Abstract
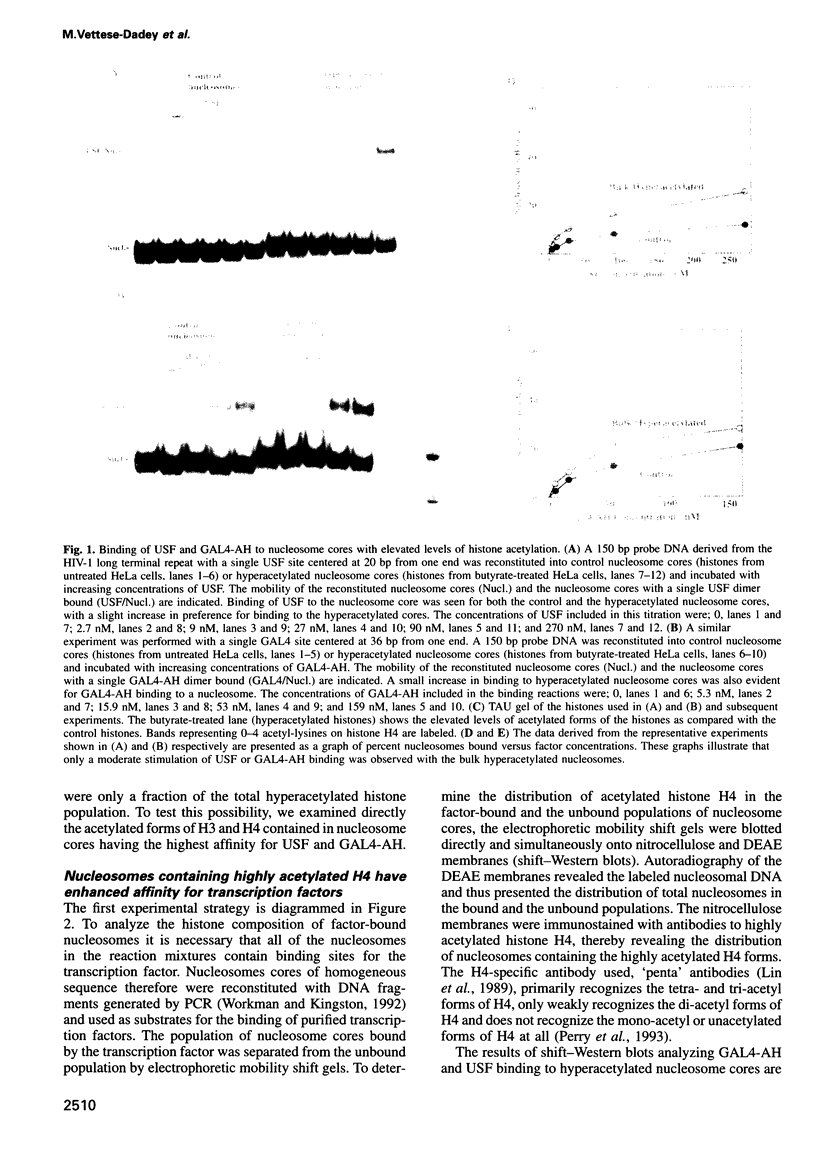
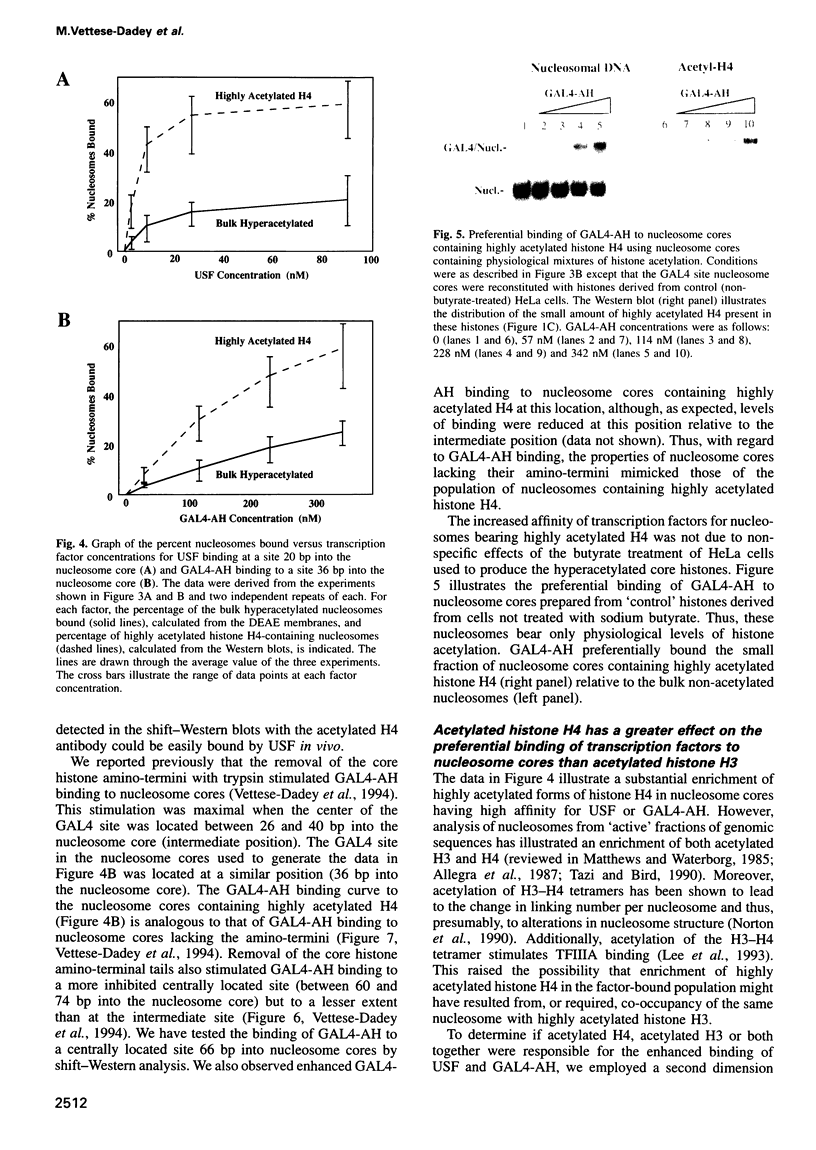
Core histones isolated from normal and butyrate-treated HeLa cells have been reconstituted into nucleosome cores in order to analyze the role of histone acetylation in enhancing transcription factor binding to recognition sites in nucleosomal DNA. Moderate stimulation of nucleosome binding was observed for the basic helix-loop-helix factor USF and the Zn cluster DNA binding domain factor GAL4-AH using heterogeneously acetylated histones. However, by coupling novel immunoblotting techniques to a gel retardation assay, we observed that nucleosome cores containing the most highly acetylated forms of histone H4 have the highest affinity for these two transcription factors. Western analysis of gel-purified USF-nucleosome and GAL4-AH-nucleosome complexes demonstrated the predominant presence of acetylated histone H4 relative to acetylated histone H3. Immunoprecipitation of USF-nucleosome complexes with anti-USF antibodies also demonstrated that these complexes were enriched preferentially in acetylated histone H4. These data show that USF and GAL4-AH preferentially interact with nucleosome cores containing highly acetylated histone H4. Acetylation of histone H4 thus appears to play a primary role in the structural changes that mediate enhanced binding of transcription factors to their recognition sites within nucleosomes.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ALLFREY V. G., FAULKNER R., MIRSKY A. E. ACETYLATION AND METHYLATION OF HISTONES AND THEIR POSSIBLE ROLE IN THE REGULATION OF RNA SYNTHESIS. Proc Natl Acad Sci U S A. 1964 May;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams C. C., Workman J. L. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol. 1995 Mar;15(3):1405–1421. doi: 10.1128/mcb.15.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J., Harborne N., Rau D. C., Gould H. Participation of core histone "tails" in the stabilization of the chromatin solenoid. J Cell Biol. 1982 May;93(2):285–297. doi: 10.1083/jcb.93.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegra P., Sterner R., Clayton D. F., Allfrey V. G. Affinity chromatographic purification of nucleosomes containing transcriptionally active DNA sequences. J Mol Biol. 1987 Jul 20;196(2):379–388. doi: 10.1016/0022-2836(87)90698-x. [DOI] [PubMed] [Google Scholar]
- Annunziato A. T., Seale R. L. Histone deacetylation is required for the maturation of newly replicated chromatin. J Biol Chem. 1983 Oct 25;258(20):12675–12684. [PubMed] [Google Scholar]
- Bauer W. R., Hayes J. J., White J. H., Wolffe A. P. Nucleosome structural changes due to acetylation. J Mol Biol. 1994 Feb 25;236(3):685–690. doi: 10.1006/jmbi.1994.1180. [DOI] [PubMed] [Google Scholar]
- Becker P. B. The establishment of active promoters in chromatin. Bioessays. 1994 Aug;16(8):541–547. doi: 10.1002/bies.950160807. [DOI] [PubMed] [Google Scholar]
- Boffa L. C., Walker J., Chen T. A., Sterner R., Mariani M. R., Allfrey V. G. Factors affecting nucleosome structure in transcriptionally active chromatin. Histone acetylation, nascent RNA and inhibitors of RNA synthesis. Eur J Biochem. 1990 Dec 27;194(3):811–823. doi: 10.1111/j.1432-1033.1990.tb19474.x. [DOI] [PubMed] [Google Scholar]
