Abstract
The CUB and sushi multiple domains 1 (CSMD1) gene harbors signals provided by clusters of nearby SNPs with 10-2 > p > 10-8 associations in genome wide association (GWAS) studies of addiction-related phenotypes. A CSMD1 intron 3 SNP displays p < 10-8 association with schizophrenia and more modest associations with individual differences in performance on tests of cognitive abilities. CSDM1 encodes a cell adhesion molecule likely to influence development, connections and plasticity of brain circuits in which it is expressed. We tested association between CSMD1 genotypes and expression of its mRNA in postmortem human brains (n = 181). Expression of CSMD1 mRNA in human postmortem cerebral cortical samples differs 15–25%, in individuals with different alleles of simple sequence length and SNP polymorphisms located in the gene’s third/fifth introns, providing nominal though not Bonferroni-corrected significance. These data support mice with altered CSMD1 expression as models for common human CSMD1 allelic variation. We tested baseline and/or cocaine-evoked addiction, emotion, motor and memory-related behaviors in +/- and -/- csmd1 knockout mice on mixed and on C57-backcrossed genetic backgrounds. Initial csmd1 knockout mice on mixed genetic backgrounds displayed a variety of coat colors and sizable individual differences in responses during behavioral testing. Backcrossed mice displayed uniform black coat colors. Cocaine conditioned place preference testing revealed significant influences of genotype (p = 0.02). Homozygote knockouts displayed poorer performance on aspects of the Morris water maze task. They displayed increased locomotion in some, though not all, environments. The combined data thus support roles for common level-of-expression CSMD1 variation in a drug reward phenotype relevant to addiction and in cognitive differences that might be relevant to schizophrenia. Mouse model results can complement data from human association findings of modest magnitude that identify likely polygenic influences.
Introduction
The gene that encodes the CUB and sushi multiple domains 1 protein (CSMD1) has been identified in case vs control genome-wide association (GWAS) samples for addiction- related phenotypes that include extensive use of an addictive substance, vulnerability to develop dependence on an addictive substance, ability to quit smoking and time from initiation of tobacco use to development of nicotine dependence. The modest association signals identified in these studies come from clusters of nearby single nucleotide polymorphisms (SNPs) with 10−2 > p > 10−8 association that often do not survive conservative Bonferroni corrections [1,2,3,4,5,6,7,8,9,10,11,12,13]. The distribution of these signals in several parts of the CSMD1 gene is consistent with substantial disease-related allelic heterogeneity.
CSMD1 has also been identified by several analyses of GWAS data and/or candidate gene studies for vulnerability to schizophrenia and clusters of schizophrenic symptoms [14,15,16,17,18,19]. Association with the CSMD1 intron 3 SNP rs10503253 is among the most studied. This or nearby SNPs have also been associated, at least modestly, with individual differences in cognitive abilities in normal and in schizophrenic samples [20,21,22,23,24]. Nominal p values were 3 x 10−3–1 x 10−4 for tests of IQ, strategy formation, planning, set shifting and problem solving in Greek military conscripts [20], 0.01–0.04 for verbal IQ, performance IQ and/or mnemonic measures in Irish and/or German schizophrenics and controls [23] and 2×10−6 with “memory cognition” in Chinese schizophrenics and controls [24].Three common CSMD1 missense variants with minor allele frequencies > 0.03 are listed in databases. However, all are distant from intron 3 (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?geneId=64478). Increased frequencies of CSMD1 copy number variants are reported in individuals with mild cognitive impairment and with dementia [25]. However, little reported data links specific human CSMD1 genomic variants to specific molecular or behavioral phenotypes relevant to particular aspects of addiction or schizophrenia.
Features of CSMD1’s neurobiology make its variants attractive candidates to contribute to individual differences in vulnerability to addiction and in mnemonic processes. The CSMD1 gene encodes a single transmembrane domain protein likely to alter the development and maintenance of connections between expressing neurons. Abundant CSMD1 immunoreactivity is found at the growth cones of cultured neurons [26]. Ventral midbrain neurons implicated in reward, likely to be dopaminergic, prominently express CSMD1 mRNA, as do hippocampal neurons implicated in mnemonic processes (http://mouse.brain-map.org/experiment/ivt?id=69608130&popup=true). Results from studies of csmd1 knockouts on mixed genetic backgrounds (see below) also support interesting phenotypes [21,27].
These neurobiologic, genetic and genomic data nominate CSMD1 as a candidate for studies that seek: a) influences of common human allelic variation on CSMD1 expression, b) influences of variation in CSMD1 expression on responses to rewarding addictive substances in mouse models, c) influences of variation in CSMD1 expression on cognitive phenotypes that may model features relevant to schizophrenia and d) comparisons with other physiological and behavioral differences in mice with altered csmd1 expression.
We now report studies of CSMD1 expression in human postmortem cerebral cortical samples that identify nominally-significant associations between levels of CSMD1 expression and CSMD1 genomic markers, including those that lie near the schizophrenia-associated SNP rs10503253, though these associations do not survive conservative Bonferroni corrections. We describe our initial data from the csmd1 knockouts on mixed genetic backgrounds and report the variability that differences in genetic background among these mice appears to provide. We then describe more extensive results from “csmd1” knockout mice backcrossed onto a C57 background for 5 generations. We study results of tests that include cocaine conditioned place preference (CPP), one of the most commonly used and heavily validated mouse tests for drug reward/reinforcement [28]. We demonstrate that mice with altered CSMD1 expression display overall differences in cocaine CPP, although their locomotion is influenced by modest to moderate doses of cocaine in ways similar to those of wildtype mice. There is modestly increased locomotion in homozygous knockouts. We identify alterations in Morris water maze testing in homozygous backcrossed csmd1 knockouts, and discuss the potential implications of these data for the CSMD1 associations with cognitive differences in schizophrenia, in normal populations, and for our CPP data from heterozygous and homozygous mice. We note ways in which these data enhance our confidence that CSMD1 variation and thus the neuronal properties and connections that CSMD1 modulates play roles in addiction phenotypes and in cognition-related phenotypes that are of likely relevance for schizophrenia.
