [Google Scholar]
- 3.Denny BJ, Wheelhouse RT, Stevens MF, Tsang LL, Slack JA. NMR and molecular modeling investigation of the mechanism of activation of the antitumor drug temozolomide and its interaction with DNA. Biochemistry. 1994;33:9045–9051. doi: 10.1021/bi00197a003. [DOI] [PubMed] [Google Scholar]
- 4.Chakravarti A, Erkkinen MG, Nestler U, Stupp R, Mehta M, Aldape K, Gilbert MR, Black PM, Loeffler JS. Temozolomide-mediated radiation enhancement in glioblastoma: A report on underlying mechanisms. Clin Cancer Res. 2006;12:4738–4746. doi: 10.1158/1078-0432.CCR-06-0596. [DOI] [PubMed] [Google Scholar]
- 5.Liu L, Gerson SL. Targeted modulation of MGMT: Clinical implications. Clin Cancer Res. 2006;12:328–331. doi: 10.1158/1078-0432.CCR-05-2543. [DOI] [PubMed] [Google Scholar]
- 6.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 7.Rydberg B, Spurr N, Karran P. cDNA cloning and chromosomal assignment of the human O6-methylguanine-DNA methyltransferase. cDNA expression in Escherichia coli and gene expression in human cells. J Biol Chem. 1990;265:9563–9569. [PubMed] [Google Scholar]
- 8.Costello JF, Futscher BW, Kroes RA, Pieper RO. Methylation-related chromatin structure is associated with exclusion of transcription factors from and suppressed expression of the O-6-methylguanine DNA methyltransferase gene in human glioma cell lines. Mol Cell Biol. 1994;14:6515–6521. doi: 10.1128/mcb.14.10.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavon I, Fuchs D, Zrihan D, Efroni G, Zelikovitch B, Fellig Y, Siegal T. Novel mechanism whereby nuclear factor kappaB mediates DNA damage repair through regulation of O(6)-methylguanine-DNA-methyltransferase. Cancer Res. 2007;67:8952–8959. doi: 10.1158/0008-5472.CAN-06-3820. [DOI] [PubMed] [Google Scholar]
- 10.Bhakat KK, Mitra S. Regulation of the human O(6)-methylguanine-DNA methyltransferase gene by transcriptional coactivators cAMP response element-binding protein-binding protein and p300. J Biol Chem. 2000;275:34197–34204. doi: 10.1074/jbc.M005447200. [DOI] [PubMed] [Google Scholar]
- 11.Boldogh I, Ramana CV, Chen Z, Biswas T, Hazra TK, Grösch S, Grombacher T, Mitra S, Kaina B. Regulation of expression of the DNA repair gene O6-methylguanine-DNA methyltransferase via protein kinase C-mediated signaling. Cancer Res. 1998;58:3950–3956. [PubMed] [Google Scholar]
- 12.Rolhion C, Penault-Llorca F, Kemeny JL, Kwiatkowski F, Lemaire JJ, Chollet P, Finat-Duclos F, Verrelle P. O(6)-methylguanine-DNA methyltransferase gene (MGMT) expression in human glioblastomas in relation to patient characteristics and p53 accumulation. Int J Cancer. 1999;84:416–420. doi: 10.1002/(SICI)1097-0215(19990820)84:4<416::AID-IJC15>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 13.Bocangel D, Sengupta S, Mitra S, Bhakat KK. p53-Mediated down-regulation of the human DNA repair gene O6-methylguanine-DNA methyltransferase (MGMT) via interaction with Sp1 transcription factor. Anticancer Res. 2009;29:3741–3750. [PMC free article] [PubMed] [Google Scholar]
- 14.Pistollato F, Abbadi S, Rampazzo E, Persano L, Della Puppa A, Frasson C, Sarto E, Scienza R, D’avella D, Basso G. Intratumoral hypoxic gradient drives stem cells distribution and MGMT expression in glioblastoma. Stem Cells. 2010;28:851–862. doi: 10.1002/stem.415. [DOI] [PubMed] [Google Scholar]
- 15.Della Puppa A, Persano L, Masi G, Rampazzo E, Sinigaglia A, Pistollato F, Denaro L, Barzon L, Palù G, Basso G, et al. MGMT expression and promoter methylation status may depend on the site of surgical sample collection within glioblastoma: A possible pitfall in stratification of patients? J Neurooncol. 2012;106:33–41. doi: 10.1007/s11060-011-0639-9. [DOI] [PubMed] [Google Scholar]
- 16.Persano L, Pistollato F, Rampazzo E, Della Puppa A, Abbadi S, Frasson C, Volpin F, Indraccolo S, Scienza R, Basso G. BMP2 sensitizes glioblastoma stem-like cells to Temozolomide by affecting HIF-1α stability and MGMT expression. Cell Death Dis. 2012;3:e412. doi: 10.1038/cddis.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiler M, Blaes J, Pusch S, Sahm F, Czabanka M, Luger S, Bunse L, Solecki G, Eichwald V, Jugold M, et al. mTOR target NDRG1 confers MGMT-dependent resistance to alkylating chemotherapy. Proc Natl Acad Sci USA. 2014;111:409–414. doi: 10.1073/pnas.1314469111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakagawachi T, Soejima H, Urano T, Zhao W, Higashimoto K, Satoh Y, Matsukura S, Kudo S, Kitajima Y, Harada H, et al. Silencing effect of CpG island hypermethylation and histone modifications on O6-methylguanine-DNA methyltransferase (MGMT) gene expression in human cancer. Oncogene. 2003;22:8835–8844. doi: 10.1038/sj.onc.1207183. [DOI] [PubMed] [Google Scholar]
- 19.Danam RP, Howell SR, Brent TP, Harris LC. Epigenetic regulation of O6-methylguanine-DNA methyltransferase gene expression by histone acetylation and methyl-CpG binding proteins. Mol Cancer Ther. 2005;4:61–69. [PubMed] [Google Scholar]
- 20.Zhao W, Soejima H, Higashimoto K, Nakagawachi T, Urano T, Kudo S, Matsukura S, Matsuo S, Joh K, Mukai T. The essential role of histone H3 Lys9 di-methylation and MeCP2 binding in MGMT silencing with poor DNA methylation of the promoter CpG island. J Biochem. 2005;137:431–440. doi: 10.1093/jb/mvi048. [DOI] [PubMed] [Google Scholar]
- 21.Meng CF, Zhu XJ, Peng G, Dai DQ. Role of histone modifications and DNA methylation in the regulation of O6-methylguanine-DNA methyltransferase gene expression in human stomach cancer cells. Cancer Invest. 2010;28:331–339. doi: 10.3109/07357900903179633. [DOI] [PubMed] [Google Scholar]
- 22.Kitange GJ, Mladek AC, Carlson BL, Schroeder MA, Pokorny JL, Cen L, Decker PA, Wu W, Lomberk GA, Gupta SK, et al. Inhibition of histone deacetylation potentiates the evolution of acquired temozolomide resistance linked to MGMT upregulation in glioblastoma xenografts. Clin Cancer Res. 2012;18:4070–4079. doi: 10.1158/1078-0432.CCR-12-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi EJ, Cho BJ, Lee DJ, Hwang YH, Chun SH, Kim HH, Kim IA. Enhanced cytotoxic effect of radiation and temozolomide in malignant glioma cells: Targeting PI3K-AKT-mTOR signaling, HSP90 and histone deacetylases. BMC Cancer. 2014;14:17. doi: 10.1186/1471-2407-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng W, Li M, Cai J, Wang K, Zhang C, Bao Z, Liu Y, Wu A. HDAC4, a prognostic and chromosomal instability marker, refines the predictive value of MGMT promoter methylation. J Neurooncol. 2015 Jan 4; doi: 10.1007/s11060-014-1709-6. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 26.Dolan ME, Stine L, Mitchell RB, Moschel RC, Pegg AE. Modulation of mammalian O6-alkylguanine-DNA alkyltransferase in vivo by O6-benzylguanine and its effect on the sensitivity of a human glioma tumor to 1-(2-chloroethyl)-3-(4-methylcyclohexyl)-1-nitrosourea. Cancer Commun. 1990;2:371–377. doi: 10.3727/095535490820873985. [DOI] [PubMed] [Google Scholar]
