SUMMARY
Chikungunya virus (CHIKV) is a mosquito-transmitted RNA virus that causes acute febrile infection associated with polyarthralgia in humans. Mechanisms of protective immunity against CHIKV are poorly understood, and no effective therapeutics or vaccines are available. We isolated and characterized human monoclonal antibodies (mAbs) that neutralize CHIKV infectivity. Among the 30 mAbs isolated, 13 had broad and ultrapotent neutralizing activity (IC50 < 10 ng/mL), and all of these mapped to domain A of the E2 envelope protein. Potent inhibitory mAbs blocked post-attachment steps required for CHIKV membrane fusion, and several were protective in a lethal challenge model in immunocompromised mice, even when administered at late time points after infection. These highly protective mAbs could be considered for prevention or treatment of CHIKV infection, and their epitope location in domain A of E2 could be targeted for rational structure-based vaccine development.
Introduction
Chikungunya virus (CHIKV) is an enveloped, positive-sense RNA virus in the Alphavirus genus of the Togaviridae family and is transmitted by Aedes species mosquitoes. The mature CHIKV virion contains two glycoproteins, the E1 fusion protein and the E2 attachment protein, which are generated from a precursor polyprotein, p62-E1, by proteolytic cleavage.. In humans, CHIKV infection causes fever and joint pain, which may be severe and last in some cases for years (Schilte et al., 2013; Sissoko et al., 2009; Staples et al., 2009). CHIKV has caused outbreaks in most regions of sub-Saharan Africa and also in parts of Asia, Europe, and the Indian and Pacific Oceans. In December 2013, the first transmission of CHIKV in the Western Hemisphere occurred, with autochthonous cases identified in St. Martin (CDC 2013). The virus spread rapidly to many islands in the Caribbean as well as Central, South, and North America. In less than one year, over a million suspected CHIKV cases in the Western Hemisphere were reported, and endemic transmission in more than 40 countries, including the United States was documented (CDC, 2014). At present, there is no licensed vaccine or antiviral therapy to prevent or treat CHIKV infection.
Although mechanisms of protective immunity to CHIKV infection in humans are not fully understood, the humoral response controls infection and limits tissue injury (Chu et al., 2013; Hallengard et al., 2014; Hawman et al., 2013; Kam et al., 2012b; Lum et al., 2013; Pal et al., 2013). Immune human γ-globulin neutralizes infectivity in cultured cells and prevents morbidity in mice when administered up to 24 h after viral inoculation (Couderc et al., 2009). Several murine monoclonal antibodies (mAbs) that neutralize CHIKV infection have been described (Brehin et al., 2008; Goh et al., 2013; Masrinoul et al., 2014; Pal et al., 2013; Pal et al., 2014), including some with efficacy when used in combination to treat mice or nonhuman primates following CHIKV challenge (Pal et al., 2013; Pal et al., 2014). In comparison, a limited number of human CHIKV mAbs have been reported, the vast majority of which exhibit modest neutralizing activity (Fong et al., 2014; Fric et al., 2013; Lee et al., 2011; Selvarajah et al., 2013; Warter et al., 2011).
We isolated a large panel of human mAbs that neutralize CHIKV infectivity in cell culture and successfully treated immunodeficient Ifnar−/− mice (lacking type I interferon receptors) inoculated with a lethal dose of CHIKV, even when administered as late as 60 h after infection. We identified the A domain of E2 as the major antigenic site for recognition by human mAbs that broadly neutralize CHIKV infection with ultrapotent activity and showed that the principal mechanism of inhibition is to prevent fusion.
Results
Isolation of CHIKV-specific human mAbs
We isolated a panel of mAbs from a single individual who acquired CHIKV infection in Sri Lanka in 2006 and presented with fever, arthralgias, and rash (Fig. S1). We transformed B cells in two separate experiments from a single blood sample collected from the donor five and a half years following natural infection. We observed a virus-specific B cell frequency of ~ 0.1% of total B cells and established 30 stable hybridomas from B cell lines secreting antibodies that bound to virus. The mAb panel contained IgGs of multiple subclasses, with 24 IgG1, 3 IgG2, and 2 IgG3; one was not determined due to poor hybridoma growth (Table 1). We determined the nucleotide sequences of the antibody variable gene region using cDNA of expressed antibody mRNAs in the cloned hybridomas. Each of the clones used distinct sequences to encode the associated mAbs, except for mAbs 2B4 and 4J21, which appeared identical in the variable regions and exhibited similar functional activity.
Table 1.
Characteristics of chikungunya virus-specific human monoclonal antibodies
| mAb1 | IgG sub- class2 |
λ/κ light chain2 |
ELISA binding to E2 ectodoma in (10 μg/mL)3 |
Major antigenic site | Neutralization against CHIKV VRP (strain SL15649)6 EC50 in ng/mL7 [95% confidence interval] |
