Down-regulation of miR-24, miR-30b, and miR-142-3p during monocyte-to-macrophage and monocyte-to-dendritic cell differentiation potentiates innate immune responses.
Keywords: cytokine, TLR
Abstract
miRNAs are ubiquitous regulators of human biology. Parallel profiling of in vitro monocyte-to-Mφ and monocyte-to-DC differentiation revealed static, convergent, and divergent expression of miRNA. Bioinformatic and network analysis of differentially expressed miRNAs implicated miR-24, miR-30b, and miR-142-3p as negative regulators of intracellular signaling pathways, triggered not only by differentiation factors (M-CSF/GM-CSF/IL-4) but also from PRRs. Manipulation of miR-24, miR-30b, and miR-142-3p expression during the differentiation of mD-Mφ and mD-DC differentiation had minimal impact on the acquisition of phenotype but significantly abrogated the ability of these cells to mount inflammatory responses to pathogen-associated stimuli. Forced expression of these miRNAs, which are down-regulated during differentiation, inhibited release of inflammatory cytokines [TNF-α, IL-12(p40), IL-6] upon stimulation with LPS. Functional analysis revealed overlapping mechanisms of inhibition, including surface expression of TLR4/CD14/MD-1 and intracellular PKCα/NF-κB activation. Potential intermediary targets of the TLR4-NF-κB axis included members of the PI3K and MAPK families and PKC isoforms. These results demonstrate the requirement of miR-24, miR-30b, and miR-142-3p down-regulation for the generation of fully functional Mφs and DCs.
Introduction
miRNA-mediated regulation of gene expression has emerged as an important component of the cellular machinery that controls cell differentiation, function, and fate [1–4]. miRNAs are expressed in all cell types, including hematopoietic cells, where their expression is dynamically regulated during lineage commitment, for example, miR-181, whose expression promotes B cell populations [4]. Monocytes possess shorter lifespans (∼3 d) and limited functionality compared with their differentiated successors, primarily Mφ and DCs [5]. Differentiation can be regulated by numerous factors, such as growth factor/cytokine receptor interactions [5–12] or ligation of PRRs [13, 14], but there exists a central role for M-CSF in Mφ differentiation [9], and GM-CSF plus IL-4 in DC differentiation [6]. Of note, miRNAs have been reported to target cytokine receptor signaling pathways, as evidenced by down-regulation of M-CSFR expression by miR-22, miR-34a, and miR-155, which promote DC over Mφ differentiation [8].
In many ways the characteristics of monocyte-to-Mφ and monocyte-to-DC differentiation parallel each other, such as increased resistance to apoptosis [15, 16] and specializations in function, including enhanced uptake and killing of intracellular pathogens [17] and antigen presentation to lymphocytes [18], respectively. Monocytes, Mφ, and DCs also possess many of the same PRRs and produce many of the same inflammatory cytokines in response to pathogen; however, important differences do exist. TLR4 is highly expressed by all 3 cell types, whereas TLR2 is highly expressed on monocytes but down-regulated during Mφ and DC differentiation [19–21]. PRRs and their associated signaling components have emerged as a particularly rich group of miRNA targets. For example, miR-155 not only targets TLR4 but also MyD88, TGF-β-activated kinase 1/MAP3K7-binding protein 2, IκB kinase ε, SHIP1, suppressor of cytokine signaling 1, and TNF [22–26].
The canonical TLR4 signaling complex is comprised of TLR4/MD-2/CD14, and this signaling may be enhanced/altered by association with TLR2 via the RP105/MD-1 complex [27–31]. Recently, the recognition of LPS by the RP105/MD-1 complex has been identified as an important component of the LPS response [30]. Molecules, such as MD-1, MD-2, RP105, and Dectin-1, represent an important group of accessory molecules in innate immunity [29, 32]. Many of these are predicted targets of miRNA, such as miR-30b (implicated in MD-1 regulation) [33], but data linking miRNA expression to protein expression are currently lacking. Moreover, fewer miRNAs targeting differentiation-associated molecules in monocyte/Mφ/DCs have been identified. Whereas altered CDw93 (C1q-binding receptor) and CD68 expression has been shown under a number of conditions involving aberrant miRNA expression [34, 35], evidence of direct regulation is harder to come by; however, a few have been discovered, such as CD209 (DC-specific intercellular adhesion molecule-3-grabbing nonintegrin) regulation by miR-155 [36].
All 3 cell types produce large quantities of inflammatory cytokines in response to LPS stimulation, including TNF-α, IL-6, and IL-12, and while their production may be modulated by miRNA, in most cases, this is believed to occur indirectly [22]. LPS-induced up-regulation of antigen-presentation machinery is more prominent in DCs than Mφ, which display the highest level of MHC, CD80, and CD86 expression, as well as induced expression of CD83, a protein believed to be involved in cell–cell interaction during antigen presentation [37]. In this context, miRNAs of note include miR-148 with its ability to alter HLA-C expression [38]; however, the majority of work in this area has focused on direct miRNA regulation of lymphocytes as opposed to APC.
Here, we surveyed the miRNA profiles during differentiation of human primary monocytes to Mφ and DCs and demonstrate that down-regulation of miR-24, miR-30b, and miR-142-3p is required for generation of fully functional mD-Mφ and mD-DC.
MATERIALS AND METHODS
Primary human monocyte isolation and differentiation
Freshly prepared buffy coats were collected from healthy donors (n = 6; Sylvan N. Goldman, Oklahoma Blood Institute, Oklahoma City, OK, USA) and CD14+ monocytes, obtained by density gradient centrifugation and magnetic bead isolation. In brief, PBMCs were purified by use of Ficoll Paque (GE Healthcare, Piscataway, NJ, USA)-based density centrifugation. PBMCs were incubated with magnetically labeled CD14 beads (Miltenyi Biotec, Cologne, Germany), according to the manufacturer’s instructions. Monocyte purity and viability were >95%, as determined by flow cytometry (Supplemental Table 1). For mD-Mφ differentiation, monocytes were plated at a density of 2 × 106/ml in DMEM, supplemented with penicillin (100 U/ml), streptomycin (100 µg/ml), and gentamicin (50 µg/ml). After 2 h, the media were substituted with media containing 10% heat-inactivated FBS (Life Technologies, Grand Island, NY, USA) and rhM-CSF (50 ng/ml; PeproTech, Rocky Hill, NJ, USA). For mD-DC, monocytes were cultured in RPMI 1640, supplemented with rhGM-CSF (1000 U/ml) and rhIL-4 (500 U/ml; both from PeproTech). Media were replaced every 72 h. At day 7, cells were harvested and differentiation confirmed by flow cytometric analysis of CDw93, CD68, CD209, CD1a, CD11b, and CD11c expression.
miRNA profiling
Total RNA was isolated at 1, 4, and 12 h, and 1, 3, 5, and 7 d of differentiation by use of the miRNeasy micro kit (Qiagen, Germantown, MD, USA), following the manufacturer’s instructions. RNA integrity was assessed by use of the Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA) and 2100 Bioanalyzer (Agilent, Foster City, CA, USA). miRNA expression was performed by Exiqon Services (Vedbaek, Denmark) by use of seventh-generation microarrays (miRBase v.19). Total RNA (225 ng) was labeled by use of the miRCURY LNA microRNA Hi-Power Labeling Kit Hy3/Hy5 and subsequently hybridized onto miRCURY LNA microRNA arrays, following the procedures described by the manufacturer. Data normalization were performed by Exiqon by use of Quantile normalization. Initial analysis was performed by Exiqon by use of R/bioconductor, primarily by use of the limma package (Exiqon). Expression analysis of variance over time was performed with P values adjusted using the Benjamini-Hochberg method and identified genes subjected to the Tukey’s "honest significant difference" test. Array data were in compliance with Minimum Information About a Microarray Experiment guidelines and deposited in the Gene Expression Omnibus public database under Accession Number GSE60839.
Bioinformatic analysis
Bioinformatic analysis was performed on miRNAs, identified as significantly (FDR < 0.05) and differentially (fold change > 0.5) expressed during mD-Mφ or mD-DC differentiation. Of these, only miRNAs whose altered level of expression was maintained [>72 h from initial time point with significance (FDR > 0.05)] were selected for further analysis. We used miRWalk to predict the candidate 3′-untranslated region of genes for miRNA-binding sites with the 8 established miRNA target prediction algorithms [39]. miRNAs that possessed no predicted targets linked to differentiation/immunity/inflammation by Gene Ontology biologic terms (http://www.geneontology.org) in at least 5 of the 8 algorithms were not considered. The remaining miRNAs were then ranked according to the sum of predicted targets, with each predicted target being given a value of 1. Identification of the target by multiple algorithms resulted in a value equal to the number of predictive algorithms (i.e., if the same target was identified by 5 of the 8 algorithms, then it was given the value of 5). The top 10 ranked miRNAs were selected for further investigation and functional analysis.
Transient miRNA transfections
miScript miRNA mimics (miR-24, miR-30b, miR-101, miR-142-3p, miR-652-3p, miR-652-5p, miR-1275, miR-3656, miR-4279) and inhibitors were purchased from Qiagen. AllStars negative mimics (Qiagen) were used as controls. Transient transfections were performed by use of Lipofectamine 2000 (Life Technologies), according to the manufacturer’s instructions. Cells were transfected at day 3 or 7. Day 3-differentiating monocytes were transfected with mimics or inhibitors at a final concentration of 100 nM. Day 7-differentiated mD-Mφ and mD-DC were transfected at a final concentration of 50 nM and 100 nM, respectively. Red siGLO oligos (Thermo Fisher Scientific) were used to determine transfection efficiency (Supplemental Table 2).