- 27.Pegg AE. Mammalian O6-alkylguanine-DNA alkyltransferase: Regulation and importance in response to alkylating carcinogenic and therapeutic agents. Cancer Res. 1990;50:6119–6129. [PubMed] [Google Scholar]
- 28.Anda T, Shabani HK, Tsunoda K, Tokunaga Y, Kaminogo M, Shibata S, Hayashi T, Iseki M. Relationship between expression of O6-methylguanine-DNA methyltransferase, glutathione-S-transferase pi in glioblastoma and the survival of the patients treated with nimustine hydrochloride: An immunohistochemical analysis. Neurol Res. 2003;25:241–248. doi: 10.1179/016164103101201445. [DOI] [PubMed] [Google Scholar]
- 29.Komine C, Watanabe T, Katayama Y, Yoshino A, Yokoyama T, Fukushima T. Promoter hypermethylation of the DNA repair gene O6-methylguanine-DNA methyltransferase is an independent predictor of shortened progression free survival in patients with low-grade diffuse astrocytomas. Brain Pathol. 2003;13:176–184. doi: 10.1111/j.1750-3639.2003.tb00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamiryo T, Tada K, Shiraishi S, Shinojima N, Kochi M, Ushio Y. Correlation between promoter hypermethylation of the O6-methylguanine-deoxyribonucleic acid methyltransferase gene and prognosis in patients with high-grade astrocytic tumors treated with surgery, radiotherapy, and 1-(4-amino-2-methyl-5-pyrimidinyl) methyl-3-(2-chloroethyl)-3-nitrosourea-based chemotherapy. Neurosurgery. 2004;54:349–357. doi: 10.1227/01.NEU.0000103422.51382.99. [DOI] [PubMed] [Google Scholar]
- 31.Paz MF, Yaya-Tur R, Rojas-Marcos I, Reynes G, Pollan M, Aguirre-Cruz L, García-Lopez JL, Piquer J, Safont MJ, Balaña C, et al. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res. 2004;10:4933–4938. doi: 10.1158/1078-0432.CCR-04-0392. [DOI] [PubMed] [Google Scholar]
- 32.Blanc JL, Wager M, Guilhot J, Kusy S, Bataille B, Chantereau T, Lapierre F, Larsen CJ, Karayan-Tapon L. Correlation of clinical features and methylation status of MGMT gene promoter in glioblastomas. J Neurooncol. 2004;68:275–283. doi: 10.1023/B:NEON.0000033385.37098.85. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe T, Katayama Y, Komine C, Yoshino A, Ogino A, Ohta T, Fukushima T. O6-methylguanine-DNA methyltrans-ferase methylation and TP53 mutation in malignant astrocytomas and their relationships with clinical course. Int J Cancer. 2005;113:581–587. doi: 10.1002/ijc.20625. [DOI] [PubMed] [Google Scholar]
- 34.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 35.Brell M, Tortosa A, Verger E, Gil JM, Viñolas N, Villá S, Acebes JJ, Caral L, Pujol T, Ferrer I, et al. Prognostic significance of O6-methylguanine-DNA methyltransferase determined by promoter hypermethylation and immunohistochemical expression in anaplastic gliomas. Clin Cancer Res. 2005;11:5167–5174. doi: 10.1158/1078-0432.CCR-05-0230. [DOI] [PubMed] [Google Scholar]
- 36.Donson AM, Addo-Yobo SO, Handler MH, Gore L, Foreman NK. MGMT promoter methylation correlates with survival benefit and sensitivity to temozolomide in pediatric glioblastoma. Pediatr Blood Cancer. 2007;48:403–407. doi: 10.1002/pbc.20803. [DOI] [PubMed] [Google Scholar]
- 37.Brandes AA, Tosoni A, Cavallo G, Reni M, Franceschi E, Bonaldi L, Bertorelle R, Gardiman M, Ghimenton C, Iuzzolino P, et al. GICNO. Correlations between O6-methylguanine DNA methyltransferase promoter methylation status, 1p and 19q deletions, and response to temozolomide in anaplastic and recurrent oligodendroglioma: A prospective GICNO study. J Clin Oncol. 2006;24:4746–4753. doi: 10.1200/JCO.2006.06.3891. [DOI] [PubMed] [Google Scholar]
- 38.Herrlinger U, Rieger J, Koch D, Loeser S, Blaschke B, Kortmann RD, Steinbach JP, Hundsberger T, Wick W, Meyermann R, et al. Phase II trial of lomustine plus temozolomide chemotherapy in addition to radiotherapy in newly diagnosed glioblastoma: UKT-03. J Clin Oncol. 2006;24:4412–4417. doi: 10.1200/JCO.2006.06.9104. [DOI] [PubMed] [Google Scholar]
- 39.Everhard S, Kaloshi G, Crinière E, Benouaich-Amiel A, Lejeune J, Marie Y, Sanson M, Kujas M, Mokhtari K, Hoang-Xuan K, et al. MGMT methylation: A marker of response to temozolomide in low-grade gliomas. Ann Neurol. 2006;60:740–743. doi: 10.1002/ana.21044. [DOI] [PubMed] [Google Scholar]
- 40.Eoli M, Menghi F, Bruzzone MG, De Simone T, Valletta L, Pollo B, Bissola L, Silvani A, Bianchessi D, D’Incerti L, et al. Methylation of O6-methylguanine DNA methyltransferase and loss of heterozygosity on 19q and/or 17p are overlapping features of secondary glioblastomas with prolonged survival. Clin Cancer Res. 2007;13:2606–2613. doi: 10.1158/1078-0432.CCR-06-2184. [DOI] [PubMed] [Google Scholar]
- 41.Brandes AA, Franceschi E, Tosoni A, Blatt V, Pession A, Tallini G, Bertorelle R, Bartolini S, Calbucci F, Andreoli A, et al. MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J Clin Oncol. 2008;26:2192–2197. doi: 10.1200/JCO.2007.14.8163. [DOI] [PubMed] [Google Scholar]
- 42.Vogelbaum MA, Berkey B, Peereboom D, Macdonald D, Giannini C, Suh JH, Jenkins R, Herman J, Brown P, Blumenthal DT, et al. Phase II trial of preirradiation and concurrent temozolomide in patients with newly diagnosed anaplastic oligodendrogliomas and mixed anaplastic oligoastrocytomas: RTOG BR0131. Neuro Oncol. 2009;11:167–175. doi: 10.1215/15228517-2008-073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sadones J, Michotte A, Veld P, Chaskis C, Sciot R, Menten J, Joossens EJ, Strauven T, D’Hondt LA, Sartenaer D, et al. MGMT promoter hypermethylation correlates with a survival benefit from temozolomide in patients with recurrent anaplastic astrocytoma but not glioblastoma. Eur J Cancer. 2009;45:146–153. doi: 10.1016/j.ejca.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Mikkelsen T, Doyle T, Anderson J, Margolis J, Paleologos N, Gutierrez J, Croteau D, Hasselbach L, Avedissian R, Schultz L. Temozolomide single-agent chemotherapy for newly diagnosed anaplastic oligodendroglioma. J Neurooncol. 2009;92:57–63. doi: 10.1007/s11060-008-9735-x. [DOI] [PubMed] [Google Scholar]
- 45.Nakagawa T, Ido K, Sakuma T, Takeuchi H, Sato K, Kubota T. Prognostic significance of the immunohistochemical expression of O6-methylguanine-DNA methyltransferase, P-glycoprotein, and multidrug resistance protein-1 in glioblastomas. Neuropathology. 2009;29:379–388. doi: 10.1111/j.1440-1789.2008.00983.x. [DOI] [PubMed] [Google Scholar]