In vitro neutralizing potency and breadth of chikungunya virus-specific human mAbs | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Competition binding group for purified E2 protein 4 |
Mutagenesis mapping | Neutralization against CHKV against indicated genotype and strain* EC50, ng/mL7 [95% confidence interval] |
|||||||||
| E2 Domain5 | E2 residues for which reduced binding was noted when altered to alanine |
West African genotype NI 64 IbH 35 strain |
ECSA genotype LR2006 OPY1 (LR) strain |
Asian genotype | |||||||
| RSUI strain | 2014 Caribbean 99659 strain |
||||||||||
| 2H1 | IgG2 | λ | ++ | Low binding | E2-DA | R80, T116 | 8 [6 – 10] | 3.7 (3.3–4.3) | 5.6 (4.9–6.3) | 5.9 (5.3–6.7) | 5.5 (4.7–6.5) |
| 4N12 | IgG2 | κ | − | NT | Arch | D250 | 8 [7 – 10] | 2.5 (2.0–3.1) | 4.0 (3.3–5.0) | 6.5 (5.7–7.3) | 7.3 (5.9–9.2) |
| 2B4 | IgG1 | λ | ++ | Low binding | NoReduct ## | − | 14 [11 – 17] | 3.2 (2.8–3.7) | 5.6 (4.6–6.7) | 6.5 (5.6–7.7) | 7.0 (6.0–8.2) |
| 4J21 | IgG1 | λ | ++ | Low binding | NoReduct | − | 5 [4 – 6] | 5.2 (4.3–6.4) | 7.4 (6.6–8.3) | 7.7 (7.0–8.6) | 7.2 (5.3–9.8) |
| 5M16 | IgG1 | κ | +++ | 2 | Arch | G253 | 5 [4 – 6] | 6.0 (5.5–6.6) | 5.9 (5.0–6.8) | 8.4 (6.7–10.4) | 11.7 (9.7–14.1) |
| 9D14 | IgG1 | λ | +++ | 2 | NoReduct | − | 6 [5 – 7] | 2.1 (1.6–2.7) | 2.9 (2.3–3.7) | 6.3 (4.7–8.4) | 86.0 (31.5–235) |
| 1H12 | IgG1 | λ | +++ | 1/2 | DA/B, Arch | T58, D59, D60, R68, D71, I74, D77, T191, N193, K234 | 17 [14 – 20] | 3.0 (2.5–3.5) | 7.5 (6.7–8.4) | 11.7 (9.3–14.8) | 11.6 (8.2–16.2) |
| 8E22 | IgG1 | λ | ++ | Low binding | DA, Arch | H62, W64, R68, H99, D117, I255 | 17 [14 – 19] | 8.2 (7.0–9.7) | 7.2 (6.4–8.3) | 42.5 (30.8–58.5) | 138.9 (64.7–298) |
| 8G18 | IgG1 | λ | ++ | Low binding | DA | H62, W64, D117 | 17 [14 – 19] | 4.7 (4.1–5.3) | 7.3 (6.3–8.4) | 34.9 (24.9–48.9) | 52.4 (24.1–114) |
| 10N24 | IgG1 | κ | − | NT | DA,B | W64, D71, R80, T116, D117, I121, N187, I190 | 21 [17 – 26] | 7.9 (6.9–9.0) | 9.5 (8.2–11.0) | 15.9 (13.2–19.2) | 23.6 (18.3–30.5) |
| 8I4 | IgG1 | κ | +++ | NSF Ab | DB, Arch | M171, Q184, I190, N193, V197, R198, Y199, G209, L210, K215, K234, V242, I255 | 8 [5 – 14] | 6.9 (3.8–12.3) | 6.2 (4.5–8.4) | 153 (78–299) | > |
| 3N23 | IgG1 | κ | − | NT | DA, Arch | D60, R68, G98, H170, M171, K233, K234 | 25 [21 – 30] | 6.0 (5.0–7.2) | 10.1 (8.9–11.5) | 14.1 (11.6–17.1) | 8.7 (7.0–10.9) |
| 5O14 | IgG1 | κ | +++ | 2 | NoReduct | − | 38 [30 – 47] | 6.7 (5.5–8.3) | 12.1 (10.9–13.5) | 17.3 (14.2–21.1) | 6.2 (5.3–7.2) |
| 4J14 | IgG1 | λ | ++ | Low binding | DA/B | D63, W64, T65, R80, I121, A162, N193 | 23 [20 – 26] | 12.9 (11.2–15.0) | 17.7 (16.1–19.4) | 23.1 (20–27) | 23.0 (18.5–28.4) |
| 3E23 | IgG2 | λ | − | NT | DA | W64 | 11 [9 – 13] | 19.4 (15.2–25.0) | 18.7 (16.3–21.5) | 36.0 (30.3–42.9) | 38.0 (30.3–47.5) |
| 1L1 | IgG1 | λ | +/− | Low binding | Arch | G253 | 18 [15 – 22] | 18.6 (15.5–22.4) | 24.2 (21.3–27.5) | 34.3 (29–40.7) | N.D. |
| 3B4 | IgG3 | κ | − | NT | DB | V192, Q195 | > | 18.7 (10.7–32.8) | 29.6 (18.7–46.8) | 271 (144–511) | N.D. |
| 4B8 | IgG1 | λ | +++ | 2 | NoReduct | − | 0.6 [0.4 – 0.8] | 22.8 (12.4–41.8) | 28.1 (19.8–39.9) | 234 (142–386) | N.D. |
| 4G20 | IgG1 | λ | − | NT | DB | D174, R198, Y199, K215 | 95 [60 – 160] | 22.3 (17.3–29.0) | 34.9 (28.2–43.8) | 131.4 (88.5–195) | N.D. |
| 1O5 | IgG1 | λ | − | NT | DA | W64, T65 | 138 [110 – 170] | 30.1 (22.6–35.3) | 37.6 (32.6–43.4) | 48.9 (37.8–63.2) | N.D. |
| 1O6 | IgG3 | λ | − | 2 | DA | R80 | 5,200 [4,100 – 6,600] | 61.7 (50.8–74.8) | 57.5 (48.8–68.1) | N.D. | N.D. |
| 2L5 | NT | NT | − | NT | NoReduct | − | 4,600 [2,400 – 9,500] | 1,076 (748–1,548) | 2,361 (1,460–3,819) | 5,632 (3,904–8,128) | N.D. |
| 3A2 | IgG1 | κ | +++ | 3 | DB | I190, R198, Y199, G209, L210, T212 | 1,300 [830 – 1,900] | 1,566 (1,111–2,207) | 1,396 (952–2,046) | > | N.D. |
| 5F19 | IgG1 | λ | +++ | 4 | DA | H18 | > | > | 9,064 (2,911–28,249) | > | N.D. |
| 1M9 | IgG1 | κ | − | NT | DA, Arch | R36, H62, R80, Q146, E165, E166, N231, D250, H256 | > | > | > | 6,187 (2,795–13,709) | N.D. |
| 1I9 | IgG1 | κ | − | NT | E2 | Inconclusive | > | > | > | > | N.D. |
| 4B10 | IgG1 | κ | − | NT | NoReduct | − | > | > | > | > | N.D. |
| 2C2 | IgG1 | λ | − | NT | NoReduct | Inconclusive | > | > | > | > | N.D. |
| 2D12 | IgG1 | κ | − | NT | E2 | Inconclusive | > | > | > | > | N.D. |
| 5N23 | IgG1 | λ | +++ | 1 | DA, Arch | E24, D117, I121 | > | > | > | > | N.D. |
| murine CHK-152 | IgG2c | κ | − | NT | E2-DA, E2-DB | D59, W235, A11, M74, G194, N193, T212, H2328 | 3 [2 – 4] | ||||
Order of antibodies reflects the level of potency degree and breadth of the antibodies in neutralization assays against clinical CHIKV isolates of diverse genotypes.
Immunoglobulin isotype, subtype, and light chain use were determined by ELISA. NT indicates not tested due to poor growth of B cell line.
(−) denotes no detectable binding [OD <0.1]; (+/−) denotes weak binding [OD 0.31–0.499]; (++) denotes moderate binding [OD 0.5–0.99]; (+++) denotes strong binding [OD >1.0].