Stimulation and activation
Cells were stimulated with Escherichia coli (0111B4) LPS (Invitrogen, Carlsbad, CA, USA) at a concentration of 50 ng/ml.
Flow cytometry
The expression of surface markers was analyzed by use of antibodies specific for human CD14, CDw93, CD16, CD68, CD1a, CD209, HLA-DR, CD80, CD83, CD86, TNF-α, TLR2, and TLR4 (all from BD PharMingen, San Jose, CA, USA). TLR1 and TLR3 antibodies were purchased from eBioscience (San Diego, CA, USA). The TLR6 antibody was purchased from BioLegend (San Diego, CA, USA). MD-1 and MD-2 antibodies were purchased from Thermo Fisher Scientific. NF-κB p65 and PKCα antibodies were purchased from BD PhosFlow (BD Biosciences, San Jose, CA, USA). Corresponding fluorescence label-conjugated isotype controls were used in all experiments. Cells were washed in ice-cold PBS, supplemented with 1% (v/v) FBS and 0.05% NaN3, and cell staining was performed on ice. Staining with MitoTracker, LysoTracker, Hoescht, and L/D dyes (all from Invitrogen) was performed in the absence of FBS or NaN3. Intracellular staining for CD68 and TNF-α was performed by use of Cytofix/Cytoperm and Perm/Wash buffer and PKCα and NF-κBp65 by use of Cytofix/perm and Phosflow Perm Buffer III (all from BD PharMingen). Nonspecific staining was blocked by incubating cells with FcR-blocking reagent (Miltenyi Biotec) before antibody addition. Samples were analyzed by use of a FACScan or BD Cyan flow cytometer by use of CellQuest software (BD Biosciences). Further analysis was performed by use of FloJo software (Tree Star, Ashland, OR, USA).
Viability assays
Cell viability was determined by use of the CellTiter 96 AQueous Cell Proliferation Assay Kit (Promega, Madison, WI, USA) and by flow cytometry by use of the LIVE/DEAD Fixable Violet Dead Cell staining kit (Life Technologies; Supplemental Table 3). In brief, cells (mD-Mφ, mD-DC, and monocytes) were plated at 2 × 106/ml in 96 or 24 well plates and transfected as described above and assays performed after 24 h, according to the manufacturer’s instructions.
ELISA
Human capture and detection antibodies (Life Technologies) were used for the development of TNF-α, IL-12p40, IL-6, and IL-10 sandwich ELISAs, according to the manufacturer’s instructions. Cell culture supernatants were analyzed by use of a 96 well plate reader with absorbance at 450 nm (reference 650 nm).
Statistical analysis
For miRNA expression, P values were calculated by ANOVA. P values were adjusted by use of the Benjamini-Hochberg method. In brief, P values were sorted and ranked (smallest value = 1, second smallest = 2, largest = N), and each P value was multiplied by N and then divided by its assigned rank to give the adjusted P values (FDR). miRNAs with FDR < 0.05 were considered significant. For flow cytometric, ELISA and MTS assay data, P values were calculated by ANOVA. Statistical analysis of data was performed by use of GraphPad Prism software (GraphPad Software, La Jolla, CA, USA). Results are presented as mean ± sd or ± sem from 3 or more biologic donors. Data from each individual donor are the mean of 2 technical replicates. P < 0.05 was considered statistically significant.
RESULTS
Identification of miRNAs differentially expressed during monocyte-to-Mφ and -DC differentiation
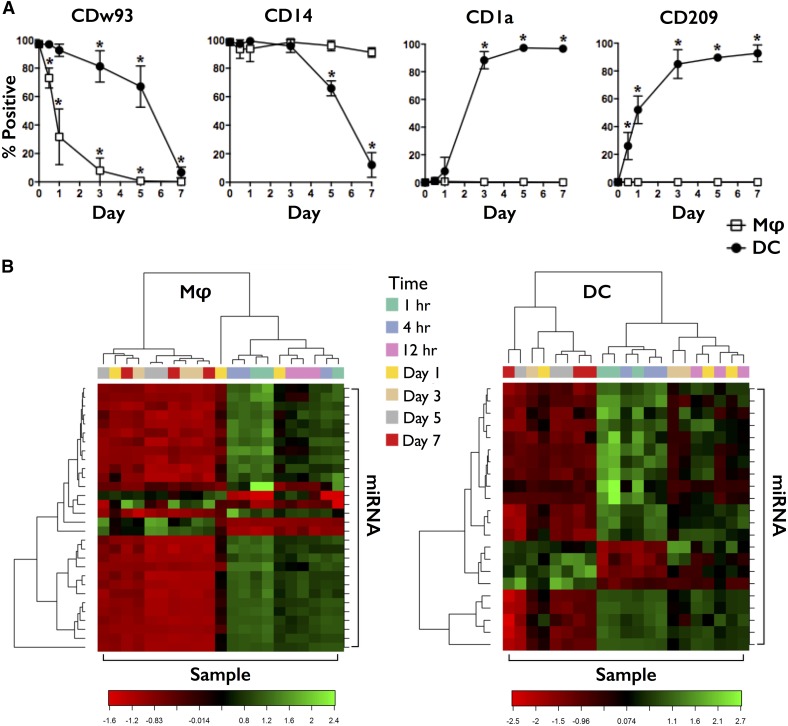
To identify miRNAs involved in the differentiation of monocytes to Mφ and DCs, we isolated peripheral blood CD14+ monocytes from normal, healthy volunteers, cultured them in media containing M-CSF or GM-CSF + IL-4, and isolated RNA at the following time points: 1, 4, and 12 h and 1, 3, 5, and 7 d. Differentiation was confirmed by microscopy and flow cytometry (Fig. 1A). Monocytes displaying a small, uniform, spherical morphology acquired the larger, heterogeneous, and adherent characteristics of Mφ or the intermediate, heterogeneous, nonadherent qualities of DCs during the differentiation process. Expression of differentiation markers also changed during this period. Monocytes were CD14+ CD16− HLA-DR+ (Supplemental Table 1) CDw93+ CD68+ CD1a− CD209− and differentiated into CDw93− CD68HI CD1a− CD209− CD14+ mD-Mφ or CDw93− CD68LO CD1a+ CD209+ CD14LO mD-DC. Further confirmation was provided by their response to LPS, with mD-DC displaying greater up-regulation of MHC (HLA-DR) and costimulatory molecule (CD86) expression, as well as exclusively up-regulating the marker of DC maturity/regulator of T cell activation, CD83 (Supplemental Table 4).
Figure 1. miRNA profiling of mD-Mφ and mD-DC.
(A) Human CD14+ monocytes were isolated and cultured in the presence of M-CSF or GM-CSF + IL-4 to promote differentiation into mD-Mφ (open square) and mD-DC (filled circle), respectively. Surface marker expression, including CDw93, CD14, CD1a, and CD209 during differentiation, was assessed by flow cytometry every 24 h up until day 7. The mean percentage of cells positive for marker expression is shown. Bars represent sd. *P < 0.05. (B) Heat map showing the scaled expression of genes and the relationship among the top 30 genes and samples in terms of genes found to be differentially expressed for the time factor relative to monocyte (0 h) expression. Green, Up-regulated; red, down-regulated.