- 46.Duffy MJ, Napieralski R, Martens JW, Span PN, Spyratos F, Sweep FC, Brunner N, Foekens JA, Schmitt M EORTC PathoBiology Group. Methylated genes as new cancer biomarkers. Eur J Cancer. 2009;45:335–346. doi: 10.1016/j.ejca.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 48.Fabi A, Metro G, Russillo M, Vidiri A, Carapella CM, Maschio M, Cognetti F, Jandolo B, Mirri MA, Sperduti I, et al. Treatment of recurrent malignant gliomas with fotemustine monotherapy: Impact of dose and correlation with MGMT promoter methylation. BMC Cancer. 2009;9:101. doi: 10.1186/1471-2407-9-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brandes AA, Franceschi E, Tosoni A, Benevento F, Scopece L, Mazzocchi V, Bacci A, Agati R, Calbucci F, Ermani M. Temozolomide concomitant and adjuvant to radiotherapy in elderly patients with glioblastoma: Correlation with MGMT promoter methylation status. Cancer. 2009;115:3512–3518. doi: 10.1002/cncr.24406. [DOI] [PubMed] [Google Scholar]
- 50.Dunn J, Baborie A, Alam F, Joyce K, Moxham M, Sibson R, Crooks D, Husband D, Shenoy A, Brodbelt A, et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009;101:124–131. doi: 10.1038/sj.bjc.6605127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Metellus P, Coulibaly B, Nanni I, Fina F, Eudes N, Giorgi R, Barrie M, Chinot O, Fuentes S, Dufour H, et al. Prognostic impact of O6-methylguanine-DNA methyltransferase silencing in patients with recurrent glioblastoma multiforme who undergo surgery and carmustine wafer implantation: A prospective patient cohort. Cancer. 2009;115:4783–4794. doi: 10.1002/cncr.24546. [DOI] [PubMed] [Google Scholar]
- 52.Weller M, Felsberg J, Hartmann C, Berger H, Steinbach JP, Schramm J, Westphal M, Schackert G, Simon M, Tonn JC, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: A prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27:5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 53.Piperi C, Themistocleous MS, Papavassiliou GA, Farmaki E, Levidou G, Korkolopoulou P, Adamopoulos C, Papavassiliou AG. High incidence of MGMT and RARbeta promoter methylation in primary glioblastomas: Association with histopathological characteristics, inflammatory mediators and clinical outcome. Mol Med. 2010;16:1–9. doi: 10.2119/molmed.2009.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao VT, Jung TY, Jung S, Jin SG, Moon KS, Kim IY, Kang SS, Park CS, Lee KH, Chae HJ. The correlation and prognostic significance of MGMT promoter methylation and MGMT protein in glioblastomas. Neurosurgery. 2009;65:866–875. doi: 10.1227/01.NEU.0000357325.90347.A1. discussion 875. [DOI] [PubMed] [Google Scholar]
- 55.Karayan-Tapon L, Quillien V, Guilhot J, Wager M, Fromont G, Saikali S, Etcheverry A, Hamlat A, Loussouarn D, Campion L, et al. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol. 2010;97:311–322. doi: 10.1007/s11060-009-0031-1. [DOI] [PubMed] [Google Scholar]
- 56.Gerstner ER, Yip S, Wang DL, Louis DN, Iafrate AJ, Batchelor TT. Mgmt methylation is a prognostic biomarker in elderly patients with newly diagnosed glioblastoma. Neurology. 2009;73:1509–1510. doi: 10.1212/WNL.0b013e3181bf9907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeAngelis LM. Anaplastic glioma: How to prognosticate outcome and choose a treatment strategy. [corrected] J Clin Oncol. 2009;27:5861–5862. doi: 10.1200/JCO.2009.24.5985. [DOI] [PubMed] [Google Scholar]
- 58.van den Bent MJ, Dubbink HJ, Sanson M, van der Lee-Haarloo CR, Hegi M, Jeuken JW, Ibdaih A, Brandes AA, Taphoorn MJ, Frenay M, et al. MGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial tumors: A report from EORTC Brain Tumor Group Study 26951. J Clin Oncol. 2009;27:5881–5886. doi: 10.1200/JCO.2009.24.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peereboom DM, Shepard DR, Ahluwalia MS, Brewer CJ, Agarwal N, Stevens GH, Suh JH, Toms SA, Vogelbaum MA, Weil RJ, et al. Phase II trial of erlotinib with temozolomide and radiation in patients with newly diagnosed glioblastoma multiforme. J Neurooncol. 2010;98:93–99. doi: 10.1007/s11060-009-0067-2. [DOI] [PubMed] [Google Scholar]
- 60.Wick W, Hartmann C, Engel C, Stoffels M, Felsberg J, Stockhammer F, Sabel MC, Koeppen S, Ketter R, et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. 2009;27:5874–5880. doi: 10.1200/JCO.2009.23.6497. [DOI] [PubMed] [Google Scholar]
- 61.Spiegl-Kreinecker S, Pirker C, Filipits M, Lötsch D, Buchroithner J, Pichler J, Silye R, Weis S, Micksche M, Fischer J, et al. O6-Methylguanine DNA methyltransferase protein expression in tumor cells predicts outcome of temozolomide therapy in glioblastoma patients. Neuro Oncol. 2010;12:28–36. doi: 10.1093/neuonc/nop003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rivera AL, Pelloski CE, Gilbert MR, Colman H, De La Cruz C, Sulman EP, Bekele BN, Aldape KD. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol. 2010;12:116–121. doi: 10.1093/neuonc/nop020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sonoda Y, Yokosawa M, Saito R, Kanamori M, Yamashita Y, Kumabe T, Watanabe M, Tominaga T. O(6)-Methylguanine DNA methyltransferase determined by promoter hypermethylation and immunohistochemical expression is correlated with progression-free survival in patients with glioblastoma. Int J Clin Oncol. 2010;15:352–358. doi: 10.1007/s10147-010-0065-6. [DOI] [PubMed] [Google Scholar]
- 64.Costa BM, Caeiro C, Guimarães I, Martinho O, Jaraquemada T, Augusto I, Castro L, Osório L, Linhares P, Honavar M, et al. Prognostic value of MGMT promoter methylation in glioblastoma patients treated with temozolomide-based chemoradiation: A Portuguese multicentre study. Oncol Rep. 2010;23:1655–1662. doi: 10.3892/or_00000808. [DOI] [PubMed] [Google Scholar]
- 65.Stupp R, Hegi ME, Neyns B, Goldbrunner R, Schlegel U, Clement PM, Grabenbauer GG, Ochsenbein AF, Simon M, Dietrich PY, et al. Phase I/IIa study of cilengitide and temozolomide with concomitant radiotherapy followed by cilengitide and temozolomide maintenance therapy in patients with newly diagnosed glioblastoma. J Clin Oncol. 2010;28:2712–2718. doi: 10.1200/JCO.2009.26.6650. [DOI] [PubMed] [Google Scholar]
- 66.Buttarelli FR, Massimino M, Antonelli M, Lauriola L, Nozza P, Donofrio V, Arcella A, Oliva MA, Di Rocco C, Giangaspero F. Evaluation status and prognostic significance of O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation in pediatric high grade gliomas. Childs Nerv Syst. 2010;26:1051–1056. doi: 10.1007/s00381-010-1191-1. [DOI] [PubMed] [Google Scholar]