Values shown represent combined data from two independent experiments. Low binding indicates incomplete mAb binding to E2 on biosensor for assessing competition. NT indicates not tested since Ab did not bind E2 ectodomain in ELISA; NSF Ab indicates insufficient supply of mAb.
NotReact indicates that the mAb did not react against the wild-type envelope proteins and could not be tested in this system. NoReduct indicates the mAb did bind to the wild-type E proteins, but no reduction was noted reproducibly for any mutant. DA indicates domain A; DB indicates domain B; Arch indicates either arch 1, arch 2, or both.
Values shown represent combined data from two or more independent experiments.
Concentration (ng/mL) at which 50% of virus was neutralized (EC50). (>) indicates EC50 value is greater than the highest mAb concentration tested (10 μg/ml).
N.D. = Not Done.
Residues identified by contacts with mAb in cryo-EM reconstruction (reference 48).
Assessment of mAb neutralization
Eighteen of the mAbs exhibited neutralizing activity against Asian CHIKV strain SL15649-GFP virus replicon particles (VRPs) with EC50 values < 40 ng/mL, with 11 exhibiting ultrapotent inhibitory activity (defined as EC50 values < 10 ng/mL, Table 1). Four mAbs possessed weak inhibitory activity (EC50 values in the 0.1 to 5 μg/mL range) and 8 of the mAbs had no inhibitory activity at the highest concentration tested (EC50 values > 10 μg/mL).
Breadth of neutralizing activity
We determined the EC50 values for each mAb against representative infectious CHIKV strains of the East/Central/South African (ECSA) genotype (LR2006 OPY1 [LR] strain), the West African genotype (NI 64 IbH 35 strain), and the Asian genotype (RSU1 and 99659 [2014 Caribbean] strains) using a high-throughput focus reduction neutralization test (FRNT) (Pal et al., 2013). Twenty-five of the mAbs exhibited neutralizing activity against at least one CHIKV strain (EC50 values < 10 μg/mL), with 8 mAbs exhibiting neutralization in a potent range (EC50 values between 10–99 ng/mL), and 13 mAbs exhibiting neutralization in an ultrapotent range (Table 1). For comparative purposes we also tested the previously reported human mAbs 5F10 and 8B10 against viruses of all three genotypes, and in every case the EC50 values were >100 ng/mL. In most cases, the mAbs we isolated exhibited relatively similar neutralizing activity against virus from all three genotypes. Six mAbs (2B4, 2H1, 4J21, 4N12, 5M16, and 9D14) inhibited viruses from all three genotypes with ultrapotent activity. These data indicate that a single individual can develop multiple CHIKV-specific antibodies that are ultrapotent and broadly neutralizing.
Binding to E2 protein
The CHIKV E2 protein is a dominant target of murine (Goh et al., 2013; Lum et al., 2013), nonhuman primate (Kam et al., 2014), and human (Fong et al., 2014; Kam et al., 2012a; Kam et al., 2012b; Selvarajah et al., 2013) humoral responses. We tested the human mAbs for binding to a monomeric form of the ectodomain of E2 protein expressed in E. coli (Pal et al., 2013). Nine mAbs bound strongly to the E2 ectodomain, 6 exhibited moderate binding, 1 bound weakly, and 14 failed to bind above background (Table 1). The capacity to bind purified E2 protein in vitro did not correlate directly with neutralizing potency (Tables 1). A subset of 17 human mAbs was tested using a surface plasmon resonance assay for binding to the p62-E1 protein derived from mammalian cells (Voss et al., 2010). All mAbs bound in the nM range, with KD values from 0.5 to 20 nM. Differences in binding kinetics did not correlate with antigenic specificity or functional activity (Table S1).
Competition-binding studies
To identify non-overlapping antigenic regions in recombinant E2 protein recognized by different neutralizing mAbs, we used a quantitative competition-binding assay. For comparison, we also evaluated 4 previously described murine mAbs (CHK-84, CHK-88, CHK-141, and CHK-265) (Pal et al., 2013) and the previously described human mAb 5F10 (Warter et al., 2011) (Figure S2). The pattern of competition was complex, but three major competition groups were evident, which we designated group 1–3. We also defined a fourth group containing the single human mAb, 5F19. These competition studies suggest that there are at least three major antigenic regions recognized by CHIKV-specific antibodies.
Epitope mapping using alanine-scanning mutagenesis
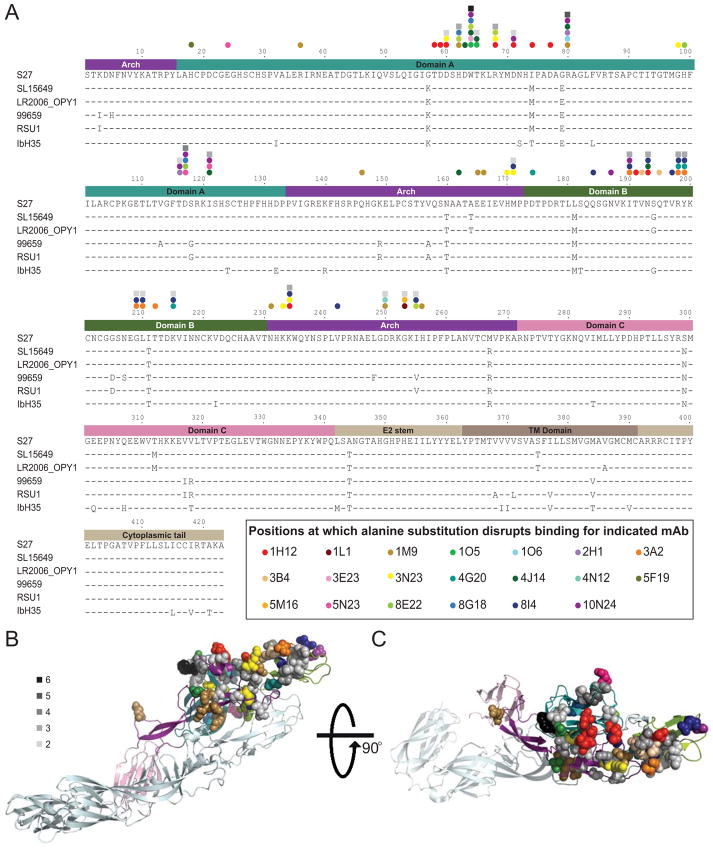
We used an alanine-scanning mutagenesis library coupled with cell-based expression and flow cytometry to identify residues in E2 and E1 proteins of CHIKV strain S27 (ECSA genotype) required for mAb binding (Fong et al., 2014) (Figure S3). Residues required for mAb binding to CHIKV glycoproteins for a subset of 20 human mAbs are listed in Table 1. Mutations affecting binding of these 20 mAbs are indicated in an alignment of the full-length E2 sequences of strain S27 and strains representing all CHIKV genotypes that were used in our study (Figure 1A). The aa in E2 that influence binding are located primarily in the solvent-exposed regions of domains A and B and arches 1 and 2 of the β-ribbon connector, which links domains A and B (Voss et al., 2010) (Figure 1A). Comparison of the antigenic sites identified by loss-of-binding experiments using alanine-scanning mutagenesis with the competition binding analysis (Figure S2) demonstrated that competition groups 1 and 2 generally corresponded to sites within domain A and the arches, whereas group 3 corresponded to regions in domain B.