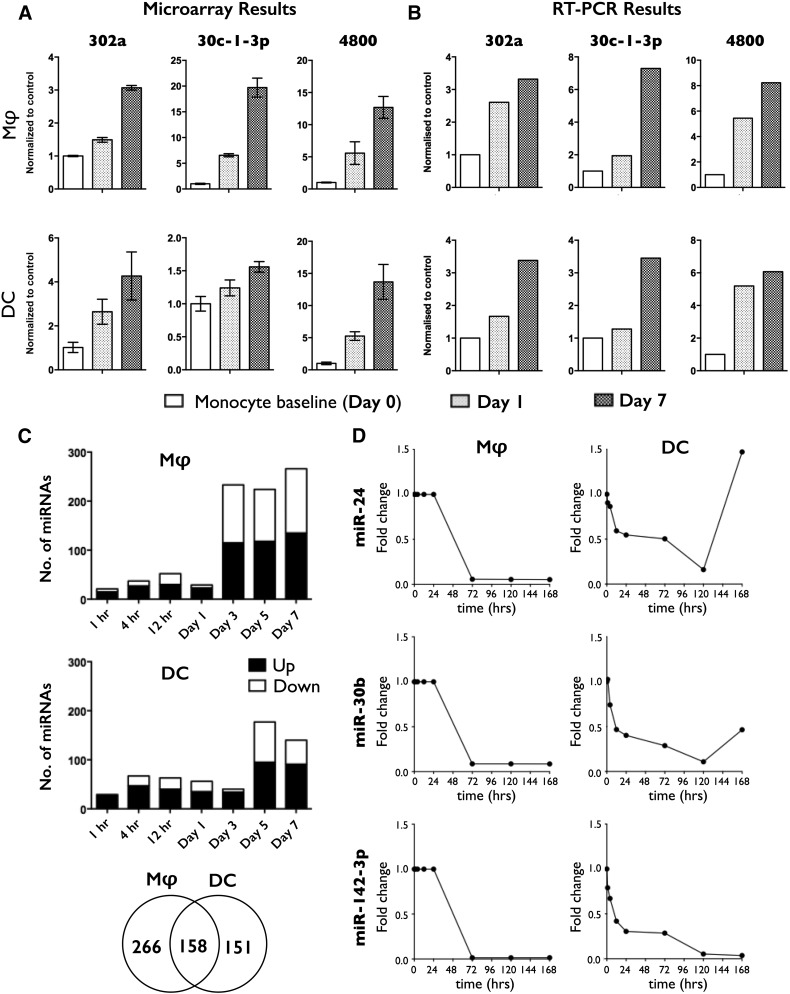
miRNA expression during the 7 d period of differentiation was monitored by use of miRNA microarrays. Data analysis revealed that time was the largest factor influencing the clustering of samples. Heat maps showing the scaled expression of genes and the relationship among the top 30 genes and samples, in terms of genes found to be differentially expressed for the time factor, are shown in Fig. 1B. miRNA array results (Fig. 2A) were validated by RT-PCR analysis of a second cohort of donors (Fig. 2B). Data analysis comparing monocyte to DC, monocyte to Mφ, and DC to Mφ revealed differentially expressed miRNAs that represented DC-specific, Mφ-specific, or common changes (Fig. 2C). Analysis of differential miRNA expression over time revealed a large number of up-regulated and down-regulated miRNAs, with the greatest number of changes occurring between days 1 and 3 for mD-Mφ (Fig. 2C, top), and days 3 and 5 for mD-DC (Fig. 2C, middle). A total of 424 and 309 miRNAs was differentially expressed during monocyte-to-Mφ and monocyte-to-DC differentiation, respectively, of which 158 were differentially expressed in both groups (Fig. 2C, bottom), albeit with differences in direction and/or magnitude. The top 20 differentially expressed miRNAs for mD-Mφ and mD-DC are presented in Supplemental Table 5. A complete list of the 158 miRNAs differentially expressed in mD-Mφ and mD-DC is presented in Supplemental Table 6. Expression of miR-24, miR-30b, and miR-142-3p was down-regulated between 24 and 72 h of Mφ differentiation and maintained until 168 h (Fig. 2D, left). By comparison, expression of miR-24, miR-30b, and miR-142-3p was down-regulated within the first 24 h of DC differentiation, continued to fall until 120 h, and was followed by up-regulation (miR-24), partial restoration (miR-30b), or maintenance (miR-142-3p; Fig. 2D, right).
Figure 2. miRNA expression during monocyte-to-Mφ and -DC differentiation.
(A) miRNA expression during monocyte-to-Mφ and monocyte-to-DC was profiled by microarray analysis. (B) Validation of microarray results by quantitative RT-PCR by use of a second cohort (n = 2) of donors. Select miRNAs, identified as being differentially expressed by microarray, are shown. (C) Number of up-regulated (filled sections of bars) and down-regulated (open sections of bars) miRNAs over time. Venn diagram displays the total number of differentially expressed miRNAs divided into mD-Mφ specific (left segment), mD-DC specific (right segment), and common (middle segment). (D) Expression of miR-24, miR-30b, and miR-142-3p by mD-Mφ (left) and mD-DC (right) during differentiation. LogFC(logarithmic fold change) > 1.5, P < 0.05 was considered to constitute significant, differential expression. Data represent 4 biologic donors.
Overexpression of miR-24, miR-30b, and miR-142-3p during mD-Mφ and mD-DC differentiation
In silico analysis of predicted miRNA-mRNA interactions was performed for miRNAs differentially expressed during monocyte-to-Mφ and monocyte-to-DC differentiation, from which candidate miRNAs were selected. Bioinformatic analysis of predicted miRNA-mRNA interactions was focused on genes with confirmed roles in cell signaling, innate immunity, and myeloid cell differentiation and function (Supplemental Table 7). miRNAs were ranked according to number of hits and number of algorithms in agreement. (miRWalk, miRDB, miRanda, TargetScan, and RNA22 were used.) miRNAs that were differentially expressed in Mφ and DC, such as miR-24, miR-30b, and miR-142-3p, were given greater consideration during the selection process for functional evaluation. Based on this criteria, miR-24, miR-30b, miR-142-3p, miR-652-3p, miR-652-5p, miR-1275, miR-3656, and miR-4279 were chosen for functional analysis. Monocytes were differentiated to mD-Mφ and mD-DC and transfected on day 3 with control mimic or miRNA mimics/inhibitors. On day 7, the expression of differentiation markers was analyzed by flow cytometry. Overexpression/knockdown of these miRNAs did not affect the normal increase in mitochondrial and lysosomal content nor did it alter autofluorescence or cell viability (Supplemental Table 8). No significant disturbance in the expression of CD45, CD1a, or CD209 (Table 1) nor CD68, CD11b, or CD11c (Supplemental Table 9) was observed in mD-Mφ or mD-DC; however, overexpression of miR-24, miR-30b, and miR-142-3p abrogated the loss of CDw93 in both cell types (Table 1). No discernible differences in cell morphology were observed. Corresponding inhibitors, control mimic, and overexpression/knockdown of miR-101-3p, miR-652-3p, miR-652-5p, miR-1275, miR-3656, and miR-4279 had no significant effect on levels of these markers (Table 1).
TABLE 1.
Overexpression of miR-24, miR-30b, and miR-142-3p during differentiation maintains CDw93 expression
| Culture | CDw93 (% +) |
CD45 (% +) |
CD1a (% +) |
CD209 (% +) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mφ | DC | Mφ | DC | Mφ | DC | Mφ | DC | |||||||||
| Untreated | 4.6 | ns | 3.7 | ns | 92.0 | ns | 96.0 | ns | 0.0 | ns | 94.1 | ns | 0.0 | ns | 97.3 | ns |
| Control mimic | 5.0 | ns | 5.8 | ns | 96.2 | ns | 98.3 | ns | 0.0 | ns | 96.3 | ns | 0.0 | ns | 92.5 | ns |
| miR-24 mimic | 29.2 | * | 20.8 | * | 92.9 | ns | 97.4 | ns | 0.0 | ns | 91.6 | ns | 0.0 | ns | 91.8 | ns |
| miR-24 inhibitor | 4.8 | ns | 4.3 | ns | 97.8 | ns | 98.0 | ns | 0.0 | ns | 93.3 | ns | 0.0 | ns | 95.5 | ns |
| miR-30b mimic | 30.5 | ** | 20.5 | * | 93.7 | ns | 99.3 | ns | 0.0 | ns | 91.0 | ns | 0.0 | ns | 95.0 | ns |
| miR-30b inhibitor | 5.5 | ns | 7.3 | ns | 95.7 | ns | 97.0 | ns | 0.0 | ns | 93.8 | ns | 0.0 | ns | 93.5 | ns |
| miR-101-3p mimic | 1.8 | ns | 7.9 | ns | 96.3 | ns | 94.4 | ns | 0.0 | ns | 88.7 | ns | 0.0 | ns | 96.2 | ns |
| miR-101-3p inhibitor | 6.0 | ns | 3.8 | ns | 99.0 | ns | 97.2 | ns | 0.0 | ns | 94.5 | ns | 0.0 | ns | 94.5 | ns |
| miR-142-3p mimic | 30.9 | * | 21.0 | * | 96.9 | ns | 98.7 | ns | 0.0 | ns | 90.5 | ns | 0.0 | ns | 93.0 | ns |
| miR-142-3p inhibitor | 4.8 | ns | 6.5 | ns | 95.4 | ns | 95.0 | ns | 0.0 | ns | 92.0 | ns | 0.0 | ns | 95.0 | ns |
| miR-652-3p mimic | 4.9 | ns | 4.1 | ns | 97.0 | ns | 96.5 | ns | 0.0 | ns | 93.1 | ns | 0.0 | ns | 94.9 | ns |
| miR-652-3p inhibitor | 8.7 | ns | 3.7 | ns | 95.6 | ns | 95.6 | ns | 0.0 | ns | 92.7 | ns | 0.0 | ns | 94.8 | ns |
| miR-652-5p mimic | 1.8 | ns | 8.6 | ns | 93.9 | ns | 93.1 | ns | 0.0 | ns | 84.5 | ns | 0.0 | ns | 91.3 | ns |
| miR-652-5p inhibitor | 2.5 | ns | 8.4 | ns | 92.2 | ns | 97.8 | ns | 0.0 | ns | 85.7 | ns | 0.0 | ns | 98.4 | ns |
| miR-1275 mimic | 8.1 | ns | 5.5 | ns | 91.8 | ns | 96.6 | ns | 0.0 | ns | 91.2 | ns | 0.0 | ns | 96.0 | ns |
| miR-1275 inhibitor | 6.8 | ns | 3.9 | ns | 96.8 | ns | 93.6 | ns | 0.0 | ns | 85.9 | ns | 0.0 | ns | 91.8 | ns |
| miR-3656 mimic | 6.8 | ns | 5.9 | ns | 92.5 | ns | 94.3 | ns | 0.0 | ns | 91.4 | ns | 0.0 | ns | 91.0 | ns |
| miR-3656 inhibitor | 6.9 | ns | 1.6 | ns | 97.7 | ns | 92.9 | ns | 0.0 | ns | 95.2 | ns | 0.0 | ns | 94.0 | ns |
| miR-4279 mimic | 5.3 | ns | 1.7 | ns | 99 | ns | 95.5 | ns | 0 | ns | 96.8 | ns | 0 | ns | 95.1 | ns |
| miR-4279 inhibitor | 5.2 | ns | 2.4 | ns | 96.0 | ns | 96.2 | ns | 0.0 | ns | 92.5 | ns | 0.0 | ns | 96.1 | ns |
mD-Mφ and mD-DC cultures were transfected on day 3 and analyzed by flow cytometry for CDw93, CD45, CD1a, and CD209 expression on day 7. Data are presented as means from 5 biologic donors.
