Abstract
Studies using the placental transcriptome to identify key molecules relevant for preeclampsia are hampered by a relatively small sample size. In addition, they use a variety of bioinformatics and statistical methods, making comparison of findings challenging. To generate a more robust preeclampsia gene expression signature, we performed a meta-analysis on the original data of 11 placenta RNA microarray experiments, representing 139 normotensive and 116 preeclamptic pregnancies. Microarray data were pre-processed and analyzed using standardized bioinformatics and statistical procedures and the effect sizes were combined using an inverse-variance random-effects model. Interactions between genes in the resulting gene expression signature were identified by pathway analysis (Ingenuity Pathway Analysis, Gene Set Enrichment Analysis, Graphite) and protein-protein associations (STRING). This approach has resulted in a comprehensive list of differentially expressed genes that led to a 388-gene meta-signature of preeclamptic placenta. Pathway analysis highlights the involvement of the previously identified hypoxia/HIF1A pathway in the establishment of the preeclamptic gene expression profile, while analysis of protein interaction networks indicates CREBBP/EP300 as a novel element central to the preeclamptic placental transcriptome. In addition, there is an apparent high incidence of preeclampsia in women carrying a child with a mutation in CREBBP/EP300 (Rubinstein-Taybi Syndrome). The 388-gene preeclampsia meta-signature offers a vital starting point for further studies into the relevance of these genes (in particular CREBBP/EP300) and their concomitant pathways as biomarkers or functional molecules in preeclampsia. This will result in a better understanding of the molecular basis of this disease and opens up the opportunity to develop rational therapies targeting the placental dysfunction causal to preeclampsia.
Introduction
Preeclampsia, defined as the development of high blood pressure in combination with proteinuria after 20 weeks of gestation, is a common (incidence 2–8%) and potentially severe pregnancy complication for both mother and child [1]. At present, apart from delivery of placenta and fetus, there is no definite treatment for preeclampsia. Although preeclampsia clinically manifests during the second half of gestation, it most likely originates at the time the early placenta develops.
The molecular mechanisms that play a role in the development of preeclampsia are largely unknown. Over the past decades, much research has been dedicated to identify preeclampsia-specific molecules that can either serve as a biomarker for prognostic/diagnostic purposes, or that play a functional role in the disease and hence are potential therapeutic targets. The main approach that has been used for this purpose is the analysis of gene expression in the placenta of normotensive and preeclamptic pregnancies. This has resulted in the identification of factors unquestionably associated with preeclampsia, such as soluble Vascular Endothelial Growth factor receptor 1 (sFLT1) [2] and soluble Endoglin [3].
We have recently performed a systematic review and meta-analysis on 30 studies reporting lists of differentially expressed genes (DEGs) in relation to preeclamptic placenta [4]. This gene list-driven approach resulted in a 40-gene preeclamptic signature that, apart from identifying most of the genes known to be associated with preeclampsia, singled out 14 genes not previously associated with preeclampsia. Although this literature-based approach resulted in an overall picture of genes differentially expressed in the preeclamptic placenta and in the identification of potential novel biomarkers, the number of genes included in the meta-signature was limited due to a high degree of between-study variability. Apart from the fact that focus of the individual research groups may have driven the reported gene lists, inconsistencies in the preeclampsia-specific gene signatures are largely due to the relatively limited sample size used in most studies, combined with differences in experimental approach, and bioinformatics and statistical methods. Recently, two papers described meta-analyses of preeclampsia RNA microarray studies. Although these studies by their nature increase the sample size and resolve some of the disadvantages of the reported gene list-driven meta-analysis, they do not, or only partly, deal with aspects like quality assessment and uniformity of data analysis [5, 6].
For the current study we retrieved the original microarray data from 11 independent placental mRNA microarray experiments. Microarray data were subjected to quality analysis, pre-processed and analyzed using standardized bioinformatics and statistical procedures. This resulted in a comprehensive preeclampsia gene signature, which upon further analysis led to the identification of a novel pathway associated with preeclampsia.
Materials and Methods
Dataset acquisition
Datasets were identified by systematic review [4], searching the NCBI Gene Expression Omnibus [7] and the EBI ArrayExpress [8] functional genomics data repositories using the term “preeclampsia” and close derivatives. Datasets from platforms that interrogated more than 10,000 genes and did not completely pool samples were selected. In addition, we generated one dataset in our laboratory [9]. If available, data was directly obtained from the data repositories. For studies identified by systematic review without publicly available data, authors were contacted to provide original data. An overview of datasets is shown in Tables 1 and 2.
Table 1. Datasets included in the meta-analysis.
| number of samples | |||||||
|---|---|---|---|---|---|---|---|
| reported | after QC | ||||||
| Data Set | Data source (first author) | NT | PE | NT | PE | Platform | probes with Gene ID |
| 1 | Authors c (Centlow) [10] | 20 | 15 | 19 | 11 | Custom, OPERON | 13010 |
| 2 | Authors$ (Enquobahrie) [11] | 18 | 18 | 15 | 16 | Custom, OPERON | 13010 |
| 3 | Authors c (Hoegh) [12] | 3 a | 3 a | 3 a | 3 a | Affymetrix HG U133A | 19955 |
| 4 | GSE54618 (Jebbink) [9] | 12 | 12 | 11 | 11 | Illumina HumanHT-12 v4 | 37463 |
| 5 | Authors c (Kivinen) [13] | 6 | 8 | 6 | 7 | Affymetrix HG U133 plus 2.0 | 41245 |
| 6 | GSE30186 (Meng) [14] | 6 | 6 | 6 | 5 | Illumina HumanHT-12 v4 | 37463 |
| 7 | GSE4707 (Nishizawa) [15] | 4 | 10 | 2 b | 7 b | Agilent Whole Genome Oligo Microarray G4112A | 29777 |
| 8 | GSE24129 (Nishizawa) [16] | 8 | 8 | 8 | 8 | Affymetrix Human Exon 1.0 ST array | 21929 |
| 9 | GSE10588 (Sitras) [17] | 26 | 17 | 24 | 16 | ABI Human Genome Survey Microarray v.2.0 | 26396 |
| 10A | GSE25906 (Tsai) [18] | 8 | 7 | 8 | 7 | Illumina Human-6 v2 | 27303 |
| 10B | 29 | 16 | 28 | 16 | |||
| 11 | GSE14722 (Winn) [19] | 11 | 12 | 9 | 9 | Affymetrix HG U133A | 19955 |
| Total | 151 | 132 | 139 | 116 | |||
GSE = GEO accession number, NT = normotensive, PE = preeclampsia, QC = Quality Control
aeach sample represents 3 pooled placentas
bsamples that are also present in dataset 8 were excluded from dataset 7
coriginal data was provided by the authors.