Materials and Methods
Common human allelic CSMD1 sequence variation was sought
Common human allelic CSMD1 sequence variation was sought by searches of dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) and the Toronto database for structural/copy number variation (http://dgvbeta.tcag.ca/dgv/app/home?ref=NCBI36/hg18). Genetic cis influences on levels of CSMD1 expression were sought by studies of CSMD1 mRNA and DNA in rapidly-frozen autopsy samples of frontal cortex of European American individuals who died without brain disease. All brain samples were supplied anonymously from tissue banks at the University of Maryland (http://medschool.umaryland.edu/btbank/) and Johns Hopkins University (http://pathology.jhu.edu/department/services/consults/neuropath.cfm). RNAs were prepared with the RNeasy lipid tissue mini kits (Qiagen), cDNA was synthesized with SuperScript III First Strand Synthesis Supermix (Invitrogen) and levels of mRNAs were assessed by quantitative RT-PCR using SybrGreen master mix (Applied Biosystems), conditions from the manufacturer’s protocol and oligonucleotide primers (sequences available from authors on request) that targeted the dominant long CSMD1 mRNA isoform (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=human&q=CSMD1) and the reference genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH), hypoxanthine phosphoribosyltransferase 1 (HPRT1) and ubiquitin C (UBC).
DNA was extracted from brain samples using Qiagen kits [29], and subjected to multiplexed SNP genotyping using Sequenom panels and oligonucleotides (S1 Table) for 38 SNPs distributed through the gene. The simple sequence repeat that is annotated as rs71534387 was amplified using oligonucleotide primers, polymerase chain reaction conditions 1X PCR Gold buffer (Invitrogen), 0.8 mM dNTP mix, 1.5 mM MgCl2, 0.4μM forward and reverse primers and 0.25 units Amplitaq Gold enzyme (Invitrogen). Amplimers and oligonucleotides provided clear peaks every 3 bp after separation using an Applied Biosystems 3730xl instrument with Liz500 size standard (performed by Genewiz, Inc.). Peaks were analyzed using Peak Scanner software and genotypes determined for each DNA sample.
Mouse models
Initial constitutive csmd1 homozygous knockout (KO), heterozygous knockout and littermate wildtype animals were produced by heterozygote x heterozygote crosses from mice that were originally created by Lexicon pharmaceuticals, distributed by Taconic Farms (TF0137) and described by others [21,27]. Embryonic stem cells derived from 129SvEvBrd mice replaced a 1,070 bp genomic sequence of the csmd1 exon 1–intron 1 junction with a LacZ/Neo selection cassette expressed in frame with the start of the CSMD1 protein-coding sequence. The “mixed background” mice derived from these ES cells were maintained on mixed genetic backgrounds that included 129 and B6 ancestries (the exact B6 substrain unknown) as reflected by varying coat colors (black, agouti, and white individuals) [21,27]. Following initial testing and identification of substantial mouse to mouse variability, these mixed background csmd1 KO mice were backcrossed to C57Bl/6J mice for 4 generations and then to Tg(Thy1-EGFP)MJrs mice (C57Bl/6J mice expressing eGFP under control of the Thy1 promoter) for the fifth generation, so that less than 2% of the initial 129 DNA was present. These backcrossed mice, termed csmd1 mice here, of both sexes were tested at 118 ± 49 days of age. All mouse breeding, care and experimentation was approved by the NIDA-IRP Animal Care and Use Committee.
Mouse behavioral studies
Motor
Muscle strength/motor persistence test: 9–10 mice of each genotype, half of them males and half females were tested to determine the time during which they could support their weight by holding onto a wire screen (5 mm hardware cloth) mounted 20 cm above and parallel to the countertop. The test was terminated at 120 sec, or when the mouse fell.
Motor coordination and learning in the same cohort of animals were measured on an accelerating rotarod cylinder on which the mice had to maintain their balance to prevent falling. The animals were tested over three consecutive days with one test per day. The starting speed of the rotating cylinder was 4 rpm and it gradually increased to 40 rpm over 5 min, which was the cut off time.
Cocaine effects
Reward: cocaine conditioned place preference was assessed in 255 naive animals, 25–26 and 13–16 subjects/genotype for 10 mg/kg and other doses, respectively, half males and half females as in [30,31]. Mice were 12 ± 3 weeks old and weighed on the pretest day to determine dose. Two 20 cm x 20 cm compartments, one with a wire-mesh floor and the other with corn cob bedding, were separated by a Plexiglas divider. In two 20 min pre-tests, subjects had access to both sides of the apparatus through a 5 cm opening in the divider. Four 20 min conditioning trials were conducted over the next 2 days during which subjects were confined to one side of the apparatus. Mice were injected with cocaine prior to confinement in the initially non-preferred compartment and with saline prior to confinement in the initially-preferred compartment. Half of the mice received cocaine/initially non preferred and half received saline/initially preferred as their first drug/environmental pairings on the first conditioning day; the order for each mouse was reversed on the second day. Control subjects received saline injections prior to separate confinements on each side. 18 hours after the last conditioning session (48 hours after the first conditioning session), subjects were again given access to both sides of the apparatus for a 20 min post-test. The preference score was calculated as the difference between time spent on the drug-paired side during the post-test and the average time spent on the drug-paired side during the pre-tests.
Locomotion was recorded: a) in 42 x 42 cm dark, sound attenuated boxes to which the mice had not been previously exposed, for 60 min trials (n = 10-11/genotype) and b) during the 20 min pretest (in both halves of the 20 x 40 conditioning apparatus), conditioning (20 x 20 cm half of the apparatus) and test (20 x 40 cm) sessions. Data was thus obtained from untreated, saline treated, and mice sampled before treatment, after one treatment and after the second treatment with cocaine. Total distance traveled was calculated from infrared beam breaks by an Optovarimax ATS System [31].
Memory and learning were evaluated in Morris water maze testing [32] in 19–24 mice/genotype of the mice previously submitted to the hanging wire, rotarod or cocaine-conditioned place preference tests. A black 90 cm diameter pool was filled with room temperature water made opaque with white tempera paint. A 9 cm diameter platform was located in the center of one quadrant, visible for the first 6 trials and then hidden 0.5 cm below the water level for subsequent trials. Each trial lasted a maximum of 60 seconds and was followed by a 15 second rest period on the platform. After two trials, mice were returned to their home cages for about 4 hours and then given an additional 2-trial session so that they received a total of 4 trials per day. For each trial, the latency to reach the platform was recorded. Three days after acquisition, defined by an average latency less than 10 seconds, the platform was removed and a 60 second probe trial was conducted. The data from the probe trial was analyzed with Ethovision software (Noldus, Netherlands), which also determined the path of the subject in the pool and the time spent in each quadrant.
Statistical analyses
Analyses of mouse data used PASW statistics 18 (SPSS) and t tests (Excel). Analyses of variance and covariance (ANOVA and ANCOVA) used between subjects factors of genotype sex, and dose, age as a cofactor, and within-subjects factors where appropriate including time for locomotion and trial day for Morris water maze. Analyses of human association data and assessment of correlations between genotypes at nearby SNPs used PLINK (pngu.mgh.harvard.edu/~purcell/plink). Normalized gene expression levels were compared between human major vs minor allele homozygotes using t tests. Bonferroni corrections were performed using (http://www.quantitativeskills.com/sisa/calculations/bonfer.php) with 0.6 and 0.8 correlations (r) between SNPs.
