Abstract
We are using induced pluripotent stem cell (iPSC) technology to study neuropsychiatric disorders associated with 22q11.2 microdeletions (del), the most common known schizophrenia (SZ)-associated genetic factor. Several genes in the region have been implicated; a promising candidate is DGCR8, which codes for a protein involved in microRNA (miRNA) biogenesis. We carried out miRNA expression profiling (miRNA-seq) on neurons generated from iPSCs derived from controls and SZ patients with 22q11.2 del. Using thresholds of p<0.01 for nominal significance and 1.5-fold differences in expression, 45 differentially expressed miRNAs were detected (13 lower in SZ and 32 higher). Of these, 6 were significantly down-regulated in patients after correcting for genome wide significance (FDR<0.05), including 4 miRNAs that map to the 22q11.2 del region. In addition, a nominally significant increase in the expression of several miRNAs was found in the 22q11.2 neurons that were previously found to be differentially expressed in autopsy samples and peripheral blood in SZ and autism spectrum disorders (e.g., miR-34, miR-4449, miR-146b-3p, and miR-23a-5p). Pathway and function analysis of predicted mRNA targets of the differentially expressed miRNAs showed enrichment for genes involved in neurological disease and psychological disorders for both up and down regulated miRNAs. Our findings suggest that: i. neurons with 22q11.2 del recapitulate the miRNA expression patterns expected of 22q11.2 haploinsufficiency, ii. differentially expressed miRNAs previously identified using autopsy samples and peripheral cells, both of which have significant methodological problems, are indeed disrupted in neuropsychiatric disorders and likely have an underlying genetic basis.
Introduction
Genome wide association studies (GWAS), copy number variation (CNV) analysis and exome sequencing show that schizophrenia (SZ) and other neuropsychiatric disorders, including bipolar disorder (BD) and autism spectrum disorders (ASD), are genetically heterogeneous complex traits. This presents a potential problem in translating molecular and genetic findings into novel therapies that would benefit a large number of patients. Consequently, investigators are applying molecular and genetic data, and bioinformatics to identify common networks onto which many seemingly disparate candidate genes converge. Identifying downstream targets of SZ and ASD candidate genes that function as transcription factors, splicing factors or chromatin remodeling complexes is one potentially useful approach. Another promising area of investigation in this regard is to characterize the role of microRNAs (miRNAs) in disease pathogenesis, considering their common mechanism of biogenesis and converging effect on genes involved in neurogenesis and synaptogenesis [1–9].
MicroRNAs regulate gene expression by inducing double-stranded RNA-mediated decay and translational arrest through base-pair specific interactions with targeted mRNAs, primarily at 3’untranslated regions [10–14]. MicroRNAs are expressed as primary molecules (pri-miRNAs) that are ~70 nucleotides in length that must be processed to form functional, mature miRNAs. The first step in their biogenesis is cleavage by a miRNA processing complex consisting of the proteins DGCR8 and DROSHA, which convert pri-miRNAs into precursor RNAs (pre-miRNAs). These are transported to the cytoplasm where cleavage by DICER occurs, ultimately yielding a single stranded ~22 base mature miRNA that’s incorporated into the RNA-induced silencing complex (RISC), which targets specific mRNAs through a complimentary seed region. Most mRNAs are regulated by more than one miRNA, and any single miRNA can potentially interact with multiple mRNAs [14,15].
Several lines of evidence support a role for miRNAs (actually, their targets) in a subgroup of SZ patients. Replicated GWAS studies, for example, show a strong association to MIR-137 [16–19]. This miRNA targets other candidate genes identified by GWAS [20,21]. Molecular studies also support a role for miRNAs in SZ. Recently, 28 miRNAs were found to be differentially expressed in the dorsolateral prefrontal cortex (DLPFC) in patients with SZ compared with controls; the mRNA targets of these miRNAs showed enrichment for genes involved in axon guidance and long-term potentiation, processes associated with neuropsychiatric disorders [22,23]. Similarly, an independent study found ~50 miRNAs that were differentially expressed in the DLPFC and superior temporal gyrus in SZ, which targeted and reciprocally down-regulated the expression of mRNAs coding for proteins involved in neurodevelopmental pathways and cell-cell signaling [24].
MicroRNAs have also been considered in the pathogenesis of SZ and other neuropsychiatric disorders that occur in a substantial proportion of patients with velocardiofacial syndrome (VCFS; DiGeorge Syndrome), which is caused by a 22q11.2 del that typically spans ~3 Mb; the DGCR8 gene maps to the deleted region [25–31]. In addition to SZ, many patients meet criteria for schizoaffective disorder (SAD), ASD, obsessive compulsive disorder (OCD), Tourette Syndrome, depression, anxiety disorder, and rapid cycling BD [32–40]. Conversely, 22q11.2 del is found in ~1% of patients with SZ, usually in the absence of the severe core clinical features characteristic of VCFS, such as cleft palate and congenital heart disease [41]. It is also found in ~4% of patients diagnosed with childhood onset SZ (COS) [42]. The T-box transcription factor TBX1 is primarily responsible for the major physical manifestations seen in 22q11.2 del [43]. However, the genes underlying the susceptibility to develop neuropsychiatric problems have not been unequivocally identified, although DGCR8 is a strong candidate [34,44–47]. Mouse Dgcr8 knockouts show down-regulation of ~25 mature miRNAs in the hippocampus and prefrontal cortex, and heterozygotes have deficits in prepulse inhibition and a spatial working memory–dependent learning task [31]. In addition, Dgcr8 knockout mice have deficits in the development of excitatory synapses and a reduction of parvalbumin interneurons in the prefrontal cortex [2,48].
In addition to viewing the role of miRNAs in 22q11.2 del from the perspective of the downstream effects of DGCR8, haploinsufficiency for individual miRNA genes that map to the deleted region have also been considered in disease pathogenesis. The most well-studied in this regard is MIR-185, which has been found to target other SZ candidate genes and influence dendritic spine density in the hippocampus [49,50]. MicroRNA-185 and its targets are also enriched in synapses [7,49,51]. MicroRNA-185 also affects immune developmental pathways, which might contribute to the immunological deficits found in a subset of patients with 22q11.2 del [52,53]. Considering the replicated genetic findings that point to the HLA locus in SZ, and the large body of epidemiological and animal studies suggesting that infectious diseases and/or autoimmune phenomena play roles in disease pathogenesis in subgroups of patients with SZ and ASD, an effect of miRNAs on immune function could potentially be of interest in neuropsychiatric disorders [54,55].
Although the 22q11.2 del mouse models have been extremely valuable, it is important to understand the molecular and genetic underpinnings in human neurons for translational research purposes. This is now possible with induced pluripotent stem cell (iPSC) technology, which we and others have been using to model neuropsychiatric disorders in vitro [56–68]. Our focus has been on 22q11.2 del syndrome because it is the most common known genetic risk factor in SZ, and one of the most penetrant as well. In order to determine if human neurons derived from patient-specific iPSCs are suitable for modeling the role of miRNAs in 22q11.2 del-associated disorders, and to identify differentially expressed miRNAs, we performed whole transcriptome miRNA sequencing and characterized the potential targets of dysregulated miRNAs.
Materials and Methods
Subjects
Controls and patients with 22q11.2 del diagnosed with a psychotic disorder (schizophrenia [SZ], childhood onset schizophrenia [COS], SAD) were recruited from two settings; the Albert Einstein College of Medicine (AECOM) and the National Institutes of Mental Health (NIMH), Child Psychiatry Branch. The study and the consent forms were approved by the AECOM Institution Review Board (IRB) and the NIMH IRB. Consents at AECOM were signed by the subjects at a time when psychotic symptoms were well-controlled with medications. For the NIMH subjects, all participants provided written assent/consent with written informed consent from a parent or legal guardian for minors. Subjects were not disadvantaged in any way if they refused to participate in the study. Consent was obtained by skilled members of the research teams who had received prior human subjects training. The procedure for obtaining informed consent was approved by the AECOM and NIMH IRBs.
The AECOM subjects were diagnosed with VCFS based on typical physical manifestations; the diagnosis was confirmed by FISH. Psychiatric diagnoses were established many years prior to recruitment by the patient’s psychiatrists using non-structured clinical interviews. Upon recruitment, a history of psychosis was confirmed by non-structured clinical interview with the patients and a parent. A more detailed clinical description for the AECOM cohort is provided in S1 Text.
Patients with childhood onset schizophrenia (COS), recruited as part of an NIMH initiative under the leadership of Dr. Judy Rapoport, met DSM-IIIR/DSM-IV criteria for SZ with documented onset of psychosis before age 13 [42]. They were subsequently interviewed for lifetime and current psychiatric disorders using structured psychiatric interviews, with diagnosis confirmed by inpatient, medication-free observation [42].
Controls in both cohorts were assessed by non-structured clinical interviews. There was no personal history of an Axis I diagnosis, and they have never been treated for a psychiatric disorder. It should be noted that we opted not to ascertain controls with 22q11.2 del who have not had psychotic episodes. This was decision was made because of the concern that a subject with 22q11.2 del who never experienced a psychotic episode could be recruited into the study as a young adult as a “control” could potentially onset later in life. Indeed, one of the subjects in this study, SZ_22q11-30, had her first psychotic episode at age 37. Consequently, controls were drawn from the general population.
A summary of the patients and controls used in this study is shown in Table 1.
Table 1. Demographics of subjects used for generating iPSCs.
| ID | age/sex | diagnosis |
|---|---|---|
| ctrl_iPSC1 | 29/F | control |
| ctrl_iPSC2 | 58/M | control |
| ctrl_iPSC5 | 32/M | control |
| ctrl_iPSC6 | 46/M | control |
| ctrl_553 | 31/M | control |
| ctrl_690 | 27/M | control |
| SZ_iPSC15 | 31/M | SAD/VCFS |
| SZ_22q11-30 | 41/F | SZ/VCFS |
| SZ_1804 | 25/F | COS |
| SZ_1220 | 31/F | COS |
| SZ_22q11-10 | 37/M | SAD/VCFS |
| SZ_22q11-60 | 25/M | SAD/VCFS |
Age refers to the age at time study was carried out. Abbreviations are Schizophrenia (SZ), Schizoaffective Disorder (SAD), Childhood Onset Schizophrenia (COS), velocardiofacial syndrome (VCFS). See S1 Text for clinical details.
Development of iPSCs from skin fibroblasts; generating iPSC lines
iPSC lines were generated from fibroblasts obtained from skin biopsies performed by board-certified physicians. The procedure for growing fibroblasts in preparation for reprogramming into iPSCs is detailed in S1 Text. Briefly, iPSC reprogramming was carried out by nucleofection. One vial of cells was thawed out and placed in a T75 flask in DMEM/F12 supplemented with 10% FBS and fed every 2 days. Cells were grown to ~50% confluence (~4–5 days), after which they were trypsinized and subjected to nucleofection (~6 x105 cells). Reprogramming was carried out using an Amaxa 4D-Nucleofector (P2 Primary Cell Kit from Lonza catalog# V4XP-2012, Program FF-135) with non-integrating plasmids containing OCT4, SOX2, KLF4, L-MYC, LIN28, and a p53 shRNA vector (Addgene Cat. # 27077, 27078, 27080), according to Okita et al., with some modifications [63,64,69]. iPSCs were maintained on Matrigel plates in mTeSR1 medium (Stem Cell Technologies) with daily feeding in 37°C/5% CO2/85% humidity.