- 67.Minniti G, Salvati M, Arcella A, Buttarelli F, D’Elia A, Lanzetta G, Esposito V, Scarpino S, Maurizi Enrici R, Giangaspero F. Correlation between O6-methylguanine-DNA methyltransferase and survival in elderly patients with glioblastoma treated with radiotherapy plus concomitant and adjuvant temozolomide. J Neurooncol. 2011;102:311–316. doi: 10.1007/s11060-010-0324-4. [DOI] [PubMed] [Google Scholar]
- 68.Shah N, Lin B, Sibenaller Z, Ryken T, Lee H, Yoon JG, Rostad S, Foltz G. Comprehensive analysis of MGMT promoter methylation: Correlation with MGMT expression and clinical response in GBM. PLoS One. 2011;6:e16146. doi: 10.1371/journal.pone.0016146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang K, Jin Q, Yan W, Zhang W, You G, Liu Y, Jiang T. Clinical correlation of MGMT protein expression and promoter methylation in Chinese glioblastoma patients. Med Oncol. 2012;29:1292–1296. doi: 10.1007/s12032-011-9901-4. [DOI] [PubMed] [Google Scholar]
- 70.Lakomy R, Sana J, Hankeova S, Fadrus P, Kren L, Lzicarova E, Svoboda M, Dolezelova H, Smrcka M, Vyzula R, et al. MiR-195, miR-196b, miR-181c, miR-21 expression levels and O-6-methylguanine-DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patients. Cancer Sci. 2011;102:2186–2190. doi: 10.1111/j.1349-7006.2011.02092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van den Bent MJ, Gravendeel LA, Gorlia T, Kros JM, Lapre L, Wesseling P, Teepen JL, Idbaih A, Sanson M, Smitt PA, et al. A hypermethylated phenotype is a better predictor of survival than MGMT methylation in anaplastic oligodendroglial brain tumors: A report from EORTC study 26951. Clin Cancer Res. 2011;17:7148–7155. doi: 10.1158/1078-0432.CCR-11-1274. [DOI] [PubMed] [Google Scholar]
- 72.Reifenberger G, Hentschel B, Felsberg J, Schackert G, Simon M, Schnell O, Westphal M, Wick W, Pietsch T, Loeffler M, et al. German Glioma Network. Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int J Cancer. 2012;131:1342–1350. doi: 10.1002/ijc.27385. [DOI] [PubMed] [Google Scholar]
- 73.Lechapt-Zalcman E, Levallet G, Dugué AE, Vital A, Diebold MD, Menei P, Colin P, Peruzzy P, Emery E, Bernaudin M, et al. O(6)-methylguanine-DNA methyltransferase (MGMT) promoter methylation and low MGMT-encoded protein expression as prognostic markers in glioblastoma patients treated with biodegradable carmustine wafer implants after initial surgery followed by radiotherapy with concomitant and adjuvant temozolomide. Cancer. 2012;118:4545–4554. doi: 10.1002/cncr.27441. [DOI] [PubMed] [Google Scholar]
- 74.Wick W, Platten M, Meisner C, Felsberg J, Tabatabai G, Simon M, Nikkhah G, Papsdorf K, Steinbach JP, Sabel M, et al. NOA-08 Study Group of Neuro-oncology Working Group (NOA) of German Cancer Society. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: The NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13:707–715. doi: 10.1016/S1470-2045(12)70164-X. [DOI] [PubMed] [Google Scholar]
- 75.Malmström A, Grønberg BH, Marosi C, Stupp R, Frappaz D, Schultz H, Abacioglu U, Tavelin B, Lhermitte B, Hegi ME, et al. Nordic Clinical Brain Tumour Study Group (NCBTSG) Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: The Nordic randomised, phase 3 trial. Lancet Oncol. 2012;13:916–926. doi: 10.1016/S1470-2045(12)70265-6. [DOI] [PubMed] [Google Scholar]
- 76.Juratli TA, Kirsch M, Geiger K, Klink B, Leipnitz E, Pinzer T, Soucek S, Schrock E, Schackert G, Krex D. The prognostic value of IDH mutations and MGMT promoter status in secondary high-grade gliomas. J Neurooncol. 2012;110:325–333. doi: 10.1007/s11060-012-0977-2. [DOI] [PubMed] [Google Scholar]
- 77.Melguizo C, Prados J, González B, Ortiz R, Concha A, Alvarez PJ, Madeddu R, Perazzoli G, Oliver JA, López R, et al. MGMT promoter methylation status and MGMT and CD133 immunohistochemical expression as prognostic markers in glioblastoma patients treated with temozolomide plus radiotherapy. J Transl Med. 2012;10:250. doi: 10.1186/1479-5876-10-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lalezari S, Chou AP, Tran A, Solis OE, Khanlou N, Chen W, Li S, Carrillo JA, Chowdhury R, Selfridge J, et al. Combined analysis of O6-methylguanine-DNA methyltransferase protein expression and promoter methylation provides optimized prognostication of glioblastoma outcome. Neuro Oncol. 2013;15:370–381. doi: 10.1093/neuonc/nos308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leu S, von Felten S, Frank S, Vassella E, Vajtai I, Taylor E, Schulz M, Hutter G, Hench J, Schucht P, et al. IDH/MGMT-driven molecular classification of low-grade glioma is a strong predictor for long-term survival. Neuro Oncol. 2013;15:469–479. doi: 10.1093/neuonc/nos317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gutenberg A, Bock HC, Brück W, Doerner L, Mehdorn HM, Roggendorf W, Westphal M, Felsberg J, Reifenberger G, Giese A. MGMT promoter methylation status and prognosis of patients with primary or recurrent glioblastoma treated with carmustine wafers. Br J Neurosurg. 2013;27:772–778. doi: 10.3109/02688697.2013.791664. [DOI] [PubMed] [Google Scholar]
- 81.van den Bent MJ, Erdem-Eraslan L, Idbaih A, de Rooi J, Eilers PH, Spliet WG, den Dunnen WF, Tijssen C, Wesseling P, Sillevis Smitt PA, et al. MGMT-STP27 methylation status as predictive marker for response to PCV in anaplastic Oligodendrogliomas and Oligoastrocytomas. A report from EORTC study 26951. Clin Cancer Res. 2013;19:5513–5522. doi: 10.1158/1078-0432.CCR-13-1157. [DOI] [PubMed] [Google Scholar]
- 82.Chen C, Huang R, MacLean A, Muzikansky A, Mukundan S, Wen PY, Norden AD. Recurrent high-grade glioma treated with bevacizumab: Prognostic value of MGMT methylation, EGFR status and pretreatment MRI in determining response and survival. J Neurooncol. 2013;115:267–276. doi: 10.1007/s11060-013-1225-0. [DOI] [PubMed] [Google Scholar]
- 83.Fiano V, Trevisan M, Trevisan E, Senetta R, Castiglione A, Sacerdote C, Gillio-Tos A, De Marco L, Grasso C, Magistrello M, et al. MGMT promoter methylation in plasma of glioma patients receiving temozolomide. J Neurooncol. 2014;117:347–357. doi: 10.1007/s11060-014-1395-4. [DOI] [PubMed] [Google Scholar]