Results
Searches for common CSMD1 variation related to addiction and/or schizophrenia
Databases contained no common CSMD1 copy number/insertion/deletion variants, though rarer copy number variants that cover much of this large gene are documented. Only three missense variants (rs28455997, rs6558702 and rs11984691), which were distant from the most schizophrenia-associated SNP rs10503253, displayed minor allele frequencies > 0.03.
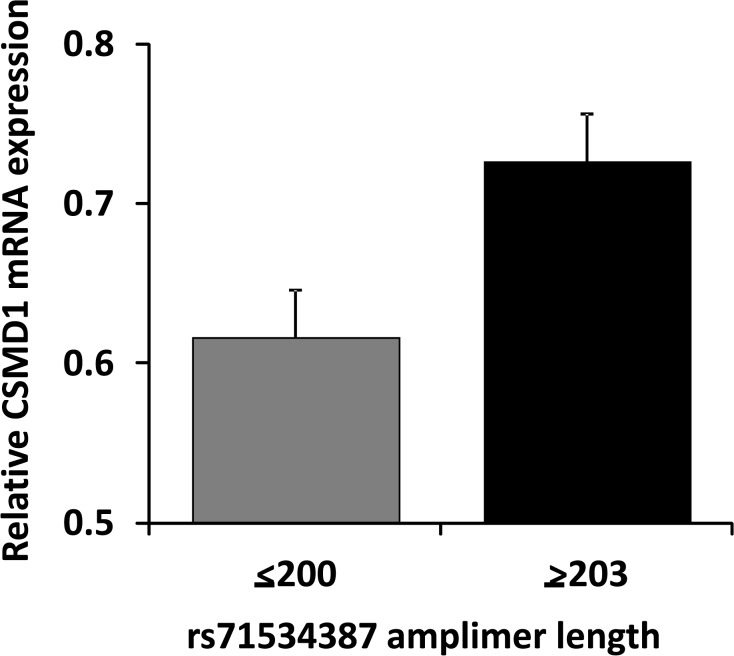
There was nominally-significant association between CSMD1 genomic markers and levels of CSMD1 mRNA expression. We detected CSMD1 mRNA in RNA extracted from cerebral cortical specimens from each of the 181 individuals tested. Data from thirty nine genomic markers that displayed minor allele frequencies > 0.03 were analyzed. The CSMD1 intron 3 trinucleotide repeat that is annotated as rs71534387 and the intron 5 SNP rs10503223 displayed the most robust associations (Fig 1; p = 0.03/0.02, respectively; t tests). The Bonferroni-corrected thresholds for two sided t tests are p < 0.012 and p < 0.024 with r = 0.6 and r = 0.8, respectively; these nominally-significant associations thus did not definitively surpass Bonferroni correction for the numbers of markers tested. These two markers displayed D’ values of 0.53 in individuals who donated the brain samples studied here. rs71534387 data comes from 104 individuals who were either hetero or homozygous for the 200 bp vs 206 bp alleles. These nominally-significant associations corresponded to 18/25% differences in levels of expression. Expression is lower in individuals with longer rs71534387 repeat lengths and in rs10503223 = G homozygotes. These differences in postmortem brain expression help to support heterozygous and/or homozygous csmd1 knockout mice as models for common CSMD1 allelic variation of relevance for addiction and, possibly, schizophrenia-related phenotypes. The schizophrenia-associated SNP rs10503253 itself did not provide the nominally-significant associations noted for the nearby rs71534387 trinucleotide repeat, however.
Fig 1. Differential expression of CSMD1 mRNA in cerebral cortical samples from individuals with different rs71534387 SSLP genotypes.
Mean +/- standard error of the mean (SEM) of relative expression of CSMD1 mRNA in cerebral cortical samples of 72 EuAm individuals with no rs71534387 allele > 203 bp vs 96 individuals with at least one rs71534387 allele > 203 bp. Triplicate RT-PCR assays. Relative expression was determined by the mean of two CSMD1 amplimers in relation to the geometric mean of three control mRNAs from the same sample.
Mice
Initial characterization of mice on mixed genetic backgrounds
Heterozygote x heterozygote crosses of mice on mixed genetic backgrounds supplied by Taconic produced offspring with genotypes in expected Mendelian ratios that displayed differences in coat colors; different mice had agouti, black or white coat colors. Initial tests of mice of these mixed genetic backgrounds revealed no significant influences of genotype on locomotion after injections of saline or 10 mg/kg cocaine doses (Table A in S2 Table; ANCOVA p = 0.729 and 0.650, respectively) or performance on the rotorod test of motor coordination/motor learning (Table B in S2 Table; ANCOVA effect of genotype p = 0.771).
Characterization of mice on C57Bl genetic backgrounds
We thus backcrossed csmd1 knockout mice of the original mixed genetic background for 5 generations to mice with C57Bl/6J genetic backgrounds as noted above, producing backcrossed csmd1 knockouts that we term csmd1 knockouts in the rest of this paper. There was significance for the modest differences in weights for the knockout males but not for the females. Weights for male/female were wildtype: 28.6/21.3 g; heterozygotes 28.6/21.5 g and homozygotes 27.1/20.7 g, n = 39–45/genotype, (Table C in S2 Table; ANCOVA with age as covariate p = 0.019 for males and 0.237 for females). Since there were no significant sex * genotype interactions, the small, sex-specific differences did not provide effects on behavioral results of the knockout.
These backcrossed mice were subjected to a number of physiologic, pharmacological and behavioral tests. The csmd1 +/- and-/- knockouts displayed no evidence for gross alterations in motor function. They were similar to wildtype littermates in screen hang time and rotarod testing (Tables D and E in S2 Table; ANCOVA p values for effects of genotype 0.346 and 0.402, respectively).
csmd1 knockout mice failed to display significant differences from their wild type siblings in the amounts of time spent in the center of an open field or their latencies before emerging from a dark box (Table F in S2 Table; ANCOVA p = 0.216 and 0.263, respectively).