Pluripotency for all iPSC lines was confirmed by immunocytochemistry using antibodies (Ab) against Tra-1-60, Tra-1-81, SSEA3 and SSEA4, which are expressed in pluripotent stem cells. In addition, the capacity to differentiate into all 3 germ layers was established by in vitro assays, as previously described [63,64]. The markers desmin (mesoderm), α-fetoprotein (endoderm), and βIII-tubulin (ectoderm) were used [70–73]. A list of the antibodies used to evaluate the iPSCs can be found in S1 Text. Karyotyping was carried out by Cell Line Genetics (Madison WI). Each iPSC line used in this study had a normal G-banded karyotype, which was used to screen for gross chromosomal changes that can occur during iPSC development. The 3 Mb deletion on 22q11.2 in the patient samples was identified by FISH using a TUPLE probe or microarray. Subjects were matched for age (control mean +/- standard deviation = 37.2+/-12.2; patients; 31.7+/-6.4, p = 0.35, Student’s t-test, two tailed). Due to technical issues there was only one female subject in the control group and three in the patient group. However, to compensate for this discrepancy, three different clones were analyzed in the control female.
Neuronal Differentiation
Neurons were generated from iPSC-derived neural progenitor cells (NPCs) as described by Marchetto et al. with slight modifications [63,65]. A detailed description of the protocol can be found S1 Text. Essentially, the protocol leads to a mixed population of glutamatergic and GABAergic neurons, from which RNA was extracted and sent for sequencing.
miRNA sequencing and data analysis
Briefly, small RNAs were extracted from day 14 neurons using miRNeasy. Libraries were constructed using NEBNext Multiplex Small RNA Library Prep Kit (Set1 for Illumina) and size selection of the Small RNA library (147 bp) was performed using Pippin Prep instrument using 3% Agarose, dye free gel with internal standards (Sage Science # CDF3010) according to the manufacturer’s instructions. MicroRNA sequencing was carried out using the Illumina HiSeq2500 Massively Parallel Sequencing platform as single end 50 bp read length.
For data analysis, 3’ adaptor sequences (TGGAATTCTCGGGTGCCAAGG) were removed from the raw miRNA-seq reads using a java script “AdRec.jar” from seqbuster [74]. Out of the total, ~7% of reads were without adaptors. 92% of processed reads after adaptor trimming were 15-35bp in length. The trimmed reads were then mapped to known pre-miRNA sequences deposited in miRBase (hsa database from miRBASE20) (http://www.mirbase.org/), allowing for one mismatch at most using bowtie ([75]. For any read to be considered as from a known mature miRNA, its 5’ and 3’ ends needed to be within 1–3 bp from the 5’ and 3’ ends of the mature miRNAs annotated in miRBase, respectively. To avoid mis-mapping, all trimmed reads were also mapped to the human genome reference sequences (hg19). Any reads initially assigned to a mature miRNA was re-assigned as non-miRNA in origin if it had a superior match outside the miRNA locus. Reads not mapped to miRNAs were also annotated and categorized based on their overlap with known gene annotations in the GENCODE (V18) (S1 Fig) [76].
To identify differentially expressed (DE) miRNAs, we applied DESeq2 to analyze the read counts of all microRNAs [77]. Specifically, DESeq2 normalized read counts across samples using size factors, estimated as the median of the ratio of a sample’s observed counts to the geometric mean of counts across samples. It then modeled the variance in miRNA read counts across replicates using the negative binomial distribution and then tested whether, for a given miRNA, the change in counts between the control and SZ/SAD samples was significantly larger as compared to the variation within each replicate group. A nominally significant p-value of < 0.01 and fold change >1.5-fold were chosen as the cutoffs for identifying differentially expressed miRNAs between SZ and controls, but a multiple comparison correction was also applied to adjust the p-values for genome-wide significance [78,79]. The total number of miRNA-seq reads for each sample is shown in S1 Table. The miRNA-seq data have been deposited in Gene Expression Omnibus (GEO; accession number GSE65367).
miRNA target gene prediction
The target genes for DE miRNAs were predicted using the Ingenuity Pathway Analysis (IPA) MicroRNA Target Filter, which combines experimentally validated targets from TarBase and miRecords, predicted targets from TargetScan, and manually collected miRNA-mRNA interactions from the peer-reviewed literature [80–83]. To reduce false positive targets, we only took into consideration the targets that were experimentally validated or predicted with high confidence. Then, IPA Core Analysis and the software DAVID were used for function analysis of the target genes [84,85]. A right-tailed Fisher’s exact test was run through IPA software and functions with p-value < 0.05 were considered significant. A modified Fisher’s exact test was run through DAVID and functional categories with p-value < 0.05 were considered significant [84,85].
Visualization of functional connections of predicted miRNA targets
To better understand how the significantly altered miRNAs could impact neural and brain function and development, we first used the DAVID analysis to detect biological process GO terms enriched (p-value < 0.05) in the targets of each differentially expressed miRNA. The neuron/brain-related GO terms were selected and the miRNA targets in these terms were then used to determine pairwise overlap coefficients [86]. A coefficient > 0.5 was used to connect two GO terms, resulting in a network. To illustrate the relationship between miRNAs and the GO term network, an edge was subsequently added between a specific miRNA and a GO term enriched among the predicted target of this miRNA. We further organized the GO terms into functional groups with reference to QuickGO [87] (http://www.ebi.ac.uk/QuickGO/). The final network was generated in Cytoscape 3.2.0 and exported for further editing of colors and labels in Adobe Illustrator [88].
Validation of mature miRNAs and DGCR8 by real time quantitative PCR (qPCR)
Expression levels of selected miRNAs were validated using Mercury LNA Universal RT microRNAs (Exiqon, Woburn, MA). PCR was carried out using an ABI-7900 HT Thermocycler in the presence of ROX (300 nM), which was used as a passive reference. Samples were analyzed in triplicate using the 2-ΔΔCt relative expression method normalized with two small RNAs; SNORD48 and U6. The mean expression values from 4–6 patient and control samples each were determined using both controls as normalizers. To measure the relative expression of DGCR8, standard qPCR was carried using beta-2 microglobulin as the normalizing control, as we have previously described [64,84]. For the statistical analysis, the control and SZ relative expression levels were pooled and the mean fold difference (controls vs SZ) was determined. A Student’s t-test was used to determine statistical significance.
Results
microRNA sequencing
MicroRNA-seq was carried out on day 14 neurons obtained from iPSCs derived from six controls and six patients with SZ or SAD. For one control sample (ctrl_iPSC1), three independent clones were analyzed, and for another control (ctrl_iPSC2), two different clones were analyzed, resulting in a total of 9 control samples. Among the 22q11.2 del samples, two different clones for SZ_iPSC15 were analyzed resulting in a total of 7 samples. After the miRNA-seq reads were analyzed and annotated, we found that the percentages of small RNA reads originating from miRNAs were, overall, similar among samples, but the SZ samples showed slightly reduced percentages (S1 Fig). The Spearman correlation coefficient was high (>0.8) across the patient samples and controls (S2 Fig).
After normalizing the counts, each sample showed high levels of expression of miRNAs associated with neurogenesis (miR-9, miR-124, miR-125a, miR-125b, miR-181a, miR-219, and let-7; for example), validating the neuronal nature of our samples. More modest levels of expression for other miRNAs involved in neurogenesis were detected, including miR-17, miR-184, miR-132, miR-324-5p, miR-326, and the SZ candidate miR-137. However, there were no significant differences in expression between patients and controls for these particular miRNAs (S2 Table).
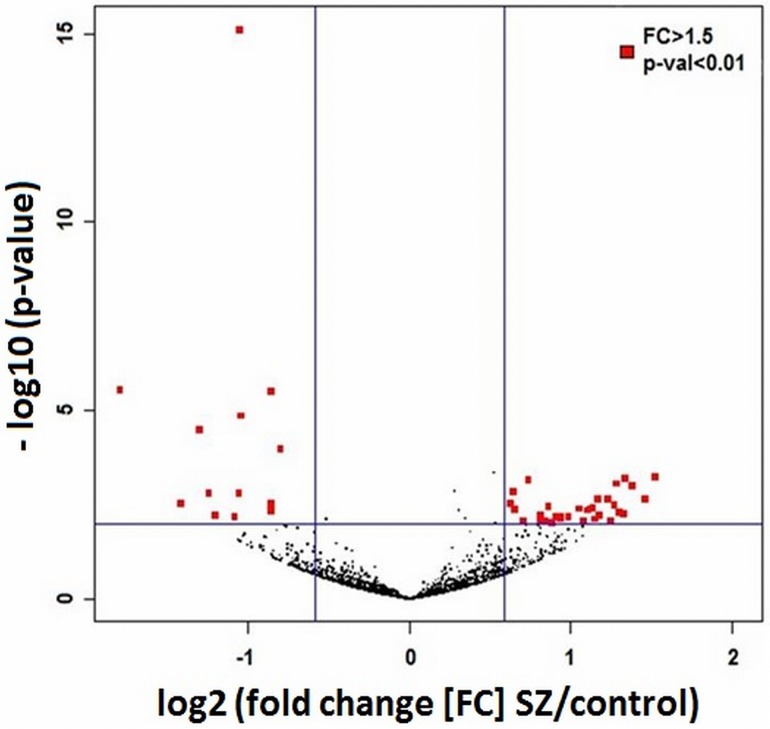
Using a p-value <0.01 and >1.5-fold differences in expression as limits, there were 45 differentially expressed miRNAs (13 lower in SZ; 32 higher; Fig 1, Table 2). Of these, 6 were significantly down-regulated in the 22q11.2 del neurons after correcting for genome wide significance (FDR<0.05), including 4 miRNAs that map to the 22q11.2 del region (miR-1306-3p, miR-1286, miR-1306-5p and miR-185-5p), and two that do not (miR-3175, miR-3158-3p). Two of the down-regulated miRNAs (miR-185 and miR-491) overlapped with the 25 that were found to be similarly down-regulated in the hippocampus and prefrontal cortex in Dgcr8 knockout mice [31,50].
Fig 1. Volcano plot showing statistical significance (-log10 of the p-values) on the y-axis vs fold change of all expressed miRNAs.
The 13 miRNAs that were significantly down-regulated in SZ are shown at the left (red squares), while the 32 that were significantly up-regulated in SZ are shown at the right (p < 0.05 and FC > 1.5).