P < 0.05 is considered significant; **P < 0.005; ns, not significant.
Overexpression of miR-24, miR-30b, and miR-142-3p impairs the inflammatory cytokine response
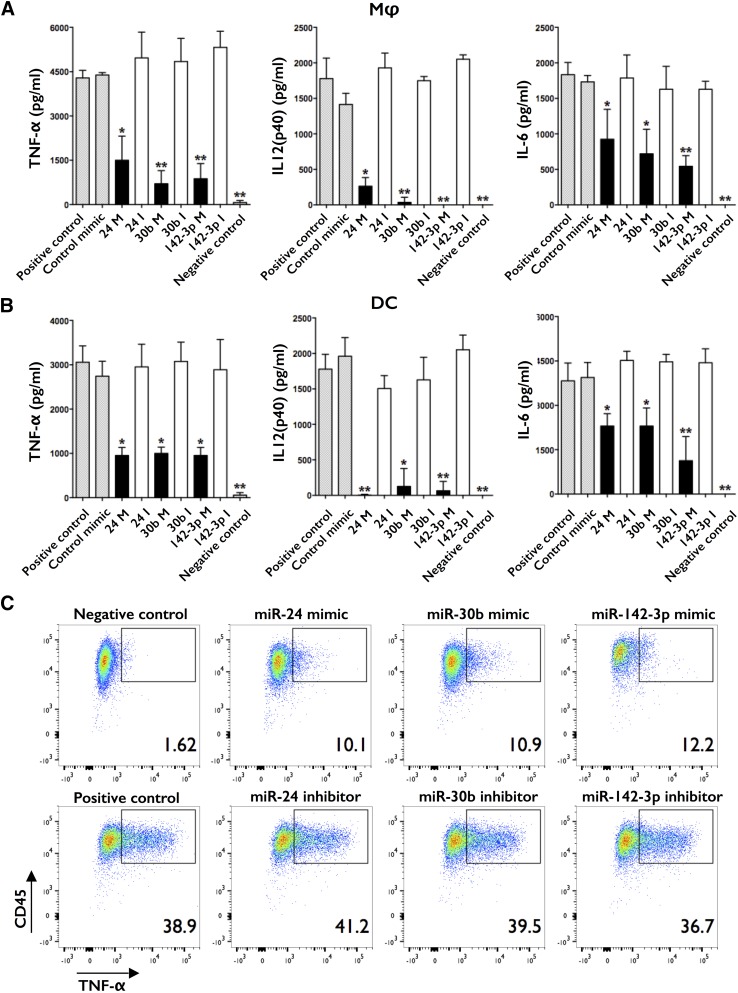
To further explore the effects of miR-24, miR-30b, and miR-142-3p, monocytes were differentiated to Mφ and DC and transfected on day 3 with control mimic or their mimics/inhibitors. Cells were stimulated with LPS on day 7 and supernatant levels of TNF-α, IL-6, IL-12(p40), and IL-10 assayed by ELISA and intracellular levels of TNF-α assayed by flow cytometry after 18 h. Significantly lower levels of TNF-α, IL-6, and IL-12(p40) were noted in supernatants of mD-Mφ and mD-DC transfected with miR-24, miR-30b, or miR-142-3p mimics (Fig. 3A and B), whereas their inhibitors had no significant effect compared with positive control (untransfected cells) or control mimic. No significant differences in IL-10 production were detected at this time point (data not shown). Interestingly, there was a trend for higher supernatant levels of TNF-α and IL-12(p40) in mD-Mφ and IL-6 in mD-DC and lower levels of IL-12(p40) in mD-DC in the presence of inhibitors, although these did not reach statistical significance (P > 0.05). Intracellular staining for TNF-α corroborated with our ELISA data, as evident from decreased intracellular levels of TNF-α in mimic-transfected cells compared with inhibitor-transfected cells (Fig. 3C; mD-DC data shown).
Figure 3. Overexpression of miR-24, miR-30b, and miR-142-3p impairs the inflammatory cytokine response.
(A) mD-Mφ and (B) mD-DC were transfected on day 3 with mimics (M)/inhibitors (I) or control mimic, stimulated with LPS on day 7 and supernatant levels of TNF-α, IL-12(p40), and IL-6 assayed by ELISA after 18 h. (C) Intracellular levels of TNF-α in mD-DC assayed by flow cytometry after 4 h of LPS stimulation. No significant differences in viability, as assayed by MTS assay and LIVE/DEAD Fixable Violet Dead Cell-positive staining kit, respectively, were observed. Data are presented as means and sd or representative images from 5 biologic donors. *P < 0.05 is considered significant; **P < 0.005.
miR-30b and miR-142-3p regulate TLR4 expression
mD-Mφ and mD-DC were transfected on day 3 and expression of pathogen recognition/response molecules assayed on day 7 by flow cytometry. Overexpression of miR-30b or miR-142-3p but not miR-24 mimics resulted in a significant decrease in TLR4 expression in mD-Mφ and mD-DC (Table 2). Next, transfected mD-Mφ and mD-DC were challenged with LPS on day 7 and TLR4 levels assayed on day 8. In mD-Mφ, LPS-induced down-regulation of TLR4 was enhanced significantly by transfection with miR-30b and miR-142-3p mimics but ameliorated by their corresponding inhibitors (Table 2). In mD-DC, transfection with miR-30b and miR-142-3p inhibitors ameliorated the LPS-induced decrease in TLR4. The corresponding mimics had no significant effect.
TABLE 2.
miR-24, miR-30b, and miR-142-3p regulate components of the TLR signaling complex
| Culture | TLR4 (% +) |
MD-1 (% +) |
TLR4 (% +) + LPS |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mφ | DC | Mφ | DC | Mφ | DC | |||||||
| Untreated | 97.5 | ns | 57.5 | ns | 90.9 | ns | 86.6 | ns | 38.9 | ns | 22.6 | ns |
| Control mimic | 96.7 | ns | 59.3 | ns | 92.0 | ns | 88.4 | ns | 29.3 | ns | 19.4 | ns |
| miR-24 mimic | 84.0 | ns | 57.6 | ns | 71.0 | ns | 77.9 | ns | 35.5 | ns | 27.4 | ns |
| miR-24 inhibitor | 83.4 | ns | 64.8 | ns | 82.9 | ns | 94.5 | ns | 40.3 | ns | 29.0 | ns |
| miR-30b mimic | 64.9 | * | 42.2 | * | 47.5 | * | 70.9 | * | 22.1 | * | 18.1 | ns |
| miR-30b inhibitor | 94.9 | ns | 69.2 | ns | 75.4 | ns | 90.9 | ns | 88.0 | * | 42.2 | * |
| miR-142-3p mimic | 63.8 | * | 38.0 | * | 28.3 | ** | 65.3 | * | 18.2 | ** | 28.5 | ns |
| miR-142-3p inhibitor | 93.4 | ns | 62.4 | ns | 79.4 | ns | 95.3 | ns | 91.0 | * | 55.3 | * |
mD-Mφ and mD-DC cultures were transfected on day 3 and analyzed by flow cytometry for TLR4 and MD-1 expression on day 7 or 8, with LPS stimulation on day 7. Data are presented as means from 5 biologic donors.
P < 0.05 is considered significant; **P < 0.005.
miR-30b and miR-142-3p regulate MD-1 expression
Cells were transfected at day 3 and levels of MD-1 and MD-2 assayed at day 7. Transfection with miR-30b and miR-142-3p but not miR-24 mimics decreased MD-1 expression during the differentiation of mD-DC and mD-Mφ (Table 2). This effect was most pronounced in cells transfected with miR-142-3p. No alteration in MD-2 expression was observed in mD-DC or mD-Mφ (data not shown).
miR-24, miR-30b, and miR-142-3p regulate CD14 expression
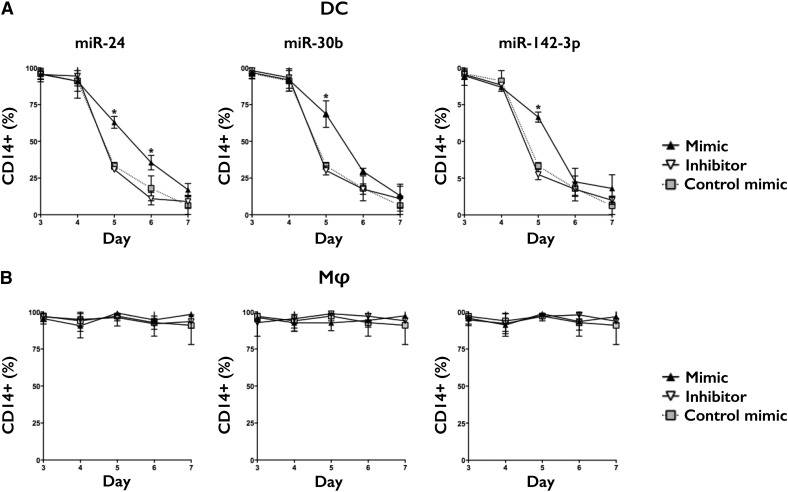
mD-Mφ and mD-DC were transfected on day 3 and expression of CD14 followed up to day 7 (Fig. 4). In mD-DC, overexpression of miR-24, miR-30b, and miR-142-3p mimics but not inhibitors significantly impeded the normal decrease in CD14 expression observed on days 5 and 6, although levels were comparable with control by day 7. In mD-Mφ, transfection with mimics or inhibitors had no significant effect on CD14 expression.