- 84.Wiestler B, Capper D, Hovestadt V, Sill M, Jones DT, Hartmann C, Felsberg J, Platten M, Feiden W, Keyvani K, et al. Assessing CpG island methylator phenotype, 1p/19q codeletion, and MGMT promoter methylation from epigenome-wide data in the biomarker cohort of the NOA-04 trial. Neuro Oncol. 2014;16:1630–1638. doi: 10.1093/neuonc/nou138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stupp R, Hegi ME, Gorlia T, Erridge SC, Perry J, Hong YK, Aldape KD, Lhermitte B, Pietsch T, Grujicic D, et al. European Organisation for Research and Treatment of Cancer (EORTC); Canadian Brain Tumor Consortium; CENTRIC study team. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): A multi-centre, randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1100–1108. doi: 10.1016/S1470-2045(14)70379-1. [DOI] [PubMed] [Google Scholar]
- 87.Weller M, Pfister SM, Wick W, Hegi ME, Reifenberger G, Stupp R. Molecular neuro-oncology in clinical practice: A new horizon. Lancet Oncol. 2013;14:e370–e379. doi: 10.1016/S1470-2045(13)70168-2. [DOI] [PubMed] [Google Scholar]
- 88.Hamilton MG, Roldán G, Magliocco A, McIntyre JB, Parney I, Easaw JC. Determination of the methylation status of MGMT in different regions within glioblastoma multiforme. J Neurooncol. 2011;102:255–260. doi: 10.1007/s11060-010-0307-5. [DOI] [PubMed] [Google Scholar]
- 89.Kreth S, Thon N, Eigenbrod S, Lutz J, Ledderose C, Egensperger R, Tonn JC, Kretzschmar HA, Hinske LC, Kreth FW. O-methylguanine-DNA methyltransferase (MGMT) mRNA expression predicts outcome in malignant glioma independent of MGMT promoter methylation. PLoS One. 2011;6:e17156. doi: 10.1371/journal.pone.0017156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 91.Sontheimer EJ, Carthew RW. Silence from within: Endogenous siRNAs and miRNAs. Cell. 2005;122:9–12. doi: 10.1016/j.cell.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 92.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 93.Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nat Rev Genet. 2013;14:535–548. doi: 10.1038/nrg3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ramakrishnan V, Kushwaha D, Koay DC, Reddy H, Mao Y, Zhou L, Ng K, Zinn P, Carter B, Chen CC. Post-transcriptional regulation of O(6)-methylguanine-DNA methyltransferase MGMT in glioblastomas. Cancer Biomark. 2011–2012;10:185–193. doi: 10.3233/CBM-2012-0245. [DOI] [PubMed] [Google Scholar]
- 95.Slaby O, Lakomy R, Fadrus P, Hrstka R, Kren L, Lzicarova E, Smrcka M, Svoboda M, Dolezalova H, Novakova J, et al. MicroRNA-181 family predicts response to concomitant chemoradiotherapy with temozolomide in glioblastoma patients. Neoplasma. 2010;57:264–269. doi: 10.4149/neo_2010_03_264. [DOI] [PubMed] [Google Scholar]
- 96.Ujifuku K, Mitsutake N, Takakura S, Matsuse M, Saenko V, Suzuki K, Hayashi K, Matsuo T, Kamada K, Nagata I, et al. miR-195, miR-455-3p and miR-10a(*) are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer Lett. 2010;296:241–248. doi: 10.1016/j.canlet.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 97.Shi L, Chen J, Yang J, Pan T, Zhang S, Wang Z. MiR-21 protected human glioblastoma U87MG cells from chemo-therapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Res. 2010;1352:255–264. doi: 10.1016/j.brainres.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 98.Shi L, Zhang S, Feng K, Wu F, Wan Y, Wang Z, Zhang J, Wang Y, Yan W, Fu Z, et al. MicroRNA-125b-2 confers human glioblastoma stem cells resistance to temozolomide through the mitochondrial pathway of apoptosis. Int J Oncol. 2012;40:119–129. doi: 10.3892/ijo.2011.1179. [DOI] [PubMed] [Google Scholar]
- 99.Zhang S, Wan Y, Pan T, Gu X, Qian C, Sun G, Sun L, Xiang Y, Wang Z, Shi L. MicroRNA-21 inhibitor sensitizes human glioblastoma U251 stem cells to chemotherapeutic drug temozolomide. J Mol Neurosci. 2012;47:346–356. doi: 10.1007/s12031-012-9759-8. [DOI] [PubMed] [Google Scholar]
- 100.Zhang W, Zhang J, Hoadley K, Kushwaha D, Ramakrishnan V, Li S, Kang C, You Y, Jiang C, Song SW, et al. miR-181d: A predictive glioblastoma biomarker that downregulates MGMT expression. Neuro Oncol. 2012;14:712–719. doi: 10.1093/neuonc/nos089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tunca B, Tezcan G, Cecener G, Egeli U, Ak S, Malyer H, Tumen G, Bilir A. Olea europaea leaf extract alters microRNA expression in human glioblastoma cells. J Cancer Res Clin Oncol. 2012;138:1831–1844. doi: 10.1007/s00432-012-1261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wong ST, Zhang XQ, Zhuang JT, Chan HL, Li CH, Leung GK. MicroRNA-21 inhibition enhances in vitro chemosensitivity of temozolomide-resistant glioblastoma cells. Anticancer Res. 2012;32:2835–2841. [PubMed] [Google Scholar]
- 103.Asuthkar S, Velpula KK, Chetty C, Gorantla B, Rao JS. Epigenetic regulation of miRNA-211 by MMP-9 governs glioma cell apoptosis, chemosensitivity and radiosensitivity. Oncotarget. 2012;3:1439–1454. doi: 10.18632/oncotarget.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Costa PM, Cardoso AL, Nóbrega C, Pereira de Almeida LF, Bruce JN, Canoll P, Pedroso de Lima MC. MicroRNA-21 silencing enhances the cytotoxic effect of the antiangiogenic drug sunitinib in glioblastoma. Hum Mol Genet. 2013;22:904–918. doi: 10.1093/hmg/dds496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ohno M, Ohkuri T, Kosaka A, Tanahashi K, June CH, Natsume A, Okada H. Expression of miR-17–92 enhances anti-tumor activity of T-cells transduced with the anti-EGFRvIII chimeric antigen receptor in mice bearing human GBM xenografts. J Immunother Cancer. 2013;1:21. doi: 10.1186/2051-1426-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kreth S, Limbeck E, Hinske LC, Schütz SV, Thon N, Hoefig K, Egensperger R, Kreth FW. In human glioblastomas transcript elongation by alternative polyadenylation and miRNA targeting is a potent mechanism of MGMT silencing. Acta Neuropathol. 2013;125:671–681. doi: 10.1007/s00401-013-1081-1. [DOI] [PubMed] [Google Scholar]
- 107.Tarassishin L, Lee SC. Interferon regulatory factor 3 alters glioma inflammatory and invasive properties. J Neurooncol. 2013;113:185–194. doi: 10.1007/s11060-013-1109-3. [DOI] [PubMed] [Google Scholar]
- 108.Li RY, Chen LC, Zhang HY, Du WZ, Feng Y, Wang HB, Wen JQ, Liu X, Li XF, Sun Y, et al. MiR-139 inhibits Mcl-1 expression and potentiates TMZ-induced apoptosis in glioma. CNS Neurosci Ther. 2013;19:477–483. doi: 10.1111/cns.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Comincini S, Allavena G, Palumbo S, Morini M, Durando F, Angeletti F, Pirtoli L, Miracco C. microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol Ther. 2013;14:574–586. doi: 10.4161/cbt.24597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Siebzehnrubl FA, Silver DJ, Tugertimur B, Deleyrolle LP, Siebzehnrubl D, Sarkisian MR, Devers KG, Yachnis AT, Kupper MD, Neal D, et al. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol Med. 2013;5:1196–1212. doi: 10.1002/emmm.201302827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wan Y, Sun G, Zhang S, Wang Z, Shi L. MicroRNA-125b inhibitor sensitizes human primary glioblastoma cells to chemotherapeutic drug temozolomide on invasion. In Vitro Cell Dev Biol Anim. 2013;49:599–607. doi: 10.1007/s11626-013-9644-y. [DOI] [PubMed] [Google Scholar]