Table 2. Differentially expressed miRNAs.
| Down regulated | coordinates | log2 FC | p-value | p-adjusted |
|---|---|---|---|---|
| miR-1306-3p | chr22:20073635_20073652 | 1.05 | 7.52E-16 | 1.34E-12 |
| miR-1286 | chr22:20236668_20236688 | 1.79 | 2.89E-06 | 1.84E-03 |
| miR-1306-5p | chr22:20073595_20073616 | 0.85 | 3.09E-06 | 1.84E-03 |
| miR-185-5p* | chr22:20020676_20020697 | 1.04 | 1.37E-05 | 6.11E-03 |
| miR-3175 | chr15:93447638_93447659 | 1.30 | 3.12E-05 | 1.12E-02 |
| miR-3158-3p | chr10:103361184_103361205,chr10:103361223_103361244 | 0.80 | 9.97E-05 | 2.97E-02 |
| miR-185-3p* | chr22:20020711_20020732 | 1.06 | 1.49E-03 | 1.69E-01 |
| miR-486-3p* | chr8:41517961_41517981,chr8:41518004_41518024 | 1.24 | 1.51E-03 | 1.69E-01 |
| miR-1249 | chr22:45596839_45596860 | 0.86 | 2.96E-03 | 2.44E-01 |
| miR-6840-5p | chr7:99954279_99954302 | 1.41 | 3.00E-03 | 2.44E-01 |
| miR-491-5p* | chr9:20716119_20716140 | 0.85 | 4.63E-03 | 2.67E-01 |
| miR-4804-5p | chr5:72174427_72174447 | 1.20 | 5.88E-03 | 2.93E-01 |
| miR-767-3p | chrX:151561919_151561941 | 1.08 | 6.32E-03 | 2.93E-01 |
| Up regulated | coordinates | |||
| miR-34b-3p* | chr11:111383712_111383733 | -1.52 | 5.81E-04 | 1.21E-01 |
| miR-34c-5p* | chr11:111384176_111384198 | -1.33 | 6.29E-04 | 1.21E-01 |
| miR-26b-5p* | chr2:219267380_219267400 | -0.74 | 6.75E-04 | 1.21E-01 |
| miR-146b-3p* | chr10:104196313_104196334 | -1.28 | 8.59E-04 | 1.40E-01 |
| miR-23a-5p* | chr19:13947444_13947465 | -1.38 | 9.86E-04 | 1.47E-01 |
| miR-296-3p* | chr20:57392681_57392702 | -0.64 | 1.44E-03 | 1.69E-01 |
| miR-4449* | chr4:53578887_53578908 | -1.46 | 2.14E-03 | 2.09E-01 |
| miR-4792 | chr3:24562903_24562920 | -1.22 | 2.21E-03 | 2.09E-01 |
| miR-148a-3p | chr7:25989542_25989563 | -1.17 | 2.22E-03 | 2.09E-01 |
| miR-320b | chr1:117214409_117214430,chr1:224444751_224444772 | -0.63 | 2.90E-03 | 2.44E-01 |
| miR-3609 | chr7:98479323_98479346 | -1.27 | 3.26E-03 | 2.54E-01 |
| miR-320c | chr18:19263520_19263539,chr18:21901680_21901699 | -0.86 | 3.52E-03 | 2.61E-01 |
| miR-126-3p* | chr9:139565105_139565126 | -1.13 | 3.87E-03 | 2.61E-01 |
| miR-320e | chr19:47212551_47212568 | -1.05 | 3.94E-03 | 2.61E-01 |
| miR-7704 | chr2:177053571_177053589 | -1.12 | 3.94E-03 | 2.61E-01 |
| miR-181b-5p* | chr1:198828054_198828076,chr9:127456004_127456026 | -0.65 | 4.08E-03 | 2.61E-01 |
| miR-146a-5p* | chr5:159912379_159912400 | -1.10 | 4.37E-03 | 2.66E-01 |
| miR-6757-5p | chr12:53450733_53450754 | -1.30 | 5.12E-03 | 2.86E-01 |
| miR-4682 | chr10:121718034_121718056 | -1.32 | 5.59E-03 | 2.93E-01 |
| miR-26a-5p* | chr12:58218441_58218462,chr3:38010904_38010925 | -0.81 | 6.08E-03 | 2.93E-01 |
| miR-3195 | chr20:60639868_60639884 | -1.18 | 6.16E-03 | 2.93E-01 |
| miR-126-5p* | chr9:139565068_139565088 | -0.94 | 6.31E-03 | 2.93E-01 |
| miR-125a-5p | chr19:52196521_52196544 | -0.91 | 6.39E-03 | 2.93E-01 |
| miR-548q | chr10:12767324_12767345 | -0.98 | 6.66E-03 | 2.98E-01 |
| miR-320d | chr13:41301964_41301982,chrX:140008337_140008355 | -0.94 | 7.37E-03 | 3.10E-01 |
| miR-4497 | chr12:110271155_110271171 | -1.15 | 7.47E-03 | 3.10E-01 |
| miR-27a-3p* | chr19:13947261_13947281 | -0.84 | 8.11E-03 | 3.17E-01 |
| miR-455-5p | chr9:116971729_116971750 | -0.70 | 8.25E-03 | 3.17E-01 |
| miR-7113-5p | chr11:67800332_67800352 | -0.81 | 8.33E-03 | 3.17E-01 |
| miR-6842-5p | chr8:27290892_27290913 | -1.25 | 8.64E-03 | 3.18E-01 |
| miR-146b-5p | chr10:104196277_104196298 | -1.07 | 8.70E-03 | 3.18E-01 |
| miR-6852-5p | chr9:35710713_35710733 | -0.88 | 9.21E-03 | 3.29E-01 |
Asterisk (*) shows miRNAs that have also been found to be differentially expressed in autopsy samples or peripheral cells in neuropsychiatric disorders (see text for references). Abbreviation: FC (fold change)
A total of 7 known miRNA genes map to the large 22q11.2 del region, producing 10 different mature miRNAs, 5 of which are expressed at relatively high levels in our differentiating neurons (miR-1306-3p, miR-1286-3p, and miR-1306-5p, miR-185-5p and miR-185-3p) (S2 Table). Three other mature miRNAs are expressed at relatively low levels (normalized RPM <1; miR-6816-3p, miR-4761-3p and miR-4761-5p), and two are not expressed at all in our neurons: miR-649 and miR-3168, the latter of which is involved in cardiovascular development. Of the 5 mature, highly expressed miRNAs, all showed a significant ~50% decrease in the 22q11.2 del samples compared with controls (miR-1306-3p, miR-1286, miR-1306-5p, miR-185-5p and miR-185-3p; miR-185-5p); four miRNAs were differentially expressed at the genome-wide significance including, miR-185, as noted above (Table 1). By contrast, 41 mature miRNAs that map to chromosome 22 outside of the deleted region are expressed in the neurons (out of total of 67 miRNA genes on chromosome 22). Of these, only one (miR-1249) showed a nominally significant difference compared to control neurons. The difference in differentially expressed mature miRNAs in the 22q11.2 del region that showed nominally significant differences in expression compared with mature miRNAs generated from the remainder of miRNA genes on chromosome 22 is highly significant (Fisher exact test, p = 2E-06). This shows that neurons derived from iPSCs that carry the 22q11.2 del recapitulate the miRNA expression patterns expected of 22q11.2 haploinsufficiency.
There were no differentially expressed up-regulated miRNAs that achieved genome wide significance. The most significant were miR-34b-3p and miR-34c-5p, which are members of the miR‑34 family. These miRNAs regulate the mitotic cell cycle, cell migration and apoptosis, and one member, miR-34a, has been found to be differentially expressed in patients with SZ and ASD (see discussion) [89–96]. It should be noted that miR-34a-5p showed a 1.6-fold increase in the SZ neurons in our study (S2 Table). However the results did not reach our threshold for nominal significance (p = 0.11).
Other miRNAs that show the largest fold increases in the SZ samples were miR-4449, miR-146b-3p, and miR-23a-5p. These are all of interest in neuropsychiatric disorders as described in greater detail in the Discussion section [97–102]. MicroRNA-146 also affects IL-6 expression and regulates inflammatory responses and innate immunity, factors associated with SZ and ASD risk [98,103–108]. These findings show that in addition to down-regulated miRNAs caused by haploinsufficiency for DGCR8 and the miRNA genes that map to the 22q11.2 del region, increased expression of some miRNAs may also play a role in the pathogenesis of neuropsychiatric disorders associated with 22q11.2 del.
Validation
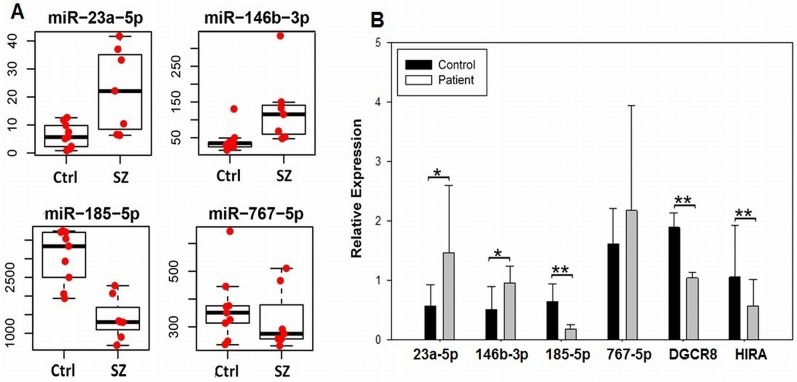
The expression of four miRNAs that showed either increased or decreased levels, or no change, in the SZ samples relative to controls was validated by qPCR, as described in the methods section. As shown in Fig 2, fold differences in expression were similar to that seen in the miRNA-seq experiments. In addition, we used qPCR to evaluate the expression of DGCR8 mRNA in our patient vs control samples. As seen in the figure, the qPCR results support the miRNA-seq findings; a significant decrease in DGCR8 expression is seen in the patient samples, as expected of a 22q11.2 del disorder. A significant decrease in the expression of HIRA, which also maps to the 22q11.2 del region was detected as well.
Fig 2.
A. miRNA-seq reads (y-axis) for controls (ctrl) and patients with 22q11.2 del (SZ) showing two, nominally significant up-regulated genes (miR-23a-5p and miR-146b-3p), a miRNA that showed a genome wide significant decrease in expression (miR-185-5p), and a miRNA that showed no difference in expression (miR-767-5p). B. Same miRNAs in 2A showing qPCR analysis using Mercury LNA Universal RT microRNAs assays, as described in methods. The relative expression of two coding genes that map to the 22q11.2 del region (DGCR and HIRA) was analyzed by routine qPCR. A Student’s t-test was used for statistical analysis. Error bars show standard deviation; p<0.05, one-tailed*; p<0.05, two-tailed.**
Predicted mRNA targets of differentially expressed miRNAs
In order to determine the potential impact of differentially expressed miRNAs on neuronal function, putative gene targets were predicted and functionally characterized as described in the Methods section. A total of 5,413 predicted targets were found with the high confidence setting in IPA among the up-regulated miRNAs, and 2,274 predicted targets were found for the down-regulated miRNAs (S3 Table). These high confidence targets were used in subsequent analyses because the number of experimentally validated targets was too small (240 targets for up-regulated miRNAs; 10 for the down-regulated miRNAs) for pathway analyses. According to IPA, the top two diseases/functions enriched within the predicted targets of both up and down-regulated miRNAs were neurological and psychological disorders (S4 Table; Table 3). Among the predicted targets of up-regulated miRNAs were a number of well-established SZ, ASD and BD candidates, including DISC1, GSK3β, MYT1L, TCF7L2, CNTNAP1, NRXN1, genes involved in glutamatergic transmission (GRM3, GRIN2A, GRIN2B, GRIN2D, GRIK2, GRIK3), and genes involved in GABAergic transmission (CCK, GABRA1 GRIN2B,GABBR2, GABRB2). Among the down-regulated miRNA gene targets were the candidate genes GSK3B, CNTNAP1, DAO, GRIA1, GRIN1, GRIK3, and SLC17A7 (VGLUT1).