Cocaine conditioned place preference
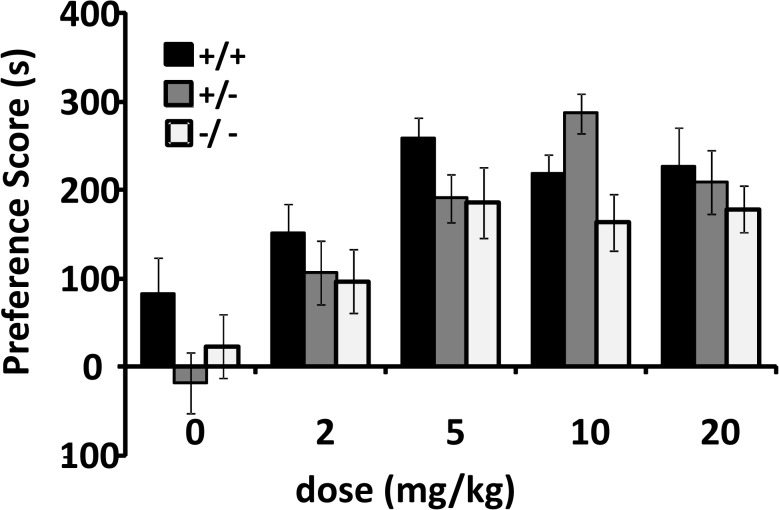
In wildtype mice, cocaine-conditioned place preference (CPP) was maximal at 5–10 mg/kg doses, as we and others have observed in studies of many other knockout mouse strains (Fig 2; Table G in S2 Table) [33]. There was a significant main effect of csmd1 genotype (p = 0.024). At most doses, mice with reduced csmd1 expression displayed numerically-reduced preferences for places where they had previously received cocaine. There was numerically-greater preference for places paired with 10 mg/kg cocaine in heterozygous knockouts. There was substantial variation among three subcohorts of mice (n = 7–11 each) that received this dose; there was p = 0.152 for genotype*dose interaction.
Fig 2. Cocaine conditioned place preference among mice with different CSMD1 genotypes.
Mean difference ± SEM in time spent on the cocaine-paired side (Y axis) before and after conditioning with different doses of cocaine for wildtype heterozygous and homozygote knockout mice (ANCOVA effect of genotype p = 0.024; n = 13–26 mice of each genotype for each dose). Data from male and female mice are combined since gender displayed no significant interaction with genotype.
Locomotion
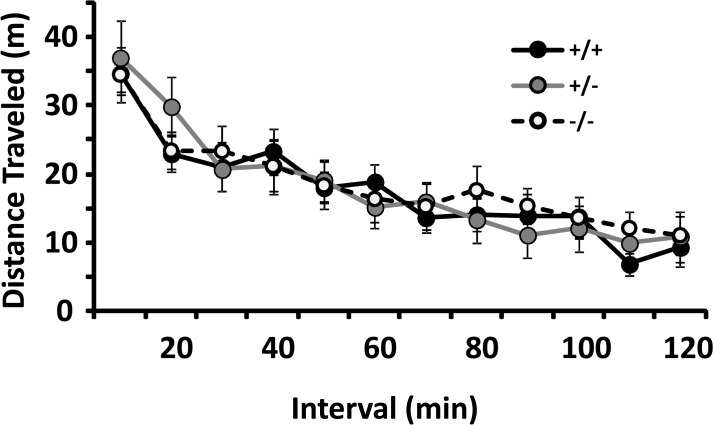
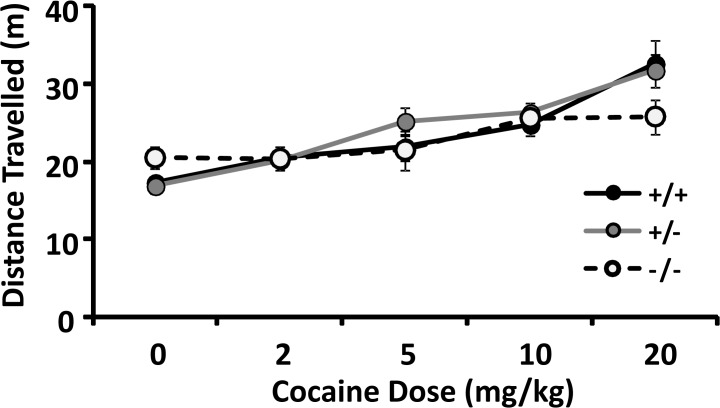
Locomotion and locomotor habituation measured in 42 x 42 cm dark and sound attenuated boxes to which the mice had not been previously exposed for 120 min trials did not identify significant influences of genotype (Fig 3; p for total distance traveled 0.967).
Fig 3. Locomotion and habituation in a novel 42 x 42 cm chamber: no significant differences between mice with different CSMD1 genotypes.
Values are mean +/- SEM of the number of meters traveled when mice were exposed for the first time to the 42 x 42 cm apparatus (n = 10/genotype, ANOVA p = 0.967).
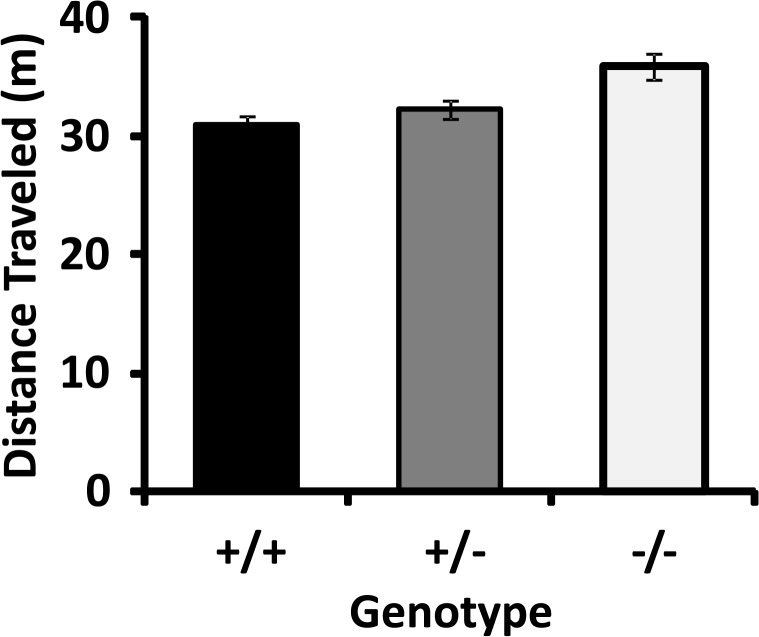
Locomotion during baseline exposures to the CPP apparatus. Mice were exposed to the CPP apparatus and allowed to explore both sides freely during the pretest that established baseline preference for each mouse. ANCOVA of data from all of the subjects revealed a highly-significant effect of genotype on locomotion during the first exposure to this smaller, more brightly lit novel environment (Fig 4; Table H in S2 Table; p = 3.7 x 10−13). Scheffe’s post-hoc testing showed striking significance for the differences between homozygotes and their wild-type siblings (p = 3.7 x 10−12) and a moderate difference between wild type and heterozygous mice (p = 0.045). There was a significant effect of gender (p = 1.0 x 10−11) but no significant genotype*sex interaction (p = 0.800).
Fig 4. CSMD1 knockouts display modestly increased locomotion in a brightly lit environment.
