Abstract
To investigate the importance of topoisomerases for transcription of the galactose induced genes, we have studied the expression of GAL1, GAL2, GAL7 and GAL10 in Saccharomyces cerevisiae cells deficient for topoisomerases I and II. We find that topoisomerases are required for transcriptional activation of the GAL genes, but are dispensable for ongoing transcription, eliminating a role of the enzymes in transcriptional elongation. Furthermore, we demonstrate that promoter chromatin remodeling of the GAL genes is unaffected in the topoisomerase deficient strain. However, the cells fail to successfully recruit RNA polymerase II due to an inability of the TATA-binding protein (TBP) to bind to the TATA box in these promoters. We therefore argue that topoisomerases are required for accurate assembly of the preinitiation complex at the promoters of the GAL genes.
Introduction
Several studies have demonstrated that transcription and DNA supercoiling are tightly coupled [1, 2]. The impact of transcription on DNA supercoiling has been explained by the “Twin-supercoiled-domain-model” [3], which implies that transcription by an RNA polymerase generates domains of positive and negative supercoiling in front of and behind the polymerase, respectively. These changes in superhelicity may eventually stop the advancing polymerase and/or perturb protein-DNA interactions if not removed or dispersed to other regions.
DNA topoisomerases solve topological problems arising during DNA metabolism. In Saccharomyces cerevisiae DNA superhelicity is primarily influenced by topoisomerase I (Top1) and topoisomerase II (Top2), encoded by the TOP1 and TOP2 gene, respectively [4]. Top1 removes helical tension by introducing a nick in one of the DNA strands, thus relieving superhelical tension by rotation of the cleaved strand around the intact strand. Top2 creates a transient double-stranded break in the DNA in order to transport another DNA duplex through the break [4]. Thus, both enzymes are able to relax supercoiled DNA, but they show differences in their substrate preferences, where Top1 is faster than Top2 in relaxation of naked DNA, whereas the opposite is the case when nucleosomal DNA is relaxed [5].
Chromatin structure adds another layer of complexity to DNA supercoiling. Approximately 80% of the genome is covered by nucleosomes in yeast [6], and nucleosomes influence transcription as they release and absorb negative superhelicity by dissociation and re-association with DNA, respectively [7]. In support of this, topoisomerases have been demonstrated to affect nucleosome dynamics. Thus, an early study showed a requirement of either Top1 or Top2 for proper chromatin assembly [8], and more recently a genome wide study demonstrated a direct requirement of Top1 for efficient nucleosome disassembly at gene promoters [9].
It has recently been suggested that chromatin is able to adapt to changes in DNA superhelicity by a slight conformational change, which is reverted upon relaxation by either Top1 or Top2 [5]. This implies that the chromatin fiber is a torsionally resilient structure, which can act as a topological buffer and facilitate dissipation of topological strain [10–12]. In addition to this, gathering evidence points to the conclusion that supercoiling is a dynamic entity, which is able to spread from the site of generation to far-reaching regions, thereby having long ranging effects [1, 12]. In eukaryotes, a change in DNA superhelicity may thus exert an additional effect on transcription via changes at the chromatin level.
Several studies have established a role of topoisomerases in transcription and transcriptional regulation. Accordingly, a genome-wide study in yeast showed a preferential localization of the enzymes to intergenic regions, i.e. promoter regions, of highly transcribed genes [13, 14], and Top1 and Top2 were found to act redundantly to enhance the recruitment of RNA polymerase II [13]. Other yeast studies have shown up- or downregulation of specific genes in the absence of either Top1 or Top2 activity, demonstrating roles of the individual enzymes in transcriptional regulation [15, 16]. Furthermore, transcription of highly expressed genes were shown to require both topoisomerase I and II in human cells, whereas genes with lower transcription managed with only topoisomerase I, demonstrating the importance of topoisomerases in gene regulation [17].
A recent study from our laboratory combined microarray gene expression analyses and single gene studies to investigate the role of topoisomerases for global gene expression [15]. Topoisomerases were found to have a major impact on transcription of a subset of genes, characterized by highly regulated transcription initiation. For the inducible PHO5 gene we demonstrated that topoisomerases were required during transcriptional activation, but not for reinitiation and transcription elongation. In the absence of topoisomerase activity, the Pho4 transcription factor failed to bind to the promoter, thus inhibiting eviction of nucleosomes from the promoter region.
In the present work we have studied transcription of the galactose inducible genes, GAL1, GAL2, GAL7, and GAL10 to investigate if topoisomerases have a similar effect on the transcriptional activation of these genes. As for the PHO5 gene, we find that topoisomerases are essential for activation of the GAL genes but not for ongoing transcription. However, we discover that nucleosome removal from the promoters is unperturbed during transcriptional activation of the GAL genes in a strain lacking functional topoisomerases, but the strain displays faulty RNA polymerase II recruitment to the GAL gene promoters. In correlation with this, we find that the TATA-binding protein (TBP) fails to bind to the TATA box in these promoters, suggesting an involvement of topoisomerases in an early step of preinitiation complex assembly. Thus, although the overall effect of topoisomerase deficiency is the same for different inducible genes, the specific step, where the enzymes exert their effect, may vary from gene to gene.
Materials and Methods
Yeast strains and growth conditions
All S. cerevisiae strains used are derivatives of W303a and are listed in Table 1. For galactose induction experiments, yeast strains were grown to a density of ~107 cells/ml at 25°C in YP with 2% raffinose and arrested in G1 with α-factor (Lipal Biochem, Zürich, Switzerland). Top2 was subsequently inhibited for 15 min at 37°C before galactose was added to a final concentration of 2% for induction of the galactose responsive genes. A sample was taken just prior to addition of galactose to serve as an uninduced control.
Table 1. S. cerevisiae strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| Ay-120 | MATa ade2-1 trp1-1 his3-11 his3-15 ura3-1 leu2-3 leu2-112 canI-100 Cir0 | R. Rothstein (W303) |
| Ay-109 | Ay-120 with top1::NAT top2-1 ts | [15] |
| Ay-127 | Ay-120 with top2-1 ts | [15] |
| Ay-161 | Ay-120 with top1::NAT | [15] |
| Ay-375 | Ay-120 with TOP1-13xcMyc-TRP | This study |
| Ay-386 | Ay-120 with TOP2-13xcMyc-TRP | This study |
| Ay-424 | Ay-120 with gal80::URA3 | This study |
| Ay-425 | Ay-120 with top1::NAT top2-1ts, gal80::URA3 | This study |
| Ay-483 | Ay-120 with 3xHA-TBP-URA3 | K. Struhl (YLK4) |
| Ay-484 | Ay-120 with top1::NAT top2-1 ts, 3xHA-TBP-URA3 | This study |
mRNA extraction and qPCR
For analysis of transcription levels by qPCR, cells were grown as described above, and samples of 2x107 cells were collected at the indicated time points. RNA was purified by use of RNeasy (Invitrogen, Carlsbad, CA), and cDNA was made by SuperScript II RT-PCR (Invitrogen, Carlsbad, CA) using oligo dT primers (Figs 1C and 2B) or by QuantiTect Reverse Transcription Kit (Qiagen) using random primers (Fig 1D). mRNA levels were quantified by Real-time PCR performed with HOT FIREPol EvaGreen qPCR Mix Plus (Solis Biodyne, Tartu, Estonia) using a Stratagene MX3000 (Agilent, Santa Clara, CA) and normalized to the ACT1 and GAPDH mRNA levels. Primer sequences are listed in Table 2.