Table 2. Studies excluded in the meta-analysis.
| number of samples | |||
|---|---|---|---|
| First author | NT | PE | Platform a |
| Ahr [20] | 4 | 3 | University Health Network Hu19k2 (cDNA) |
| Guller [21] | 6 | 3 | Affymetrix HG U133 plus 2.0 |
| Jarvenpaa [22] | 3 | 2 | Affymetrix HG U133 plus 2.0 |
| Kang [23] | 16 | 17 | Codelink Human Whole Genome Bioarrays |
| Lee [24] | 13 | 13 | Agilent Human 4X44K Oligomicroarrays Chip |
| Liu [25] | 28 | 24 | CapitalBio, OPERON |
| Mayor-Lynn [26] | 6 | 3 | Illumina HumRef-12 v3 |
| Varkonyi [27] | 10 | 12 | Agilent Human 4X44K Oligomicroarrays Chip |
| Zhou [28] | 5 | 5 | CapitalBio, OPERON |
| 91 | 82 | ||
NT = normotensive, PE = preeclampsia
aplatform names were obtained from description in cited publication.
Data processing and meta-analysis
The meta-analysis was performed according to the stepwise approach described in [29]. In brief, original data was retrieved where possible and subsequently pre-processed using state-of-the art methods for each of the platforms included. Samples of low quality were removed using the quality criteria calculated by the R package arrayQualityMetrics. Effect sizes (Hedges’ g) and their variances were calculated for each gene in each dataset. Dataset-specific effect sizes were combined using an inverse-variance random-effects model. As a final step, a leave-one-out-analysis was performed to increase the robustness of the resulting meta-signature. Details of the analysis are provided as Supplementary Methods in S1 text.
STRING 9.1 [30] was used for protein-protein association analysis. All 388 genes from the meta‐signature were used as input for STRING analysis and a network was built based on high confidence (0.8) evidence from experimental protein‐protein interaction and curated databases.
KEGG Pathway analysis was performed using Graphite [31]. The 388 gene meta-signature was used as input. Pathways were considered significantly enriched at q-values ≤ 0.05.
GSEAPreranked was used for gene set enrichment analysis [32]. All 8612 genes of the meta-analysis were ranked according to their mu-value and used as ranked gene list. Enrichment scores were calculated based on the standard GSEAPreranked settings and the curated gene set database (c2.all.v5.0.entrez.gmt, 4530 gene sets). Gene sets were considered significantly enriched at FDR q-values ≤ 0.05.
Ingenuity Pathway Analysis (Qiagen) Upstream Regulator Analysis was performed to identify upstream regulators that can explain the observed 388 gene meta-signature. Analysis was performed using standard settings. The resulting activation z-score was used to infer the activation states of the predicted regulator. A z-score of > 2 (activated) or <-2 (inhibited) was considered significant.
Gene Expression Barcode: RNA microarray data for human placentas of different gestational ages was obtained from the NCBI Gene Expression Omnibus (accession GSE9984). Samples were pre-processed and barcoded as described by the Gene Expression Barcode 3.0 using the R package frma [33]. Probesets were mapped to official gene symbols using the R package hgu133plus2.db
Results
For the current meta-analysis, we included 11 different studies representing a total of 151 normotensive and 132 preeclamptic placenta samples (Table 1). Nine studies (covering 91 normotensive and 82 preeclamptic samples) were not included (Table 2) because authors did not respond to our request for data (5 instances) or because authors did not wish to share their data (3 instances). One author was not able to recover the original data files.
Quality analysis of the microarray data resulted in the removal of 28 samples. One dataset [18] consisted of two batches, which induced systematic differences in gene expression levels and that were therefore treated as two independent datasets. Meta-analysis was performed on 12 datasets (covering placentas from 139 normotensive and 116 preeclamptic pregnancies) combining the effect sizes using an inverse-variance random-effects model. The meta-analysis was performed on the 8,612 genes available on all platforms, resulting in the identification of 688 differentially expressed genes in preeclamptic versus normotensive placenta (Fig 1). To increase the robustness of the meta-signature, an additional leave-one-out-analysis was performed which reduced the number of genes to 388 (Benjamini-Hochberg-based corrected p-value<0.05), of which 214 were overexpressed in preeclamptic placentas compared to placentas from normotensive pregnancies. A detailed list of the 388 genes and the corresponding statistics is shown in S1 Table, corresponding forest plots are shown in S1 Fig.
Fig 1. Volcano plot of DEGs.
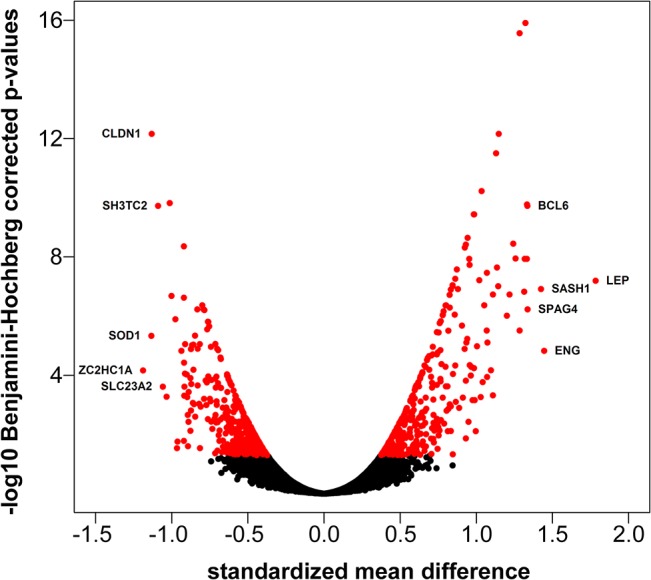
Volcano plot showing the standardized mean difference and the adjusted p-value for the 8,612 genes included on all platforms. The red dots represent genes differentially expressed (adjusted P<0.05) in the preeclamptic versus normotensive placenta. The symbol-marked dots indicate the five genes with the largest negative or positive standardized mean difference.