Values are mean +/- SEM of the number of cm traveled during the first pretest, in which mice were exposed for the first time to the CPP apparatus and allowed to explore both sides (n = 48/genotype, ANCOVA p = 3.7 x 10−13).
Locomotion during the first 20 min conditioning session was assessed when mice were confined to 20 x 20 cm portions of the conditioned place preference boxes after receiving their first saline or cocaine injections. Saline-injected csmd1 knockouts displayed significantly increased locomotion (Fig 5; Table I in S2 Table; p = 0.002) in comparison to mice of other genotypes. Significant effects of cocaine dose and gender were identified in ANCOVA of data from cocaine-injected mice of all genotypes (p = 5.5 x 10−15 and 0.009, respectively). There was a trend towards genotype*dose interaction (p = 0.054). Cocaine injections increased locomotion in wild type and heterozygous mice (p = 4.7 x 10−6 and 4.9 x 10−8, respectively), but did not significantly increase locomotion in the homozygotes (p = 0.078).
Fig 5. CSMD1 knockouts display differences in locomotion during the 20 min conditioning sessions when confined to 20 x 20 cm portions of the conditioned place preference boxes after receiving their first saline or cocaine injections.
Saline-injected CSMD1 knockouts displayed significantly increased locomotion (p = 0.002) in comparison to mice of other genotypes. Significant effects of cocaine dose and gender were identified in ANCOVA of data from cocaine-injected mice of all genotypes (p = 5.5 x 10−15 and 0.009, respectively). There was a trend towards genotype * dose interaction (p = 0.054). Values are mean +/- SEM of the number of m traveled.
Locomotor sensitization was sought by comparing locomotion during the second vs first cocaine conditioning sessions. Mice of different genotypes failed to display significant overall differences between the quantities of locomotion that they displayed during these two sessions (repeated measures ANCOVA effect of genotype (Table J in S2 Table; p = 0.432,). By contrast, there was a highly significant main effect of dose (p = 3.5 x 10−26), significant genotype*dose interactions (p = 0.007) and significant dose*session interactions (p = 2.0 x 10−13).
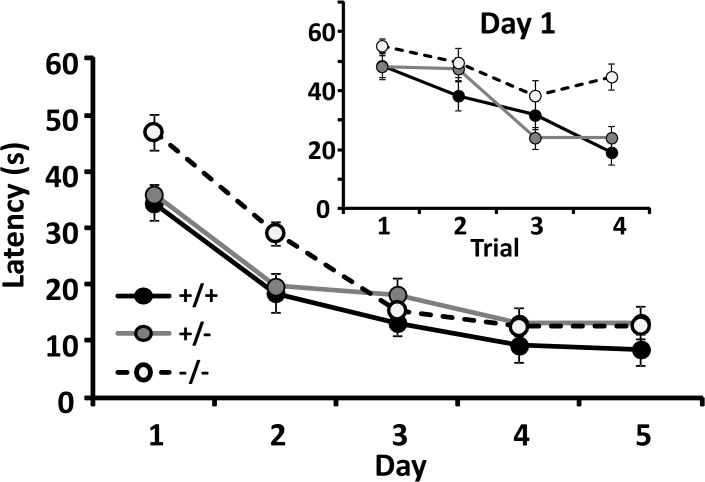
Memory: Morris water maze: Mice of all csmd1 genotypes swam through the Morris water maze apparatus with similar velocities (Table M in S2 Table; p = 0.106). Averaged daily latencies to reach the platform during the learning phase of the task were significantly longer in the homozygous csmd1 knockouts (Fig 6; Table K in S2 Table; repeated measures ANOVA genotype*day interaction p = 0.004). Effects were especially notable during the first day of training (Fig 6 inset; Table L in S2 Table). Latencies during the first trial on this first day were similar across genotypes. Wild type and heterozygote knockouts improved performance during the day, but homozygote knockout mice failed to improve performance. These differences reached the margin of statistical significance (repeated measures ANOVA genotype * trial interaction p = 0.058). These results are consistent with at least some results from some of the studies of cognitive associations with human CSMD1 variants [20,23,24] and cognitive tests of mixed-background csmd1 knockouts reported from other laboratories [21,27]. Despite the longer latencies to find the hidden platform during the acquisition phase of testing, the homozygous csmd1 knockouts did not display significantly worse performance during probe trials in which the platform was removed (Table M in S2 Table; distance from platform location p = 0.854, time in target quadrant p = 0.909).
Fig 6. Morris water maze performance: time to reach platform in mice with different CSMD1 genotypes.
Main Fig: Averaged daily latencies ± SEM to reach the Morris water maze platform for wildtype, heterozygous and homozygous CSMD1 knockout mice (repeated measures ANOVA genotype * day interaction p = 0.004 for learning. There were no significant differences in swimming speed (see text). Inset: More detailed data showing latencies to reach the (visible) platform during the first four trials (day 1). The trend for poorer performance in the homozygous knockouts reached the margin of statistical significance (p = 0.058).
Discussion
Our current results document 15–25% differences in levels of expression of CSMD1 in postmortem brains of individuals with common CSMD1 genotype marker alleles that lie near CSMD1 markers that have been associated with substance dependence, ability to quit smoking and vulnerability to developing schizophrenia. These nominally-significant associations do not reach Bonferroni-corrected statistical significance. In csmd1 knockout mice with reduced CSMD1 mRNA expression, there is an overall alteration in preference for places associated with exposure to effects of cocaine. Taken together with data that describe association between human CSMD1 variants and addiction-related phenotypes, these combined results support likely contributions of moderate individual differences in levels of CSMD1 expression to individual differences in levels of responses to rewarding and addictive drugs.
Conditioned place preference tests combinations of drug reward and the memory processes that are triggered by prior rewarding drug-associated experiences. It is thus conceivable that the reduced CPP in homozygous knockouts could receive contributions from the observed reductions in performance on mnemonic testing. Others’ results in which csmd1 knockouts on mixed genetic backgrounds fail to display substantially altered memory in tests of novel object recognition or differences in sucrose preference fail to provide evidence for large confounding influences in these mice [21,27]. Alterations in cocaine-induced place preference in the heterozygous csmd1 knockout mice that express little difference in mnemonic testing suggest that we cannot ascribe the CPP findings in these mice to mnemonic influences of more moderately-altered levels of csmd1 expression, however.