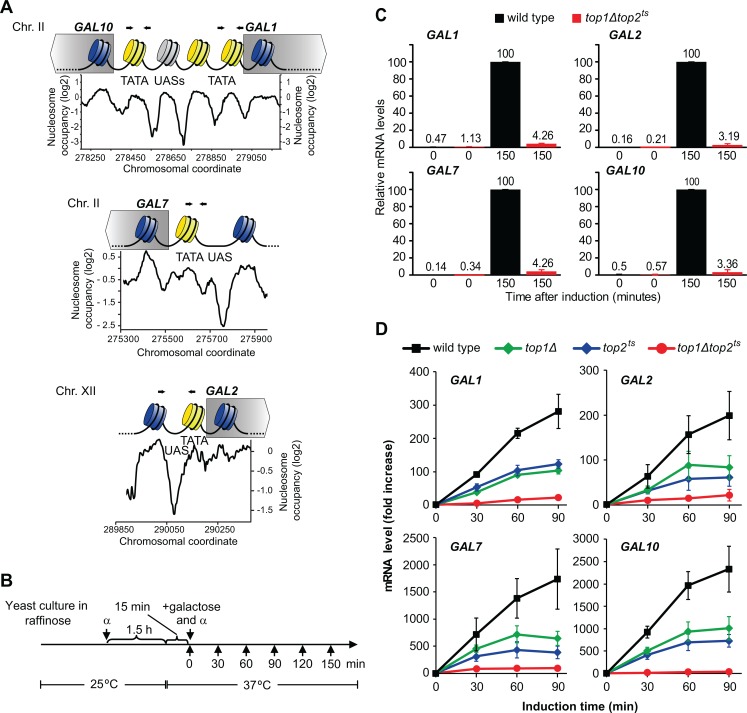
Fig 1. GAL gene transcription requires topoisomerase activity.
(A) Organization of the GAL genes. GAL1 and GAL10 are located on Chromosome II (Chr. II) and share a promoter with two UASs and three nucleosomes. The GAL7 promoter, located immediately after the GAL10 open reading frame on Chr. II, contains a single UAS and a single nucleosome. The GAL2 promoter is similar to the GAL7 promoter, but is located on Chr. XII. Promoter nucleosomes are illustrated in yellow. A promoter nucleosome occupancy profile [6] is shown below each gene (black line) indicating chromosomal coordinates of nucleosomes. Primers used in ChIP experiments are indicated by black arrows. UAS, Upstream Activating Sequence. TATA, TATA box. (B) Experimental setup. Cells were grown at 25°C in raffinose media (de-repressive conditions), and α-factor was added to arrest cells in G1. After 1.5 hours, cells were shifted to the restrictive temperature. After Top2 inactivation for 15 minutes, cells were treated with galactose to induce the GAL genes, α-factor was added again to keep cells in G1, and samples were collected at the indicated time points. α, α-factor. (C) Induction of GAL1, GAL2, GAL7, and GAL10 in wild type and top1Δtop2 ts cells. Cells were treated as illustrated in (B), and samples were collected 0 and 150 minutes after galactose treatment. mRNA was isolated, and the levels of the individual GAL genes and two control genes (GAPDH and ACT1) were quantified by qPCR. mRNA levels of the GAL genes were calculated relative to the mean of the mRNA levels for GAPDH and ACT1 for each time point and the value obtained in wild type at the latest time point was set to 100. Numbers indicate relative mRNA levels. Averages from two individual experiments are shown with error bars representing ± one standard deviation. (D) Induction of GAL1, GAL2, GAL7, and GAL10 in wild type, top1Δ, top2 ts, and top1Δtop2 ts cells. Cells were treated as shown in (B), and samples were collected at the indicated time points for qPCR measurements of GAL gene mRNA levels. mRNA levels are presented as fold increase relative to the level at time point 0. Averages from three individual experiments are shown with error bars representing ± one standard deviation.
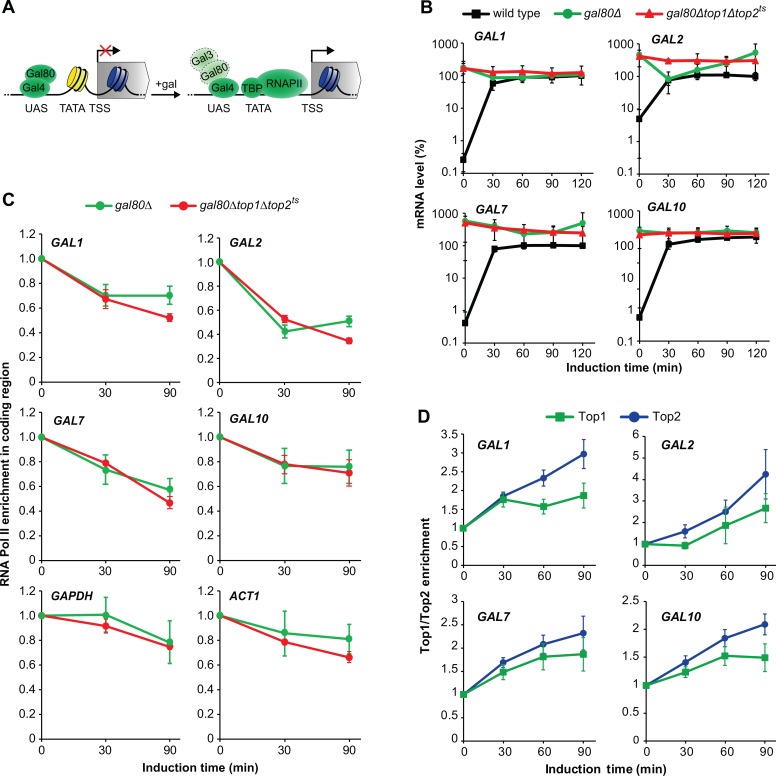
Fig 2. Topoisomerase activity has a direct role in GAL gene activation but is not required for transcriptional elongation and reinitiation.