Figure 4. miR-24, miR-30b, and miR-142-3p regulate CD14 expression during DC differentiation.
(A) mD-DC and (B) mD-Mφ were transfected on day 3 and CD14 expression assessed by flow cytometry over 7 d of differentiation. Data are presented as means and sd from 5 biologic donors. *P < 0.05 is considered significant.
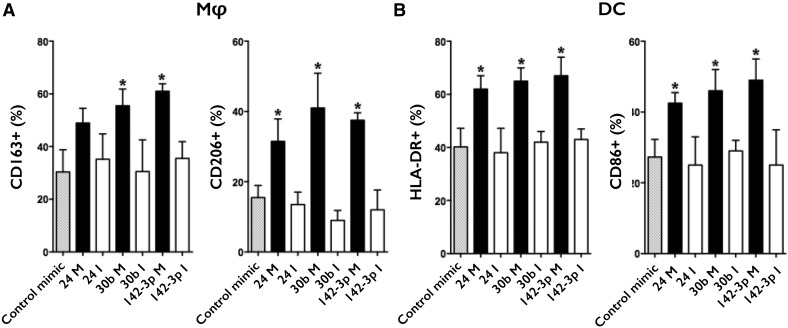
miR-24, miR-30b, and miR-142-3p regulate CD163, CD206, HLA-DR, and CD86 expression
In mD-Mφ, transfection with miR-30b and miR-142-3p mimics resulted in significantly higher expression of CD163 (Fig. 5A). Although there was a trend for elevated levels of CD163 in cells transfected with miR-24, this did not reach statistical significance. All 3 miRNA mimics maintained elevated levels of CD206 in mD-Mφ. No disturbance in CD163 or CD206 expression was detected in mD-DC (Supplemental Table 10A). In mD-DC, overexpression of miR-24, miR-30b, and miR-142-3p mimics increased HLA-DR and CD86 expression (Fig. 5B). No disturbance in HLA-DR or CD86 expression was observed in mD-Mφ (Supplemental Table 10B).
Figure 5. miR-24, miR-30b, and miR-142-3p regulate CD163 and CD206 expression in Mφ and HLA-DR and CD86 expression in DC.
(A) mD-Mφ transfected on day 3 and analyzed for CD163 and CD206 expression on day 7 by flow cytometry. (B) mD-DC transfected on day 3 and analyzed for HLA-DR and CD86 expression on day 7 by flow cytometry. Data are presented as means and sd from 5 biologic donors. *P < 0.05 is considered significant.
miR-24, miR-30b, and miR-142-3p target components of M-CSF/GM-CSF receptor signaling, including PKCα
PKC activation is a component of DC and Mφ differentiation and also PRR-mediated activation [40]. We assayed the effect of altered miR-24, miR-30b, and miR-142-3p expression on PKCα activation during differentiation. In mD-Mφ, overexpression of miR-24, miR-30b, and miR-142-3p mimics but not inhibitors decreased PKCα activation relative to controls. In mD-DC, miR-30b, and miR-142-3p but not miR-24 decreased PKCα activation relative to controls (Table 3).
TABLE 3.
miR-24, miR-30b, and miR-142-3p target PKCα and NF-κB activation
| Culture | PKCα (% +) |
NF-κBp65 (% +) |
||||||
|---|---|---|---|---|---|---|---|---|
| Mφ | DC | Mφ | DC | |||||
| Untreated | 89.1 | ns | 80.0 | ns | 90.9 | ns | 91.2 | ns |
| Control mimic | 87.3 | ns | 85.8 | ns | 92.1 | ns | 90.9 | ns |
| miR-24 mimic | 63.3 | * | 81.4 | ns | 63.7 | * | 48.6 | * |
| miR-24 inhibitor | 86.5 | ns | 83.3 | ns | 93.5 | ns | 92.9 | ns |
| miR-30b mimic | 60.8 | * | 49.2 | * | 64.0 | * | 29.0 | * |
| miR-30b inhibitor | 89.0 | ns | 83.7 | ns | 91.8 | ns | 88.6 | ns |
| miR-142-3p mimic | 56.3 | * | 33.2 | * | 31.5 | ** | 4.4 | * |
| miR-142-3p inhibitor | 84.2 | ns | 87.5 | ns | 90.7 | ns | 93.2 | ns |
mD-Mφ and mD-DC cultures were transfected on day 3 and PKCα activation analyzed by flow cytometry on day 5. Data are presented as means and sd from 3 biologic donors. mD-Mφ and mD-DC cultures were transfected on day 3, stimulated with LPS on day 7, and analyzed for NF-κBp65 expression after 4 h by flow cytometry. Data are presented as means from 5 biologic donors.
P < 0.05 is considered significant; **P < 0.005.
miR-24, miR-30b, and miR-142-3p target multiple components of PRR signaling to inhibit NF-κB activation
Previously, we reported that transfection of mD-Mφ and mD-DC with miR-24, miR-30b, and miR-142-3p mimics or inhibitors altered mRNA levels of numerous genes, including PKCα, CD163, and CD206 in mD-Mφ and PKCα, CD14, TLR4, and MD-1 in mD-DC [41], which mostly correlated with protein levels observed in this study. In these data, we also observed decreased gene expression of multiple signaling components involved in the processes of Mφ and DC differentiation and innate function, including PI3K, MAPK, PKB, and PKC isoforms. To investigate the possibility that miR-24-, miR-30b-, and miR-142-3p-mediated inhibition of the LPS response converged at the point of NF-κB, we transfected mD-Mφ and mD-DC at day 3, stimulated with LPS on day 7, and NF-κBp65 activation assayed after 4 h. We noted that overexpression of miR-24, miR-30b, and miR-142-3p mimics decreased NF-κBp65 activation in mD-Mφ and mD-DC (Table 3). Similar to effects on MD-1, transfection with miR-142-3p had the most profound effects on NF-κBp65 levels. Their corresponding inhibitors had no significant effect.
DISCUSSION
Our miRNA-profiling data of in vitro monocyte-to-Mφ and monocyte-to-DC differentiation identified numerous miRNAs that were differentially expressed over the course of 7 d. Notably, the largest change was seen between days 1 and 3 during mD-Mφ differentiation and days 3 and 5 during mD-DC differentiation. A core set of miRNAs (158) was differentially expressed in both groups, suggesting that they regulate functions that are common to mD-Mφ and mD-DC function (Supplemental Table 6). This list includes many miRNAs with known roles in innate immunity, such as let-7 family members (let-7a-5p, let-7c, let-7d-5p, let-7f-5p, let-7g-5p, let-7i-5p), miR-155 (with 140 confirmed gene targets with roles in myelopoiesis and leukemogenesis, inflammation, and innate immunity) [42], and miR-146a (one of the first miRNAs whose expression was identified as NF-κB dependent) [43]. At first glance, the high number (>50%) of miRNAs that was found to be differentially expressed during monocyte-to-Mφ and monocyte-to-DC differentiation would infer an equal (or larger) number of genes whose expression directs monocyte differentiation independently of Mφ or DC specialization; however, further analysis of the characteristics of these miRNAs on the basis of cell type (e.g., direction and magnitude of regulation, time of induction, and kinetics) has revealed differences that may allow for specific or bipolar regulation of Mφ and DC by manipulation of a single miRNA. The latter scenario would be of particular interest for miRNA-based therapy of inflammatory disorders, wherein the goal would entail inhibition of the classic activation of Mφ, while maintaining APC functionality of DC.
Expression of miR-24, miR-30b, and miR-142-3p was down-regulated significantly during mD-Mφ differentiation and miR-30b and miR-142-3p during mD-DC differentiation. Forced expression of these miRNAs had no effect on the general morphology or cytochemistry of the cells, as indicated by mitochondrial, lysosomal, and nuclear staining, and gain in autofluorescence (representative of combined size, granularity, and organelle composition/complexity). Of the differentiation markers analyzed, only CDw93, found on monocytes but not Mφ or DC, was altered by disturbance of miR-24, miR-30b, and miR-142-3p expression. This and other alterations were not a result of a global disturbance of expression, as multiple markers, including CD45, CD45RA, CD68, CD11b, CD11c, CD1a, and CD209, remained at control levels. We noted that miR-30b, miR-101, and miR-652-3p expression was down-regulated, whereas miR-652-5p, miR-3656, and miR-4279 were up-regulated in both groups. The alteration of the expression of these miRNAs during differentiation did not significantly affect the phenotype or function under the parameters tested; however, we do not exclude the possibility, or indeed likelihood, that these miRNAs are actively involved in other aspects of myeloid cell biology.