The modest changes in Morris water maze performance in the csmd1 homozygous knockout mice with virtual elimination of CSMD1 expression are consistent with other data from mice and humans. These results accord with the prominent CSMD1 expression in memory-associated brain regions, especially the hippocampus. They are consistent with a report of more frequent CSMD1 copy number variants in individuals with mild cognitive impairment or dementia, when compared to controls [25]. The overall data, and contrasting data for missense or other common variation, support the idea that variation in levels of CSMD1 expression attributable to 5’ haplotypes in this gene provides a significant molecular source of common functional human individual difference at the CSMD1 locus. This positive set of translational data, some of which achieve only nominal statistical significance, contrasts with others’ inabilities to detect significant differences in prepulse inhibition in csmd1 knockout animals with mixed genetic backgrounds [21,27]. Taken together, these data support the possibility that human CSMD1 variation might play disproportionate roles in schizophrenia vulnerabilities due to influences on the cognitive difficulties that are commonly experienced by individuals with this diagnosis [34], though recent analyses also point to roles for CSMD1 variation in core schizophrenic psychotic features and in disease severity, providing a caution to accepting this simple explanation [16].
The intron 5 SNP rs10503223 and the intron 3 trinucleotide repeat annotated as rs71534387 are each associated with modest-to-moderate individual differences in levels of expression of CSMD1 mRNA that provide results at the margins of Bonferroni-corrected statistical significance. Variants in 5’ aspects of many genes are associated with their levels of expression [35]. The relatively large set of brains available here for these “level of expression” association studies, the relatively 5’ location of the genomic markers that display nominally-significant association and the lack of common missense variants in the 5’ exons of this gene provide support for common level of expression variation in CSMD1. There are also features that suggest caution. As noted above, the observed SNP and SSLP associations with levels of expression fail to surpass Bonferroni corrections for the numbers of correlated markers tested. The SNP whose variants are most studied for association with schizophrenia and individual differences in cognitive function, rs10503253, fails to display significant association with levels of CSMD1 mRNA expression. Though rs10503253 lies only ca 35 kb from the rs71534387 SSLP marker that does display associations with levels of CSMD1 expression, there is only modest linkage disequilibrium (D’ = 0.1) between these markers in the individuals whose brains we sampled here. Indeed, there is substantial variation in the patterns and extents of linkage disequilibrium across the 5’ aspects of the CSMD1 gene in samples from different populations (http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap3r2_B36/#search), supporting the idea that use of the markers described here will provide differing patterns of information about nearby variations in samples from distinct populations.
The differences in expression associated with these markers’ alleles suggest that heterozygous and/or homozygous knockout mice with reduced expression provide valid models for common variation in CSMD1 in ways that accord nicely with their manifestation of phenotypes relevant to addiction and cognitive abilities. Based on the human associations with addiction phenotypes, it is thus of interest both that there are significant differences in cocaine conditioned place preference in the heterozygous knockout mice and that these differences display some evidence for specificity. Heterozygous CSMD1 knockout mice fail to display reductions in tests of locomotion or swimming speed that would provide confounding explanations for altered performance in tests of preference for places paired with cocaine or the Morris water maze. There is no evidence that heterozygous knockout alters the conditioned locomotion that can be exerted by cocaine during conditioned place preference testing. There is no enhanced lethality or other profound physiological alteration noted with the lifelong reductions in CSMD1 expression found in heterozygous knockouts. Others have reported only modest changes in expression of genes other than CSMD1 in initial csmd1 knockouts [21]. In csmd1 mice, we have found only modest (25–30%) increases in cerebral cortical levels of expression of mRNAs for the two CSMD1 family members, CSMD2 and CSMD3 (ANOVA p = 0.015 and < 0.001, respectively), providing additional evidence for the modest magnitude of the adaptive brain changes caused by lifelong reductions in CSMD1 expression (JD and GRU, unpublished observations, 2013).
The current results add to data from studies of mice with altered expression of other cell adhesion molecules (NrCAM [36], CDH13, PTPRD (JD, GRU et al, submitted) and receptors (nAChR5 [37]), providing increasingly-robust validation for conditioned place preference as a model for human allelic variation associated with addiction vulnerability phenotypes. The conditioned place preference results from studies of heterozygous csmd1 knockouts add significant value in confirming and extending modest-magnitude human associations that might otherwise be difficult to confirm in any other manner. These data support the working hypothesis that differences in neuronal connections, including those mediated by differences in cell adhesion molecules, are likely to help underpin individual differences in addiction and cognitive phenotypes [38].
Supporting Information
(XLSX)
Table A: Original spreadsheet and statistical analysis of the 60 min locomotion in the original CSMD1 line. Mice were injected with saline or 10 mg/kg s.c. cocaine, as indicated and immediately placed into dark, sound-attenuating locomotor boxes (42 x 42 cm). Total distance traveled was calculated from infrared beam breaks by an Optovarimax ATS System. Table B: Original spreadsheet and statistical analysis of the Rotarod test in the original CSMD1 line. Latencies (s) to fall off on each day are presented. Table C: Original spreadsheet and statistical analysis of the weights of the back-crossed CSMD1 mice. Table D: Original spreadsheet and statistical analysis of the hanging wire test of the back-crossed CSMD1 mice. Table E: Original spreadsheet and statistical analysis of the Rotarod test of the back-crossed CSMD1 mice. Latencies (s) to fall off on each day are presented. Table F: Original spreadsheet and statistical analysis of the Dark box emergence and the open field tests of the back-crossed CSMD1 mice. For the Dark box emergence test, the testing cage (18 x 36 cm) consisted of two compartments: a dark chamber (18 x 18 cm) with black walls and a small opening (5 cm) leading to a Plexiglas compartment. An animal was placed into the dark chamber. The latency to emerge and the time spent outside the dark chamber during the 10 min trial were measured using the Optovarimax system (Columbus Instruments, OH). For the Open Field test, mice were placed singly in an open field (42 x 42 cm) for ten minutes. The time each animal spent in the central quadrant was recorded. Table G: Original spreadsheet and statistical analysis of the Conditioned Place preference (CPP) test of the back-crossed CSMD1 mice. Times spent on the cocaine-paired side during two pre-tests (Wire-T1, Wire-T2) and post-test (Wire-T3) and the difference (Ave ∆T) between the post-test and average of the two pretests (Ave T 1–2) are presented. Table H: Original spreadsheet and statistical analysis of the locomotion recorded during the CPP pre-test (T1- Loco) of the back-crossed CSMD1 mice. Table I: Original spreadsheet and statistical analysis of the locomotion recorded during the first CPP conditioning session (1st COC Loco) with the back-crossed CSMD1 mice. Table J: Original spreadsheet and statistical analysis of the locomotor sensitiziation of the back-crossed CSMD1 mice that occurred during the two CPP conditioning sessions (1st COC, 2nd COC). Table K: Original spreadsheet and statistical analysis of the Morris water maze (MWM) learning curves of the back-crossed CSMD1 mice. Values for each day are averages of four trials performed on that day. Table L: Original spreadsheet and statistical analysis of the Morris water maze (MWM) Latencies to reach the platform during each trial on day 1 are presented. Table M: Original spreadsheet and statistical analysis of the results from the (MWM) probe trial.