Table 3. Ingenuity Pathway Analysis (IPA): Diseases/Functions of mRNA targets of differentially expressed miRNAs.
| Diseases/Functions: targets of up-regulated miRNAs | p-value |
| Neurological Disease | 6.74E-04-4.49E-02 |
| Psychological Disorders | 6.74E-04-4.49E-02 |
| Metabolic Disease | 7.39E-04-4.49E-02 |
| Cell Death and Survival | 2.18E-03-4.76E-02 |
| Cellular Growth and Proliferation | 2.26E-03-3.42E-02 |
| Cellular Development | 3.38E-03-4.49E-02 |
| Nervous System Development and Function | 3.38E-03-4.49E-02 |
| Tissue Development | 3.38E-03-4.49E-02 |
| Tissue Morphology | 6.1E-03-4.49E-02 |
| Cellular Assembly and Organization | 1.49E-02-3.42E-02 |
| Disease/Functions: targets of down-regulated miRNAs | p-value |
| Neurological Disease | 1.42E-02-2.95E-02 |
| Psychological Disorders | 1.42E-02-2.95E-02 |
| Metabolic Disease | 1.66E-02-1.96E-02 |
| Cell Morphology | 1.89E-02-1.89E-02 |
| Cellular Function and Maintenance | 1.89E-02-1.89E-02 |
| Nervous System Development and Function | 1.89E-02-1.89E-02 |
Ingenuity Pathway Analysis (IPA), Diseases/Functions category of mRNA targets of differentially expressed miRNAs. Table shows top IPA diseases and functions for predicted gene targets.
Similarly, using the DAVID functional annotation tool, SZ was the top disease category (Table 4) for up-regulated miRNA target genes, when the predicted targets were analyzed for enrichment of genes associated with genetic disorders. In addition, other neuropsychiatric disorders previously reported in patients with 22q11.2 del were also among the top diseases, including BD, Tourette Syndrome and SAD, as well as cleft palate, one of the most common physical anomalies associated with VCFS [32–34,36,109,110]. Interestingly, target genes involved in type 2 diabetes were also somewhat enriched. This suggests that there may be some common genetic factors involved in the development of type 2 diabetes, which is seen as part of the “metabolic syndrome” that occurs in a substantial subgroup of SZ patients treated with anti-psychotic medications, consistent with some published reports [111,112].
Table 4. DAVID functional annotation; Top Diseases for predicted targets of differentially expressed miRNAs.
| Top Diseases for predicted targets of up-regulated miRNAs | |||
| Term | Count | % | P-Value |
| schizophrenia | 116 | 2.29 | 3.30E-04 |
| Alzheimer's Disease | 91 | 1.79 | 0.01 |
| colon cancer rectal cancer | 6 | 0.12 | 0.01 |
| schizophrenia; schizoaffective disorder; bipolar disorder | 10 | 0.20 | 0.02 |
| prostate cancer | 66 | 1.30 | 0.02 |
| bipolar affective disorder | 8 | 0.16 | 0.02 |
| sleep disorders | 8 | 0.16 | 0.02 |
| cleft lip with cleft palate cleft lip without cleft palate cleft palate | 16 | 0.32 | 0.03 |
| colorectal cancer; Tourette syndrome; bone density; pregnancy loss, recurrent; cleft lip without cleft palate; juvenile polyposis; cleft palate | 6 | 0.12 | 0.03 |
| diabetes, type 2 triglycerides | 6 | 0.12 | 0.03 |
| Top Diseases for predicted targets of down-regulated miRNAs | |||
| Term | Count | % | P-Value |
| myelopathy, HTLV-1 associated | 5 | 0.24 | 0.003 |
| narcolepsy | 7 | 0.33 | 0.004 |
| diabetes, type 1 | 38 | 1.80 | 0.004 |
| pulmonary hypertension | 4 | 0.19 | 0.02 |
| sclerosis, systemic | 10 | 0.47 | 0.03 |
| HTLV-1 infection | 5 | 0.24 | 0.04 |
| malaria; schistosomiasis | 3 | 0.14 | 0.04 |
| migraine; migraine with aura | 4 | 0.19 | 0.05 |
| normal variation | 7 | 0.33 | 0.05 |
| diabetes mellitus | 5 | 0.24 | 0.05 |
| Graves' disease | 9 | 0.43 | 0.06 |
| bipolar disorder | 20 | 0.95 | 0.06 |
| schizophrenia | 45 | 2.13 | 0.07 |
DAVID functional annotation; Top Diseases for predicted targets of differentially expressed miRNAs.
Genes involved in SZ and BD were also found among the predicted targets of down-regulated miRNAs, although the p-values were modest (p = 0.06 and 0.07, respectively, Table 4). A number of infectious and autoimmune disorders were among the top diseases in this category as well, consistent with the immune problems found in patients with 22q11.2 del. Whether this also influences the underlying autoimmune and/or infectious disease etiology believed to play a role in the pathogenesis of SZ and ASD in a subgroup of patients remains to be determined [54,55,113–116].
The predicted target genes were also characterized by Gene Ontology (GO). As seen in Table 5, the most enriched GO terms in the cellular component (CC) assessment were synapse and neuron projection for predicted targets of up-regulated miRNAs; these were also among the top hits for predicted targets of down-regulated miRNAs (see S5 Table and S6 Table for details). Abnormal synaptogenesis and neuron projection are primary pathogenic features of both SZ and ASD [117–125].
Table 5. DAVID functional annotation; Gene Ontology (GO) categories (CC).
| GO categories | Count | % | p-value | FE | Bonferroni | Benjamini | FDR |
| GO:0045202~synapse | 172 | 3.39 | 1.23E-18 | 1.81 | 9.56E-16 | 9.56E-16 | 1.88E-15 |
| GO:0043005~neuron projection | 167 | 3.29 | 1.54E-18 | 1.82 | 1.20E-15 | 6.02E-16 | 2.36E-15 |
| GO:0005794~Golgi apparatus | 332 | 6.55 | 4.56E-14 | 1.42 | 3.56E-11 | 1.19E-11 | 6.98E-11 |
| GO:0042995~cell projection | 269 | 5.30 | 2.87E-12 | 1.44 | 2.24E-09 | 5.60E-10 | 4.39E-09 |
| GO:0005626~insoluble fraction | 313 | 6.17 | 4.99E-12 | 1.39 | 3.89E-09 | 7.79E-10 | 7.64E-09 |
| GO:0005624~membrane fraction | 302 | 5.95 | 1.15E-11 | 1.39 | 8.95E-09 | 1.49E-09 | 1.76E-08 |
| GO:0030424~axon | 82 | 1.62 | 6.01E-11 | 1.93 | 4.69E-08 | 6.70E-09 | 9.20E-08 |
| GO:0000267~cell fraction | 383 | 7.55 | 6.93E-11 | 1.32 | 5.40E-08 | 6.75E-09 | 1.06E-07 |
| GO:0019898~extrinsic to membrane | 197 | 3.88 | 1.31E-10 | 1.49 | 1.02E-07 | 1.13E-08 | 2.00E-07 |
| GO:0044459~plasma membrane part | 711 | 14.02 | 1.95E-10 | 1.21 | 1.52E-07 | 1.52E-08 | 2.98E-07 |
| GO categories | Count | % | p-value | FE | Bonferroni | Benjamini | FDR |
| GO:0044459~plasma membrane part | 356 | 16.84 | 9.98E-13 | 1.40 | 5.97E-10 | 5.97E-10 | 1.47E-09 |
| GO:0005886~plasma membrane | 519 | 24.55 | 7.08E-07 | 1.19 | 4.23E-04 | 2.12E-04 | 1.05E-03 |
| GO:0045202~synapse | 73 | 3.45 | 1.11E-06 | 1.78 | 6.61E-04 | 2.21E-04 | 1.63E-03 |
| GO:0042995~cell projection | 121 | 5.72 | 3.54E-06 | 1.50 | 0.00 | 5.29E-04 | 0.01 |
| GO:0043005~neuron projection | 69 | 3.26 | 4.52E-06 | 1.74 | 0.00 | 5.40E-04 | 0.01 |
| GO:0045121~membrane raft | 36 | 1.70 | 1.12E-05 | 2.18 | 0.01 | 0.00 | 0.02 |
| GO:0031226~intrinsic to plasma membrane | 184 | 8.70 | 6.27E-05 | 1.31 | 0.04 | 0.01 | 0.09 |
| GO:0005887~integral to plasma membrane | 180 | 8.51 | 7.38E-05 | 1.31 | 0.04 | 0.01 | 0.11 |
| GO:0005624~membrane fraction | 129 | 6.10 | 1.06E-04 | 1.38 | 0.06 | 0.01 | 0.16 |
Network analysis
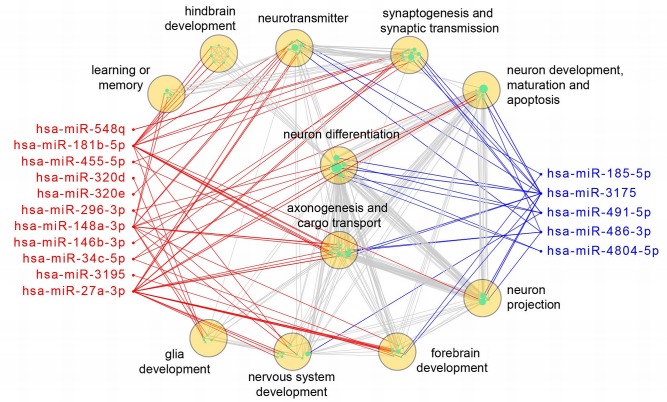
To better illustrate how the differentially expressed miRNAs may independently or cooperatively affect brain development and function, we constructed a network of differentially expressed miRNAs and the enriched neural GO terms of their putative targets, as described in the Methods section [86,87] (Fig 3). The data show that 11 up-regulated and 5 down-regulated miRNAs can regulate a broad range of brain related genes, with miR-181b-5p, miR148a-3p, miR-27a-3p, and miR-3175 potentially having the largest impact. The result also indicates that genes involved in neurotransmitter function, synaptogenesis, and neuronal differentiation will likely be affected most by the disruption of the miRNAs found in our 22q11.2 del neurons. Also, while forebrain development can be affected by both up- and down-regulated miRNAs in SZ, hindbrain development seems be affected more by the up-regulated miRNAs. The figure reinforces our finding that in differentiating neurons, up-regulated, as well as down-regulated miRNAs and their targets are affected by 22q11.2 del, and consequently can affect multiple processes in brain development and function.
Fig 3. A network view of neurological function of significantly altered miRNAs.
Red nodes stand for up-regulated miRNAs, blue for down-regulated miRNAs, and cyan for enriched GO terms (n = 91, with sizes proportional to the numbers of predicted miRNA targets). GO terms were further classified into broader function groups (yellow circles) by QuickGO. A red or blue edge indicates a GO term was enriched in the predicted targets of up-regulated or down-regulated miRNAs, respectively. Grey edges link GO terms with overlapped targets (overlap coefficient > 0.5). This network was created by the Cytoscape 3.2.0.