- 112.Shi L, Wan Y, Sun G, Zhang S, Wang Z, Zeng Y. miR-125b inhibitor may enhance the invasion-prevention activity of temozolomide in glioblastoma stem cells by targeting PIAS3. BioDrugs. 2014;28:41–54. doi: 10.1007/s40259-013-0053-2. [DOI] [PubMed] [Google Scholar]
- 113.Munoz JL, Bliss SA, Greco SJ, Ramkissoon SH, Ligon KL, Rameshwar P. Delivery of functional anti-miR-9 by mesenchymal stem cell-derived exosomes to glioblastoma multiforme cells conferred chemosensitivity. Mol Ther Nucleic Acids. 2013;2:e126. doi: 10.1038/mtna.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Quintavalle C, Mangani D, Roscigno G, Romano G, Diaz-Lagares A, Iaboni M, Donnarumma E, Fiore D, De Marinis P, Soini Y, et al. MiR-221/222 target the DNA methyltransferase MGMT in glioma cells. PLoS One. 2013;8:e74466. doi: 10.1371/journal.pone.0074466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Alrfaei BM, Vemuganti R, Kuo JS. microRNA-100 targets SMRT/NCOR2, reduces proliferation, and improves survival in glioblastoma animal models. PLoS One. 2013;8:e80865. doi: 10.1371/journal.pone.0080865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Parker NR, Correia N, Crossley B, Buckland ME, Howell VM, Wheeler HR. Correlation of microRNA 132 up-regulation with an unfavorable clinical outcome in patients with primary glioblastoma multiforme treated with radiotherapy plus concomitant and adjuvant temozolomide chemotherapy. Transl Oncol. 2013;6:742–748. doi: 10.1593/tlo.13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.She X, Yu Z, Cui Y, Lei Q, Wang Z, Xu G, Luo Z, Li G, Wu M. miR-181 subunits enhance the chemosensitivity of temozolomide by Rap1B-mediated cytoskeleton remodeling in glioblastoma cells. Med Oncol. 2014;31:892. doi: 10.1007/s12032-014-0892-9. [DOI] [PubMed] [Google Scholar]
- 118.Chen J, Fu X, Wan Y, Wang Z, Jiang D, Shi L. miR-125b inhibitor enhance the chemosensitivity of glioblastoma stem cells to temozolomide by targeting Bak1. Tumour Biol. 2014;35:6293–6302. doi: 10.1007/s13277-014-1821-4. [DOI] [PubMed] [Google Scholar]
- 119.Tezcan G, Tunca B, Bekar A, Preusser M, Berghoff AS, Egeli U, Cecener G, Ricken G, Budak F, Taskapılıoglu MO, et al. microRNA expression pattern modulates temozolomide response in GBM tumors with cancer stem cells. Cell Mol Neurobiol. 2014;34:679–692. doi: 10.1007/s10571-014-0050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Berthois Y, Delfino C, Metellus P, Fina F, Nanni-Metellus I, Al Aswy H, Pirisi V, Ouafik L, Boudouresque F. Differential expression of miR200a-3p and miR21 in grade II–III and grade IV gliomas: Evidence that miR200a-3p is regulated by O6-methylguanine methyltransferase and promotes temozolomide responsiveness. Cancer Biol Ther. 2014;15:938–950. doi: 10.4161/cbt.28920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Haemmig S, Baumgartner U, Glück A, Zbinden S, Tschan MP, Kappeler A, Mariani L, Vajtai I, Vassella E. miR-125b controls apoptosis and temozolomide resistance by targeting TNFAIP3 and NKIRAS2 in glioblastomas. Cell Death Dis. 2014;5:e1279. doi: 10.1038/cddis.2014.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kushwaha D, Ramakrishnan V, Ng K, Steed T, Nguyen T, Futalan D, Akers JC, Sarkaria J, Jiang T, Chowdhury D, et al. A genome-wide miRNA screen revealed miR-603 as a MGMT-regulating miRNA in glioblastomas. Oncotarget. 2014;5:4026–4039. doi: 10.18632/oncotarget.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.She X, Yu Z, Cui Y, Lei Q, Wang Z, Xu G, Xiang J, Wu M, Li G. miR-128 and miR-149 enhance the chemosensitivity of temozolomide by Rap1B-mediated cytoskeletal remodeling in glioblastoma. Oncol Rep. 2014;32:957–964. doi: 10.3892/or.2014.3318. [DOI] [PubMed] [Google Scholar]
- 124.Tezcan G, Tunca B, Bekar A, Budak F, Sahin S, Cecener G, Egeli U, Taskapılıoglu MO, Kocaeli H, Tolunay S, et al. Olea europaea leaf extract improves the treatment response of GBM stem cells by modulating miRNA expression. Am J Cancer Res. 2014;4:572–590. [PMC free article] [PubMed] [Google Scholar]
- 125.Liu Q, Zou R, Zhou R, Gong C, Wang Z, Cai T, Tan C, Fang J. miR-155 Regulates glioma cells invasion and chemosensitivity by p38 isforms in vitro. J Cell Biochem. 2014 Dec 23; doi: 10.1002/jcb.25073. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 126.Munoz JL, Rodriguez-Cruz V, Ramkissoon SH, Ligon KL, Greco SJ, Rameshwar P. Temozolomide resistance in glioblastoma occurs by miRNA-9-targeted PTCH1, independent of sonic hedgehog level. Oncotarget. 2015;6:1190–1201. doi: 10.18632/oncotarget.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang Z, Yang J, Xu G, Wang W, Liu C, Yang H, Yu Z, Lei Q, Xiao L, Xiong J, et al. Targeting miR-381-NEFL axis sensitizes glioblastoma cells to temozolomide by regulating stemness factors and multidrug resistance factors. Oncotarget. 2014 Dec 18; doi: 10.18632/oncotarget.3061. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bleeker FE, Molenaar RJ, Leenstra S. Recent advances in the molecular understanding of glioblastoma. J Neurooncol. 2012;108:11–27. doi: 10.1007/s11060-011-0793-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brognara E, Fabbri E, Bazzoli E, Montagner G, Ghimenton C, Eccher A, Cantù C, Manicardi A, Bianchi N, Finotti A, et al. Uptake by human glioma cell lines and biological effects of a peptide-nucleic acids targeting miR-221. J Neurooncol. 2014;118:19–28. doi: 10.1007/s11060-014-1405-6. [DOI] [PubMed] [Google Scholar]
- 131.Chen L, Zhang J, Han L, Zhang A, Zhang C, Zheng Y, Jiang T, Pu P, Jiang C, Kang C. Downregulation of miR-221/222 sensitizes glioma cells to temozolomide by regulating apoptosis independently of p53 status. Oncol Rep. 2012;27:854–860. doi: 10.3892/or.2011.1535. [DOI] [PubMed] [Google Scholar]
- 132.Li W, Guo F, Wang P, Hong S, Zhang C. miR-221/222 confers radioresistance in glioblastoma cells through activating Akt independent of PTEN status. Curr Mol Med. 2014;14:185–195. doi: 10.2174/1566524013666131203103147. [DOI] [PubMed] [Google Scholar]
- 133.Xie Q, Yan Y, Huang Z, Zhong X, Huang L. MicroRNA-221 targeting PI3-K/Akt signaling axis induces cell proliferation and BCNU resistance in human glioblastoma. Neuropathology. 2014;34:455–464. doi: 10.1111/neup.12129. [DOI] [PubMed] [Google Scholar]