(XLSX)
Acknowledgments
This work was supported by the National Institutes of Health (NIH)–Intramural Research Program, NIDA, DHHS (Dr. Uhl). We are grateful for access to brain samples from the University of Maryland Brain Tissue Bank.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Funding was provided by National Institutes on Drug Abuse Intramural Research Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE, et al. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Archives of general psychiatry 2008;65: 683–693. 10.1001/archpsyc.65.6.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Uhl GR, Drgon T, Johnson C, Fatusin OO, Liu QR, Contoreggi C, et al. "Higher order" addiction molecular genetics: convergent data from genome-wide association in humans and mice. Biochemical pharmacology 2008;75: 98–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Drgon T, Zhang PW, Johnson C, Walther D, Hess J, Nino M, et al. Genome wide association for addiction: replicated results and comparisons of two analytic approaches. PloS one 2010;5: e8832 10.1371/journal.pone.0008832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Drgon T, Johnson CA, Nino M, Drgonova J, Walther DM, Uhl GR. "Replicated" genome wide association for dependence on illegal substances: genomic regions identified by overlapping clusters of nominally positive SNPs. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics 2011;156: 125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Drgon T, Johnson C, Walther D, Albino AP, Rose JE, Uhl GR. Genome-wide association for smoking cessation success: participants in a trial with adjunctive denicotinized cigarettes. Molecular medicine 2009;15: 268–274. 10.2119/molmed.2009.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson C, Drgon T, Liu QR, Zhang PW, Walther D, Li C- Y, et al. Genome wide association for substance dependence: convergent results from epidemiologic and research volunteer samples. BMC medical genetics 2008;9: 113 10.1186/1471-2350-9-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson C, Drgon T, Walther D, Uhl GR. Genomic regions identified by overlapping clusters of nominally-positive SNPs from genome-wide studies of alcohol and illegal substance dependence. PloS one 2011;6: e19210 10.1371/journal.pone.0019210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Drgon T, Montoya I, Johnson C, Liu QR, Walther D, Hamer D, et al. Genome-wide association for nicotine dependence and smoking cessation success in NIH research volunteers. Molecular medicine 2009;15: 21–27. 10.2119/molmed.2008.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Uhl GR, Drgon T, Johnson C, Walther D, David SP, Murphy M, et al. Genome-wide association for smoking cessation success: participants in the Patch in Practice trial of nicotine replacement. Pharmacogenomics 2010;11: 357–367. 10.2217/pgs.09.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uhl GR, Drgon T, Johnson C, Ramoni MF, Behm FM, Rose JE. Genome-wide association for smoking cessation success in a trial of precessation nicotine replacement. Molecular medicine 2010;16: 513–526. 10.2119/molmed.2010.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Uhl GR, Walther D, Musci R, Fisher C, Anthony JC, Storr CL, et al. Smoking quit success genotype score predicts quit success and distinct patterns of developmental involvement with common addictive substances. Molecular psychiatry 2014;19: 50–54. 10.1038/mp.2012.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li M, Gardiner JC, Breslau N, Anthony JC, Lu Q. A non-parametric approach for detecting gene-gene interactions associated with age-at-onset outcomes. BMC genetics 2014;15: 79 10.1186/1471-2156-15-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McGue M, Zhang Y, Miller MB, Basu S, Vrieze S, Hicks B, et al. A genome-wide association study of behavioral disinhibition. Behavior genetics 2013;43: 363–373. 10.1007/s10519-013-9606-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Havik B, Le Hellard S, Rietschel M, Lybaek H, Djurovic S, Mattheisen M, et al. The complement control-related genes CSMD1 and CSMD2 associate to schizophrenia. Biological psychiatry 2011;70: 35–42. 10.1016/j.biopsych.2011.01.030 [DOI] [PubMed] [Google Scholar]
- 15. Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nature genetics 2011;43: 969–976. 10.1038/ng.940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arnedo J, Svrakic DM, Del Val C, Romero-Zaliz R, Hernandez-Cuervo H, Molecular Genetics of Schizophrenia Consortium, et al. Uncovering the Hidden Risk Architecture of the Schizophrenias: Confirmation in Three Independent Genome-Wide Association Studies. The American journal of psychiatry 2015;172:139–153. 10.1176/appi.ajp.2014.14040435 [DOI] [PubMed] [Google Scholar]
- 17. Bergen SE, O'Dushlaine CT, Ripke S, Lee PH, Ruderfer DM, Akterin S, et al. Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Molecular psychiatry 2012;17: 880–886. 10.1038/mp.2012.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nature genetics 2013;45: 1150–1159. 10.1038/ng.2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sleiman P, Wang D, Glessner J, Hadley D, Gur RE, Cohen N, et al. GWAS meta analysis identifies TSNARE1 as a novel Schizophrenia / Bipolar susceptibility locus. Scientific reports 2013;3: 3075 10.1038/srep03075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koiliari E, Roussos P, Pasparakis E, Lencz T, Malhotra A, Siever LJ, et al. The CSMD1 genome-wide associated schizophrenia risk variant rs10503253 affects general cognitive ability and executive function in healthy males. Schizophrenia research 2014;154: 42–47. 10.1016/j.schres.2014.02.017 [DOI] [PubMed] [Google Scholar]
- 21. Steen VM, Nepal C, Ersland KM, Holdhus R, Naevdal M, Ratvik SM, et al. Neuropsychological deficits in mice depleted of the schizophrenia susceptibility gene CSMD1. PloS one 2013;8: e79501 10.1371/journal.pone.0079501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rose EJ, Morris DW, Hargreaves A, Fahey C, Greene C, Garavan H, et al. Neural effects of the CSMD1 genome-wide associated schizophrenia risk variant rs10503253. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics 2013;162B: 530–537. [DOI] [PubMed] [Google Scholar]
- 23. Donohoe G, Walters J, Hargreaves A, Rose EJ, Morris DW, Fahey C, et al. Neuropsychological effects of the CSMD1 genome-wide associated schizophrenia risk variant rs10503253. Genes, brain, and behavior 2013;12: 203–209. 10.1111/gbb.12016 [DOI] [PubMed] [Google Scholar]
- 24. Xiang B, Wu JY, Ma XH, Wang YC, Deng W, Chen ZF, et al. [Genome-wide association study with memory measures as a quantitative trait locus for schizophrenia]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. = Chinese journal of medical genetics 2012;29: 255–259. [DOI] [PubMed] [Google Scholar]