Figure 1. Structural analysis of E2 residues important for mAb binding.
(A) Sequence alignment of E2 from the CHIKV strains (indicated on the left) used in this study. The numbers above the sequence correspond to the aa position in the mature E2 protein. Amino acids identical to strain S27 are indicated by a dash. Domains of E2 determined from the crystal structure of the E2/E1 heterodimer (Voss et al., 2010) are depicted in the diagram above the alignment and are color-coded (cyan: domain A, purple: β-ribbon connector, green: domain B, pink: domain C, taupe shades: regions not present in the crystal structure). The position of residues at which alanine substitution disrupts mAb binding, as determined by alanine-scanning mutagenesis, are designated by color-coded dots for each specific mAb. Residues that influence the binding of multiple antibodies are indicated by squares shaded in gray, with the darker the shade of gray, the greater number of antibodies influenced by substitution at that residue (legend in panel (B). (B) Location of residues required for mAb binding mapped onto the crystal structure of the mature envelope glycoprotein complex (PDB ID 3N41). A side view of a ribbon trace of a single heterodimer of E1/E2 is shown with E1 in light cyan and the domains of E2 colored as in panel A. The side chains of the aa required for mAb binding are shown as space-filling forms and color-coded for each of the 20 individual antibodies according to the legend in panel A.. (C) A top view of the E1/E2 heterodimer, rotated 90° from the structure in panel B. Also see Figures S3 and S4.
Structural analysis of antigenic regions
A large and diverse number of the surface residues in domains A and B and the arches are contacted by at least 1 of the mAbs (Figure 1B and 1C). Two principal antigenic regions in E2 accounted for the binding of multiple mAbs. The first region is located in domain A, between residues 58 and 80, and contains the putative receptor-binding domain (RBD) (Sun et al., 2014; Voss et al., 2010). The second region is located in domain B, between residues 190 and 215. Both sequence regions project away from the viral envelope and are located near the E2 trimer apex (Figure S3 and S4).
Mechanism of neutralization
We conducted pre- and post-attachment neutralization assays using mAbs displaying a range of inhibitory activities. As expected, all 5 mAbs tested neutralized infection efficiently when pre-incubated with VRPs (Figure 2A). However, mAb 4B8 did not neutralize VRPs completely even at high concentrations, suggesting the presence of a fraction of CHIKV virions resistant to this mAb; this pattern also was observed in assays using viable CHIKV strains corresponding to the three distinct CHIKV genotypes. In contrast, mAbs 3E23, 4J21, 5M16, and 9D14 completely neutralized infection when administered before attachment. All five human mAbs also neutralized CHIKV infection when added following attachment, but we observed three different patterns of activity (Figure 2A). MAb 4B8 was incapable of complete neutralization when added post-attachment, and the fraction of resistant virions was larger compared with that observed following pre-attachment neutralization. MAb 9D14 neutralized VRPs with comparable efficiency whether added before or after attachment. MAbs 3E23, 4J21, and 5M16 displayed complete neutralization of VRPs, but the efficiency of neutralization post-attachment was lower than that following pre-attachment. The mAbs 2H1 and 4N12 also efficiently neutralized VRPs when added prior to or after attachment (data not shown).
Figure 2. Mechanism of neutralization by human anti-CHIKV mAbs.
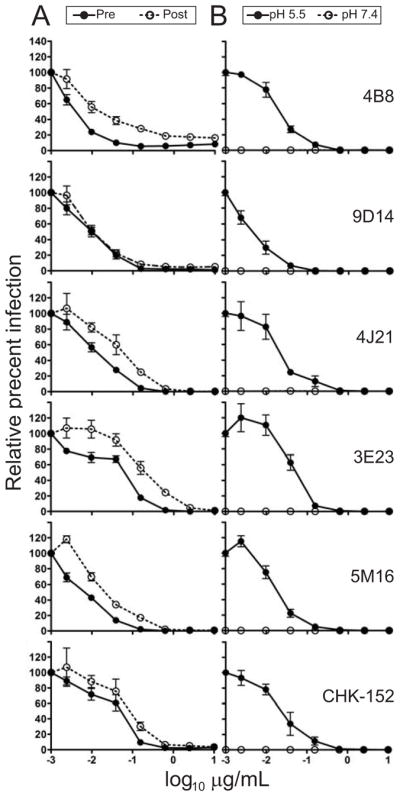
(A) Pre- and post-attachment neutralization assays. SL15649 VRPs were (i) incubated with the mAbs shown (including CHK-152, a positive control mAb) prior to addition to pre-chilled Vero cells, followed by removal of unbound virus by three washes (pre-attachment; filled circle) or (ii) allowed to adsorb to pre-chilled Vero cells followed by addition of the indicated mAbs (post- attachment; open circles). (B) FFWO assay. SL15649 VRPs were adsorbed to pre-chilled Vero cells, followed by addition of the mAbs shown (including CHK-152, a positive control murine mAb). Unbound virus was removed, and cells were exposed to low (pH 5.5 to trigger viral fusion at the plasma membrane; filled circles) or neutral (pH 7.4 as a control; open circles) pH medium at 37°C for 2 min.. For both (A) and (B) cells were incubated at 37°C until 18 h after infection, and GFP-positive cells were quantified using fluorescence microscopy. The data are combined from two independent experiments, each performed in triplicate, and represented as mean +/− SEM.
Fusion-from-without (FFWO) assay testing (Edwards and Brown, 1986) of five of the ultrapotently neutralizing mAbs (3E23, 4B8, 4J21, 5M16, or 9D14) revealed that all inhibited fusion. In the absence of antibody treatment, a short pulse of acidic pH-buffered medium resulted in infected cells, indicating fusion between the viral envelope and plasma membrane, whereas a pulse of neutral pH resulted in little to no infection as expected (Figure 2B). Notably, all 5 human mAbs inhibited plasma membrane fusion and infection, with mAb 9D14 exhibiting the greatest potency in this assay. These studies suggest that ultrapotently neutralizing mAbs block CHIKV fusion.