(A) Overview of promoter changes during transcriptional induction of the GAL genes. (left) In raffinose, the GAL gene promoter is covered by nucleosomes except at the UAS, which binds Gal4 having its activation domain blocked by Gal80. (right) Upon galactose addition, Gal3 binds Gal80, leaving the activation domain of Gal4 free to bind chromatin remodelers. Subsequent removal of promoter nucleosomes allows recruitment of TBP and RNA polymerase II. UAS, Upstream Activating Sequence. TATA, TATA box. TSS, Transcription Start Site. gal, galactose. Light coloring and dashed borders of Gal3 and Gal80 indicate that the enzymes do not block the Gal4 activation domain, either due to dissociation or rearrangement of the complex. (B) Time course experiment of GAL1, GAL2, GAL7, and GAL10 transcription in wild type, gal80Δ, and gal80Δtop1Δtop2 ts cells. Cells were treated as in Fig 1B, and mRNA levels of the individual genes were quantified by qPCR, normalized to the wild type level at the latest time point (set to 100%), and presented on a log10-scale. The average from two individual experiments is shown, and error bars represent ± one standard deviation. (C) Time course experiment with ChIP analysis of RNA polymerase II enrichment in the coding regions of the GAL genes and two control genes, GAPDH and ACTI, following transcriptional activation. Cells were treated as in Fig 1B, and ChIP was performed with antibodies recognizing the C-terminal domain of the Rpb1 subunit of RNA polymerase II. RNA polymerase II binding levels were normalized relative to the binding at the 0 min time point (set to 1). (D) Time course experiment with ChIP analysis of Top1 and Top2 enrichment in the promoters of the GAL genes following transcriptional activation. Cells expressing the endogenous Top1 or Top2 enzymes fused to a cMyc tag were treated as described in Fig 1B, and ChIP was performed with antibodies recognizing the cMyc tag. Top1 and Top2 binding levels were normalized as in (C). In (C) and (D) averages from three individual experiments are shown, and error bars represent ± one standard deviation. Positions of primers used in the ChIP experiments for the individual GAL genes are indicated with arrows in Fig 1A and presented in Table 2.
Table 2. Primers used in this study.
| Name | Sequence | Comment |
|---|---|---|
| AP 487 | CACCAACTGTTTGGCTCCAT | RT-qPCR of TDH1 (GAPDH) |
| AP 488 | TAGCAGCACCGGTAGAGGAT | RT-qPCR of TDH1 (GAPDH) |
| AP 1820 | CCATAATGCCTCCTATATTTAGCCTTT | ChIP at TEL06R (normalization) |
| AP 1821 | TCCGAACGCTATTCCAGAAAGT | ChIP at TEL06R (normalization) |
| AP 2109 | GCCTTCTACGTTTCCATCCA | RT-qPCR of ACT1 |
| AP 2110 | GGCCAAATCGATTCTCAAAA | RT-qPCR of ACT1 |
| AP 2161 | AACAAACATTTCGCAGGCTA | ChIP in the promoter of GAL2 |
| AP 2162 | TATTCTTGATGATAATTGAA | ChIP in the promoter of GAL2 |
| AP 2165 | TTCCGACCTGCTTTTATATC | ChIP in the promoter of GAL7 |
| AP 2166 | ACAGTGTTCACAAAATAGCC | ChIP in the promoter of GAL7 |
| AP 2190 | AGCTGCATAACCACTTTAAC | ChIP in the promoter of GAL1 |
| AP 2191 | GACGTTAAAGTATAGAGGTA | ChIP in the promoter of GAL1 |
| AP 2192 | GGCATTACCACCATATACAT | ChIP in the promoter of GAL10 |
| AP 2193 | GAAAGTTCCAAAGAGAAGGT | ChIP in the promoter of GAL10 |
| AP 2359 | CGTTGCTTTAGCTGTTGTT | RT-qPCR of GAL1 |
| AP 2360 | CTGATCCATACCGCCATT | RT-qPCR of GAL1 |
| AP 2361 | TTGGCCTGGATGATTCCT | RT-qPCR of GAL2 |
| AP 2362 | AGCGCCCAAAAGTAAACA | RT-qPCR of GAL2 |
| AP 2363 | CCCAGTATGGAACAACAAC | RT-qPCR of GAL7 |
| AP 2364 | CTGATTTGTTTGCCGATTAC | RT-qPCR of GAL7 |
| AP 2365 | ACCAGAAGCTTTGCAGAA | RT-qPCR of GAL10 |
| AP 2366 | AAGGTTTGTGTCGTGAGT | RT-qPCR of GAL10 |
ChIP and Western Blot
ChIP was performed with 2.5x108 cells as described previously [15] with minor modifications. Thus, wash of antibody-coupled beads after incubation with extract was performed two times with Lysis buffer (50 mM Hepes, pH 7.5, 140 mM NaCl, 1 mM EDTA, pH 8, 1% Triton X-100, 0.1% Natriumdeoxycholate and protease inhibitors), one time with Wash buffer (10 mM Tris-HCl, pH 8, 500 mM NaCl, 1 mM EDTA, pH 8, 0.5% NP-40, 0.5% Natriumdeoxycholate and protease inhibitors), and one time with TE buffer (Tris-HCl, pH 7.5, mM EDTA). Histone H3 was precipitated with monoclonal antibodies recognizing the H3 C-terminal tail (ab1791 available from Abcam, Cambridge, UK), Gal4 was precipitated with a polyclonal anti-Gal4 antibody (ab1396 available from Abcam, Cambridge, UK) and RNA polymerase II was precipitated using a monoclonal antibody against the C-terminal domain of the RPB1 subunit (ab5408 available from Abcam, Cambridge, UK). For ChIP of RNA polymerase II, an extra washing step was included with Lysis500 (Lysis buffer containing 500 mM NaCl) after before wash with the Wash buffer. For ChIP of 3xHA-tagged TBP, monoclonal antibodies against the HA epitope tag were used (Santa Cruz), whereas ChIP of cMyc tagged Top1 or Top2 was performed with monoclonal antibodies targeting the cMyc epitope tag (Santa Cruz). Enrichment was calculated as 2(CT IP – CT beads)/CT Input, and was normalized to the enrichment in a telomeric region (TEL06R). The 0 min time point was set to 1. Sequences of the primers used in the ChIP experiments are listed in Table 2. For each gene, the same primer set was used for ChIP of H3, the RPB1 subunit of RNA polymerase II, TBP, Top1, Top2, and Gal4. The resolution of the individual ChIP assays was approximately 500 bp. For Western blotting of TBP, proteins were precipitated from wild type and top1Δtop2ts cells expressing HA tagged TBP by trichloroacetic acid. Proteins were subjected to sodium dodecyl sulphate polyacrylamide gel electrophoresis, and western blotting was performed with antibodies targeting the HA epitope tag on TBP (Santa Cruz) and anti Mcm2 antibodies (Santa Cruz).