Comparison of the current 388-gene meta-signature with the gene list-driven signature of 40 genes we previously obtained from published gene lists reported as being differentially expressed in preeclamptic placenta [4] shows that most of the literature-based genes are also included in the 388-gene signature (S2 Table). In total, 12 genes from the gene list-driven signature were not included in the meta-signature from our current analysis. Six of these were not represented by a gene-specific probe on some platforms (INHBA, CGB, F5, HTRA4, PVRL4, RDH13). Calculation of the standardized mean difference (mu) based on the studies that did contain a gene-specific probe for these six genes shows that all are differentially expressed. Two of the 12 genes (HSD17B1, VIM) were among the 688 differentially expressed genes in the initial meta-signature, but were removed in the leave-one-out-analysis, and four genes (CGA, IGFBP1, PGF, TNFSF10) were not significantly differentially expressed in our meta-analysis on the original microarray data. For those genes in the gene list-driven signature that were also differentially expressed in the current analysis, the direction of differential expression was identical except for SOD1, SPP1 and VIM. SOD1 and VIM, which have been described in the literature both as being upregulated and downregulated in the preeclamptic placenta, are definitely downregulated in the current analysis. In fact, SOD1 is one of the most strongly downregulated genes in the preeclamptic placenta, as shown in S1 Table. The only genuine discrepancy is the direction of SPP1. Although reported as being upregulated in the preeclamptic placenta in three different publications, our current meta-analysis identifies SPP1 as a gene downregulated in preeclampsia.
Two recent studies also performed a meta-analysis on microarray data exclusively. Vaiman et al [6] analyzed six datasets (corresponding to dataset 6, 7, 8, 9, 10 and 11 in this paper) and used vote counting to identify genes that were consistently upregulated or downregulated in the majority of studies. Moslehi et al [5] analyzed four datasets (corresponding to dataset 7, 8, 9, and 11 in this paper) and performed their meta-analysis by averaging p-values and fold differences. After removal of the genes not included in our 8,612 gene universe, 77% (57 out of 74) of the DEGs reported by Vaiman et al are included in our current 388-gene meta-signature, while the overlap with the Moslehi study is 44% (123 out of 281) as shown in Fig 2.
Fig 2. Overlapping genes with previous meta-analyses.
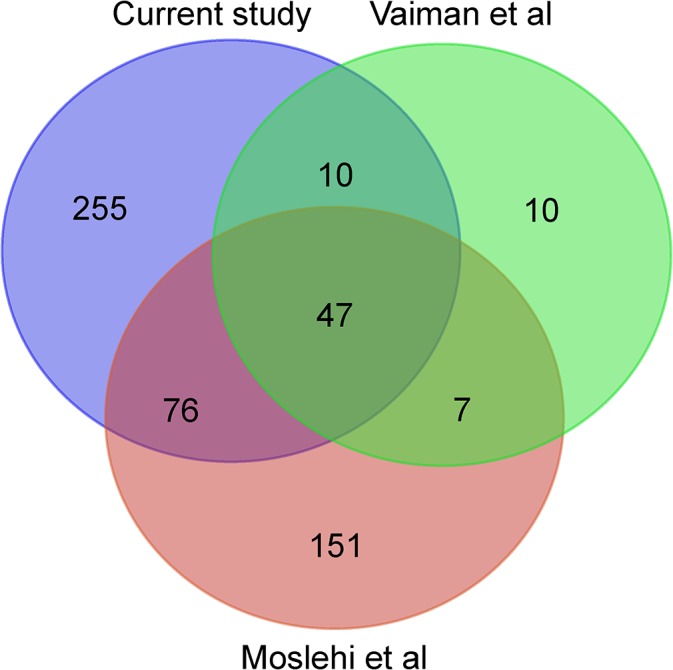
Venn diagram of the DEGs from the current study and the previously published studies. DEGs were extracted from the publications [5, 6] and only genes within the gene universe of our study were included in the comparison.
To further identify preeclampsia-related pathways or networks within our 388-gene meta-signature, the STRING network of both physical and functional interactions between proteins was used [30]. By employing a stringent confidence score of 0.8 and experiments plus curated databases as active prediction methods, 112 interactions between proteins encoded by meta-signature genes were observed (S2 Fig). CREBBP is a hub in this network, with 13 direct protein interactions and 23 interactions in total (Fig 3).
Fig 3. Protein interaction network analysis indicates a central role for CREBBP in the preeclamptic placenta.
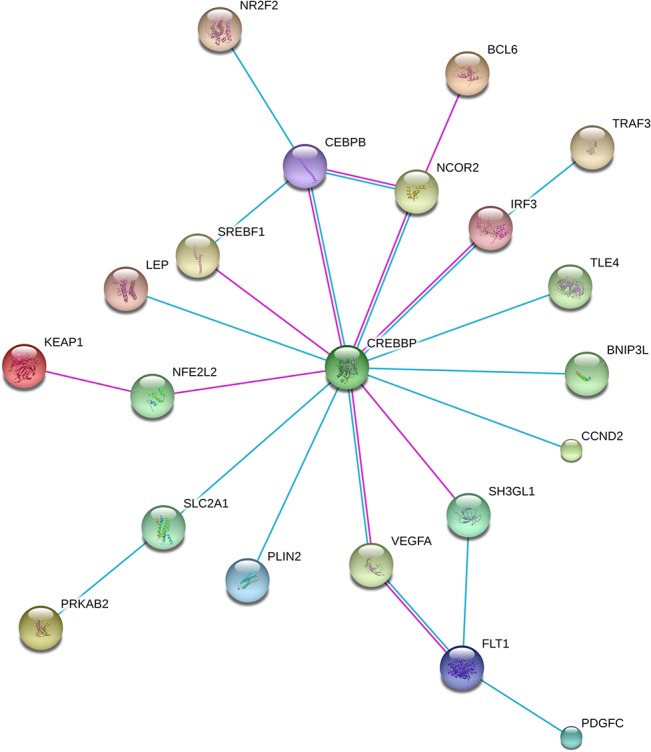
All 388 genes were used as input for STRING analysis and a network was built (S2 Fig). Shown are the details of CREBBP interactions based on high confidence (0.8) evidence from experimental protein-protein interaction (blue lines) and curated (purple lines) databases. Proteins are indicated by nodes labeled with the encoding gene symbol. The network is enriched in interactions (p = 2.17E-13) [30].