MAb prophylaxis in vivo
We tested a subset of mAbs exhibiting diverse levels of neutralizing activity (Table 1) in a lethal infection model with 6-week-old, highly immunodeficient Ifnar−/− mice. Mice were pre-treated with a single 50 μg dose (~ 3 mg/kg) of human anti-CHIKV mAbs or a West Nile virus (WNV)-specific isotype control mAb (WNV hE16) 24 h before subcutaneous injection with a lethal dose of CHIKV-LR2006. All mice treated with the isotype control mAb succumbed to infection by 4 d post-inoculation. Pretreatment with mAbs 4B8, 4J21, or 5M16 completely protected Ifnar−/− mice, whereas treatment with mAbs 3E23 or 9D14 partially protected the infected animals, with 50% and 67% survival rates, respectively (Fig. 3A). Surprisingly, mAb 2D12, which weakly neutralized in vitro, protected 80% of the animals.
Figure 3. Human mAb prophylaxis and therapy against lethal CHIKV infection in Ifnar−/− mice.
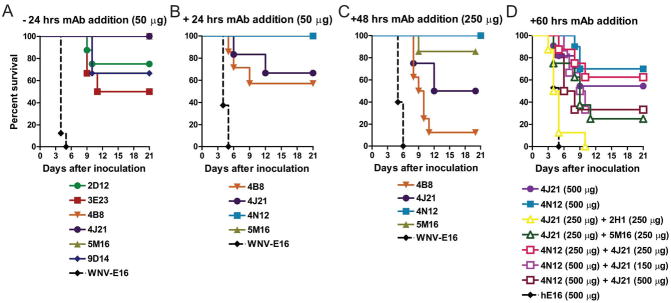
Mice were administered either 50 or 250 μg of indicated CHIKV-specific or control mAb by intraperitoneal injection 24 h before (A; n = 6 to 8 mice per mAb tested) or 24 h (B; n = 5 to 8 mice per mAb tested) or 48 h after (C; n = 7 to 10 mice per mAb tested) a lethal challenge of CHIKV (D) Mice were administered 150, 250 or 500 μg of indicated CHIKV-specific mAbs in combination by intraperitoneal injection 60 h after a lethal challenge of CHIKV (n = 6 to 13 mice per mAb combination tested). For monotherapy with 4J21, 4N12 or hE16 (negative control), a single dose of 500 μg was given (n = 10 to 17 mice per mAb tested). All data in this figure were pooled from at least two independent experiments. The following statistical analysis was performed using the Mantel-Cox log rank test: 4N12 versus 4J21, P = 0.39; 4N12 (500 μg) versus 4N12 (250 μg) + 4J21 (250 μg), P = 0.69; 4N12 (500 μg) versus 4N12 (500 μg) + 4J21 (150 μg), P = 0.13; 4N12 (500 μg) versus 4N12 (500 μg) + 4J21 (500 μg), P = 0.06. All Ab administrations with the exception of 4J21 (250 μg) + 2H1 (250 μg) differed significantly from the hE16 control (P < 0.002).
MAb post-exposure therapy in vivo
Ifnar−/− mice were inoculated with a lethal dose of CHIKV-LR2006 and then administered a single 50 μg (~ 3 mg/kg) dose of representative mAbs 24 h following virus inoculation. Therapeutic administration of mAb 4N12 or 5M16 mAbs provided complete protection, whereas the isotype-control mAb provided no protection, and others provided partial protection (Figure 3B). To define further the therapeutic window of efficacy, Ifnar−/− mice were administered a single 250 μg (~ 14 mg/kg) dose of representative mAbs 48 h after challenge with CHIKV-LR2006. Treatment with 4N12, 5M16, 4J21, and 4B8 protected 100%, 85%, 50%, and 12.5% of the animals, respectively (Figure 3C). Remarkably, monotherapy with 4N12 or 4J21 at the later time point of 60 h protected 70% and 55% of animals when used at a dose of 500 μg, [~ 28 mg/kg]) (Figure 3D). The observed differences in efficacy of the mAbs are likely not due to varying in vivo half-life in mice, as there was no appreciable difference in the rate of clearance in the serum for mAbs 4B8, 5M16, 4N12, and 4J21 (data not shown). These data establish that human mAbs can protect against CHIKV-induced death, even at intervals well after infection is established.
Combination mAb therapy in vivo
Given the possibility of resistance selection in vivo in animals treated with a single anti-CHIKV mAb (Pal et al., 2013), we tested whether a combination of two anti-CHIKV human mAbs could protect mice against lethal challenge. We chose pairs of neutralizing mAbs based on the potency of individual mAbs in vitro as well as protective activity in vivo as monotherapy. Ifnar−/− mice were administered a single combination antibody treatment dose of the most effective mAbs 60 h after inoculation. None of the combinations tested at varying doses ([4J21 + 2H1], [4J21 + 5M16], or [4J21 + 4N12]) provided superior protection to 4J21 or 4N12 monotherapy.
Discussion
We report the isolation of a diverse panel of naturally-occurring human mAbs from a single individual, the majority of which recognize the CHIKV E2 protein and display remarkable neutralizing activity in vitro and therapeutic efficacy in vivo. As a class, the most inhibitory antibodies also exhibited broad activity, neutralizing viruses from all 3 CHIKV genotypes, including a strain currently circulating in the Caribbean. The majority of human CHIKV-specific mAbs isolated in this study neutralized the virus at concentrations < 100 ng/mL, and many exhibited inhibitory activity at < 10 ng/mL. This activity is greater than we have observed in our previous studies of human mAbs against other pathogenic human viruses, including H1, H2, H3, or H5 influenza viruses (Hong et al., 2013; Krause et al., 2012; Krause et al., 2011a; Krause et al., 2011b; Krause et al., 2010; Thornburg et al., 2013; Yu et al., 2008), dengue viruses (Messer et al., 2014; Smith et al., 2013a; Smith et al., 2014; Smith et al., 2013b; Smith et al., 2012), and others. The potency of many human CHIKV mAbs is comparable to or exceeds that of best-in-class murine neutralizing CHIKV mAbs (Fong et al., 2014; Fric et al., 2013; Pal et al., 2013; Warter et al., 2011), which were generated after iterative boosting and affinity maturation. Most other neutralizing human mAbs against CHIKV are substantially less potent (Fong et al., 2014; Selvarajah et al., 2013; Warter et al., 2011). A single previously reported human CHIKV-specific mAb (IM-CKV063) displays activity comparable to the ultrapotent neutralizing mAbs reported here (Fong et al., 2014).