Results
Topoisomerase activity is required for GAL gene transcription
The GAL genes are a group of genes, which is induced when galactose is used as the carbon source in S. cerevisiae. To investigate, how topoisomerase deficiency influences transcription of this group of genes, we have studied transcription of GAL1, GAL2, GAL7 and GAL10 in an S. cerevisiae strain having a deletion of the TOP1 gene and a temperature-sensitive mutation in the TOP2 gene (top10Δtop2 ts). A schematic presentation of the individual GAL genes is shown in Fig 1A. In an earlier study we found that absence of topoisomerases dramatically inhibited the transcription of inducible genes, including the GAL genes [15]. To establish if lack of topoisomerases causes a true inhibition of GAL gene transcription or merely a kinetic delay, we performed an experiment with wild type and top1Δtop2 ts cells as outlined in Fig 1B, where transcription was followed for an extensive period of time. To avoid genome-wide effects of topological challenges caused by replication [14, 18] as well as abortive mitosis due to lack of Top2 activity, the cells were kept in G1 throughout the experiment by treatment with α-factor. After inactivation of Top2 by transfer of cells to the restrictive temperature of 37°C, the GAL genes were activated by addition of galactose. Cells were collected before transfer to inducible conditions (“0”) and after an induction period of 150 min, and mRNA levels were determined by qPCR. In top1Δtop2 ts cells the induction levels of all four GAL genes remained very low even after 150 minutes under inducing conditions relative to the levels obtained in wild type cells (Fig 1C). Thus, the absence of topoisomerases does not result in a kinetic delay in GAL gene transcription. Rather, the enzymes are required for transcription of the genes per se.
The observed lack of GAL gene transcription in top1Δtop2 ts cells suggests that topoisomerase relaxation activity is required for transcription. Alternatively, one of the enzymes could play a more specific, although essential role for GAL gene transcription, in which case no transcription would be observed in cells lacking this enzyme. To differentiate between these two possibilities, we compared GAL gene transcription in top1Δ, top2 ts and top1Δtop2 ts cells using the experimental setup outlined in Fig 1B. Although top1Δ and top2 ts cells showed reduced GAL gene transcription relative to wild type cells, the level in each single mutant was significantly increased compared to the level in the double mutant (Fig 1D). This demonstrates that GAL gene transcription is sensitive to topoisomerase dosage. The result therefore strongly suggests that GAL gene transcription requires DNA relaxation activity, which is the only common activity of Top1 and Top2.
Topoisomerases are directly required for GAL gene activation but are dispensable for transcriptional elongation and reinitiation
When yeast cells are grown in media containing raffinose, the GAL genes are in a “primed” or de-repressed state, ready to undergo rapid induction upon galactose addition [19]. In this state, the transcription factor Gal4 is bound to the Upstream Activating Sequence (UAS) in the promoter region. However, the protein domain responsible for recruitment of factors involved in transcriptional activation is blocked by Gal80 through protein-protein interactions (Fig 2A, left) [20, 21]. When galactose is added to the media, the activation domain on Gal4 becomes accessible for binding of chromatin remodeling factors. This has been suggested to take place either by a direct dissociation of Gal80 due to interaction with Gal3 [22, 23] or by formation of a complex between Gal4, Gal80 and Gal3, which leaves the domain accessible for other interactions [24, 25]. Binding of chromatin remodeling factors ensures nucleosome eviction from the promoter region and exposure of the TATA box, thus paving the way for transcriptional activation (Fig 2A, right) [24, 26]. To investigate, whether topoisomerases were required for transcriptional activation of the GAL genes or for continued transcription, we took advantage of the fact that deletion of GAL80 leads to constitutive expression of the GAL genes due to elimination of the need for external stimuli. Wild type, gal80Δ, as well as gal80Δtop1Δtop2 ts cells were therefore cultured as shown in Fig 1B, and samples were withdrawn at the indicated time points and analyzed for GAL gene mRNA levels. In contrast to wild type cells gal80Δ and gal80Δtop1Δtop2 ts cells displayed high mRNA levels of the GAL genes under de-repressive conditions at the “0” minutes time points (reflecting the situation at the permissive temperature for top2 ts) (Fig 2B). Interestingly, following transfer to inducible conditions at the restrictive temperature, gal80Δtop1Δtop2 ts cells still accumulated mRNA at a level comparable to gal80Δ cells and similar to fully induced wild type cells. This demonstrates that once activation has taken place, continued transcription including transcription elongation is independent of topoisomerase activity.
To verify that transcription elongation took place to the same extent in gal80Δ and gal80Δtop1Δtop2 ts cells we investigated the level of RNA polymerase II in the coding region of the GAL genes in the two strains. This was performed by chromatin immunoprecipitation (ChIP) with antibodies against the C-terminal domain of the RPBI subunit of RNA polymerase II (Fig 2C). As expected, the level of RNA polymerase II was similar in the coding regions of the GAL genes in the two strains, consistent with a topoisomerase independency of GAL gene transcription elongation. A drop in RNA polymerase II enrichment of about 50% was seen in both strains during the experiment. A drop was also seen in two control genes, GAPDH and ACTI, and probably reflects the transfer of cells to 37°C. Taken together, the experiments demonstrate that topoisomerases are required during the initial activation of the GAL genes, whereas transcription elongation and re-initiation can occur in the absence of topoisomerase activity, as was observed earlier for the PHO5 gene [15].
The observed requirement of topoisomerase activity during GAL gene activation can be caused by a direct need for DNA relaxation activity in the GAL gene promoters. However, relaxation activity could also be required indirectly in a separate step essential for GAL gene activation. If the enzymes exert a direct function to relax promoter regions during GAL gene activation, we expect a recruitment of Top1 and Top2 to the promoter regions during activation. To investigate if this took place, yeast strains expressing either Top1 or Top2 fused to a cMyc epitope tag were generated, and chromatin immunoprecipitation (ChIP) was performed with antibodies targeting the cMyc epitope tag. Top1 and Top2 were enriched approximately 2 and 3 fold, respectively, in the individual GAL gene promoters (Fig 2D), demonstrating a recruitment of both topoisomerases to the promoter regions upon induction. Taken together, the results suggest that Top1 and Top2 directly bind to the promoter regions to ensure that the topological state is optimal for efficient GAL gene activation.