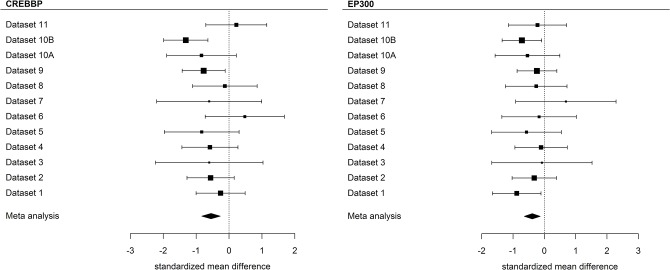
As shown in Fig 4, CREBBP (adjusted p-value = 0.004, mu = -0.549) and its functionally similar paralogue EP300 (adjusted p-value = 0.038, mu = -0.388; present in the 688 differentially expressed gene list, but removed in leave-one-out-analysis) [34], are consistently downregulated in almost all individual datasets.
Fig 4. CREBBP/EP300 expression in individual studies.
Forest plots for CREBBP and EP300. Squares are proportional to weights used in the meta-analysis, and the lines represent the 95% confidence interval. The diamond represents the standardized mean difference (log2 scale).
Parallel approaches using pathway analysis resulted in a large number of pathways that are enriched in the 388-gene meta-signature with a consistent enrichment for the hypoxia/HIF1A pathway in Ingenuity Pathway Analysis, Graphite, and Gene Set Enrichment Analysis (Table 3).
Table 3. Pathway analysis.
| Ingenuity Upstream Regulator Analysis a | ||||
| Upstream Regulator | Molecule Type | Predicted Activation State | Activation z-score | |
| HIF1A | transcription regulator | Activated | 4.163 | |
| PDGF BB | complex | Activated | 3.400 | |
| CREB1 | transcription regulator | Activated | 3.392 | |
| PPARG | ligand-dependent nuclear receptor | Activated | 3.085 | |
| CSF3 | cytokine | Activated | 2.961 | |
| Graphite Pathway Analysis | ||||
| Pathway | q-value | |||
| HIF-1 signaling pathway | 0.0000 | |||
| Fc gamma R-mediated phagocytosis | 0.0003 | |||
| Gene Set Enrichment Analysis (top of the ranked gene list) a | ||||
| Gene Set Name | Size b | NES c | FDR q-val | |
| SEMENZA_HIF1_TARGETS | 31 | 2.517 | 0.000 | |
| ELVIDGE_HYPOXIA_BY_DMOG_UP | 109 | 2.470 | 0.000 | |
| MENSE_HYPOXIA_UP | 70 | 2.461 | 0.000 | |
| LEONARD_HYPOXIA | 41 | 2.450 | 0.000 | |
| ELVIDGE_HIF1A_AND_HIF2A_TARGETS_DN | 83 | 2.437 | 0.000 | |
| ELVIDGE_HYPOXIA_UP | 140 | 2.398 | 0.000 | |
| ELVIDGE_HIF1A_TARGETS_DN | 72 | 2.301 | 0.000 | |
| WANG_ADIPOGENIC_GENES_REPRESSED_BY_SIRT1 | 17 | 2.253 | 0.000 | |
| WINTER_HYPOXIA_METAGENE | 187 | 2.240 | 0.001 | |
| FARDIN_HYPOXIA_11 | 25 | 2.212 | 0.001 | |
a For the Graphite Pathway Analysis all the significant pathways are shown. For Ingenuity Upstream Regulator Analysis the top 5 activated upstream regulators, and for Gene Set Enrichment Analysis the top 10 gene sets overrepresented at the top of the meta-signature are shown. A full list of all significant results is presented in S3 Table.
bSize: Number of genes in the gene set after filtering out those genes not in the expression dataset.
cNES: Normalized enrichment score.
To obtain information if the genes listed in the meta-signature are also expressed earlier during gestation, we performed a Gene Expression Barcode analysis on a microarray data set including 1st and 2nd trimester placentas [35]. As shown in S4 Table most of the meta-signature genes are expressed above background (z-score > 3 in 2nd, and even 1st trimester placenta.
Discussion
We established a robust 388-gene meta-signature in what is presently the most complete meta-analysis of the preeclamptic placental transcriptome. Our meta-analysis not only greatly increases the number of samples included as compared to the individual studies, but also largely resolves the between-study variation in bioinformatics and statistical analysis, which was one of the major inconsistencies reported in our previous gene list-driven meta-analysis [4].
Recently, several other approaches to summarize placental mRNA expression data in relation to preeclampsia have been reported. As mentioned above, we previously performed a systematic review and vote-counting-based meta-analysis on published preeclampsia lists of DEGs from both genome-wide microarray and smaller scale mRNA expression studies [4]. Most genes in the resulting gene list-driven meta-signature are also included in our current 388-gene meta-signature. Genes not included in our current meta-signature had a lower number of votes and mainly originate from older studies. The current meta-analysis clearly generates many additional DEGs compared with the literature-based approach.
The 388-gene meta-signature we currently present shows considerable overlap (77%; odds ratio = 82.86, P<2.2·10−16, Fisher’s exact test) with the genes in the meta-signature of Vaiman and colleagues, and with the meta-signature reported by Moslehi and colleagues (44%; odds ratio = 23.67, P<2.2·10−16, Fisher’s exact test). However, our current approach has generated many more DEGs. Possible reasons for this are multiple and include (i) an increase in statistical power by the larger number of studies included in our meta-analysis, (ii) renormalization of the original data using an uniform set of state-of-the-art methods whenever possible, and (iii) use of stringent quality control criteria to detect and discard low-quality arrays. Differences in meta-analysis techniques used also impact the meta-signatures obtained. Combining effect sizes using an inverse-variance random-effects model has several advantages compared to the meta-analysis techniques used by the other two studies [29]. Particularly important is the incorporation of between-study heterogeneity into the study weights, since the 11 studies included in our meta-analysis are geographically (eight different countries) and technically (eight different platforms, 10 different laboratories) diverse.