We observed a diversity of epitope recognition patterns in E2 by the different neutralizing CHIKV mAbs tested. Fine epitope mapping with alanine-substituted CHIKV glycoproteins showed that recognition of three structural regions in E2 is associated with mAb-mediated neutralization: domain A, which contains the putative RBD (Sun et al., 2013; Voss et al., 2010), domain B, which contacts and shields the fusion loop in E1 (Voss et al., 2010), and arches 1 and 2 of the β-ribbon connector, which contains an acid-sensitive region and links domains A and B (Voss et al., 2010). Of the antibodies mapped to epitopes in E2, the bulk (those in competition groups 1 and 2) preferentially recognized sites in domain A and arches 1 and 2, whereas a smaller group (in competition group 3) recognized sites in domain B. These data suggest that surface-exposed regions in domain A and the arches are dominant antigenic sites that elicit human neutralizing antibody responses. We conclude that the highly conserved region in domain A and arch 2 might elicit a broadly protective immune response and serve as an attractive candidate for epitope-focused vaccine design.
Remarkably, almost a quarter of surface-exposed residues in the critical E2 domains appear to be engaged by one or more mAbs from a single individual. The existence of functionally diverse binding modes on the major antigenic sites is implied by two observations: (a) some mAbs bound to similar epitopes but exhibited inhibitory activity that varied by several orders of magnitude and (b) there was little correlation between neutralization capacity and affinity of binding to E2 protein. Seven of the most potently neutralizing human mAbs (2H1, 3E23, 4B8, 4J21, 4N12, 5M16, and 9D14) inhibited CHIKV infection at a step following attachment, likely via prevention of pH-dependent structural changes, which prevents nucleocapsid penetration into the cytoplasm (Kielian et al., 2010).
As therapeutic efficacy in mice appears to predict treatment outcomes in experimentally-induced infection and arthritis in nonhuman primates (Pal et al., 2013; Pal et al., 2014), the data here suggest that prophylaxis of humans with CHIKV-specific human mAbs would prevent infection. Given concerns about selection of resistant variants with monotherapy (Pal et al., 2013), combination therapy using ultrapotent neutralizing antibodies that target different regions of E2 may be desirable. Unexpectedly, we did not observe a superior therapeutic effect for combinations of mAbs compared with monotherapy at late time points in these studies with immunodeficient mice. In fact, the survival in most groups treated with combination therapy trended toward less protection than that of the groups treated with 4J21 or 4N12 alone. Although further study is warranted, the lack of enhanced therapeutic benefit with the particular mAb combinations tested could be due to competition or structural hindrance of binding of individual antibody molecules to adjacent epitopes on E2 proteins on the icosahedral virion surface. In comparison, a prior study with anti-E2 (CHK-152) and anti-E1 (CHK-166) mouse MAbs did show advantage as combination therapy (Pal et al., 2013). Regardless, our data suggest that patient populations at markedly increased risk of severe disease could be targeted for prophylaxis or treatment with human anti-CHIKV mAbs during outbreaks, including those with serious underlying medical conditions (e.g., late-term pregnant women, the immunocompromised, and the elderly). Further clinical testing is planned to determine whether neutralizing human mAbs can prevent or ameliorate established joint disease in humans.
Experimental Procedures
Isolation of human mAbs
PBMCs were obtained from a human ~ 5.5 years after documented symptomatic CHKV infection in Sri Lanka. B cells were transformed with EBV in the presence of CpG. The supernatants from the resulting B cell lymphoblastic cells lines were screened for CHKV-neutralizing activity using SL15649 VRPS. Positive wells were further selected for the presence of human CHKV-specific binding antibodies by ELISA using live CHIKV vaccine strain 181/25 virus as antigen. Transformed B cells were collected and fused to a myeloma cell line, distributed into culture plates and expansion, and selected by growth in hypoxanthine-aminopterin-thymidine medium containing ouabain. Hybridomas were cloned by single-cell sorting. Supernatants from cloned hybridomas growing in serum-free medium were collected, purified, and concentrated from clarified medium by protein G chromatography.
Neutralization assays
Purified IgG mAb proteins were tested for neutralizing activity using CHKV VRPs or fully infectious CHIKV. VRPs were incubated with serial dilutions of mAbs then inoculated onto Vero 81 cell monolayers for 18 hrs; infected cells and total cells (identified with a nuclear marker) were identified with a fluorescence imaging system. Neutralizing activity for four infectious virus strains was determined in a focus reduction neutralization test (Pal et al., 2013). Serial dilutions of mAbs were incubated with 100 focus-forming units of CHIKV and then added to Vero cells. Foci were detected with a mouse anti-CHIKV mAb after cell fixation using immunoperoxidase detection and quantified using an ImmunoSpot 5.0.37 macroanalyzer (Cellular Technologies Ltd).
E2 ELISA
Recombinant CHIKV E2 ectodomain protein (corresponding to the CHIKV-LR2006 strain) was generated in E. coli (Pal et al., 2013) and adsorbed to microtiter plates. Human mAbs were applied, and bound CHKV-specific mAbs were detected with biotin-conjugated goat anti-human IgG.
Competition binding assay
We identified groups of antibodies binding to the same major antigenic site by competing pairs of antibodies for binding to CHIKV-LR2006 E2 ectodomain protein containing a polyhistidine-tag attached to an Anti-Penta-His biosensor tip (ForteBio #18-5077) in an Octet Red biosensor (ForteBio).
Alanine scanning mutagenesis for epitope mapping
A CHIKV envelope protein expression construct (strain S27, Uniprot Reference #Q8JUX5) with a C-terminal V5 tag was subjected to alanine-scanning mutagenesis to generate a comprehensive mutation library. Primers were designed to mutate each residue within the E2, 6K, and E1 regions of the envelope proteins (residues Y326 to H1248 in the structural polyprotein) to alanine; alanine codons were mutated to serine. In total, 910 CHIKV envelope protein mutants were generated. Loss of binding of mAbs to each construct was determined using an immunofluorescence binding assay, using cellular fluorescence detected with a high-throughput flow cytometer.