Eviction of nucleosomes from the GAL gene promoters during activation occurs independent of topoisomerases
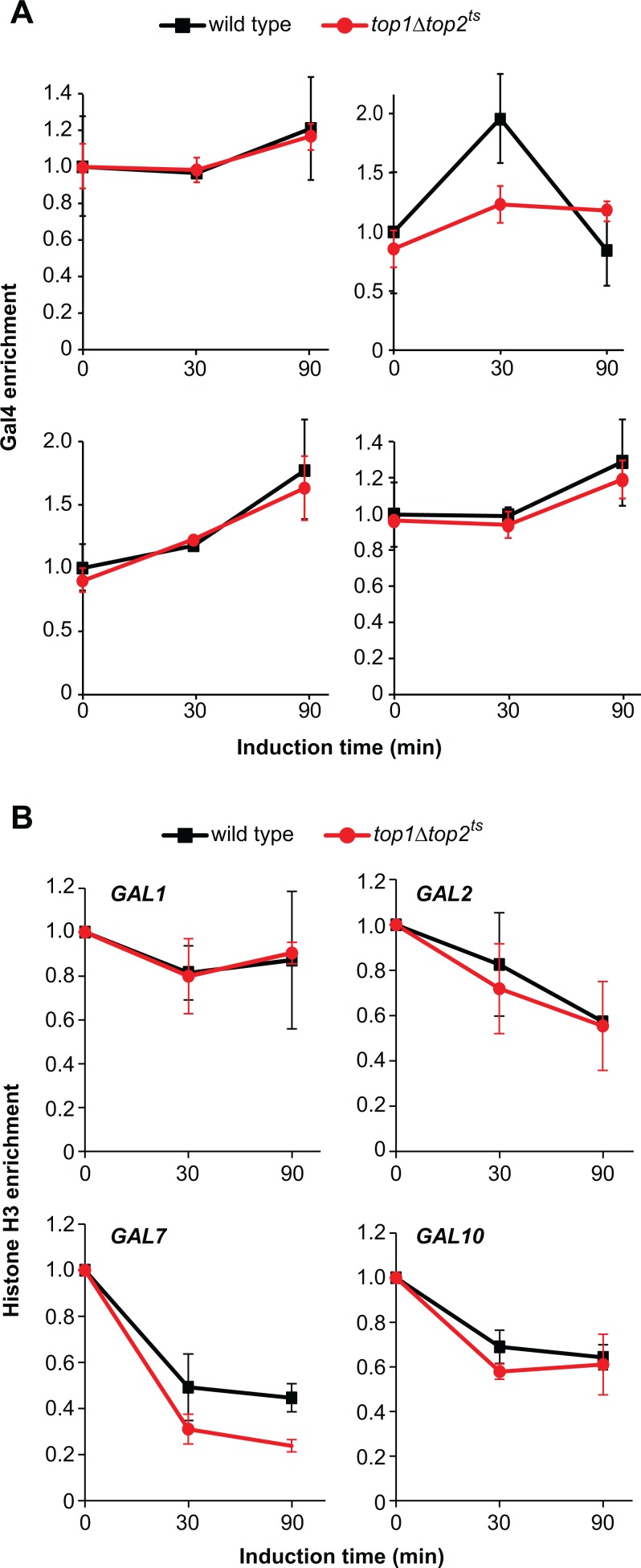
We next addressed, during which step in the activation process topoisomerases were required. A prerequisite for induction of the galactose responsive genes is removal of nucleosomes from the promoter regions to allow access of essential transcription factors (Figs 1A and 2A) [27]. Since the presence of Gal4 in the promoter region is essential for this step, we first used ChIP with antibodies against Gal4 to investigate whether or not this factor remained bound in top1Δtop2 ts cells. For all GAL genes, the level of Gal4 in the promoter region was similar in wild type and top1Δtop2 ts cells (Fig 3A). Thus, Gal4 remains bound in the absence of topoisomerases leaving the fundament for chromatin remodeling intact in top1Δtop2 ts cells.
Fig 3. Promoter chromatin remodeling is not affected by lack of topoisomerases.
(A) ChIP analysis of Gal4 binding in the GAL gene promoters of wild type and top1Δtop2 ts cells following transcriptional activation. Experimental setup was as described for Fig 1B, and ChIP was performed using antibodies against Gal4. Gal4 binding levels in the GAL gene promoters were normalized to the binding under de-repressed conditions in wild type at the 0 min time point (set to 1). (B) ChIP analysis of nucleosome removal from GAL gene promoters of wild type and top1Δtop2 ts cells following transcriptional activation. ChIP was performed using antibodies targeting histone H3. H3 binding levels in the GAL gene promoters were normalized relative to the binding under uninduced conditions at the 0 min time point (set to 1). Averages from three (A) or two (B) individual experiments are shown, and error bars represent ± one standard deviation. Positions of primers used in the ChIP experiments for the individual GAL genes are indicated with arrows in Fig 1A and presented in Table 2.
To investigate, whether nucleosome removal per se was perturbed in top1Δtop2 ts cells, we next used ChIP with antibodies targeting histone H3 to measure the nucleosomal occupancy in the promoter regions of the GAL genes during galactose induction. Nucleosomes were removed with equal kinetics and to a similar extent in both wild type and top1Δtop2 ts cells (Fig 3B). Thus, in contrast to our earlier observation with the inducible PHO5 gene [15], nucleosome removal from the GAL gene promoters occurs independent of topoisomerase activity.
GAL gene activation requires topoisomerases for preinitiation complex assembly
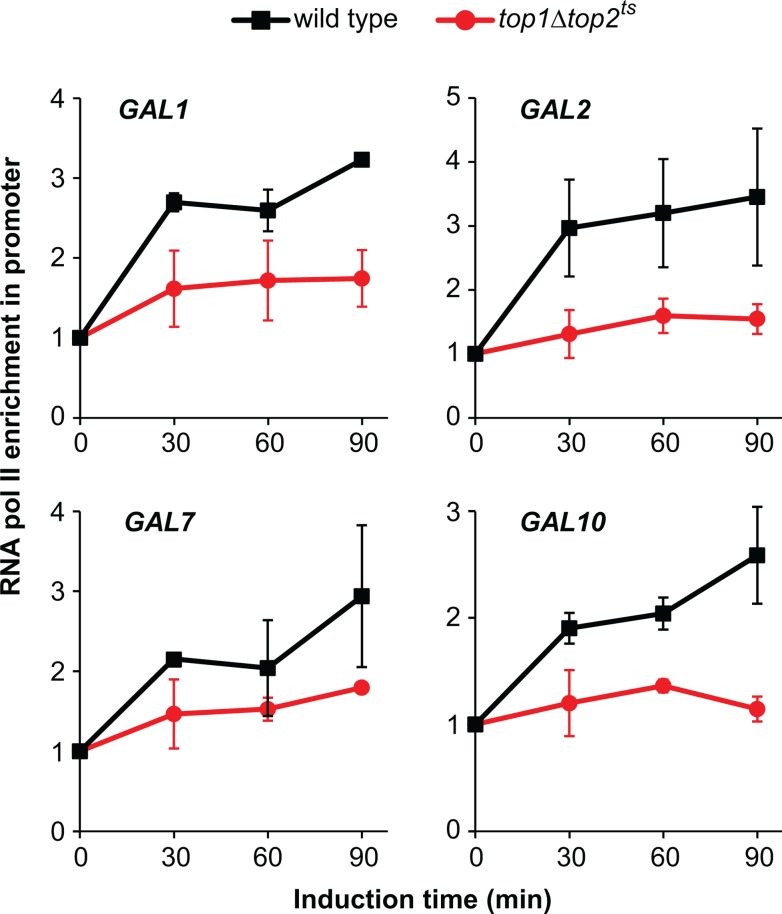
The step following eviction of nucleosomes from the promoters in transcriptional activation of the GAL genes is assembly of the preinitiation complex [19]. A principal part of this assembly is the recruitment of RNA polymerase II. In order to study the recruitment of RNA polymerase II to the promoters of the GAL genes, ChIP was performed using antibodies against the C-terminal domain of the RPB1 subunit of RNA polymerase II. As expected, an increase in RNA polymerase II occupancy in the promoter regions of the GAL genes was observed in wild type cells (Fig 4). Conversely, very little or no RNA polymerase II enrichment was seen in the GAL gene promoters in top1Δtop2 ts cells, demonstrating a requirement of topoisomerases for the recruitment of RNA polymerase II to the GAL gene promoters.