- Bone J. R., Lavender J., Richman R., Palmer M. J., Turner B. M., Kuroda M. I. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 1994 Jan;8(1):96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Rose A. B., Holmes S. G., Allis C. D., Broach J. R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993 Apr;7(4):592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- Carthew R. W., Chodosh L. A., Sharp P. A. The major late transcription factor binds to and activates the mouse metallothionein I promoter. Genes Dev. 1987 Nov;1(9):973–980. doi: 10.1101/gad.1.9.973. [DOI] [PubMed] [Google Scholar]
- Chodosh L. A., Carthew R. W., Morgan J. G., Crabtree G. R., Sharp P. A. The adenovirus major late transcription factor activates the rat gamma-fibrinogen promoter. Science. 1987 Oct 30;238(4827):684–688. doi: 10.1126/science.3672119. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Tullius T. D., Klug A. Mode of interaction of the zinc finger protein TFIIIA with a 5S RNA gene of Xenopus. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5528–5532. doi: 10.1073/pnas.87.14.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton A. L., Hebbes T. R., Thorne A. W., Crane-Robinson C. Histone acetylation and gene induction in human cells. FEBS Lett. 1993 Dec 20;336(1):23–26. doi: 10.1016/0014-5793(93)81601-u. [DOI] [PubMed] [Google Scholar]
- Csordas A. On the biological role of histone acetylation. Biochem J. 1990 Jan 1;265(1):23–38. doi: 10.1042/bj2650023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demczuk S., Harbers M., Vennström B. Identification and analysis of all components of a gel retardation assay by combination with immunoblotting. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2574–2578. doi: 10.1073/pnas.90.7.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrin L. K., Mann R. K., Kayne P. S., Grunstein M. Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. Cell. 1991 Jun 14;65(6):1023–1031. doi: 10.1016/0092-8674(91)90554-c. [DOI] [PubMed] [Google Scholar]
- Fedor M. J., Lue N. F., Kornberg R. D. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988 Nov 5;204(1):109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A. R., Pognonec P., Roeder R. G., Burley S. K. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994 Jan 1;13(1):180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher-Adams G., Grunstein M. Yeast histone H4 and H3 N-termini have different effects on the chromatin structure of the GAL1 promoter. EMBO J. 1995 Apr 3;14(7):1468–1477. doi: 10.1002/j.1460-2075.1995.tb07133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovsky M. A., Pleger G. L., Keevert J. B., Johmann C. A. Studies on histone fraction F2A1 in macro- and micronuclei of Tetrahymena pyriformis. J Cell Biol. 1973 Jun;57(3):773–781. doi: 10.1083/jcb.57.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes T. R., Clayton A. L., Thorne A. W., Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994 Apr 15;13(8):1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes T. R., Thorne A. W., Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988 May;7(5):1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes T. R., Turner C. H., Thorne A. W., Crane-Robinson C. A "minimal epitope" anti-protein antibody that recognises a single modified amino acid. Mol Immunol. 1989 Sep;26(9):865–873. doi: 10.1016/0161-5890(89)90143-0. [DOI] [PubMed] [Google Scholar]