Discussion
Research on the potential role of miRNAs in 22q11.2 del-associated neuropsychiatric disorders has primarily focused on analyzing the effects of DGCR8 on miRNA expression and behavior, which stands to reason considering the effect it has on miRNA biogenesis [31,48,117]. More recently, there have been studies showing the effects of specific miRNAs that map to the deleted region, most notably MIR-185, which may influence neuronal function and synaptogenesis independently of DGCR8-mediated biogenesis [7,49,51,117]. Overall, the bioinformatics analysis of predicted targets of miRNAs that were found to be down-regulated in our 22q11.2 del samples, which included miR-185, is consistent with the mouse studies in that there was enrichment for genes involved in neuropsychiatric disorders, synaptogenesis and neuron projection. However, without direct functional studies, the relative contribution of DGCR8, miR-185 and other down-regulated miRNAs, some of which are affected by reduced levels of DGCR8, remains uncertain.
There were two miRNAs that were significantly down-regulated in our 22q11.2 neurons that overlapped with the 25 that were found in Dgcr8 knockouts; miR-185 and miR-491[31]. The reason for the small degree of overlap is not clear, but could be due to species differences, cellular heterogeneity in the neuroanatomical structures analyzed in mice, which could potentially amplify an effect in non-neuronal cells, and the relative immaturity and spatio-temporal differences of the neurons used in our analysis. Although miR-185 has established effects on neuronal function, as noted above, less is known about miR-491 in the context of neuronal function and disease pathogenesis. However, in cancer cells, expression is correlated with an increase in apoptosis through activation of intrinsic mitochondrial apoptotic pathways, and cell growth is affected by the inhibitory effect of miR-491 on PI3K/Akt signaling [126]. Abnormalities in apoptosis and PI3K/Akt signaling have been described in SZ and ASD [127–131].
As for other down-regulated miRNAs that map to 22q11.2, there are no reported functional studies, so their potential role in neuropsychiatric disorders is uncertain. The same can be said for the down-regulated miRNAs detected in our study that map to other chromosomes, with the exception of miR-486 and miR-491. miR-486 was the top hit in an analysis of differentially expressed genes carried out in discordant siblings for ASD using lymphoblastoid cell lines (although expression was higher in the ASD subjects, and lower in our 22q11.2 del neurons) [132]. MicroRNA-486 is known to regulate Akt signaling by targeting PTEN, the latter of which is a well-established ASD candidate gene [133–135]. MicroRNA-491 is one of several miRNAs that may affect impulsivity and co-morbid traits associated with synaptic plasticity in the mouse amygdala [136]. This could be of translational interest because amygdala size has been reported to be abnormal in 22q11.2 del [137,138]. Functional abnormalities in the amygdala probably plays some role in the high rate of anxiety disorder seen in patients with 22q11.2 del, as well as their tendency towards impulsivity [39,40,139]. Down-regulation of these miRNAs is likely not due to DGCR8 haploinsufficiency, at least not entirely, since they were not identified as such in Dgcr8 knockout mice [31].
Thus, in 22q11.2 del, some down-regulated miRNAs that may play a role in disease pathogenesis are regulated independently of DGCR8, their expression affected perhaps by one or more of the transcriptional and chromatin regulators that map to this region of the genome.
An effect on miRNA expression in 22q11.2 del that is independent of DGCR8 is supported by the finding that 32 differentially expressed miRNAs were up-regulated in the 22q11.2 del samples, rather than down-regulated, a number of which have previously been connected to neuropsychiatric disorders. The miR-34 family of related miRNAs is an example. A key member of the miR-34 family is miR-34a, which plays a role in neural stem cell differentiation [93]. Most interestingly, miR-34a is expressed at higher levels in the peripheral blood mononuclear cells and in the prefrontal cortex in SZ patients compared to controls [92,94,95]. Similarly, miR-34a, 34b, and 34c are upregulated in the hippocampus of Fmr1 KO mice: a CGG trinucleotide repeat in the FMR1 gene causes Fragile X syndrome [140]. A number of ASD, SZ and BD candidate genes are predicted targets of miR-34c, including CNTNAP1, CNTNAP2, GABRA3, RELN, FOXP2, NRXN2, and ANK3 (S3 Table).
Finally miR-34a was identified as a hub molecule in a bioinformatics analysis of CNVs in ASD [96].
Another up-regulated miRNA of interest is miR-4449; a significant increase in expression was detected in Brodmann area 46 in SZ autopsy samples, and a trend towards a significant increase was found in the hair follicles of living patients [97]. Others are three members of the miR-146 family and miR-296. MicroRNA-146a and miR-146b were two of the top differentially expressed miRNAs in an animal model of Rett syndrome, although it is lower in the Mecp2 knockouts, while miR-296 is expressed at significantly higher levels, similar to our SZ neuronal samples [141]. MicroRNA-146 expression is modulated by neuronal activity, and affects IL-6 expression, inflammatory responses and innate immunity, factors associated with SZ and ASD risk [98,103–108]. This is consistent with the finding that predicted targets of both miR-146a-5p and miR-146b-3p are multiple members of the interleukin signaling cascade (S3 Table).
Another suggestive up-regulated miRNA is miR-23a, the expression of which is increased in the cerebellum and transformed lymphoblasts of patients with ASD ([99,100]. Expression is also increased (as is miR-146a) in the hippocampus of patients with epilepsy [102]. Epilepsy is found in a subgroup of patients with 22q11.2 del and shares common genetic risk factors with SZ and ASD at multiple loci [142–147]. Finally, the expression of miR-26a, miR-27a-3p, miR-181b and miR-26b is higher in the DLPFC in SZ [101]. A number of SZ, ASD and bipolar disorder candidates are also high confidence predicted targets of these genes, including JARID2, RGS4, TCF7L2, GSK3B, GABRB3, FOXP2, CACNA1C, NRXN1, ANK3, CNTNAP2, among others (S3 Table).
Identifying differentially expressed miRNAs in our in vitro model for 22q11.2 del-associated neuropsychiatric disorders is extremely important from the standpoint of interpreting miRNA expression data uncovered using autopsy specimens. Although informative findings have emerged from the analysis of mRNAs and miRNAs using this resource, it is an imperfect system from a variety of perspectives. In addition to the technical challenges associated with using autopsy samples (e.g., brain pH, premorbid medical issues, cause of death, postmortem delay, cellular heterogeneity), there are pathophysiological considerations as well. Patients with SZ, for example, are exposed to a number of environmental factors that can each influence gene expression, such as medications, alcohol and nicotine use, and illicit drugs. The differential expression of mRNAs and miRNAs found in expression profiling studies could easily be influenced by one or more of these factors. A similar set of circumstances could also influence the interpretation of gene expression studies in ASD autopsy samples. In addition, the neurodevelopmental underpinnings of both SZ and ASD predate gene expression changes found in autopsy samples by decades. The finding that a number of differentially expressed miRNAs we detected in iPSC-derived neurons grown under controlled conditions overlap with those found in autopsy samples, supports their involvement in neuropsychiatric disorders. This provides a strong rationale for their more extensive analysis in animal and in vitro models.
The same principle applies to the use of peripheral, non-neuronal cells, which are exposed to the same confounding environmental factors, and suffer from the additional shortcoming of being a very imperfect proxy for explaining pathophysiological processes involved in neurodevelopmental disorders. However, peripheral cells are excellent sources of potential molecular biomarkers, as long as a connection can be established between those biomarkers and neuronal function and/or behavior. The finding of common differentially expressed miRNAs in peripheral cells and our cultured neurons, such as the miR-34 family, miR-4449 and miR-23a, supports the idea that they should be evaluated as potential biomarkers to assess clinically relevant phenotypes.
Finally, the finding of common differentially expressed miRNAs in neurons containing a 22q11.2 del and clinical/autopsy samples drawn from the general population of SZ and ASD patients suggests that there are underlying molecular genetic networks shared by 22q11.2 del and candidate genes at other loci.
One caveat to our findings that needs to be discussed is that our analysis was carried out using a mix of phenotypes that included both COS and patients who developed psychotic symptoms during adolescence and adulthood. In addition, we used independent replicate clones for two of the controls and one 22q11.2 del subject. Consequently, we repeated the differential expression analysis without the COS subjects, and again, without the independent clones. Using our threshold for nominal significance (p-value of < 0.01 and fold change >1.5-fold), 26 out of 45 differentially expressed miRNAs fell below the level of significance when the data were analyzed without the COS samples (S7 Table). Similarly, without the replicate samples, 13 differentially miRNAs fell below the threshold. However, the dropouts are due to a small increase in p-values. In the COS samples, 23/26 remained below 0.05 (that is, between 0.01–0.05), and the highest was 0.08. In addition, the fold changes comparing patients vs controls are largely unaffected. Indeed, 17 of the miRNAs that dropped below the threshold showed an increase in the fold change—although p-values increased because of higher sample to sample variability. For the 13 differentially expressed miRNAs that fell below the significance threshold when replicates were removed, all but 2 had p-values between 0.01 and 0.05. In addition, the fold change increased in 10 samples, although, as in the COS samples, the p-values increased because of sample to sample variability. This reanalysis indicates that omitting the COS and replicate samples does not significantly change the major conclusions drawn from our study, and that we need to include all samples to maximize statistical power.
To summarize, we show that iPSCs derived from patients with SZ who have 22q11.2 del is a good model system to study the neuropsychiatric manifestations of this condition in vitro. Furthermore, expression profiling shows changes in the expression of several miRNAs that are similar to those found in clinical samples, supporting a role in SZ and ASD pathogenesis and their utility as peripheral biomarkers.
Supporting Information
(TIF)
(TIF)
(DOCX)
(XLSX)
(XLSX)
(XLS)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Acknowledgments
We thank Dr. Robert Shprintzen, President and Chairman of the Board of the Virtual Center for Velo-Cardio-Facial Syndrome, Inc for patient referrals, Dr. Judith Rapoport for supplying fibroblasts from patients with childhood onset schizophrenia, Dr. Kwangmi Ahn for providing genotyping data, Dr. Shahina Maqbool and her staff at the Einstein Epigenomics Shared Facility for miRNA-sequencing. We also want to thank participating subjects and their families.
Data Availability
All relevant data are within the paper and its Supporting Information files. The miRNA-seq data have been deposited in Gene Expression Omnibus (GEO; accession number GSE65367).