Mechanism of neutralization
MAbs were interacted with VRPs before or after attachment to Vero 81 cells, and then cells were stained, imaged, and analyzed as described for VRP neutralization assays to determine at what stage mAbs exerted the antiviral effect. Fusion from without assays were performed as detailed in Supplemental Experimental Procedures.
In vivo protection studies in mice
Ifnar−/− mice were bred in pathogen-free animal facilities and infection experiments were performed in A-BSL3 facilities. Footpad injections were performed under anesthesia. For prophylaxis studies, human mAbs were administered by intraperitoneal injection to 6 week-old Ifnar−/− mice 1 d prior to subcutaneous inoculation in the footpad with 10 FFU of CHIKV-LR. For therapeutic studies, 10 FFU of CHIKV-LR was delivered 24, 48, or 60 h prior to administration of a single dose of individual or combinations of human mAbs at specified doses.
Supplementary Material
Highlights.
A panel of 30 chikungunya virus-specific antibodies (Abs) isolated from a single donor
13 Abs exhibited broad and potent neutralizing activity with IC50 < 10 ng/mL
Potently neutralizing Abs bind to the E2 envelope protein and block viral fusion
Several Abs exhibited prophylactic and therapeutic activity in a mouse model
Acknowledgments
We thank Aravinda de Silva (UNC Chapel Hill) for assistance with acquisition of the donor sample, Frances Smith-House at Vanderbilt University for excellent laboratory management support, Melissa Edeling and Katie O’Brien at WUSTL for generating E2 proteins and performing some of the initial mAb binding experiments, Edgar Davidson, Andrew Ettenger, Johnathan Guest, Trevor Barnes, Surabhi Srinivasan, and Bernard Lieberman at Integral Molecular for help with epitope mapping, and Chris Slaughter for assistance with biostatistical analysis of VRP neutralization data.
Funding: This work was supported by U.S. National Institutes of Health grants K08 AI103038 (S.A.S), F32 AI096833 (L.A.S.), T32 HL007751 (A.W.A.), T32 5T32AI007151-33 (C.E.M.) U54 AI057157 (T.S.D.), R01 AI104545 (M.S.D.), and NIH contract HHSN272200900055C (B.J.D.). The work also received support from the Elizabeth B. Lamb Center for Pediatric Research (T.S.D.), Infectious Diseases Society of America Education and Research Foundation (S.A.S), and National Foundation for Infectious Diseases Young Investigator Award in Vaccine Development sponsored by Pfizer (S.A.S). The project described was supported by the National Center for Research Resources, Grant UL1 RR024975-01 and is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06.
Footnotes
Author Contributions: S.A.S and L.A.S performed initial screening and isolation of antibodies. S.A.S., N.K., and G.S. isolated hybridomas, purified antibodies, and sequenced mAb clones. C.E.M. and M.T.H. devised and executed construction of SL15649 replicon plasmids. L.A.S., S.K., and A.W.A. devised and conducted VRP neutralization and mechanistic assays. J.M.F. and P.P. and M.S.D. devised and performed FRNT assays with infectious virus. S.K.A. and M.S.D. devised and conducted surface plasmon resonance studies. A.F. and J.E.C. devised and performed Octet-based competition binding assays and provided associated data. K.M.K., R.H.F., S.S., and B.J.D. devised and performed alanine-scanning mutagenesis. L.A.S. performed structural analysis of epitope residues. J.M.F., J.D.B., and M.S.D. devised and performed mouse studies. S.A.S., L.A.S., M.S.D, T.S.D., and J.E.C. prepared the manuscript. All authors revised and approved the final version of the manuscript.
The content is solely the responsibility of the authors and does not represent the official views of the NIH.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brehin AC, Rubrecht L, Navarro-Sanchez ME, Marechal V, Frenkiel MP, Lapalud P, Laune D, Sall AA, Despres P. Production and characterization of mouse monoclonal antibodies reactive to Chikungunya envelope E2 glycoprotein. Virology. 2008;371:185–195. doi: 10.1016/j.virol.2007.09.028. [DOI] [PubMed] [Google Scholar]
- CDC. Chikungunya virus. Atlanta, GA: US Department of Health and Human Services; 2013. http://www.cdc.gov/media/releases/2013/p1218-chikungunyas.html. [Google Scholar]
- CDC. Chikungunya in the Americas. Atlanta, GA: US Department of Health and Human Services; 2014. http://www.cdc.gov/chikungunya/geo/americas.html. [Google Scholar]
- Chu H, Das SC, Fuchs JF, Suresh M, Weaver SC, Stinchcomb DT, Partidos CD, Osorio JE. Deciphering the protective role of adaptive immunity to CHIKV/IRES a novel candidate vaccine against Chikungunya in the A129 mouse model. Vaccine. 2013;31:3353–3360. doi: 10.1016/j.vaccine.2013.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couderc T, Khandoudi N, Grandadam M, Visse C, Gangneux N, Bagot S, Prost JF, Lecuit M. Prophylaxis and therapy for Chikungunya virus infection. J Infect Dis. 2009;200:516–523. doi: 10.1086/600381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J, Brown DT. Sindbis virus-mediated cell fusion from without is a two-step event. J Gen Virol. 1986;67(Pt 2):377–380. doi: 10.1099/0022-1317-67-2-377. [DOI] [PubMed] [Google Scholar]
- Fong RH, et al. Exposure of epitope residues on the outer face of the chikungunya virus envelope trimer determines antibody neutralizing efficacy. J Virol. 2014;88:14364–14379. doi: 10.1128/JVI.01943-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fric J, Bertin-Maghit S, Wang CI, Nardin A, Warter L. Use of human monoclonal antibodies to treat Chikungunya virus infection. J Infect Dis. 2013;207:319–322. doi: 10.1093/infdis/jis674. [DOI] [PubMed] [Google Scholar]
- Goh LY, Hobson-Peters J, Prow NA, Gardner J, Bielefeldt-Ohmann H, Pyke AT, Suhrbier A, Hall RA. Neutralizing monoclonal antibodies to the E2 protein of chikungunya virus protects against disease in a mouse model. Clin Immunol. 2013;149:487–497. doi: 10.1016/j.clim.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Hallengard D, et al. Prime-boost immunization strategies against Chikungunya virus. J Virol. 2014;88:13333–13343. doi: 10.1128/JVI.01926-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawman DW, Stoermer KA, Montgomery SA, Pal P, Oko L, Diamond MS, Morrison TE. Chronic joint disease caused by persistent Chikungunya virus infection is controlled by the adaptive immune response. J Virol. 2013;87:13878–13888. doi: 10.1128/JVI.02666-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, et al. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol. 2013;87:12471–12480. doi: 10.1128/JVI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam YW, et al. Longitudinal analysis of the human antibody response to Chikungunya virus infection: implications for serodiagnosis and vaccine development. J Virol. 