- Hecht A., Laroche T., Strahl-Bolsinger S., Gasser S. M., Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995 Feb 24;80(4):583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hong L., Schroth G. P., Matthews H. R., Yau P., Bradbury E. M. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 "tail" to DNA. J Biol Chem. 1993 Jan 5;268(1):305–314. [PubMed] [Google Scholar]
- Ip Y. T., Jackson V., Meier J., Chalkley R. The separation of transcriptionally engaged genes. J Biol Chem. 1988 Oct 5;263(28):14044–14052. [PubMed] [Google Scholar]
- Jeppesen P., Turner B. M. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993 Jul 30;74(2):281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- Johnson L. M., Kayne P. S., Kahn E. S., Grunstein M. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan L. J., Utley R. T., Adams C. C., Vettese-Dadey M., Workman J. L. Differential repression of transcription factor binding by histone H1 is regulated by the core histone amino termini. EMBO J. 1994 Dec 15;13(24):6031–6040. doi: 10.1002/j.1460-2075.1994.tb06949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayne P. S., Kim U. J., Han M., Mullen J. R., Yoshizaki F., Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988 Oct 7;55(1):27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- Kleff S., Andrulis E. D., Anderson C. W., Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995 Oct 20;270(42):24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- Lee D. Y., Hayes J. J., Pruss D., Wolffe A. P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993 Jan 15;72(1):73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- Lin R., Leone J. W., Cook R. G., Allis C. D. Antibodies specific to acetylated histones document the existence of deposition- and transcription-related histone acetylation in Tetrahymena. J Cell Biol. 1989 May;108(5):1577–1588. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl P. Histone acetylation: facts and questions. Chromosoma. 1994 Dec;103(7):441–449. doi: 10.1007/BF00337382. [DOI] [PubMed] [Google Scholar]
- Mann R. K., Grunstein M. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 1992 Sep;11(9):3297–3306. doi: 10.1002/j.1460-2075.1992.tb05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R., Carey M., Ptashne M., Harrison S. C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992 Apr 2;356(6368):408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Nickol J. M., Felsenfeld G., Rau D. C. Histone hyperacetylation has little effect on the higher order folding of chromatin. Nucleic Acids Res. 1983 Jun 25;11(12):4065–4075. doi: 10.1093/nar/11.12.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H. Nucleosome disruption by transcription factor binding in yeast. Science. 1993 Dec 3;262(5139):1563–1566. doi: 10.1126/science.8248805. [DOI] [PubMed] [Google Scholar]