Fig 4. Topoisomerase activity is required for RNA polymerase II recruitment.
ChIP analysis of RNA polymerase II enrichment in GAL gene promoters of wild type and top1Δtop2 ts cells following transcriptional activation. Experimental setup was as described for Fig 1B, and ChIP was performed using antibodies targeting RNA polymerase II. RNA polymerase II binding levels were normalized relative to the binding under uninduced conditions at the 0 min time point (set to 1). Averages from two individual experiments are shown, and error bars represent ± one standard deviation. Positions of primers used in the ChIP experiments for the individual GAL genes are indicated with arrows in Fig 1A.
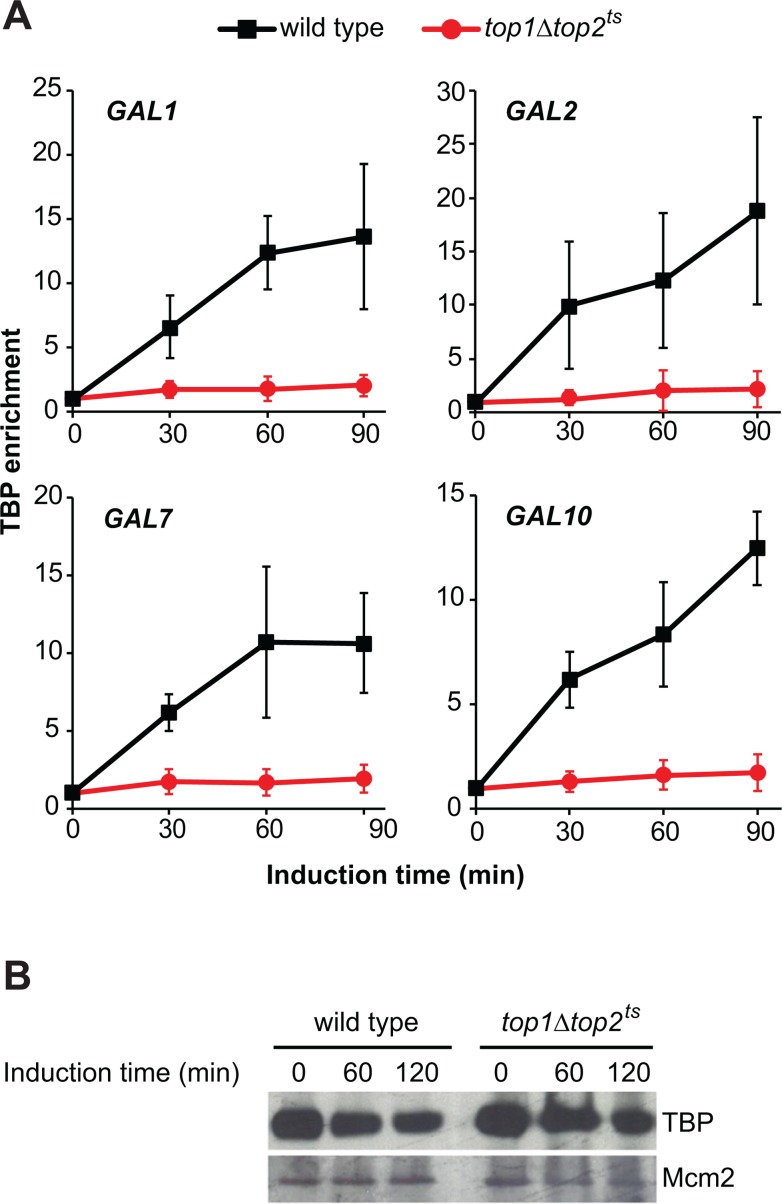
Binding of the TATA-binding protein (TBP) to the TATA box is the first step in the assembly of the preinitiation complex [28]. In order to investigate the role of topoisomerases in the binding of TBP to the TATA box in the promoter regions of the GAL genes, ChIP was again performed, this time with antibodies targeting an HA-tag N-terminally fused to TBP. As expected, an increase in TBP enrichment was observed in the promoters of the GAL genes in wild type cells (Fig 5A). However, no significant enrichment was seen in top1Δtop2 ts cells. Taken together with the observation that the cellular level of TBP was similar in wild type and topoisomerase deficient cells, (Fig 5B), this demonstrates a requirement of topoisomerases for TBP binding. Based on our results, we conclude that GAL gene activation requires topoisomerases either directly for TBP binding to the TATA box, or in a step between nucleosome eviction and TBP binding.
Fig 5. TBP binding to the TATA box requires topoisomerase activity.
(A) ChIP analysis of TBP enrichment in GAL gene promoters of wild type and top1Δtop2 ts cells following transcriptional activation. Experimental setup was as described for Fig 1B. ChIP was performed on cells having the endogenous TBP protein fused to a HA epitope tag using anti-HA antibodies. TBP binding levels were normalized relative to the binding under uninduced conditions at the 0 min time point (set to 1). Averages from three individual experiments are shown, and error bars represent ± one standard deviation. Positions of primers used in the ChIP experiments for the individual GAL genes are indicated with arrows in Fig 1A. (B) TBP protein levels in wild type and top1Δtop2 ts cells. Cells used in (A) were treated as in Fig 1B, and samples taken at the indicated time points were processed for Western Blot analysis with antibodies targeting the HA tag on TBP. Mcm2 was used as loading control.
Discussion
Our findings have revealed that topoisomerase activity is required for transcriptional activation of the GAL genes, whereas ongoing transcription can occur in the absence of Top1 and Top2. Furthermore, we demonstrate that topoisomerases are required for TBP binding to the TATA box, or in a step downstream of nucleosome eviction, but upstream of TBP binding. We have previously shown that topoisomerases are required for transcriptional activation of the phosphate regulated PHO5 gene, where the enzymes are necessary for binding of the transcription factor Pho4 to the promoter region [15]. In the present study, lack of topoisomerase activity does not influence the GAL gene induction pathway, as wild type and top1Δtop2 ts cells display equal nucleosome promoter clearing (Fig 3). The disparities in topoisomerase requirement between the PHO5 gene and the GAL genes are probably reflected in the dissimilarities in the induction pathways. In the case of PHO5, the Pho4 transcription factor has to bind the PHO5 promoter in order for nucleosome remodeling to occur [29]. In contrast, the corresponding Gal4 transcription factor, which is required for chromatin remodeling in the GAL system, is already bound to the promoter, but its chromatin remodeler recruiting domain is blocked by Gal80 [21]. Thus, this step in chromatin remodeling does not rely on novel protein-DNA interactions as it does in the PHO5 system.