A central, perhaps defining, aspect of monocyte-to-Mφ or -DC differentiation is the acquisition of function. Therefore, we analyzed the effects of altered miR-24, miR-30b, and miR-142-3p expression on the ability of these cells to recognize and respond to innate stimuli. We found that overexpression of miR-30b and miR-142-3p during differentiation resulted in decreased expression of TLR4 and MD-1 (Table 2), and in mD-DC, this was accompanied by greater retention of CD14 expression (Fig. 4). To test the functional significance of these findings, we stimulated cells transfected with miR-24, miR-30b, and miR-142-3p mimics with LPS and observed significant inhibition of inflammatory cytokines TNF-α, IL-12 (p40), and IL-6, whose physiologic roles include (but are not limited to) mediation of inflammation, cell activation, and Th1/17 differentiation. No differences in IL-10 production were detected at this time point, a finding that is mirrored in similar experiments by use of inactivated E. coli as the stimulus [41]. These data imply that miR-24/30b/142-3p down-regulation is not required for phenotype acquisition but is necessary for acquisition of function. This indicates that down-regulation of these miRNAs is necessary for differentiation of fully functional Mφ and DC.
We observed reciprocal regulation for TLR4 and MD-1 expression, whereby mimics decreased expression during differentiation, and inhibitors prevented down-regulation following LPS stimulation. TLR4 and MD-1 are present on the cell surface in complexes comprising TLR4/MD-2/CD14 and MD-1/RP105, with both complexes recognizing LPS to trigger signaling pathways that include MyD88, PI3K, MAPK, PKC, PKB, and NF-κB. LPS-induced proinflammatory cytokine production was inhibited by transfection with miR-24, miR-30b, and miR-142-3p mimics in mD-Mφ and mD-DC, suggesting that a common mode of action was at play. TLR4 and MD-1 expression was decreased in mD-DC and mD-Mφ by miR-30b and miR-142-3p mimics.
Bioinformatic analysis of predicted mRNA targets for miR-24, miR-30b, and miR-142-3p identified several signaling molecules of relevance, including members of the PI3K and MAPK families, and validation of these targets would help to elucidate shared/exclusive facets of the miRNA-mediated inhibition. These targets exist upstream and downstream of PKCα, a molecule that was down-regulated by all 3 mimics in mD-Mφ and miR-30b and miR-142-3p mimics in mD-DC. Whereas miR-24 mimic did not reduce PKCα expression in mD-DC, decreased NF-κB activation was observed by all 3 mimics in both cell types. This observation, in the context of our related findings, suggests that miR-24, miR-30b, and miR-142-3p target multiple components of a common signaling network, with each miRNA possessing a distinctive pattern of targets. Importantly, these miRNAs demonstrate a focused inhibition of the innate response to pathogen-associated stimuli by the gatekeepers of immunity: Mφ and DC. Based on our current understanding, we hypothesize that down-regulation of miR-24, miR-30b, and miR-142-3p during Mφ and DC differentiation is part of a preparatory program, in which via increased expression of pathogen-recognition molecules, cells are sensitized to innate stimuli. Furthermore, this may render the cell permissive for increased miR-24, miR-30b, and miR-142-3p expression upon stimulation, with the down-regulation of their targets a potential mechanism of negative feedback to prevent excessive and prolonged inflammatory activation.
During normal monocyte-to-Mφ and monocyte-to-DC differentiation, PKCα expression increases, and we have found this increase to be impeded by forced expression of miR-24, miR-30b, and miR-142-3p. MD-1, in complex with RP105, can also transmit powerful survival signals [30], so the observation that down-regulation of miR-30b and miR-142-3p during differentiation is required for MD-1 up-regulation further points to a role for these miRNAs in blocking monocyte-to-Mφ/DC transition. These observations, combined with the lack of CDw93 (also known as C1qR) down-regulation in the presence of mimics, are consistent with the idea that these miRNAs function as "handbrakes," and loss of expression is required to gain traction on the road toward Mφ/DC differentiation. In mD-DC, we noted that altered expression of these miRNAs impacted the expression of the antigen presentation and costimulatory molecules HLA-DR and CD86. These alterations are likely to have knock-on effects on lymphocyte proliferation and differentiation and the effective initiation of adaptive immune responses.
CD163 and CD206, also known as the scavenger and mannose receptors, are important mediators (and markers) of Mφ function, and their altered expression would be predicted to impact the generation or effectiveness of alternatively activated Mφ [44–46]. CDw93 binds the first component, the complement pathway (C1), as well as mannose-binding lectin and pulmonary surfactant protein A [47, 48]. Through these interactions, CDw93 expression may enhance phagocytosis, and its modulation by miR-24, miR-30b, and miR-142-3p would be predicted to decrease phagocytic capacity. Furthermore, it has not escaped our notice that FcR expression can impact differentiation via alterations in PKC isoform expression [12, 49], and this may represent an additional level of miR-24-, miR-30b-, and miR-142-3p-mediated alteration in Mφ/DC development.
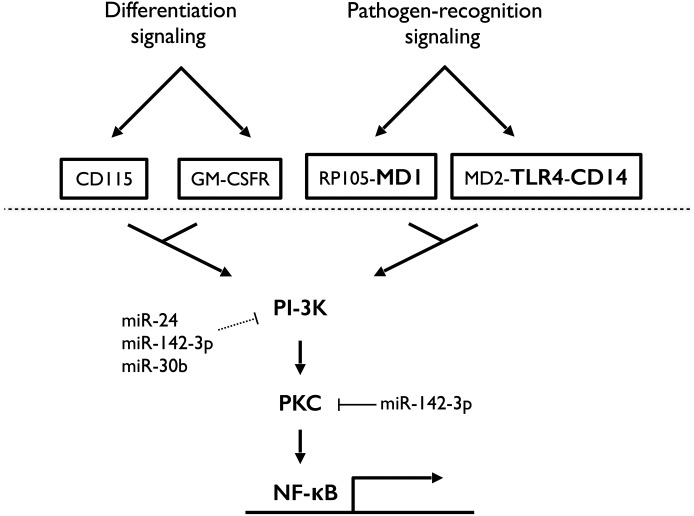
We have identified 2 mechanistic aspects of these miRNAs, 1 during differentiation and 1 during activation, which may explain the observed phenotype. The binding of M-CSF and GM-CSF to their receptors on monocytes induces sequential activation of, amongst others, PI3K, PKC isoforms, and NF-κB, resulting in increased survival and differentiation [13, 40, 49–53]. We have shown previously that overexpression of miR-24, miR-30b, and miR-142-3p alters the expression of several of these genes [41], and here, we show decreased PKCα expression at the level of protein. Up-regulation of PKCα during Mφ and DC differentiation is an important contributor to their increased resistance to apoptosis, but PKC expression has also been shown to potentiate TLR signaling and mediate NF-κB activation in monocytes, Mφ and DC [54]. The convergence of intracellular signaling between differentiation factors and PRRs is highlighted in Fig. 6. Furthermore, the use of PKC inhibitors blocks LPS-induced cytokine secretion [55, 56]. These facts are consistent with our observed results and support our finding of miR-30b and miR-142-3p to be regulators of PKC expression.
Figure 6. Schematic diagram highlighting the role of miR-24, miR-30b, and miR-142-3p on convergent signaling triggered by differentiation factors (M-CSF and GM-CSF) and pathogen-recognition (LPS).
Unbroken lines represent confirmed target interactions. Broken lines represented putative or indirect target interactions.
No disturbance in NF-κB activation was observed when miR-24, miR-30b, and miR-142-3p expression was altered during differentiation; however, reduced NF-κB activation in response to LPS was observed. Overexpression of these miRNAs resulted in decreased activation of NF-κB, as indicated by reduced detection of the (phosphorylated) NF-κB p65 subunit (availability is inversely correlated to the rate of IκB/NF-κB complex formation). In mD-DC, this may be partially explained by reduced TLR4 expression; however, reduced NF-κB activation was also observed in mD-Mφ, where TLR4 expression was only moderately decreased. Overexpression of miR-30b and miR-142-3p resulted in lower MD-1 expression, a molecule that associates with RP105 to enhance TLR4/MD-2-mediated signaling [27], and this mechanism may contribute to the decreased LPS responsiveness that we observed in mD-Mφ and mD-DC. MD-1 expression was modulated by these miRNAs in mD-Mφ and mD-DC. MD-1 associates with RP105 to stabilize its expression and contributes to TLR4-dependent signaling and survival [30–32, 57]. RP105/MD-1 also mediates TLR4/MD-2-independent, inflammatory responses via NF-κB activation and another, currently unknown, signaling pathway. It has been suggested that the TLR4/MD-2-independent ligand for RP105/MD-1 is a fatty acid released during adipose tissue inflammation [58].