Funding Statement
This work was supported by National Institute of Mental Health grants MH087840, MH099427, MH093869 and MH099452. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Liu J, Githinji J, Mclaughlin B, Wilczek K, Nolta J. Role of miRNAs in neuronal differentiation from human embryonic stem cell-derived neural stem cells. Stem Cell Rev. 2012;8: 1129–1137. 10.1007/s12015-012-9411-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hsu R, Schofield CM, Dela Cruz CG, Jones-Davis DM, Blelloch R, Ullian EM. Loss of microRNAs in pyramidal neurons leads to specific changes in inhibitory synaptic transmission in the prefrontal cortex. Mol Cell Neurosci. 2012;50: 283–292. 10.1016/j.mcn.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jimenez-Mateos EM, Engel T, Merino-Serrais P, McKiernan RC, Tanaka K, Mouri G, et al. Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat Med. 2012;18: 1087–1094. 10.1038/nm.2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Konopka W, Kiryk A, Novak M, Herwerth M, Parkitna JR, Wawrzyniak M, et al. MicroRNA loss enhances learning and memory in mice. J Neurosci. 2010;30: 14835–14842. 10.1523/JNEUROSCI.3030-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lambert TJ, Storm DR, Sullivan JM. MicroRNA132 modulates short-term synaptic plasticity but not basal release probability in hippocampal neurons. PLoS One. 2010;5: e15182 10.1371/journal.pone.0015182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9: 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smalheiser NR, Lugli G. microRNA regulation of synaptic plasticity. Neuromolecular Med. 2009;11: 133–140. 10.1007/s12017-009-8065-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5: R13 10.1186/gb-2004-5-3-r13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meza-Sosa KF, Pedraza-Alva G, Perez-Martinez L. microRNAs: Key triggers of neuronal cell fate. Front Cell Neurosci. 2014;8: 175 10.3389/fncel.2014.00175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136: 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11: 441–450. 10.1016/j.devcel.2006.09.009 [DOI] [PubMed] [Google Scholar]
- 12. Chitwood DH, Timmermans MC. Small RNAs are on the move. Nature. 2010;467: 415–419. 10.1038/nature09351 [DOI] [PubMed] [Google Scholar]
- 13. Slezak-Prochazka I, Durmus S, Kroesen BJ, van den Berg A. MicroRNAs, macrocontrol: Regulation of miRNA processing. RNA. 2010;16: 1087–1095. 10.1261/rna.1804410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15: 509–524. 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- 15. Stappert L, Roese-Koerner B, Brustle O. The role of microRNAs in human neural stem cells, neuronal differentiation and subtype specification. Cell Tissue Res. 2015;359: 47–64. 10.1007/s00441-014-1981-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43: 969–976. 10.1038/ng.940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lett TA, Chakavarty MM, Felsky D, Brandl EJ, Tiwari AK, Goncalves VF, et al. The genome-wide supported microRNA-137 variant predicts phenotypic heterogeneity within schizophrenia. Mol Psychiatry. 2013. 10.1038/mp.2013.17 [DOI] [PubMed]
- 18.Green MJ, Cairns MJ, Wu J, Dragovic M, Jablensky A, Tooney PA, et al. Genome-wide supported variant MIR137 and severe negative symptoms predict membership of an impaired cognitive subtype of schizophrenia. Mol Psychiatry. 2012. 10.1038/mp.2012.84 [DOI] [PubMed]
- 19. Das S, Chang C. Regulation of early xenopus embryogenesis by smad ubiquitination regulatory factor 2. Dev Dyn. 2012;241: 1260–1273. 10.1002/dvdy.23811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kwon E, Wang W, Tsai LH. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Mol Psychiatry. 2013;18: 11–12. 10.1038/mp.2011.170 [DOI] [PubMed] [Google Scholar]
- 21. Kim AH, Parker EK, Williamson V, McMichael GO, Fanous AH, Vladimirov VI. Experimental validation of candidate schizophrenia gene ZNF804A as target for hsa-miR-137. Schizophr Res. 2012;141: 60–64. 10.1016/j.schres.2012.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex brodmann area 46 in schizophrenia. Biol Psychiatry. 2011;69: 180–187. 10.1016/j.biopsych.2010.09.030 [DOI] [PubMed] [Google Scholar]
- 23. Moreau MP, Bruse SE, David-Rus R, Buyske S, Brzustowicz LM. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol Psychiatry. 2011;69: 188–193. 10.1016/j.biopsych.2010.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beveridge NJ, Santarelli DM, Wang X, Tooney PA, Webster MJ, Weickert CS, et al. Maturation of the human dorsolateral prefrontal cortex coincides with a dynamic shift in MicroRNA expression. Schizophr Bull. 2013. 10.1093/schbul/sbs198 [DOI] [PMC free article] [PubMed]
- 25. Scambler PJ, Kelly D, Lindsay E, Williamson R, Goldberg R, Shprintzen R, et al. Velo-cardio-facial syndrome associated with chromosome 22 deletions encompassing the DiGeorge locus. Lancet. 1992;339: 1138–1139. [DOI] [PubMed] [Google Scholar]
- 26. Carlson C, Papolos D, Pandita RK, Faedda GL, Veit S, Goldberg R, et al. Molecular analysis of velo-cardio-facial syndrome patients with psychiatric disorders. Am J Hum Genet. 1997;60: 851–859. [PMC free article] [PubMed] [Google Scholar]
- 27. Carlson C, Sirotkin H, Pandita R, Goldberg R, McKie J, Wadey R, et al. Molecular definition of 22q11 deletions in 151 velo-cardio-facial syndrome patients. Am J Hum Genet. 1997;61: 620–629. 10.1086/515508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Driscoll DA, Spinner NB, Budarf ML, McDonald-McGinn DM, Zackai EH, Goldberg RB, et al. Deletions and microdeletions of 22q11.2 in velo-cardio-facial syndrome. Am J Med Genet. 1992;44: 261–268. 10.1002/ajmg.1320440237 [DOI] [PubMed] [Google Scholar]
- 29. Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, et al. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet. 1999;8: 1157–1167. [DOI] [PubMed] [Google Scholar]
- 30. Brzustowicz LM, Bassett AS. miRNA-mediated risk for schizophrenia in 22q11.2 deletion syndrome. Front Genet. 2012;3: 291 10.3389/fgene.2012.00291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40: 751–760. 10.1038/ng.138 [DOI] [PubMed] [Google Scholar]
- 32. Shprintzen RJ, Goldberg RB, Lewin ML, Sidoti EJ, Berkman MD, Argamaso RV, et al. A new syndrome involving cleft palate, cardiac anomalies, typical facies, and learning disabilities: Velo-cardio-facial syndrome. Cleft Palate J. 1978;15: 56–62. [PubMed] [Google Scholar]
- 33. Papolos DF, Veit S, Faedda GL, Saito T, Lachman HM. Ultra-ultra rapid cycling bipolar disorder is associated with the low activity catecholamine-O-methyltransferase allele. Mol Psychiatry. 1998;3: 346–349. [DOI] [PubMed] [Google Scholar]
- 34. Lachman HM, Morrow B, Shprintzen R, Veit S, Parsia SS, Faedda G, et al. Association of codon 108/158 catechol-O-methyltransferase gene polymorphism with the psychiatric manifestations of velo-cardio-facial syndrome. Am J Med Genet. 1996;67: 468–472. doi: 2-G. [DOI] [PubMed] [Google Scholar]
- 35. Murphy KC, Owen MJ. Velo-cardio-facial syndrome: A model for understanding the genetics and pathogenesis of schizophrenia. Br J Psychiatry. 2001;179: 397–402. [DOI] [PubMed] [Google Scholar]
- 36. Aneja A, Fremont WP, Antshel KM, Faraone SV, AbdulSabur N, Higgins AM, et al. Manic symptoms and behavioral dysregulation in youth with velocardiofacial syndrome (22q11.2 deletion syndrome). J Child Adolesc Psychopharmacol. 2007;17: 105–114. 10.1089/cap.2006.0023 [DOI] [PubMed] [Google Scholar]
- 37. Kates WR, Antshel KM, Fremont WP, Shprintzen RJ, Strunge LA, Burnette CP, et al. Comparing phenotypes in patients with idiopathic autism to patients with velocardiofacial syndrome (22q11 DS) with and without autism. Am J Med Genet A. 2007;143A: 2642–2650. 10.1002/ajmg.a.32012 [DOI] [PubMed] [Google Scholar]
- 38. Ivanov D, Kirov G, Norton N, Williams HJ, Williams NM, Nikolov I, et al. Chromosome 22q11 deletions, velo-cardio-facial syndrome and early-onset psychosis. molecular genetic study. Br J Psychiatry. 2003;183: 409–413. [DOI] [PubMed] [Google Scholar]
- 39. Schneider M, Debbane M, Bassett AS, Chow EW, Fung WL, van den Bree M, et al. Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: Results from the international consortium on brain and behavior in 22q11.2 deletion syndrome. Am J Psychiatry. 2014;171: 627–639. doi: 1838527 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gothelf D, Schneider M, Green T, Debbane M, Frisch A, Glaser B, et al. Risk factors and the evolution of psychosis in 22q11.2 deletion syndrome: A longitudinal 2-site study. J Am Acad Child Adolesc Psychiatry. 2013;52: 1192–1203.e3. 10.1016/j.jaac.2013.08.008 [DOI] [PubMed] [Google Scholar]
- 41. Bassett AS, Chow EW. Schizophrenia and 22q11.2 deletion syndrome. Curr Psychiatry Rep. 2008;10: 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ahn K, Gotay N, Andersen TM, Anvari AA, Gochman P, Lee Y, et al. High rate of disease-related copy number variations in childhood onset schizophrenia. Mol Psychiatry. 2014;19: 568–572. 10.1038/mp.2013.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104: 619–629. [DOI] [PubMed] [Google Scholar]
- 44. Hiramoto T, Kang G, Suzuki G, Satoh Y, Kucherlapati R, Watanabe Y, et al. Tbx1: Identification of a 22q11.2 gene as a risk factor for autism spectrum disorder in a mouse model. Hum Mol Genet. 2011;20: 4775–4785. 10.1093/hmg/ddr404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43: 864–868. 10.1038/ng.902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li T, Ma X, Hu X, Wang Y, Yan C, Meng H, et al. PRODH gene is associated with executive function in schizophrenic families. Am J Med Genet B Neuropsychiatr Genet. 2007. 10.1002/ajmg.b.30648 [DOI] [PubMed]
- 47. Karayiorgou M, Gogos JA. The molecular genetics of the 22q11-associated schizophrenia. Brain Res Mol Brain Res. 2004;132: 95–104. 10.1016/j.molbrainres.2004.09.029 [DOI] [PubMed] [Google Scholar]