Interestingly, no RNA polymerase II or TBP enrichment is observed in the promoters of the GAL genes in top1Δtop2 ts cells during transcriptional activation (Figs 4 and 5), whereas topoisomerases become dispensable once the GAL genes are activated (Fig 2). This indicates that transcriptional activation is fully dependent on topoisomerase activity although reinitiation and transcription elongation can take place in the already activated genes independent of topoisomerases. One explanation for this discrepancy could be that the half-lives of the mRNAs in question are very high, making it impossible to accurately assess, whether transcription in the gal80∆top1Δtop2 ts mutant is deregulated upon transfer to the restrictive temperature. However, as the half-lives of the mRNA’s from GAL1, GAL2, GAL7, and GAL10 are 18, 49, 27, and 20 minutes, respectively [30], and we measure transcript levels 135 min after transfer of the cells to the restrictive temperature for top2 ts, we find this explanation unlikely. A more plausible explanation is that after the RNA polymerase has escaped from the preinitiation complex, some of the general transcription factors remain in the promoter region, creating a reinitiation scaffold [31], which can persist even under topological conditions, where one or more of the factors would be unable to bind individually. Furthermore, several GAL genes have been shown to exhibit gene looping [32, 33], where the promoter and termination regions are brought into close proximity. Gene looping allows for easy RNA polymerase shuttling from the termination region to the promoter, and is believed to keep the promoter chromatin free and primed for reinitiation, which may eliminate the requirement for topoisomerases.
Transcriptional activation of a gene is a complicated process involving a plethora of proteins. The DNA superhelical changes occurring in topoisomerase deficient strains will most likely affect the activation process in a step involving protein-DNA interactions. As the nucleosomes bound at the GAL gene promoters are removed with equal kinetics in both wild type and top1Δtop2 ts cells (Fig 3), but TBP is unable to bind to the promoter in topoisomerase deficient cells, the requirement for topoisomerase activity has to be found between these two steps. Following nucleosome depletion, different co-activators, including Mediator and SAGA, bind at GAL gene promoters through interaction with Gal4 [34, 35]. These co-activators then assist in the recruitment of TBP to the TATA box through direct interactions [20]. The step requiring topoisomerase activity can therefore either be binding of TBP or one of its co-activators. However, we find it unlikely that binding of Mediator and SAGA is influenced by lack of topoisomerase activity, since these co-activators have been suggested to act as bridging proteins between Gal4 and TBP [20]. Rather, we expect that the DNA interaction of TBP will be impeded in topoisomerase deficient strains, where the global superhelical level is shifted towards a more positive state [12, 15, 36]. To this end, earlier studies have demonstrated that TBP binding is supercoiling sensitive [37–39]. This is supported by the earlier finding that TBP binding extensively distorts the DNA helix by introducing a 90° kink in the DNA, causing a slight underwinding of the DNA [39, 40]. Furthermore, an in vitro study has demonstrated that negative superhelicity results in a more active transcription of the immunoglobulin heavy chain gene when only TBP, TFIIB and RNA polymerase II are present [38].
If lack of topoisomerases affects TBP binding, one may speculate, how reinitiation can occur in gal80Δtop1Δtop2 ts cells. In these cells, TBP is already bound to the DNA template before cells are exposed to the positive supercoiling, which follows the transfer of cells to the restrictive temperature for top2 ts, and although TBP binding may be sensitive to DNA superhelicity, its dissociation may not necessarily be affected. Alternatively, the transient negative supercoiling generated due to polymerase movement during elongation could maintain binding of TBP, as has been suggested earlier [39].
In conclusion, we show that topoisomerase activity is essential for GAL gene activation, being required for TBP binding downstream of nucleosome eviction. This is in contrast to our earlier finding with the PHO5 inducible gene system, where topoisomerase activity was found to be required for binding of the transcription factor Pho4 prior to nucleosome eviction [15]. Topoisomerases can therefore influence different steps in the activation of inducible genes both upstream and downstream of promoter nucleosome clearance. Due to the multitude of protein-DNA interactions required for activation of inducible genes, these genes are thus highly vulnerable to changes in promoter topology.
Acknowledgments
We are grateful to Kevin Struhl for providing yeast strain Ay-483 containing the HA-tagged TBP (originally YLK4) and to Noriko Hansen for skillful technical assistance.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the Danish Natural Science Research Council, the Novo Nordisk Foundation, the Karen Elise Jensen Foundation, the Gangsted Foundation, the Simon Fougner Hartmanns Foundation, the Dagmar Marshalls Foundation, the Astrid Thaysens Foundation, and the Villum Kann Rasmussen Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15(2):146–54. Epub 2008/01/15. 10.1038/nsmb.1372 nsmb.1372 [pii]. . [DOI] [PubMed] [Google Scholar]
- 2. Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326(6111):414–6. Epub 1987/03/01. 10.1038/326414a0 . [DOI] [PubMed] [Google Scholar]
- 3. Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(20):7024–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nature reviews Molecular cell biology. 2002;3(6):430–40. 10.1038/nrm831 . [DOI] [PubMed] [Google Scholar]
- 5. Salceda J, Fernandez X, Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25(11):2575–83. Epub 2006/05/20. doi: 7601142 [pii] 10.1038/sj.emboj.7601142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nature genetics. 2007;39(10):1235–44. 10.1038/ng2117 . [DOI] [PubMed] [Google Scholar]
- 7. Clark DJ, Felsenfeld G. Formation of nucleosomes on positively supercoiled DNA. EMBO J. 1991;10(2):387–95. Epub 1991/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garinther WI, Schultz MC. Topoisomerase function during replication-independent chromatin assembly in yeast. Mol Cell Biol. 1997;17(7):3520–6. Epub 1997/07/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Durand-Dubief M, Persson J, Norman U, Hartsuiker E, Ekwall K. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J. 2010;29(13):2126–34. Epub 2010/06/08. 10.1038/emboj.2010.109 emboj2010109 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hamiche A, Carot V, Alilat M, De Lucia F, O'Donohue MF, Revet B, et al. Interaction of the histone (H3-H4)2 tetramer of the nucleosome with positively supercoiled DNA minicircles: Potential flipping of the protein from a left- to a right-handed superhelical form. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(15):7588–93. Epub 1996/07/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bancaud A, Conde e Silva N, Barbi M, Wagner G, Allemand JF, Mozziconacci J, et al. Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat Struct Mol Biol. 2006;13(5):444–50. Epub 2006/04/20. doi: nsmb1087 [pii] 10.1038/nsmb1087 . [DOI] [PubMed] [Google Scholar]