When taken together, these data support a model in which miR-24, miR-30b, and miR-142-3p down-regulation promotes innate immunity via the acquisition of enhanced and specialized functions of Mφ and DC over monocytes. These data also highlight the significance of timing in cell-signaling pathways, whereby activation of a pathway with 1 set of ligands (i.e., differentiation factors) confers phenotype acquisition and also primes the same pathway for subsequent activation with a different set of ligands (i.e., PRRs). In this sense, miRNA provides short-term immunological memory to enhance the efficiency and variety of cellular responses in a way that incorporates information from prior signaling events into current signaling events. The commonalities in M-CSF/GM-CSF- and TLR4/CD14/MD-1-mediated signaling and miR-24, miR-30b, and miR-142-3p targeting are presented in schematic form in Fig. 6. Simultaneously, the presence of miRNAs, differentially expressed in monocyte-to-Mφ and monocyte-to-DC differentiation, and miRNAs that are unchanged, that is, present in monocytes, mD-Mφ and mD-DC, highlight the similarity of these cells from a miRNA perspective. These points should be important considerations in the development of molecular therapies based on the manipulation of their expression, as unwanted activation/suppression of different facets of innate immunity may be avoided with careful selection.
We recognize that the in vitro system used, while being well-established in the relevant body of scientific literature, represents a cytokine-specific mode of differentiation, one that may be considered minimalistic when compared with the complexity of differentiation factors found in vivo. We are actively pursuing several alternative models of Mφ/DC differentiation and thus far, have demonstrated the suppressive effects of miR-24, miR-30b, and miR-142-3p overexpression in Mφ differentiated from monocytes by use of GM-CSF (published as Supplemental data; Naqvi et al. [41]). In conjunction with an in vivo model of differentiation, we expect further characterization of these alternative modes to cast light on the true, physiologically defined impact of these miRNAs. At present, we highlight the importance of these miRNAs in the acquisition of Mφ- and DC-specific functionality from monocytes responding to the important differentiation factors M-CSF and GM-CSF + IL-4, respectively. We also highlight the potential of miR-24, miR-30b, and miR-142-3p mimics for the treatment of deleterious, proinflammatory responses to gram-negative bacteria. As such, future endeavors should include an evaluation of the local overexpression of miR-24, miR-30b, or miR-142-3p, with the goal of ameliorating tissue destruction as a result of immunopathology.
AUTHORSHIP
J.B.F., A.R.N., and S.N. made the initial discovery, designed the experiments, analyzed the data, and wrote the manuscript. J.B.F. and A.R.N. performed the experiments.
Acknowledgments
This study was supported by the National Institute of Dental and Craniofacial Research of the U.S. National Institutes of Health (R01DE021052).
Glossary
- DC
dendritic cell
- FDR
false discovery rate
- Mφ
macrophage
- MD-1
lymphocyte antigen 86
- MD2
lymphocyte antigen 96
- mD-DC
monocyte-derived dendritic cell
- mD-Mφ
monocyte-derived macrophage
- miRNA/miR
microRNA
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- PKB/C
protein kinase B/C
- PRR
pathogen recognition receptor
- rh
recombinant human
Footnotes
The online version of this paper, found at www.jleukbio.org, includes supplemental information.
DISCLOSURES
The authors do not have any conflicts of interest to declare.
REFERENCES
- 1.Lee R. C., Feinbaum R. L., Ambros V. (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- 2.Wightman B., Ha I., Ruvkun G. (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. [DOI] [PubMed] [Google Scholar]
- 3.Androulidaki A., Iliopoulos D., Arranz A., Doxaki C., Schworer S., Zacharioudaki V., Margioris A. N., Tsichlis P. N., Tsatsanis C. (2009) The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity 31, 220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C. Z., Li L., Lodish H. F., Bartel D. P. (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303, 83–86. [DOI] [PubMed] [Google Scholar]
- 5.Fogg D. K., Sibon C., Miled C., Jung S., Aucouturier P., Littman D. R., Cumano A., Geissmann F. (2006) A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science 311, 83–87. [DOI] [PubMed] [Google Scholar]
- 6.Sallusto F., Lanzavecchia A. (1994) Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 179, 1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiktor-Jedrzejczak W., Gordon S. (1996) Cytokine regulation of the macrophage (M phi) system studied using the colony stimulating factor-1-deficient op/op mouse. Physiol. Rev. 76, 927–947. [DOI] [PubMed] [Google Scholar]
- 8.Riepsaame J., van Oudenaren A., den Broeder B. J., van Ijcken W. F., Pothof J., Leenen P. J. (2013) MicroRNA-mediated down-regulation of M-CSF receptor contributes to maturation of mouse monocyte-derived dendritic cells. Front. Immunol. 4, 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stanley E. R., Cifone M., Heard P. M., Defendi V. (1976) Factors regulating macrophage production and growth: identity of colony-stimulating factor and macrophage growth factor. J. Exp. Med. 143, 631–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hashimoto S., Suzuki T., Dong H. Y., Yamazaki N., Matsushima K. (1999) Serial analysis of gene expression in human monocytes and macrophages. Blood 94, 837–844. [PubMed] [Google Scholar]
- 11.Dai X. M., Ryan G. R., Hapel A. J., Dominguez M. G., Russell R. G., Kapp S., Sylvestre V., Stanley E. R. (2002) Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood 99, 111–120. [DOI] [PubMed] [Google Scholar]
- 12.Dai X., Jayapal M., Tay H. K., Reghunathan R., Lin G., Too C. T., Lim Y. T., Chan S. H., Kemeny D. M., Floto R. A., Smith K. G., Melendez A. J., MacAry P. A. (2009) Differential signal transduction, membrane trafficking, and immune effector functions mediated by FcgammaRI versus FcgammaRIIa. Blood 114, 318–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palucka K. A., Taquet N., Sanchez-Chapuis F., Gluckman J. C. (1999) Lipopolysaccharide can block the potential of monocytes to differentiate into dendritic cells. J. Leukoc. Biol. 65, 232–240. [DOI] [PubMed] [Google Scholar]
- 14.Krutzik S. R., Tan B., Li H., Ochoa M. T., Liu P. T., Sharfstein S. E., Graeber T. G., Sieling P. A., Liu Y. J., Rea T. H., Bloom B. R., Modlin R. L. (2005) TLR activation triggers the rapid differentiation of monocytes into macrophages and dendritic cells. Nat. Med. 11, 653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munn D. H., Beall A. C., Song D., Wrenn R. W., Throckmorton D. C. (1995) Activation-induced apoptosis in human macrophages: developmental regulation of a novel cell death pathway by macrophage colony-stimulating factor and interferon gamma. J. Exp. Med. 181, 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fahy R. J., Doseff A. I., Wewers M. D. (1999) Spontaneous human monocyte apoptosis utilizes a caspase-3-dependent pathway that is blocked by endotoxin and is independent of caspase-1. J. Immunol. 163, 1755–1762. [PubMed] [Google Scholar]
- 17.Roilides E., Lyman C. A., Sein T., Petraitiene R., Walsh T. J. (2003) Macrophage colony-stimulating factor enhances phagocytosis and oxidative burst of mononuclear phagocytes against Penicillium marneffei conidia. FEMS Immunol. Med. Microbiol. 36, 19–26. [DOI] [PubMed] [Google Scholar]
- 18.Steinman R. M. (1991) The dendritic cell system and its role in immunogenicity. Annu. Rev. Immunol. 9, 271–296. [DOI] [PubMed] [Google Scholar]
- 19.Lien E., Sellati T. J., Yoshimura A., Flo T. H., Rawadi G., Finberg R. W., Carroll J. D., Espevik T., Ingalls R. R., Radolf J. D., Golenbock D. T. (1999) Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J. Biol. Chem. 274, 33419–33425. [DOI] [PubMed] [Google Scholar]
- 20.Thoma-Uszynski S., Kiertscher S. M., Ochoa M. T., Bouis D. A., Norgard M. V., Miyake K., Godowski P. J., Roth M. D., Modlin R. L. (2000) Activation of Toll-like receptor 2 on human dendritic cells triggers induction of IL-12, but not IL-10. J. Immunol. 165, 3804–3810. [DOI] [PubMed] [Google Scholar]
- 21.Juarez E., Nuñez C., Sada E., Ellner J. J., Schwander S. K., Torres M. (2010) Differential expression of Toll-like receptors on human alveolar macrophages and autologous peripheral monocytes. Respir. Res. 11, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Neill L. A., Sheedy F. J., McCoy C. E. (2011) MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 11, 163–175. [DOI] [PubMed] [Google Scholar]