Higher statistical power of the current meta-analysis compared to the individual studies leads to discovery of novel genes/pathways as illustrated by CREBBP (Fig 4). CREBBP is downregulated in preeclamptic placenta in most studies, but with wide confidence intervals. However, its association with preeclampsia is exposed by combining all studies in the current meta-analysis.
There are two obvious issues that have limited the full potential of the current analysis. (i) Of the 20 suitable studies identified, only seven had submitted their data to a publicly available repository; the data of four additional studies were obtained directly from the authors. Although the acquired original data represents the majority of samples, it is disappointing that from almost half of the studies data did not become available.
(ii) In order to obtain summary statistics and p-values based on data from all studies, only the genes present on all platforms were taken into account. This results in a large decrease of the gene universe in the current analysis (8,612 genes) compared to the more than 20,000 genes located on most of the microarray platforms included. In particular, the inclusion of the Operon platforms (representing approximately 13,000 Entrez gene IDs) dramatically decreases the number of genes in the intersection.
A shortcoming of the current meta-analysis (but also for all preeclampsia placental gene expression studies) in relation to both function of gene products and their potential role as predictors in preeclampsia is the timing of the gene expression pattern. All mRNA microarray data included in the current meta-analysis are generated from placentas at delivery, while the initial defects leading to preeclampsia occur earlier during pregnancy [1, 36]. Based on data from one of the few microarray data sets on early gestation placenta, we show that the majority of the meta-signature genes is expressed during the 1st and 2nd trimester. CREBBP shows clear placental expression from 1st trimester onwards, but the level of EP300 mRNA expression in the early placenta is less pronounced. Novel developments allowing the quantification of placental RNAs in maternal blood offers the enticing opportunity to study placental gene expression in ongoing pregnancy [37].
A further asset would be the coupling of individual gene expression data to more extensive clinical parameters. The method and time of delivery and the use of antihypertensive drugs, among other things, are likely to affect placental gene expression. Preeclampsia, as a heterogeneous condition consists of several subtypes with different etiology and degree of placental contribution [38]. Unfortunately, most of the datasets available for the current study are not accompanied by sufficient individual clinical information to allow more detailed placental transcriptome to clinical phenotype coupling.
Identification of functionally relevant pathways from the large number of genes in the preeclampsia-placental signature is a major challenge. Building networks based on protein-protein interactions can help to identify these functionally relevant pathways. Even when using a rather conservative approach, a protein-protein interaction network within our preeclampsia meta-signature emerges with CREBBP as key protein within this network. CREBBP interacts with several proteins well known to be associated with preeclampsia, such as NFE2L2 [39, 40], LEP [41] and VEGF/FLT1 [42], but also with proteins not-yet related to preeclampsia. CREBBP and EP300 are transcriptional/epigenetic regulators involved in many physiological and pathological processes [43]. Although their functions appear to be largely redundant, differences have been reported [34]. Even though CREBBP/EP300 were not identified as DEGs in the other two published meta-analyses, their involvement in preeclampsia was suggested based on association with differentially expressed transcription factors by Vaiman and colleagues [6].
Mutations in CREBBP and EP300 are causative for Rubinstein-Taybi syndrome (RSTS, OMIM 180849 and 613684). RSTS is a rare (approximately 1 in 100,000 newborns) autosomal dominant congenital disorder characterized by mental retardation, dysmorphic facial features, postnatal growth deficiency, and skeletal abnormalities [44]. Almost all mutations occur de novo and range from large deletions to point mutations. There is no clear genotype-phenotype correlation, but loss of the CREBBP/EP300 histone acetyltransferase activity appears to be essential for the development of RSTS [45]. Interestingly, there have been frequent reports of preeclampsia-complicated pregnancies in mothers giving birth to a RSTS affected child [46–51]. Thus, the fetal genotype not only causes RSTS in the neonate but also influences placenta function resulting in an increased incidence of preeclampsia in the mother. If we translate these observations and our meta-analysis results to non-RSTS women, it appears that decreased CEBBP/EP300 levels are detrimental for placenta functioning, resulting in an increased probability for the mother to develop preeclampsia.
Analysis of canonical pathways and gene expression patterns clearly indicated a prominent role for hypoxia in the establishment of the preeclamptic gene meta-signature, a conclusion that was also drawn by Moslehi et al, based on the DEGs from their meta-analysis [5].
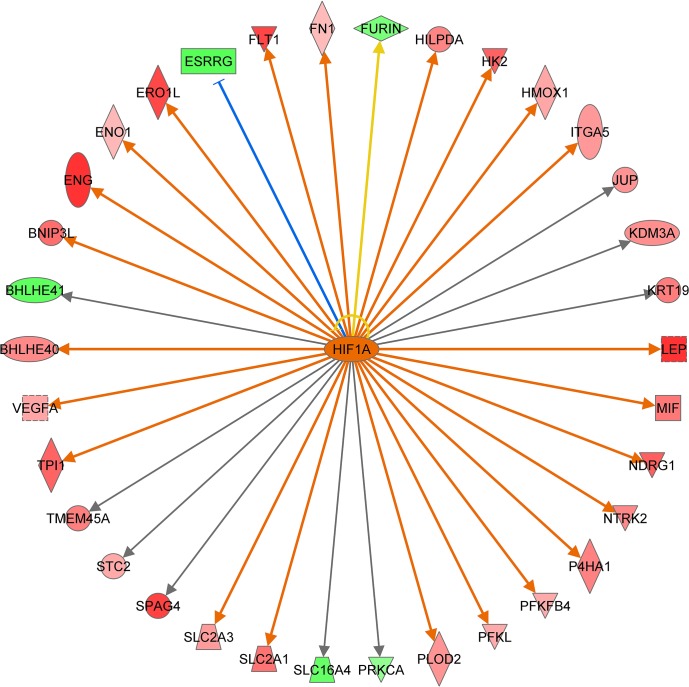
Preeclamptic placentas are compromised by hypoxia/ischemia, and the effect on gene expression is largely mediated via HIF-1α [52]. More than 10% of the upregulated meta-signature genes are direct targets of HIF-1α (Fig 5). CREBBP/EP300 is an important transcriptional coactivator of HIF-1α [53–56]. As a result, decreased CREBBP/EP300 levels may impair the placenta’s capacity to respond to low oxygen, resulting in a further decline of its (preeclamptic) condition.