- 134.Weller M, Stupp R, Reifenberger G, Brandes AA, van den Bent MJ, Wick W, Hegi ME. MGMT promoter methylation in malignant gliomas: Ready for personalized medicine? Nat Rev Neurol. 2010;6:39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- 135.Silber JR, Bobola MS, Ghatan S, Blank A, Kolstoe DD, Berger MS. O6-methylguanine-DNA methyltransferase activity in adult gliomas: Relation to patient and tumor characteristics. Cancer Res. 1998;58:1068–1073. [PubMed] [Google Scholar]
- 136.Silber JR, Blank A, Bobola MS, Ghatan S, Kolstoe DD, Berger MS. O6-methylguanine-DNA methyltransferase-deficient phenotype in human gliomas: Frequency and time to tumor progression after alkylating agent-based chemotherapy. Clin Cancer Res. 1999;5:807–814. [PubMed] [Google Scholar]
- 137.Citron M, White A, Decker R, Wasserman P, Li B, Randall T, Guerra D, Belanich M, Yarosh D. O6-methylguanine-DNA methyltransferase in human brain tumors detected by activity assay and monoclonal antibodies. Oncol Res. 1995;7:49–55. [PubMed] [Google Scholar]
- 138.Jaeckle KA, Eyre HJ, Townsend JJ, Schulman S, Knudson HM, Belanich M, Yarosh DB, Bearman SI, Giroux DJ, Schold SC. Correlation of tumor O6 methylguanine-DNA methyltransferase levels with survival of malignant astrocytoma patients treated with bis-chloroethylnitrosourea: A Southwest Oncology Group study. J Clin Oncol. 1998;16:3310–3315. doi: 10.1200/JCO.1998.16.10.3310. [DOI] [PubMed] [Google Scholar]
- 139.Chinot OL, Barrié M, Fuentes S, Eudes N, Lancelot S, Metellus P, Muracciole X, Braguer D, Ouafik L, Martin PM, et al. Correlation between O6-methylguanine-DNA methyltransferase and survival in inoperable newly diagnosed glioblastoma patients treated with neoadjuvant temozolomide. J Clin Oncol. 2007;25:1470–1475. doi: 10.1200/JCO.2006.07.4807. [DOI] [PubMed] [Google Scholar]
- 140.Levin N, Lavon I, Zelikovitsh B, Fuchs D, Bokstein F, Fellig Y, Siegal T. Progressive low-grade oligodendrogliomas: Response to temozolomide and correlation between genetic profile and O6-methylguanine DNA methyltransferase protein expression. Cancer. 2006;106:1759–1765. doi: 10.1002/cncr.21809. [DOI] [PubMed] [Google Scholar]
- 141.Preusser M, Charles Janzer R, Felsberg J, Reifenberger G, Hamou MF, Diserens AC, Stupp R, Gorlia T, Marosi C, Heinzl H, et al. Anti-O6-methylguanine-methyltransferase (MGMT) immunohistochemistry in glioblastoma multiforme: Observer variability and lack of association with patient survival impede its use as clinical biomarker. Brain Pathol. 2008;18:520–532. doi: 10.1111/j.1750-3639.2008.00153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rodriguez FJ, Thibodeau SN, Jenkins RB, Schowalter KV, Caron BL, O’neill BP, James CD, Passe S, Slezak J, Giannini C. MGMT immunohistochemical expression and promoter methylation in human glioblastoma. Appl Immunohistochem Mol Morphol. 2008;16:59–65. doi: 10.1097/PAI.0b013e31802fac2f. [DOI] [PubMed] [Google Scholar]
- 143.Mineura K, Yanagisawa T, Watanabe K, Kowada M, Yasui N. Human brain tumor O(6)-methylguanine-DNA methyltransferase mRNA and its significance as an indicator of selective chloroethylnitrosourea chemotherapy. Int J Cancer. 1996;69:420–425. doi: 10.1002/(SICI)1097-0215(19961021)69:5<420::AID-IJC12>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 144.Tanaka S, Kobayashi I, Utsuki S, Oka H, Fujii K, Watanabe T, Nagashima T, Hori T. O6-methylguanine-DNA methyl-transpherase gene expression in gliomas by means of real-time quantitative RT-PCR and clinical response to nitrosoureas. Int J Cancer. 2003;103:67–72. doi: 10.1002/ijc.10757. [DOI] [PubMed] [Google Scholar]
- 145.Ohe N, Saio M, Kijima M, Tamakawa N, Suwa T, Kojima Y, Yano H, Kaku Y, Iwama T, Shinoda J, et al. In situ detection of O6-methylguanine-DNA methyltransferase messenger RNA in paraffin-embedded human astrocytic tumor tissues by nested in situ RT-PCR is useful in predicting chemotherapy-resistance of tumors. Int J Oncol. 2003;22:543–549. [PubMed] [Google Scholar]
- 146.Tanaka S, Oka H, Fujii K, Watanabe K, Nagao K, Kakimoto A. Quantitation of O6-methylguanine-DNA methyltransferase gene messenger RNA in gliomas by means of real-time RT-PCR and clinical response to nitrosoureas. Cell Mol Neurobiol. 2005;25:1067–1071. doi: 10.1007/s10571-005-8475-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tanaka S, Akimoto J, Kobayashi I, Oka H, Ujiie H. Individual adjuvant therapy for malignant gliomas based on O6-methylguanine-DNA methyltransferase messenger RNA quantitation by real-time reverse-transcription polymerase chain-reaction. Oncol Rep. 2008;20:165–171. [PubMed] [Google Scholar]
- 148.Everhard S, Tost J, El Abdalaoui H, Crinière E, Busato F, Marie Y, Gut IG, Sanson M, Mokhtari K, Laigle-Donadey F, et al. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol. 2009;11:348–356. doi: 10.1215/15228517-2009-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yoshino A, Ogino A, Yachi K, Ohta T, Fukushima T, Watanabe T, Katayama Y, Okamoto Y, Naruse N, Sano E, et al. Gene expression profiling predicts response to temozolomide in malignant gliomas. Int J Oncol. 2010;36:1367–1377. doi: 10.3892/ijo_00000621. [DOI] [PubMed] [Google Scholar]
- 150.Tanaka S, Akimoto J, Narita Y, Oka H, Tashiro T. Is the absolute value of O(6)-methylguanine-DNA methyltransferase gene messenger RNA a prognostic factor, and does it predict the results of treatment of glioblastoma with temozolomide? J Neurosurg. 2014;121:818–826. doi: 10.3171/2014.6.JNS132535. [DOI] [PubMed] [Google Scholar]
- 151.Hegi ME, Diserens AC, Godard S, Dietrich PY, Regli L, Ostermann S, Otten P, Van Melle G, de Tribolet N, Stupp R. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10:1871–1874. doi: 10.1158/1078-0432.CCR-03-0384. [DOI] [PubMed] [Google Scholar]
- 152.Crinière E, Kaloshi G, Laigle-Donadey F, Lejeune J, Auger N, Benouaich-Amiel A, Everhard S, Mokhtari K, Polivka M, Delattre JY, et al. MGMT prognostic impact on glioblastoma is dependent on therapeutic modalities. J Neurooncol. 2007;83:173–179. doi: 10.1007/s11060-006-9320-0. [DOI] [PubMed] [Google Scholar]
- 153.Brandes AA, Tosoni A, Franceschi E, Sotti G, Frezza G, Amistà P, Morandi L, Spagnolli F, Ermani M. Recurrence pattern after temozolomide concomitant with and adjuvant to radiotherapy in newly diagnosed patients with glioblastoma: Correlation with MGMT promoter methylation status. J Clin Oncol. 2009;27:1275–1279. doi: 10.1200/JCO.2008.19.4969. [DOI] [PubMed] [Google Scholar]