2012a;86:13005–13015. doi: 10.1128/JVI.01780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam YW, Lee WW, Simarmata D, Le Grand R, Tolou H, Merits A, Roques P, Ng LF. Unique epitopes recognized by antibodies induced in Chikungunya virus-infected non-human primates: implications for the study of immunopathology and vaccine development. PLoS One. 2014;9:e95647. doi: 10.1371/journal.pone.0095647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam YW, et al. Early neutralizing IgG response to Chikungunya virus in infected patients targets a dominant linear epitope on the E2 glycoprotein. EMBO Mol Med. 2012b;4:330–343. doi: 10.1002/emmm.201200213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M, Chanel-Vos C, Liao M. Alphavirus entry and membrane fusion. Viruses. 2010;2:796–825. doi: 10.3390/v2040796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause JC, et al. Human monoclonal antibodies to pandemic 1957 H2N2 and pandemic 1968 H3N2 influenza viruses. J Virol. 2012;86:6334–6340. doi: 10.1128/JVI.07158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE., Jr A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol. 2011a;85:10905–10908. doi: 10.1128/JVI.00700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Briney BS, Smith SA, Basler CF, Crowe JE., Jr Epitope-specific human influenza antibody repertoires diversify by B cell intraclonal sequence divergence and interclonal convergence. J Immunol. 2011b;187:3704–3711. doi: 10.4049/jimmunol.1101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause JC, Tumpey TM, Huffman CJ, McGraw PA, Pearce MB, Tsibane T, Hai R, Basler CF, Crowe JE., Jr Naturally occurring human monoclonal antibodies neutralize both 1918 and 2009 pandemic influenza A (H1N1) viruses. J Virol. 2010;84:3127–3130. doi: 10.1128/JVI.02184-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, et al. Chikungunya virus neutralization antigens and direct cell-to-cell transmission are revealed by human antibody-escape mutants. PLoS Pathog. 2011;7:e1002390. doi: 10.1371/journal.ppat.1002390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum FM, Teo TH, Lee WW, Kam YW, Renia L, Ng LF. An essential role of antibodies in the control of Chikungunya virus infection. J Immunol. 2013;190:6295–6302. doi: 10.4049/jimmunol.1300304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masrinoul P, Puiprom O, Tanaka A, Kuwahara M, Chaichana P, Ikuta K, Ramasoota P, Okabayashi T. Monoclonal antibody targeting chikungunya virus envelope 1 protein inhibits virus release. Virology. 2014;464–465:111–117. doi: 10.1016/j.virol.2014.05.038. [DOI] [PubMed] [Google Scholar]
- Messer WB, et al. Dengue virus envelope protein domain I/II hinge determines long-lived serotype-specific dengue immunity. Proc Natl Acad Sci U S A. 2014;111:1939–1944. doi: 10.1073/pnas.1317350111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pal P, et al. Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS Pathog. 2013;9:e1003312. doi: 10.1371/journal.ppat.1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal P, et al. Chikungunya viruses that escape monoclonal antibody therapy are clinically attenuated, stable, and not purified in mosquitoes. J Virol. 2014;88:8213–8226. doi: 10.1128/JVI.01032-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilte C, Staikowsky F, Couderc T, Madec Y, Carpentier F, Kassab S, Albert ML, Lecuit M, Michault A. Chikungunya virus-associated long-term arthralgia: a 36-month prospective longitudinal study. PLoS Negl Trop Dis. 2013;7:e2137. doi: 10.1371/journal.pntd.0002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvarajah S, et al. A neutralizing monoclonal antibody targeting the acid-sensitive region in chikungunya virus E2 protects from disease. PLoS Negl Trop Dis. 2013;7:e2423. doi: 10.1371/journal.pntd.0002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sissoko D, Malvy D, Ezzedine K, Renault P, Moscetti F, Ledrans M, Pierre V. Post-epidemic Chikungunya disease on Reunion Island: course of rheumatic manifestations and associated factors over a 15-month period. PLoS Negl Trop Dis. 2009;3:e389. doi: 10.1371/journal.pntd.0000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, et al. The potent and broadly neutralizing human dengue virus-specific monoclonal antibody 1C19 reveals a unique cross-reactive epitope on the bc loop of domain II of the envelope protein. MBio. 2013a;4:e00873–00813. doi: 10.1128/mBio.00873-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, de Alwis AR, Kose N, Jadi RS, de Silva AM, Crowe JE., Jr Isolation of dengue virus-specific memory B cells with live virus antigen from human subjects following natural infection reveals the presence of diverse novel functional groups of antibody clones. J Virol. 2014;88:12233–12241. doi: 10.1128/JVI.00247-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, de Alwis R, Kose N, Durbin AP, Whitehead SS, de Silva AM, Crowe JE., Jr Human monoclonal antibodies derived from memory B cells following live attenuated dengue virus vaccination or natural infection exhibit similar characteristics. J Infect Dis. 2013b;207:1898–1908. doi: 10.1093/infdis/jit119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Zhou Y, Olivarez NP, Broadwater AH, de Silva AM, Crowe JE., Jr Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection. J Virol. 2012;86:2665–2675. doi: 10.1128/JVI.06335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples JE, Breiman RF, Powers AM. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin Infect Dis. 2009;49:942–948. doi: 10.1086/605496. [DOI] [PubMed] [Google Scholar]
- Sun C, Gardner CL, Watson AM, Ryman KD, Klimstra WB. Stable, high-level expression of reporter proteins from improved alphavirus expression vectors to track replication and dissemination during encephalitic and arthritogenic disease. J Virol. 2014;88:2035–2046. doi: 10.1128/JVI.02990-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Xiang Y, Akahata W, Holdaway H, Pal P, Zhang X, Diamond MS, Nabel GJ, Rossmann MG. Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. Elife. 2013;2:e00435. doi: 10.7554/eLife.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg NJ, et al. Human antibodies that neutralize respiratory droplet transmissible H5N1 influenza viruses. J Clin Invest. 2013;123:4405–4409. doi: 10.1172/JCI69377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- Warter L, et al. Chikungunya virus envelope-specific human monoclonal antibodies with broad neutralization potency. J Immunol. 2011;186:3258–3264. doi: 10.4049/jimmunol.1003139. [DOI] [PubMed] [Google Scholar]
- Yu X, et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455:532–536. doi: 10.1038/nature07231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.