Fig 5. Ingenuity Pathway Analysis identifies HIF1A as the most prominently activated upstream regulator.
Thirty four out of 388 genes are direct HIF1A downstream targets according to the Ingenuity Knowledge Database. Arrows indicate the predicted relationship: orange = leads to activation (23 genes), blue = leads to inhibition (1 gene), grey = effect not predicted (9 genes), yellow = findings inconsistent with the state of downstream molecule (1 gene). Red symbols indicate genes upregulated in preeclamptic placenta, green symbols indicate genes downregulated in preeclamptic placenta. Intensity of the coloring is related to the level of up/downregulation.
Conclusions
The robust 388-gene signature of differentially expressed genes in the preeclamptic placenta serves as a starting point to investigate their function in the development of preeclampsia. This will result in a better understanding of the molecular basis of this disease and opens up the opportunity to develop rational therapies targeting the placental dysfunction causal to preeclampsia.
Supporting Information
Forest plots of the 388 differentially expressed genes in the preeclamptic placenta ordered on the absolute value of the standardized mean difference (see S1 Table). Squares are proportional to weights used in the meta-analysis, and the lines represent the 95% confidence interval. The diamond represents the standardized mean difference (log2 scale).
(PDF)
All 388 genes from the meta‐signature were used as input for STRING analysis and a network was built based on high confidence (0.8) evidence from experimental protein‐protein interaction (blue lines) and curated (purple lines) databases. Proteins are indicated by nodes labeled with the encoding gene symbol. Two genes (CEBPA and SEPP1) are not present in the used STRING version 9.1. The network is enriched in interactions (p = 0.007) using the intersection of 8,612 genes present on all platforms as background. Additional cluster analysis was used to color the nodes of the interaction networks. Yellow nodes represent proteins that have no protein‐protein interaction at this confidence setting.
(PDF)
(XLSX)
(XLSX)
(XLSX)
Indicated microarray samples were analyzed by the Gene Expression Barcode 3.0 and the resulting z-scores are shown. The z-score is the number of standard deviations above the mean expression signal of the unexpressed null distribution. An average z-score of ≥ 3 for a gene-specific probe has been labeled as being expressed in that trimester.
(XLSX)
(DOCX)
Acknowledgments
The authors appreciate the contribution of Aldo Jongejan for improving the ABI probe annotation and Fulco van der Veen for critically reading the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study has been supported by funds from the Swedish Research Council (www.vr.se), Academy of Finland (http://www.aka.fi), Sigrid Juselius Foundation (http://www.sigridjuselius.fi), Finnish Medical Foundation (http://www.laaketieteensaatio.fi), Helsinki University Central Hospital Research Funds (http://www.med.helsinki.fi), Helsinki University Research Funds (http://www.helsinki.fi), Päivikki and Sakari Sohlberg Foundation (http://www.pss-saatio.fi) and Uusimaa Regional Fund of the Finnish Cultural Foundation (www.skr.fi) (H.L.); National Iinstitute of Health (www.nih.gov) grants R01HD32562, R01HD055566, and K01HL103174 (D.A.E.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Steegers EA, von Dadelszen P, Duvekot JJ, Pijnenborg R. Pre-eclampsia. Lancet. 2010;376(9741):631–44. 10.1016/S0140-6736(10)60279-6 [DOI] [PubMed] [Google Scholar]
- 2. Maynard SE, Min JY, Merchan J, Lim KH, Li J, Mondal S, et al. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J Clin Invest. 2003;111(5):649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Venkatesha S, Toporsian M, Lam C, Hanai J-i, Mammoto T, Kim YM, et al. Soluble endoglin contributes to the pathogenesis of preeclampsia. Nat Med. 2006;12(6):642–9. [DOI] [PubMed] [Google Scholar]
- 4. Kleinrouweler CE, van Uitert M, Moerland PD, Ris-Stalpers C, van der Post JA, Afink GB. Differentially expressed genes in the pre-eclamptic placenta: a systematic review and meta-analysis. PLoS One. 2013;8(7):e68991 10.1371/journal.pone.0068991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moslehi R, Mills JL, Signore C, Kumar A, Ambroggio X, Dzutsev A. Integrative transcriptome analysis reveals dysregulation of canonical cancer molecular pathways in placenta leading to preeclampsia. Scientific reports. 2013;3:2407 10.1038/srep02407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vaiman D, Calicchio R, Miralles F. Landscape of transcriptional deregulations in the preeclamptic placenta. PLoS One. 2013;8(6):e65498 10.1371/journal.pone.0065498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic acids research. 2013;41(Database issue):D991–5. 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rustici G, Kolesnikov N, Brandizi M, Burdett T, Dylag M, Emam I, et al. ArrayExpress update—trends in database growth and links to data analysis tools. Nucleic acids research. 2013;41(Database issue):D987–90. 10.1093/nar/gks1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jebbink JM, Boot RG, Keijser R, Moerland PD, Aten J, Veenboer GJ, et al. Increased glucocerebrosidase expression and activity in preeclamptic placenta. Placenta. 2015;36(2):160–9. 10.1016/j.placenta.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 10. Centlow M, Wingren C, Borrebaeck C, Brownstein MJ, Hansson SR. Differential gene expression analysis of placentas with increased vascular resistance and pre-eclampsia using whole-genome microarrays. Journal of pregnancy. 2011;2011:472354 10.1155/2011/472354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Enquobahrie DA, Meller M, Rice K, Psaty BM, Siscovick DS, Williams MA. Differential placental gene expression in preeclampsia. American journal of obstetrics and gynecology. 2008;199(5):566.e1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoegh AM, Borup R, Nielsen FC, Sorensen S, Hviid TV. Gene expression profiling of placentas affected by pre-eclampsia. Journal of biomedicine & biotechnology. 2010;2010:787545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kivinen K, Peterson H, Hiltunen L, Laivuori H, Heino S, Tiala I, et al. Evaluation of STOX1 as a preeclampsia candidate gene in a population-wide sample. European journal of human genetics: EJHG. 2007;15(4):494–7. [DOI] [PubMed] [Google Scholar]