- 154.Zawlik I, Vaccarella S, Kita D, Mittelbronn M, Franceschi S, Ohgaki H. Promoter methylation and polymorphisms of the MGMT gene in glioblastomas: A population-based study. Neuroepidemiology. 2009;32:21–29. doi: 10.1159/000170088. [DOI] [PubMed] [Google Scholar]
- 155.Prados MD, Chang SM, Butowski N, DeBoer R, Parvataneni R, Carliner H, Kabuubi P, Ayers-Ringler J, Rabbitt J, Page M, et al. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol. 2009;27:579–584. doi: 10.1200/JCO.2008.18.9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Weiler M, Hartmann C, Wiewrodt D, Herrlinger U, Gorlia T, Bähr O, Meyermann R, Bamberg M, Tatagiba M, von Deimling A, et al. Chemoradiotherapy of newly diagnosed glioblastoma with intensified temozolomide. Int J Radiat Oncol Biol Phys. 2010;77:670–676. doi: 10.1016/j.ijrobp.2009.05.031. [DOI] [PubMed] [Google Scholar]
- 157.Clarke JL, Iwamoto FM, Sul J, Panageas K, Lassman AB, DeAngelis LM, Hormigo A, Nolan CP, Gavrilovic I, Karimi S, et al. Randomized phase II trial of chemoradiotherapy followed by either dose-dense or metronomic temozolomide for newly diagnosed glioblastoma. J Clin Oncol. 2009;27:3861–3867. doi: 10.1200/JCO.2008.20.7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mikeska T, Bock C, El-Maarri O, Hübner A, Ehrentraut D, Schramm J, Felsberg J, Kahl P, Büttner R, Pietsch T, et al. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J Mol Diagn. 2007;9:368–381. doi: 10.2353/jmoldx.2007.060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vlassenbroeck I, Califice S, Diserens AC, Migliavacca E, Straub J, Di Stefano I, Moreau F, Hamou MF, Renard I, Delorenzi M, et al. Validation of real-time methylation-specific PCR to determine O6-methylguanine-DNA methyltransferase gene promoter methylation in glioma. J Mol Diagn. 2008;10:332–337. doi: 10.2353/jmoldx.2008.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Preusser M, Elezi L, Hainfellner JA. Reliability and reproducibility of PCR-based testing of O6-methylguanine-DNA methyltransferase gene (MGMT) promoter methylation status in formalin-fixed and paraffin-embedded neurosurgical biopsy specimens. Clin Neuropathol. 2008;27:388–390. doi: 10.5414/NPP27388. [DOI] [PubMed] [Google Scholar]
- 161.Håvik AB, Brandal P, Honne H, Dahlback HS, Scheie D, Hektoen M, Meling TR, Helseth E, Heim S, Lothe RA, et al. MGMT promoter methylation in gliomas-assessment by pyrosequencing and quantitative methylation-specific PCR. J Transl Med. 2012;10:36. doi: 10.1186/1479-5876-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Quillien V, Lavenu A, Karayan-Tapon L, Carpentier C, Labussière M, Lesimple T, Chinot O, Wager M, Honnorat J, Saikali S, et al. Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, MethyLight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer. 2012;118:4201–4211. doi: 10.1002/cncr.27392. [DOI] [PubMed] [Google Scholar]
- 163.Lattanzio L, Borgognone M, Mocellini C, Giordano F, Favata E, Fasano G, Vivenza D, Monteverde M, Tonissi F, Ghiglia A, et al. MGMT promoter methylation and glioblastoma: A comparison of analytical methods and of tumor specimens. Int J Biol Markers. 2014;0 doi: 10.5301/jbm.5000126. [DOI] [PubMed] [Google Scholar]
- 164.Preusser M, Berghoff AS, Manzl C, Filipits M, Weinhäusel A, Pulverer W, Dieckmann K, Widhalm G, Wöhrer A, Knosp E, et al. Clinical Neuropathology practice news 1–2014: Pyrosequencing meets clinical and analytical performance criteria for routine testing of MGMT promoter methylation status in glioblastoma. Clin Neuropathol. 2014;33:6–14. doi: 10.5414/NP300730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 166.Alexander BM, Galanis E, Yung WK, Ballman KV, Boyett JM, Cloughesy TF, Degroot JF, Huse JT, Mann B, Mason W, et al. Brain Malignancy Steering Committee clinical trials planning workshop: Report from the Targeted Therapies Working Group. Neuro Oncol. 2015;17:180–188. doi: 10.1093/neuonc/nou154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, Carpentier AF, Hoang-Xuan K, Kavan P, Cernea D, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370:709–722. doi: 10.1056/NEJMoa1308345. [DOI] [PubMed] [Google Scholar]
- 168.Lu KV, Chang JP, Parachoniak CA, Pandika MM, Aghi MK, Meyronet D, Isachenko N, Fouse SD, Phillips JJ, Cheresh DA, et al. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell. 2012;22:21–35. doi: 10.1016/j.ccr.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Atkins RJ, Ng W, Stylli SS, Hovens CM, Kaye AH. Repair mechanisms help glioblastoma resist treatment. J Clin Neurosci. 2015;22:14–20. doi: 10.1016/j.jocn.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 170.Dolan ME, Moschel RC, Pegg AE. Depletion of mammalian O6-alkylguanine-DNA alkyltransferase activity by O6-benzylguanine provides a means to evaluate the role of this protein in protection against carcinogenic and therapeutic alkylating agents. Proc Natl Acad Sci USA. 1990;87:5368–5372. doi: 10.1073/pnas.87.14.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Quinn JA, Jiang SX, Reardon DA, Desjardins A, Vredenburgh JJ, Rich JN, Gururangan S, Friedman AH, Bigner DD, Sampson JH, et al. Phase II trial of temozolomide plus O6-benzylguanine in adults with recurrent, temozolomide-resistant malignant glioma. J Clin Oncol. 2009;27:1262–1267. doi: 10.1200/JCO.2008.18.8417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gambari R, Fabbri E, Borgatti M, Lampronti I, Finotti A, Brognara E, Bianchi N, Manicardi A, Marchelli R, Corradini R. Targeting microRNAs involved in human diseases: A novel approach for modification of gene expression and drug development. Biochem Pharmacol. 2011;82:1416–1429. doi: 10.1016/j.bcp.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 173.Piva R, Spandidos DA, Gambari R. From microRNA functions to microRNA therapeutics: Novel targets and novel drugs in breast cancer research and treatment (Review) Int J Oncol. 2013;43:985–994. doi: 10.3892/ijo.2013.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Avitabile C, Fabbri E, Bianchi N, Gambari R, Romanelli A. Inhibition of miRNA maturation by peptide nucleic acids. Methods Mol Biol. 2014;1095:157–164. doi: 10.1007/978-1-62703-703-7_13. [DOI] [PubMed] [Google Scholar]
- 175.Brognara E, Fabbri E, Bianchi N, Finotti A, Corradini R, Gambari R. Molecular methods for validation of the biological activity of peptide nucleic acids targeting microRNAs. Methods Mol Biol. 2014;1095:165–176. doi: 10.1007/978-1-62703-703-7_14. [DOI] [PubMed] [Google Scholar]
- 176.Gambari R. Peptide nucleic acids: A review on recent patents and technology transfer. Expert Opin Ther Pat. 2014;24:267–294. doi: 10.1517/13543776.2014.863874. [DOI] [PubMed] [Google Scholar]