- 25. Swaminathan S, Kim S, Shen L, Risacher SL, Foroud T, Pankratz N, et al. Genomic Copy Number Analysis in Alzheimer's Disease and Mild Cognitive Impairment: An ADNI Study. International journal of Alzheimer's disease 2011;2011: 729478 10.4061/2011/729478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kraus DM, Elliott GS, Chute H, Horan T, Pfenninger KH, Sanford SD,et al. CSMD1 is a novel multiple domain complement-regulatory protein highly expressed in the central nervous system and epithelial tissues. Journal of immunology 2006;176: 4419–4430. [DOI] [PubMed] [Google Scholar]
- 27. Distler MG, Opal MD, Dulawa SC, Palmer AA. Assessment of behaviors modeling aspects of schizophrenia in Csmd1 mutant mice. 2012;PloS one 7: e51235 10.1371/journal.pone.0051235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tzschentke TM. Measuring reward with the conditioned place preference paradigm: a comprehensive review of drug effects, recent progress and new issues. Progress in neurobiology 1998;56: 613–672. [DOI] [PubMed] [Google Scholar]
- 29. Hishimoto A, Liu QR, Drgon T, Pletnikova O, Walther D, Zhu XG, et al. Neurexin 3 polymorphisms are associated with alcohol dependence and altered expression of specific isoforms. Human Molecular Genetics 2007;16: 2880–2891. [DOI] [PubMed] [Google Scholar]
- 30. Drgonova J, Zimonjic DB, Hall FS, Uhl GR Effect of KEPI (Ppp1r14c) deletion on morphine analgesia and tolerance in mice of different genetic backgrounds: when a knockout is near a relevant quantitative trait locus. Neuroscience 2010;165: 882–895. 10.1016/j.neuroscience.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hall FS, Centeno M, Perona MT, Adair J, Dobner PR, Uhl GR. Effects of neurotensin gene knockout in mice on the behavioral effects of cocaine. Psychopharmacology 2012; 219:35–45. 10.1007/s00213-011-2370-9 [DOI] [PubMed] [Google Scholar]
- 32. Morris R Developments of a water-maze procedure for studying spatial learning in the rat. Journal of Neuroscience Methods 1984;11: 47–60. [DOI] [PubMed] [Google Scholar]
- 33. Sora I, Li B, Igari M, Hall FS, Ikeda K. Transgenic mice in the study of drug addiction and the effects of psychostimulant drugs. Annals of the New York Academy of Sciences 2010;1187: 218–246. 10.1111/j.1749-6632.2009.05276.x [DOI] [PubMed] [Google Scholar]
- 34. McCleery A, Ventura J, Kern RS, Subotnik KL, Gretchen-Doorly D, Green MF, et al. Cognitive functioning in first-episode schizophrenia: MATRICS Consensus Cognitive Battery (MCCB) Profile of Impairment. Schizophrenia Res. 2014; 157:33–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bhatia S, Kleinjan DA Disruption of long-range gene regulation in human genetic disease: a kaleidoscope of general principles, diverse mechanisms and unique phenotypic consequences. Human genetics 2014;133: 815–845. 10.1007/s00439-014-1424-6 [DOI] [PubMed] [Google Scholar]
- 36. Ishiguro H, Liu QR, Gong JP, Hall FS, Ujike H, Morales M, et al. NrCAM in addiction vulnerability: positional cloning, drug-regulation, haplotype-specific expression, and altered drug reward in knockout mice. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology 2006;31: 572–584. [DOI] [PubMed] [Google Scholar]
- 37. Jackson KJ, Marks MJ, Vann RE, Chen X, Gamage TF, Warner JA, et al. Role of alpha5 nicotinic acetylcholine receptors in pharmacological and behavioral effects of nicotine in mice. The Journal of pharmacology and experimental therapeutics 2010;334: 137–146. 10.1124/jpet.110.165738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Uhl GR, Drgon T, Johnson C, Li CY, Contoreggi C, Hess J, et al. Molecular genetics of addiction and related heritable phenotypes: genome-wide association approaches identify "connectivity constellation" and drug target genes with pleiotropic effects. Annals of the New York Academy of Sciences 2008;1141: 318–381. 10.1196/annals.1441.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Table A: Original spreadsheet and statistical analysis of the 60 min locomotion in the original CSMD1 line. Mice were injected with saline or 10 mg/kg s.c. cocaine, as indicated and immediately placed into dark, sound-attenuating locomotor boxes (42 x 42 cm). Total distance traveled was calculated from infrared beam breaks by an Optovarimax ATS System. Table B: Original spreadsheet and statistical analysis of the Rotarod test in the original CSMD1 line. Latencies (s) to fall off on each day are presented. Table C: Original spreadsheet and statistical analysis of the weights of the back-crossed CSMD1 mice. Table D: Original spreadsheet and statistical analysis of the hanging wire test of the back-crossed CSMD1 mice. Table E: Original spreadsheet and statistical analysis of the Rotarod test of the back-crossed CSMD1 mice. Latencies (s) to fall off on each day are presented. Table F: Original spreadsheet and statistical analysis of the Dark box emergence and the open field tests of the back-crossed CSMD1 mice. For the Dark box emergence test, the testing cage (18 x 36 cm) consisted of two compartments: a dark chamber (18 x 18 cm) with black walls and a small opening (5 cm) leading to a Plexiglas compartment. An animal was placed into the dark chamber. The latency to emerge and the time spent outside the dark chamber during the 10 min trial were measured using the Optovarimax system (Columbus Instruments, OH). For the Open Field test, mice were placed singly in an open field (42 x 42 cm) for ten minutes. The time each animal spent in the central quadrant was recorded. Table G: Original spreadsheet and statistical analysis of the Conditioned Place preference (CPP) test of the back-crossed CSMD1 mice. Times spent on the cocaine-paired side during two pre-tests (Wire-T1, Wire-T2) and post-test (Wire-T3) and the difference (Ave ∆T) between the post-test and average of the two pretests (Ave T 1–2) are presented. Table H: Original spreadsheet and statistical analysis of the locomotion recorded during the CPP pre-test (T1- Loco) of the back-crossed CSMD1 mice. Table I: Original spreadsheet and statistical analysis of the locomotion recorded during the first CPP conditioning session (1st COC Loco) with the back-crossed CSMD1 mice. Table J: Original spreadsheet and statistical analysis of the locomotor sensitiziation of the back-crossed CSMD1 mice that occurred during the two CPP conditioning sessions (1st COC, 2nd COC). Table K: Original spreadsheet and statistical analysis of the Morris water maze (MWM) learning curves of the back-crossed CSMD1 mice. Values for each day are averages of four trials performed on that day. Table L: Original spreadsheet and statistical analysis of the Morris water maze (MWM) Latencies to reach the platform during each trial on day 1 are presented. Table M: Original spreadsheet and statistical analysis of the results from the (MWM) probe trial.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.