- 48. Schofield CM, Hsu R, Barker AJ, Gertz CC, Blelloch R, Ullian EM. Monoallelic deletion of the microRNA biogenesis gene Dgcr8 produces deficits in the development of excitatory synaptic transmission in the prefrontal cortex. Neural Dev. 2011;6: 11-8104-6-11. 10.1186/1749-8104-6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Forstner AJ, Basmanav FB, Mattheisen M, Bohmer AC, Hollegaard MV, Janson E, et al. Investigation of the involvement of MIR185 and its target genes in the development of schizophrenia. J Psychiatry Neurosci. 2014;39: 386–396. 10.1503/jpn.130189 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xu B, Hsu PK, Stark KL, Karayiorgou M, Gogos JA. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell. 2013;152: 262–275. 10.1016/j.cell.2012.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Earls LR, Fricke RG, Yu J, Berry RB, Baldwin LT, Zakharenko SS. Age-dependent microRNA control of synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J Neurosci. 2012;32: 14132–14144. 10.1523/JNEUROSCI.1312-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Belkaya S, Murray SE, Eitson JL, de la Morena MT, Forman JA, van Oers NS. Transgenic expression of microRNA-185 causes a developmental arrest of T cells by targeting multiple genes including Mzb1. J Biol Chem. 2013;288: 30752–30762. 10.1074/jbc.M113.503532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. de la Morena MT, Eitson JL, Dozmorov IM, Belkaya S, Hoover AR, Anguiano E, et al. Signature MicroRNA expression patterns identified in humans with 22q11.2 deletion/DiGeorge syndrome. Clin Immunol. 2013;147: 11–22. 10.1016/j.clim.2013.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Irish Schizophrenia Genomics Consortium and the Wellcome Trust Case Control Consortium 2. Genome-wide association study implicates HLA-C*01:02 as a risk factor at the major histocompatibility complex locus in schizophrenia. Biol Psychiatry. 2012;72: 620–628. 10.1016/j.biopsych.2012.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511: 421–427. 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6: e26203 10.1371/journal.pone.0026203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011. 10.1038/nature09915 [DOI] [PMC free article] [PubMed]
- 58. Chamberlain SJ, Chen PF, Ng KY, Bourgois-Rocha F, Lemtiri-Chlieh F, Levine ES, et al. Induced pluripotent stem cell models of the genomic imprinting disorders angelman and prader-willi syndromes. Proc Natl Acad Sci U S A. 2010;107: 17668–17673. 10.1073/pnas.1004487107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chiang CH, Su Y, Wen Z, Yoritomo N, Ross CA, Margolis RL, et al. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol Psychiatry. 2011. 10.1038/mp.2011.13 [DOI] [PMC free article] [PubMed]
- 60. DeRosa BA, Van Baaren JM, Dubey GK, Lee JM, Cuccaro ML, Vance JM, et al. Derivation of autism spectrum disorder-specific induced pluripotent stem cells from peripheral blood mononuclear cells. Neurosci Lett. 2012;516: 9–14. 10.1016/j.neulet.2012.02.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farra N, Zhang WB, Pasceri P, Eubanks JH, Salter MW, Ellis J. Rett syndrome induced pluripotent stem cell-derived neurons reveal novel neurophysiological alterations. Mol Psychiatry. 2012. 10.1038/mp.2011.180 [DOI] [PMC free article] [PubMed]
- 62.Kim KY, Jung YW, Sullivan GJ, Chung L, Park IH. Cellular reprogramming: A novel tool for investigating autism spectrum disorders. Trends Mol Med. 2012. 10.1016/j.molmed.2012.06.002 [DOI] [PMC free article] [PubMed]
- 63. Lin M, Hrabovsky A, Pedrosa E, Wang T, Zheng D, Lachman HM. Allele-biased expression in differentiating human neurons: Implications for neuropsychiatric disorders. PLoS One. 2012;7: e44017 10.1371/journal.pone.0044017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D, et al. RNA-seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS One. 2011;6: e23356 10.1371/journal.pone.0023356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, et al. A model for neural development and treatment of rett syndrome using human induced pluripotent stem cells. Cell. 2010;143: 527–539. 10.1016/j.cell.2010.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tian Y, Voineagu I, Pasca SP, Won H, Chandran V, Horvath S, et al. Alteration in basal and depolarization induced transcriptional network in iPSC derived neurons from timothy syndrome. Genome Med. 2014;6: 75-014-0075-5. eCollection 2014. 10.1186/s13073-014-0075-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhao WN, Cheng C, Theriault KM, Sheridan SD, Tsai LH, Haggarty SJ. A high-throughput screen for Wnt/beta-catenin signaling pathway modulators in human iPSC-derived neural progenitors. J Biomol Screen. 2012;17: 1252–1263. 10.1177/1087057112456876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515: 414–418. 10.1038/nature13716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8: 409–412. 10.1038/nmeth.1591 [DOI] [PubMed] [Google Scholar]
- 70. Muenthaisong S, Ujhelly O, Polgar Z, Varga E, Ivics Z, Pirity MK, et al. Generation of mouse induced pluripotent stem cells from different genetic backgrounds using sleeping beauty transposon mediated gene transfer. Exp Cell Res. 2012;318: 2482–2489. 10.1016/j.yexcr.2012.07.014 [DOI] [PubMed] [Google Scholar]
- 71.Pal R, Mamidi MK, Das AK, Bhonde R. Comparative analysis of cardiomyocyte differentiation from human embryonic stem cells under 3-D and 2-D culture conditions. J Biosci Bioeng. 2012. 10.1016/j.jbiosc.2012.08.018 [DOI] [PubMed]
- 72. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126: 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 73. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131: 861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 74. Pantano L, Estivill X, Marti E. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res. 2010;38: e34 10.1093/nar/gkp1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: The reference human genome annotation for the ENCODE project. Genome Res. 2012;22: 1760–1774. 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15: 550. doi: s13059-014-0550-8 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9: 811–818. [DOI] [PubMed] [Google Scholar]
- 79. Green GH, Diggle PJ. On the operational characteristics of the benjamini and hochberg false discovery rate procedure. Stat Appl Genet Mol Biol. 2007;6: Article27. 10.2202/1544-6115.1302 [DOI] [PubMed] [Google Scholar]
- 80. Vlachos IS, Paraskevopoulou MD, Karagkouni D, Georgakilas G, Vergoulis T, Kanellos I, et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:MRNA interactions. Nucleic Acids Res. 2015;43: D153–9. 10.1093/nar/gku1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vergoulis T, Kanellos I, Kostoulas N, Georgakilas G, Sellis T, Hatzigeorgiou A, et al. mirPub: A database for searching microRNA publications. Bioinformatics. 2014. doi: btu819 [pii]. [DOI] [PMC free article] [PubMed]
- 82. Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: An integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37: D105–10. 10.1093/nar/gkn851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19: 92–105. 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lin M, Zhao D, Hrabovsky A, Pedrosa E, Zheng D, Lachman HM. Heat shock alters the expression of schizophrenia and autism candidate genes in an induced pluripotent stem cell model of the human telencephalon. PLoS One. 2014;9: e94968 10.1371/journal.pone.0094968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 86. Isserlin R, Merico D, Voisin V, Bader GD. Enrichment map—a cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Res. 2014;3: 141 10.12688/f1000research.4536.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Binns D, Dimmer E, Huntley R, Barrell D, O'Donovan C, Apweiler R. QuickGO: A web-based tool for gene ontology searching. Bioinformatics. 2009;25: 3045–3046. 10.1093/bioinformatics/btp536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, et al. A travel guide to cytoscape plugins. Nat Methods. 2012;9: 1069–1076. 10.1038/nmeth.2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wang L, Yu J, Xu J, Zheng C, Li X, Du J. The analysis of microRNA-34 family expression in human cancer studies comparing cancer tissues with corresponding pericarcinous tissues. Gene. 2014;554: 1–8. doi: S0378-1119(14)01192-5 [pii]. 10.1016/j.gene.2014.10.032 [DOI] [PubMed] [Google Scholar]
- 90. Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26: 731–743. doi: S1097-2765(07)00318-8 [pii]. [DOI] [PubMed] [Google Scholar]
- 91. Migliore C, Petrelli A, Ghiso E, Corso S, Capparuccia L, Eramo A, et al. MicroRNAs impair MET-mediated invasive growth. Cancer Res. 2008;68: 10128–10136. 10.1158/0008-5472.CAN-08-2148 [DOI] [PubMed] [Google Scholar]
- 92. Lai CY, Yu SL, Hsieh MH, Chen CH, Chen HY, Wen CC, et al. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS One. 2011;6: e21635 10.1371/journal.pone.0021635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morgado AL, Xavier JM, Dionisio PA, Ribeiro MF, Dias RB, Sebastiao AM, et al. MicroRNA-34a modulates neural stem cell differentiation by regulating expression of synaptic and autophagic proteins. Mol Neurobiol. 2014. 10.1007/s12035-014-8794-6 [DOI] [PubMed]
- 94. Kim AH, Reimers M, Maher B, Williamson V, McMichael O, McClay JL, et al. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr Res. 2010;124: 183–191. 10.1016/j.schres.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Smalheiser NR, Lugli G, Zhang H, Rizavi H, Cook EH, Dwivedi Y. Expression of microRNAs and other small RNAs in prefrontal cortex in schizophrenia, bipolar disorder and depressed subjects. PLoS One. 2014;9: e86469 10.1371/journal.pone.0086469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Vaishnavi V, Manikandan M, Tiwary BK, Munirajan AK. Insights on the functional impact of microRNAs present in autism-associated copy number variants. PLoS One. 2013;8: e56781 10.1371/journal.pone.0056781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maekawa M, Yamada K, Toyoshima M, Ohnishi T, Iwayama Y, Shimamoto C, et al. Utility of scalp hair follicles as a novel source of biomarker genes for psychiatric illnesses. Biol Psychiatry. 2014. doi: S0006-3223(14)00570-8 [pii]. [DOI] [PubMed]
- 98. van Spronsen M, van Battum EY, Kuijpers M, Vangoor VR, Rietman ML, Pothof J, et al. Developmental and activity-dependent miRNA expression profiling in primary hippocampal neuron cultures. PLoS One. 2013;8: e74907 10.1371/journal.pone.0074907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sarachana T, Hu VW. Genome-wide identification of transcriptional targets of RORA reveals direct regulation of multiple genes associated with autism spectrum disorder. Mol Autism. 2013;4: 14-2392-4-14. 10.1186/2040-2392-4-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Abu-Elneel K, Liu T, Gazzaniga FS, Nishimura Y, Wall DP, Geschwind DH, et al. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9: 153–161. 10.1007/s10048-008-0133-5 [DOI] [PubMed] [Google Scholar]
- 101. Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry. 2010;15: 1176–1189. 10.1038/mp.2009.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Henshall DC. MicroRNA and epilepsy: Profiling, functions and potential clinical applications. Curr Opin Neurol. 2014;27: 199–205. 10.1097/WCO.0000000000000079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Na KS, Jung HY, Kim YK. The role of pro-inflammatory cytokines in the neuroinflammation and neurogenesis of schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2014;48: 277–286. 10.1016/j.pnpbp.2012.10.022 [DOI] [PubMed] [Google Scholar]
- 104. Wei H, Chadman KK, McCloskey DP, Sheikh AM, Malik M, Brown WT, et al. Brain IL-6 elevation causes neuronal circuitry imbalances and mediates autism-like behaviors. Biochim Biophys Acta. 2012;1822: 831–842. 10.1016/j.bbadis.2012.01.011 [DOI] [PubMed] [Google Scholar]