- 14. Meng T, Chen H, Sun M, Wang H, Zhao G, Wang X. Identification of differential gene expression profiles in placentas from preeclamptic pregnancies versus normal pregnancies by DNA microarrays. Omics: a journal of integrative biology. 2012;16(6):301–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nishizawa H, Pryor-Koishi K, Kato T, Kowa H, Kurahashi H, Udagawa Y. Microarray analysis of differentially expressed fetal genes in placental tissue derived from early and late onset severe pre-eclampsia. Placenta. 2007;28(5–6):487–97. [DOI] [PubMed] [Google Scholar]
- 16. Nishizawa H, Ota S, Suzuki M, Kato T, Sekiya T, Kurahashi H, et al. Comparative gene expression profiling of placentas from patients with severe pre-eclampsia and unexplained fetal growth restriction. Reproductive biology and endocrinology: RB&E. 2011;9:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sitras V, Paulssen RH, Gronaas H, Leirvik J, Hanssen TA, Vartun A, et al. Differential placental gene expression in severe preeclampsia. Placenta. 2009;30(5):424–33. 10.1016/j.placenta.2009.01.012 [DOI] [PubMed] [Google Scholar]
- 18. Tsai S, Hardison NE, James AH, Motsinger-Reif AA, Bischoff SR, Thames BH, et al. Transcriptional profiling of human placentas from pregnancies complicated by preeclampsia reveals disregulation of sialic acid acetylesterase and immune signalling pathways. Placenta. 2011;32(2):175–82. 10.1016/j.placenta.2010.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Winn VD, Gormley M, Paquet AC, Kjaer-Sorensen K, Kramer A, Rumer KK, et al. Severe preeclampsia-related changes in gene expression at the maternal-fetal interface include sialic acid-binding immunoglobulin-like lectin-6 and pappalysin-2. Endocrinology. 2009;150(1):452–62. 10.1210/en.2008-0990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ahr A, Holtrich U, Karn T, Reitter A, Rody A, Kaufmann M, et al. [Identification of preeclampsia by cDNA-gene expression profiling in human placentas and serum—a pilot study]. Zentralblatt fur Gynakologie. 2006;128(3):138–42. [DOI] [PubMed] [Google Scholar]
- 21. Guller S, Buhimschi CS, Ma YY, Huang STJ, Yang L, Kuczynski E, et al. Placental expression of ceruloplasmin in pregnancies complicated by severe preeclampsia. Lab Invest. 2008;88(10):1057–67. 10.1038/labinvest.2008.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jarvenpaa J, Vuoristo JT, Santaniemi M, Ukkola O, Savolainen ER, Jaaskelainen M, et al. Adiponectin induced placental cell apoptosis could be mediated via the ADIPOR1-receptor in pre-eclampsia with IUGR. Journal of perinatal medicine. 2009;37(3):257–62. 10.1515/JPM.2009.046 [DOI] [PubMed] [Google Scholar]
- 23. Kang JH, Song H, Yoon JA, Park DY, Kim SH, Lee KJ, et al. Preeclampsia leads to dysregulation of various signaling pathways in placenta. Journal of hypertension. 2011;29(5):928–36. 10.1097/HJH.0b013e328344a82c [DOI] [PubMed] [Google Scholar]
- 24. Lee GS, Joe YS, Kim SJ, Shin JC. Cytokine-related genes and oxidation-related genes detected in preeclamptic placentas. Archives of gynecology and obstetrics. 2010;282(4):363–9. 10.1007/s00404-009-1222-x [DOI] [PubMed] [Google Scholar]
- 25. Liu Y, Li N, You L, Liu X, Li H, Wang X. HSP70 is associated with endothelial activation in placental vascular diseases. Molecular medicine (Cambridge, Mass). 2008;14(9–10):561–6. [DOI] [PubMed] [Google Scholar]
- 26. Mayor-Lynn K, Toloubeydokhti T, Cruz AC, Chegini N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod Sci. 2011;18(1):46–56. 10.1177/1933719110374115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Varkonyi T, Nagy B, Fule T, Tarca AL, Karaszi K, Schonleber J, et al. Microarray profiling reveals that placental transcriptomes of early-onset HELLP syndrome and preeclampsia are similar. Placenta. 2011;32 Suppl:S21–9. 10.1016/j.placenta.2010.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou R, Zhu Q, Wang Y, Ren Y, Zhang L, Zhou Y. Genomewide oligonucleotide microarray analysis on placentae of pre-eclamptic pregnancies. Gynecologic and obstetric investigation. 2006;62(2):108–14. [DOI] [PubMed] [Google Scholar]
- 29. Ramasamy A, Mondry A, Holmes CC, Altman DG. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med. 2008;5(9):e184 10.1371/journal.pmed.0050184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2013;41(D1):D808–D15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sales G, Calura E, Martini P, Romualdi C. Graphite Web: Web tool for gene set analysis exploiting pathway topology. Nucleic acids research. 2013;41(Web Server issue):W89–97. 10.1093/nar/gkt386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCall MN, Uppal K, Jaffee HA, Zilliox MJ, Irizarry RA. The Gene Expression Barcode: leveraging public data repositories to begin cataloging the human and murine transcriptomes. Nucleic acids research. 2011;39(Database issue):D1011–5. 10.1093/nar/gkq1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kalkhoven E. CBP and p300: HATs for different occasions. Biochemical pharmacology. 2004;68(6):1145–55. [DOI] [PubMed] [Google Scholar]
- 35. Mikheev AM, Nabekura T, Kaddoumi A, Bammler TK, Govindarajan R, Hebert MF, et al. Profiling gene expression in human placentae of different gestational ages: an OPRU Network and UW SCOR Study. Reprod Sci. 2008;15(9):866–77. 10.1177/1933719108322425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Redman CW, Sargent IL, Staff AC. IFPA Senior Award Lecture: making sense of pre-eclampsia—two placental causes of preeclampsia? Placenta. 2014;35 Suppl:S20–5. 10.1016/j.placenta.2013.12.008 [DOI] [PubMed] [Google Scholar]