- 23.O’Connell R. M., Chaudhuri A. A., Rao D. S., Baltimore D. (2009) Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. USA 106, 7113–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ceppi M., Pereira P. M., Dunand-Sauthier I., Barras E., Reith W., Santos M. A., Pierre P. (2009) MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. USA 106, 2735–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thai T. H., Calado D. P., Casola S., Ansel K. M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J. L., Schmidt-Supprian M., Rajewsky N., Yancopoulos G., Rao A., Rajewsky K. (2007) Regulation of the germinal center response by microRNA-155. Science 316, 604–608. [DOI] [PubMed] [Google Scholar]
- 26.Tili E., Michaille J. J., Cimino A., Costinean S., Dumitru C. D., Adair B., Fabbri M., Alder H., Liu C. G., Calin G. A., Croce C. M. (2007) Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 179, 5082–5089. [DOI] [PubMed] [Google Scholar]
- 27.Nagai Y., Yanagibashi T., Watanabe Y., Ikutani M., Kariyone A., Ohta S., Hirai Y., Kimoto M., Miyake K., Takatsu K. (2012) The RP105/MD-1 complex is indispensable for TLR4/MD-2-dependent proliferation and IgM-secreting plasma cell differentiation of marginal zone B cells. Int. Immunol. 24, 389–400. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto M., Sato S., Hemmi H., Sanjo H., Uematsu S., Kaisho T., Hoshino K., Takeuchi O., Kobayashi M., Fujita T., Takeda K., Akira S. (2002) Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature 420, 324–329. [DOI] [PubMed] [Google Scholar]
- 29.Weck M. M., Appel S., Werth D., Sinzger C., Bringmann A., Grünebach F., Brossart P. (2008) hDectin-1 is involved in uptake and cross-presentation of cellular antigens. Blood 111, 4264–4272. [DOI] [PubMed] [Google Scholar]
- 30.Kimoto M., Nagasawa K., Miyake K. (2003) Role of TLR4/MD-2 and RP105/MD-1 in innate recognition of lipopolysaccharide. Scand. J. Infect. Dis. 35, 568–572. [DOI] [PubMed] [Google Scholar]
- 31.Nagai Y., Kobayashi T., Motoi Y., Ishiguro K., Akashi S., Saitoh S., Kusumoto Y., Kaisho T., Akira S., Matsumoto M., Takatsu K., Miyake K. (2005) The radioprotective 105/MD-1 complex links TLR2 and TLR4/MD-2 in antibody response to microbial membranes. J. Immunol. 174, 7043–7049. [DOI] [PubMed] [Google Scholar]
- 32.Dziarski R., Wang Q., Miyake K., Kirschning C. J., Gupta D. (2001) MD-2 enables Toll-like receptor 2 (TLR2)-mediated responses to lipopolysaccharide and enhances TLR2-mediated responses to Gram-positive and Gram-negative bacteria and their cell wall components. J. Immunol. 166, 1938–1944. [DOI] [PubMed] [Google Scholar]
- 33.Le Guillou S., Sdassi N., Laubier J., Passet B., Vilotte M., Castille J., Laloë D., Polyte J., Bouet S., Jaffrézic F., Cribiu E., Vilotte J.L., Le Provost F. (2012) Overexpression of miR-30b in the developing mouse mammary gland causes a lactation defect and delays involution. PLoS ONE 7, e45727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poy M. N., Hausser J., Trajkovski M., Braun M., Collins S., Rorsman P., Zavolan M., Stoffel M. (2009) miR-375 maintains normal pancreatic α- and β-cell mass. Proc. Natl. Acad. Sci. USA 106, 5813–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sohn J. J., Schetter A. J., Yfantis H. G., Ridnour L. A., Horikawa I., Khan M. A., Robles A. I., Hussain S. P., Goto A., Bowman E. D., Hofseth L. J., Bartkova J., Bartek J., Wogan G. N., Wink D. A., Harris C. C. (2012) Macrophages, nitric oxide and microRNAs are associated with DNA damage response pathway and senescence in inflammatory bowel disease. PLoS ONE 7, e44156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Nunez R. T., Louafi F., Friedmann P. S., Sanchez-Elsner T. (2009) MicroRNA-155 modulates the pathogen binding ability of dendritic cells (DCs) by down-regulation of DC-specific intercellular adhesion molecule-3 grabbing non-integrin (DC-SIGN). J. Biol. Chem. 284, 16334–16342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou L. J., Tedder T. F. (1995) Human blood dendritic cells selectively express CD83, a member of the immunoglobulin superfamily. J. Immunol. 154, 3821–3835. [PubMed] [Google Scholar]
- 38.Kulkarni S., Savan R., Qi Y., Gao X., Yuki Y., Bass S. E., Martin M. P., Hunt P., Deeks S. G., Telenti A., Pereyra F., Goldstein D., Wolinsky S., Walker B., Young H. A., Carrington M. (2011) Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature 472, 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dweep H., Sticht C., Pandey P., Gretz N. (2011) miRWalk—database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 44, 839–847. [DOI] [PubMed] [Google Scholar]
- 40.Guler R., Afshar M., Arendse B., Parihar S. P., Revaz-Breton M., Leitges M., Schwegmann A., Brombacher F. (2011) PKCδ regulates IL-12p40/p70 production by macrophages and dendritic cells, driving a type 1 healer phenotype in cutaneous leishmaniasis. Eur. J. Immunol. 41, 706–715. [DOI] [PubMed] [Google Scholar]
- 41.Naqvi A. R., Fordham J. B., Nares S. (2015) miR-24, miR-30b, and miR-142-3p regulate phagocytosis in myeloid inflammatory cells. J. Immunol. 194, 1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vigorito E., Kohlhaas S., Lu D., Leyland R. (2013) miR-155: an ancient regulator of the immune system. Immunol. Rev. 253, 146–157. [DOI] [PubMed] [Google Scholar]
- 43.Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. (2006) NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 103, 12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fabriek B. O., van Bruggen R., Deng D. M., Ligtenberg A. J., Nazmi K., Schornagel K., Vloet R. P., Dijkstra C. D., van den Berg T. K. (2009) The macrophage scavenger receptor CD163 functions as an innate immune sensor for bacteria. Blood 113, 887–892. [DOI] [PubMed] [Google Scholar]
- 45.Wermeling F., Chen Y., Pikkarainen T., Scheynius A., Winqvist O., Izui S., Ravetch J. V., Tryggvason K., Karlsson M. C. I. (2007) Class A scavenger receptors regulate tolerance against apoptotic cells, and autoantibodies against these receptors are predictive of systemic lupus. J. Exp. Med. 204, 2259–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Porcheray F., Viaud S., Rimaniol A. C., Léone C., Samah B., Dereuddre-Bosquet N., Dormont D., Gras G. (2005) Macrophage activation switching: an asset for the resolution of inflammation. Clin. Exp. Immunol. 142, 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fraser D. A., Bohlson S. S., Jasinskiene N., Rawal N., Palmarini G., Ruiz S., Rochford R., Tenner A. J. (2006) C1q and MBL, components of the innate immune system, influence monocyte cytokine expression. J. Leukoc. Biol. 80, 107–116. [DOI] [PubMed] [Google Scholar]
- 48.Nepomuceno R. R., Henschen-Edman A. H., Burgess W. H., Tenner A. J. (1997) cDNA cloning and primary structure analysis of C1qR(P), the human C1q/MBL/SPA receptor that mediates enhanced phagocytosis in vitro. Immunity 6, 119–129. [DOI] [PubMed] [Google Scholar]
- 49.Melendez A. J., Harnett M. M., Allen J. M. (1999) Differentiation-dependent switch in protein kinase C isoenzyme activation by FcgammaRI, the human high-affinity receptor for immunoglobulin G. Immunology 96, 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gutcher I., Webb P. R., Anderson N. G. (2003) The isoform-specific regulation of apoptosis by protein kinase C. Cell. Mol. Life Sci. 60, 1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Folgueira L., McElhinny J. A., Bren G. D., MacMorran W. S., Diaz-Meco M. T., Moscat J., Paya C. V. (1996) Protein kinase C-zeta mediates NF-kappa B activation in human immunodeficiency virus-infected monocytes. J. Virol. 70, 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mischak H., Pierce J. H., Goodnight J., Kazanietz M. G., Blumberg P. M., Mushinski J. F. (1993) Phorbol ester-induced myeloid differentiation is mediated by protein kinase C-alpha and -delta and not by protein kinase C-beta II, -epsilon, -zeta, and -eta. J. Biol. Chem. 268, 20110–20115. [PubMed] [Google Scholar]
- 53.Mangan D. F., Welch G. R., Wahl S. M. (1991) Lipopolysaccharide, tumor necrosis factor-alpha, and IL-1 beta prevent programmed cell death (apoptosis) in human peripheral blood monocytes. J. Immunol. 146, 1541–1546. [PubMed] [Google Scholar]
- 54.Loegering D. J., Lennartz M. R. (2011) Protein kinase C and Toll-like receptor signaling. Enzyme Res. 2011, 537821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Comalada M., Xaus J., Valledor A. F., López-López C., Pennington D. J., Celada A. (2003) PKC epsilon is involved in JNK activation that mediates LPS-induced TNF-α, which induces apoptosis in macrophages. Am. J. Physiol. Cell Physiol. 285, C1235–C1245. [DOI] [PubMed] [Google Scholar]
- 56.Fronhofer V., Lennartz M. R., Loegering D. J. (2006) Role of PKC isoforms in the Fc(γ)R-mediated inhibition of LPS-stimulated IL-12 secretion by macrophages. J. Leukoc. Biol. 79, 408–415. [DOI] [PubMed] [Google Scholar]
- 57.Divanovic S., Trompette A., Atabani S. F., Madan R., Golenbock D. T., Visintin A., Finberg R. W., Tarakhovsky A., Vogel S. N., Belkaid Y., Kurt-Jones E. A., Karp C. L. (2005) Inhibition of TLR-4/MD-2 signaling by RP105/MD-1. J. Endotoxin Res. 11, 363–368. [DOI] [PubMed] [Google Scholar]
- 58.Nagai Y., Watanabe Y., Takatsu K. (2013) The TLR family protein RP105/MD-1 complex: a new player in obesity and adipose tissue inflammation. Adipocyte 2, 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]