- 105. Garbett KA, Hsiao EY, Kalman S, Patterson PH, Mirnics K. Effects of maternal immune activation on gene expression patterns in the fetal brain. Transl Psychiatry. 2012;2: e98 10.1038/tp.2012.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Schulte LN, Westermann AJ, Vogel J. Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing. Nucleic Acids Res. 2013;41: 542–553. 10.1093/nar/gks1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sheedy FJ, O'Neill LA. Adding fuel to fire: MicroRNAs as a new class of mediators of inflammation. Ann Rheum Dis. 2008;67 Suppl 3: iii50–5. 10.1136/ard.2008.100289 [DOI] [PubMed] [Google Scholar]
- 108. Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs and immunity: Novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18: 131–140. 10.1016/j.semcancer.2008.01.005 [DOI] [PubMed] [Google Scholar]
- 109. Clarke RA, Fang ZM, Diwan AD, Gilbert DL. Tourette syndrome and klippel-feil anomaly in a child with chromosome 22q11 duplication. Case Rep Med. 2009;2009: 361518 10.1155/2009/361518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Robertson MM, Shelley BP, Dalwai S, Brewer C, Critchley HD. A patient with both gilles de la tourette's syndrome and chromosome 22q11 deletion syndrome: Clue to the genetics of gilles de la tourette's syndrome? J Psychosom Res. 2006;61: 365–368. doi: S0022-3999(06)00299-6 [pii]. [DOI] [PubMed] [Google Scholar]
- 111. Liu Y, Li Z, Zhang M, Deng Y, Yi Z, Shi T. Exploring the pathogenetic association between schizophrenia and type 2 diabetes mellitus diseases based on pathway analysis. BMC Med Genomics. 2013;6 Suppl 1: S17-8794-6-S1-S17. Epub 2013 Jan 23. 10.1186/1755-8794-6-S1-S17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zhang Y, Chen M, Wu Z, Chen J, Yu S, Fang Y, et al. Association study of Val66Met polymorphism in brain-derived neurotrophic factor gene with clozapine-induced metabolic syndrome: Preliminary results. PLoS One. 2013;8: e72652 10.1371/journal.pone.0072652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zerbo O, Iosif AM, Walker C, Ozonoff S, Hansen RL, Hertz-Picciotto I. Is maternal influenza or fever during pregnancy associated with autism or developmental delays? results from the CHARGE (CHildhood autism risks from genetics and environment) study. J Autism Dev Disord. 2012. 10.1007/s10803-012-1540-x [DOI] [PMC free article] [PubMed]
- 114. Parker-Athill EC, Tan J. Maternal immune activation and autism spectrum disorder: Interleukin-6 signaling as a key mechanistic pathway. Neurosignals. 2010;18: 113–128. 10.1159/000319828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hsiao EY, McBride SW, Chow J, Mazmanian SK, Patterson PH. Modeling an autism risk factor in mice leads to permanent immune dysregulation. Proc Natl Acad Sci U S A. 2012;109: 12776–12781. 10.1073/pnas.1202556109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ghanizadeh A. Could fever and neuroinflammation play a role in the neurobiology of autism? A subject worthy of more research. Int J Hyperthermia. 2011;27: 737–738. 10.3109/02656736.2011.604665 [DOI] [PubMed] [Google Scholar]
- 117. Merico D, Costain G, Butcher NJ, Warnica W, Ogura L, Alfred SE, et al. MicroRNA dysregulation, gene networks, and risk for schizophrenia in 22q11.2 deletion syndrome. Front Neurol. 2014;5: 238 10.3389/fneur.2014.00238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Isshiki M, Tanaka S, Kuriu T, Tabuchi K, Takumi T, Okabe S. Enhanced synapse remodelling as a common phenotype in mouse models of autism. Nat Commun. 2014;5: 4742 10.1038/ncomms5742 [DOI] [PubMed] [Google Scholar]
- 119.Hall J, Trent S, Thomas KL, O'Donovan MC, Owen MJ. Genetic risk for schizophrenia: Convergence on synaptic pathways involved in plasticity. Biol Psychiatry. 2014. doi: S0006-3223(14)00519-8 [pii]. [DOI] [PubMed]
- 120. Leblond CS, Nava C, Polge A, Gauthier J, Huguet G, Lumbroso S, et al. Meta-analysis of SHANK mutations in autism spectrum disorders: A gradient of severity in cognitive impairments. PLoS Genet. 2014;10: e1004580 10.1371/journal.pgen.1004580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Penzes P, Buonanno A, Passafaro M, Sala C, Sweet RA. Developmental vulnerability of synapses and circuits associated with neuropsychiatric disorders. J Neurochem. 2013;126: 165–182. 10.1111/jnc.12261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Tsai NP, Wilkerson JR, Guo W, Maksimova MA, DeMartino GN, Cowan CW, et al. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell. 2012;151: 1581–1594. 10.1016/j.cell.2012.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Lepagnol-Bestel AM, Kvajo M, Karayiorgou M, Simonneau M, Gogos JA. A Disc1 mutation differentially affects neurites and spines in hippocampal and cortical neurons. Mol Cell Neurosci. 2013;54: 84–92. 10.1016/j.mcn.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Sudhof TC. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 2013;154: 75–88. 10.1016/j.cell.2013.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Missler M, Sudhof TC, Biederer T. Synaptic cell adhesion. Cold Spring Harb Perspect Biol. 2012;4: a005694 10.1101/cshperspect.a005694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Guo R, Wang Y, Shi WY, Liu B, Hou SQ, Liu L. MicroRNA miR-491-5p targeting both TP53 and bcl-XL induces cell apoptosis in SW1990 pancreatic cancer cells through mitochondria mediated pathway. Molecules. 2012;17: 14733–14747. 10.3390/molecules171214733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kumarasinghe N, Beveridge NJ, Gardiner E, Scott RJ, Yasawardene S, Perera A, et al. Gene expression profiling in treatment-naive schizophrenia patients identifies abnormalities in biological pathways involving AKT1 that are corrected by antipsychotic medication. Int J Neuropsychopharmacol. 2013: 1–21. 10.1017/S1461145713000035 [DOI] [PubMed]
- 128. Yao JJ, Sun J, Zhao QR, Wang CY, Mei YA. Neuregulin-1/ErbB4 signaling regulates Kv4.2-mediated transient outward K+ current through the Akt/mTOR pathway. Am J Physiol Cell Physiol. 2013;305: C197–206. 10.1152/ajpcell.00041.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Porteous D, Millar K. How DISC1 regulates postnatal brain development: Girdin gets in on the AKT. Neuron. 2009;63: 711–713. 10.1016/j.neuron.2009.09.017 [DOI] [PubMed] [Google Scholar]
- 130. Gasso P, Mas S, Molina O, Lafuente A, Bernardo M, Parellada E. Increased susceptibility to apoptosis in cultured fibroblasts from antipsychotic-naive first-episode schizophrenia patients. J Psychiatr Res. 2014;48: 94–101. 10.1016/j.jpsychires.2013.09.017 [DOI] [PubMed] [Google Scholar]
- 131. Drerup CM, Wiora HM, Topczewski J, Morris JA. Disc1 regulates foxd3 and sox10 expression, affecting neural crest migration and differentiation. Development. 2009;136: 2623–2632. 10.1242/dev.030577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ghahramani Seno MM, Hu P, Gwadry FG, Pinto D, Marshall CR, Casallo G, et al. Gene and miRNA expression profiles in autism spectrum disorders. Brain Res. 2011;1380: 85–97. 10.1016/j.brainres.2010.09.046 [DOI] [PubMed] [Google Scholar]
- 133.Clipperton-Allen AE, Page DT. Pten haploinsufficient mice show broad brain overgrowth but selective impairments in autism-relevant behavioral tests. Hum Mol Genet. 2014. 10.1093/hmg/ddu057 [DOI] [PubMed]
- 134. Lv JW, Cheng TL, Qiu ZL, Zhou WH. Role of the PTEN signaling pathway in autism spectrum disorder. Neurosci Bull. 2013;29: 773–778. 10.1007/s12264-013-1382-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shaham L, Vendramini E, Ge Y, Goren Y, Birger Y, Tijssen MR, et al. MicroRNA-486-5p is an erythroid oncomiR of the myeloid leukemias of down syndrome. Blood. 2014. doi: blood-2014-06-581892 [pii]. [DOI] [PMC free article] [PubMed]
- 136. Pietrzykowski AZ, Spijker S. Impulsivity and comorbid traits: A multi-step approach for finding putative responsible microRNAs in the amygdala. Front Neurosci. 2014;8: 389 10.3389/fnins.2014.00389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Antshel KM, Aneja A, Strunge L, Peebles J, Fremont WP, Stallone K, et al. Autistic spectrum disorders in velo-cardio facial syndrome (22q11.2 deletion). J Autism Dev Disord. 2007;37: 1776–1786. 10.1007/s10803-006-0308-6 [DOI] [PubMed] [Google Scholar]
- 138. Kates WR, Miller AM, Abdulsabur N, Antshel KM, Conchelos J, Fremont W, et al. Temporal lobe anatomy and psychiatric symptoms in velocardiofacial syndrome (22q11.2 deletion syndrome). J Am Acad Child Adolesc Psychiatry. 2006;45: 587–595. 10.1097/01.chi.0000205704.33077.4a [DOI] [PubMed] [Google Scholar]
- 139.Montojo CA, Jalbrzikowski M, Congdon E, Domicoli S, Chow C, Dawson C, et al. Neural substrates of inhibitory control deficits in 22q11.2 deletion syndrome. Cereb Cortex. 2013. doi: bht304 [pii]. [DOI] [PMC free article] [PubMed]
- 140.Liu T, Wan RP, Tang LJ, Liu SJ, Li HJ, Zhao QH, et al. A MicroRNA profile in Fmr1 knockout mice reveals MicroRNA expression alterations with possible roles in fragile X syndrome. Mol Neurobiol. 2014. 10.1007/s12035-014-8770-1 [DOI] [PubMed]
- 141. Urdinguio RG, Fernandez AF, Lopez-Nieva P, Rossi S, Huertas D, Kulis M, et al. Disrupted microRNA expression caused by Mecp2 loss in a mouse model of rett syndrome. Epigenetics. 2010;5: 656–663. 10.4161/epi.5.7.13055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Simao I, Lourenco T, Lopes L, Ramos MP. First seizure as late presentation of velo-cardio-facial syndrome. J Pediatr Endocrinol Metab. 2013;26: 381–383. 10.1515/jpem-2012-0348 [DOI] [PubMed] [Google Scholar]
- 143. Cheung EN, George SR, Andrade DM, Chow EW, Silversides CK, Bassett AS. Neonatal hypocalcemia, neonatal seizures, and intellectual disability in 22q11.2 deletion syndrome. Genet Med. 2014;16: 40–44. 10.1038/gim.2013.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Lionel AC, Vaags AK, Sato D, Gazzellone MJ, Mitchell EB, Chen HY, et al. Rare exonic deletions implicate the synaptic organizer gephyrin (GPHN) in risk for autism, schizophrenia and seizures. Hum Mol Genet. 2013;22: 2055–2066. 10.1093/hmg/ddt056 [DOI] [PubMed] [Google Scholar]
- 145. Clarke MC, Tanskanen A, Huttunen MO, Clancy M, Cotter DR, Cannon M. Evidence for shared susceptibility to epilepsy and psychosis: A population-based family study. Biol Psychiatry. 2012;71: 836–839. 10.1016/j.biopsych.2012.01.011 [DOI] [PubMed] [Google Scholar]
- 146. Murdoch JD, State MW. Recent developments in the genetics of autism spectrum disorders. Curr Opin Genet Dev. 2013;23: 310–315. 10.1016/j.gde.2013.02.003 [DOI] [PubMed] [Google Scholar]
- 147. Doherty JL, O'Donovan MC, Owen MJ. Recent genomic advances in schizophrenia. Clin Genet. 2012;81: 103–109. 10.1111/j.1399-0004.2011.01773.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(DOCX)
(XLSX)
(XLSX)
(XLS)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. The miRNA-seq data have been deposited in Gene Expression Omnibus (GEO; accession number GSE65367).