- 12. Joshi RS, Pina B, Roca J. Positional dependence of transcriptional inhibition by DNA torsional stress in yeast chromosomes. EMBO J. 2010;29(4):740–8. Epub 2010/01/09. 10.1038/emboj.2009.391 emboj2009391 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sperling AS, Jeong KS, Kitada T, Grunstein M. Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(31):12693–8. Epub 2011/07/21. 10.1073/pnas.1106834108 1106834108 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bermejo R, Capra T, Gonzalez-Huici V, Fachinetti D, Cocito A, Natoli G, et al. Genome-organizing factors Top2 and Hmo1 prevent chromosome fragility at sites of S phase transcription. Cell. 2009;138(5):870–84. Epub 2009/09/10. 10.1016/j.cell.2009.06.022 S0092-8674(09)00722-3 [pii]. 19737516. [DOI] [PubMed] [Google Scholar]
- 15. Pedersen JM, Fredsoe J, Roedgaard M, Andreasen L, Mundbjerg K, Kruhoffer M, et al. DNA Topoisomerases maintain promoters in a state competent for transcriptional activation in Saccharomyces cerevisiae. PLoS genetics. 2012;8(12):e1003128 10.1371/journal.pgen.1003128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nikolaou C, Bermudez I, Manichanh C, Garcia-Martinez J, Guigo R, Perez-Ortin JE, et al. Topoisomerase II regulates yeast genes with singular chromatin architectures. Nucleic Acids Res. 2013;41(20):9243–56. 10.1093/nar/gkt707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kouzine F, Gupta A, Baranello L, Wojtowicz D, Ben-Aissa K, Liu J, et al. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol. 2013;20(3):396–403. Epub 2013/02/19. 10.1038/nsmb.2517 nsmb.2517 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bermejo R, Doksani Y, Capra T, Katou YM, Tanaka H, Shirahige K, et al. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 2007;21(15):1921–36. Epub 2007/08/03. doi: 21/15/1921 [pii] 10.1101/gad.432107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sellick CA, Campbell RN, Reece RJ. Galactose metabolism in yeast-structure and regulation of the leloir pathway enzymes and the genes encoding them. International review of cell and molecular biology. 2008;269:111–50. 10.1016/S1937-6448(08)01003-4 . [DOI] [PubMed] [Google Scholar]
- 20. Traven A, Jelicic B, Sopta M. Yeast Gal4: a transcriptional paradigm revisited. EMBO Rep. 2006;7(5):496–9. Epub 2006/05/04. doi: 7400679 [pii] 10.1038/sj.embor.7400679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lue NF, Chasman DI, Buchman AR, Kornberg RD. Interaction of GAL4 and GAL80 gene regulatory proteins in vitro. Molecular and cellular biology. 1987;7(10):3446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sil AK, Alam S, Xin P, Ma L, Morgan M, Lebo CM, et al. The Gal3p-Gal80p-Gal4p transcription switch of yeast: Gal3p destabilizes the Gal80p-Gal4p complex in response to galactose and ATP. Mol Cell Biol. 1999;19(11):7828–40. Epub 1999/10/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang FL, Frey BR, Evans ML, Friel JC, Hopper JE. Gene Activation by Dissociation of an Inhibitor from a Transcriptional Activation Domain. Molecular and Cellular Biology. 2009;29(20):5604–10. 10.1128/Mcb.00632-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Platt A, Reece RJ. The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. EMBO J. 1998;17(14):4086–91. Epub 1998/07/22. 10.1093/emboj/17.14.4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abramczyk D, Holden S, Page CJ, Reece RJ. Interplay of a ligand sensor and an enzyme in controlling expression of the Saccharomyces cerevisiae GAL genes. Eukaryot Cell. 2012;11(3):334–42. Epub 2012/01/03. 10.1128/EC.05294-11 EC.05294-11 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pilauri V, Bewley M, Diep C, Hopper J. Gal80 dimerization and the yeast GAL gene switch. Genetics. 2005;169(4):1903–14. 10.1534/genetics.104.036723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lohr D, Venkov P, Zlatanova J. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 1995;9(9):777–87. . [DOI] [PubMed] [Google Scholar]
- 28. Martinez E. Multi-protein complexes in eukaryotic gene transcription. Plant Mol Biol. 2002;50(6):925–47. Epub 2003/01/09. . [DOI] [PubMed] [Google Scholar]
- 29. Svaren J, Schmitz J, Horz W. The transactivation domain of Pho4 is required for nucleosome disruption at the PHO5 promoter. EMBO J. 1994;13(20):4856–62. Epub 1994/10/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(9):5860–5. Epub 2002/04/25. 10.1073/pnas.092538799 092538799 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sarge KD, Park-Sarge OK. Gene bookmarking: keeping the pages open. Trends Biochem Sci. 2005;30(11):605–10. Epub 2005/09/29. doi: S0968-0004(05)00270-7 [pii] 10.1016/j.tibs.2005.09.004 . [DOI] [PubMed] [Google Scholar]
- 32. Laine JP, Singh BN, Krishnamurthy S, Hampsey M. A physiological role for gene loops in yeast. Genes Dev. 2009;23(22):2604–9. Epub 2009/11/26. 10.1101/gad.1823609 23/22/2604 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Greger IH, Proudfoot NJ. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 1998;17(16):4771–9. Epub 1998/08/26. 10.1093/emboj/17.16.4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bryant GO, Prabhu V, Floer M, Wang X, Spagna D, Schreiber D, et al. Activator control of nucleosome occupancy in activation and repression of transcription. PLoS Biol. 2008;6(12):2928–39. Epub 2008/12/26. 10.1371/journal.pbio.0060317 08-PLBI-RA-3549 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15(15):1946–56. Epub 2001/08/04. 10.1101/gad.911501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Joshi RS, Pina B, Roca J. Topoisomerase II is required for the production of long Pol II gene transcripts in yeast. Nucleic acids research. 2012;40(16):7907–15. Epub 2012/06/22. 10.1093/nar/gks626 gks626 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tabuchi H, Handa H, Hirose S. Underwinding of DNA on binding of yeast TFIID to the TATA element. Biochem Biophys Res Commun. 1993;192(3):1432–8. Epub 1993/05/14. doi: S0006-291X(83)71576-7 [pii] 10.1006/bbrc.1993.1576 . [DOI] [PubMed] [Google Scholar]
- 38. Parvin JD, Sharp PA. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73(3):533–40. . [DOI] [PubMed] [Google Scholar]
- 39. Kahn JD. Topological effects of the TATA box binding protein on minicircle DNA and a possible thermodynamic linkage to chromatin remodeling. Biochemistry. 2000;39(13):3520–4. Epub 2000/03/29. doi: bi992263f [pii]. . [DOI] [PubMed] [Google Scholar]
- 40. Nikolov DB, Chen H, Halay ED, Usheva AA, Hisatake K, Lee DK, et al. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377(6545):119–28. Epub 1995/09/14. 10.1038/377119a0 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.