- 37. Hahn S, Rusterholz C, Hösli I, Lapaire O. Cell-free Nucleic Acids as Potential Markers for Preeclampsia. Placenta. 2011;32, Supplement 1(0):S17–S20. [DOI] [PubMed] [Google Scholar]
- 38. Myatt L, Redman CW, Staff AC, Hansson S, Wilson ML, Laivuori H, et al. Strategy for standardization of preeclampsia research study design. Hypertension. 2014;63(6):1293–301. 10.1161/HYPERTENSIONAHA.113.02664 [DOI] [PubMed] [Google Scholar]
- 39. Chigusa Y, Tatsumi K, Kondoh E, Fujita K, Nishimura F, Mogami H, et al. Decreased lectin-like oxidized LDL receptor 1 (LOX-1) and low Nrf2 activation in placenta are involved in preeclampsia. J Clin Endocrinol Metab. 2012;97(10):E1862–70. 10.1210/jc.2012-1268 [DOI] [PubMed] [Google Scholar]
- 40. Kweider N, Huppertz B, Kadyrov M, Rath W, Pufe T, Wruck CJ. A possible protective role of Nrf2 in preeclampsia. Annals of anatomy = Anatomischer Anzeiger: official organ of the Anatomische Gesellschaft. 2014;196(5):268–77. [DOI] [PubMed] [Google Scholar]
- 41. Henson MC, Castracane VD. Leptin in pregnancy: an update. Biology of reproduction. 2006;74(2):218–29. [DOI] [PubMed] [Google Scholar]
- 42. Andraweera PH, Dekker GA, Roberts CT. The vascular endothelial growth factor family in adverse pregnancy outcomes. Hum Reprod Update. 2012;18(4):436–57. 10.1093/humupd/dms011 [DOI] [PubMed] [Google Scholar]
- 43. Wang F, Marshall CB, Ikura M. Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cellular and molecular life sciences: CMLS. 2013;70(21):3989–4008. 10.1007/s00018-012-1254-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hennekam RC. Rubinstein-Taybi syndrome. European journal of human genetics: EJHG. 2006;14(9):981–5. [DOI] [PubMed] [Google Scholar]
- 45. Roelfsema JH, Peters DJ. Rubinstein-Taybi syndrome: clinical and molecular overview. Expert reviews in molecular medicine. 2007;9(23):1–16. [DOI] [PubMed] [Google Scholar]
- 46. Negri G, Milani D, Colapietro P, Forzano F, Della Monica M, Rusconi D, et al. Clinical and molecular characterization of Rubinstein-Taybi syndrome patients carrying distinct novel mutations of the EP300 gene. Clinical genetics. 2015;87(2):148–54. 10.1111/cge.12348 [DOI] [PubMed] [Google Scholar]
- 47. Tsai AC, Dossett CJ, Walton CS, Cramer AE, Eng PA, Nowakowska BA, et al. Exon deletions of the EP300 and CREBBP genes in two children with Rubinstein-Taybi syndrome detected by aCGH. European journal of human genetics: EJHG. 2011;19(1):43–9. 10.1038/ejhg.2010.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wieczorek D, Bartsch O, Lechno S, Kohlhase J, Peters DJ, Dauwerse H, et al. Two adults with Rubinstein-Taybi syndrome with mild mental retardation, glaucoma, normal growth and skull circumference, and camptodactyly of third fingers. American journal of medical genetics Part A. 2009;149a(12):2849–54. 10.1002/ajmg.a.33129 [DOI] [PubMed] [Google Scholar]
- 49. Foley P, Bunyan D, Stratton J, Dillon M, Lynch SA. Further case of Rubinstein-Taybi syndrome due to a deletion in EP300. American journal of medical genetics Part A. 2009;149a(5):997–1000. 10.1002/ajmg.a.32771 [DOI] [PubMed] [Google Scholar]
- 50. Parsley L, Bellus G, Handler M, Tsai AC. Identical twin sisters with Rubinstein-Taybi syndrome associated with Chiari malformations and syrinx. American journal of medical genetics Part A. 2011;155a(11):2766–70. 10.1002/ajmg.a.34227 [DOI] [PubMed] [Google Scholar]
- 51. Bartholdi D, Roelfsema JH, Papadia F, Breuning MH, Niedrist D, Hennekam RC, et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: delineation of the phenotype of the first patients carrying mutations in EP300. J Med Genet. 2007;44(5):327–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tal R. The role of hypoxia and hypoxia-inducible factor-1alpha in preeclampsia pathogenesis. Biology of reproduction. 2012;87(6):134 10.1095/biolreprod.112.102723 [DOI] [PubMed] [Google Scholar]
- 53. Carrero P, Okamoto K, Coumailleau P, O'Brien S, Tanaka H, Poellinger L. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1alpha. Molecular and cellular biology. 2000;20(1):402–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kallio PJ, Okamoto K, O'Brien S, Carrero P, Makino Y, Tanaka H, et al. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. The EMBO journal. 1998;17(22):6573–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science (New York, NY). 2002;295(5556):858–61. [DOI] [PubMed] [Google Scholar]
- 56. Arany Z, Huang LE, Eckner R, Bhattacharya S, Jiang C, Goldberg MA, et al. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci U S A. 1996;93(23):12969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Forest plots of the 388 differentially expressed genes in the preeclamptic placenta ordered on the absolute value of the standardized mean difference (see S1 Table). Squares are proportional to weights used in the meta-analysis, and the lines represent the 95% confidence interval. The diamond represents the standardized mean difference (log2 scale).
(PDF)
All 388 genes from the meta‐signature were used as input for STRING analysis and a network was built based on high confidence (0.8) evidence from experimental protein‐protein interaction (blue lines) and curated (purple lines) databases. Proteins are indicated by nodes labeled with the encoding gene symbol. Two genes (CEBPA and SEPP1) are not present in the used STRING version 9.1. The network is enriched in interactions (p = 0.007) using the intersection of 8,612 genes present on all platforms as background. Additional cluster analysis was used to color the nodes of the interaction networks. Yellow nodes represent proteins that have no protein‐protein interaction at this confidence setting.
(PDF)
(XLSX)
(XLSX)
(XLSX)
Indicated microarray samples were analyzed by the Gene Expression Barcode 3.0 and the resulting z-scores are shown. The z-score is the number of standard deviations above the mean expression signal of the unexpressed null distribution. An average z-score of ≥ 3 for a gene-specific probe has been labeled as being expressed in that trimester.
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.