- Mueller P. R., Salser S. J., Wold B. Constitutive and metal-inducible protein:DNA interactions at the mouse metallothionein I promoter examined by in vivo and in vitro footprinting. Genes Dev. 1988 Apr;2(4):412–427. doi: 10.1101/gad.2.4.412. [DOI] [PubMed] [Google Scholar]
- Navankasattusas S., Sawadogo M., van Bilsen M., Dang C. V., Chien K. R. The basic helix-loop-helix protein upstream stimulating factor regulates the cardiac ventricular myosin light-chain 2 gene via independent cis regulatory elements. Mol Cell Biol. 1994 Nov;14(11):7331–7339. doi: 10.1128/mcb.14.11.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton V. G., Imai B. S., Yau P., Bradbury E. M. Histone acetylation reduces nucleosome core particle linking number change. Cell. 1989 May 5;57(3):449–457. doi: 10.1016/0092-8674(89)90920-3. [DOI] [PubMed] [Google Scholar]
- Norton V. G., Marvin K. W., Yau P., Bradbury E. M. Nucleosome linking number change controlled by acetylation of histones H3 and H4. J Biol Chem. 1990 Nov 15;265(32):19848–19852. [PubMed] [Google Scholar]
- O'Neill L. P., Turner B. M. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 1995 Aug 15;14(16):3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen-Hughes T., Workman J. L. Experimental analysis of chromatin function in transcription control. Crit Rev Eukaryot Gene Expr. 1994;4(4):403–441. [PubMed] [Google Scholar]
- Park E. C., Szostak J. W. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol Cell Biol. 1990 Sep;10(9):4932–4934. doi: 10.1128/mcb.10.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry C. A., Annunziato A. T. Histone acetylation reduces H1-mediated nucleosome interactions during chromatin assembly. Exp Cell Res. 1991 Oct;196(2):337–345. doi: 10.1016/0014-4827(91)90269-z. [DOI] [PubMed] [Google Scholar]
- Perry C. A., Annunziato A. T. Influence of histone acetylation on the solubility, H1 content and DNase I sensitivity of newly assembled chromatin. Nucleic Acids Res. 1989 Jun 12;17(11):4275–4291. doi: 10.1093/nar/17.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry C. A., Dadd C. A., Allis C. D., Annunziato A. T. Analysis of nucleosome assembly and histone exchange using antibodies specific for acetylated H4. Biochemistry. 1993 Dec 14;32(49):13605–13614. doi: 10.1021/bi00212a028. [DOI] [PubMed] [Google Scholar]
- Pfeffer U., Vidali G. Histone acetylation: recent approaches to a basic mechanism of genome organization. Int J Biochem. 1991;23(3):277–285. doi: 10.1016/0020-711x(91)90107-x. [DOI] [PubMed] [Google Scholar]
- Pognonec P., Kato H., Sumimoto H., Kretzschmar M., Roeder R. G. A quick procedure for purification of functional recombinant proteins over-expressed in E.coli. Nucleic Acids Res. 1991 Dec 11;19(23):6650–6650. doi: 10.1093/nar/19.23.6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pognonec P., Roeder R. G. Recombinant 43-kDa USF binds to DNA and activates transcription in a manner indistinguishable from that of natural 43/44-kDa USF. Mol Cell Biol. 1991 Oct;11(10):5125–5136. doi: 10.1128/mcb.11.10.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridsdale J. A., Davie J. R. Chicken erythrocyte polynucleosomes which are soluble at physiological ionic strength and contain linker histones are highly enriched in beta-globin gene sequences. Nucleic Acids Res. 1987 Feb 11;15(3):1081–1096. doi: 10.1093/nar/15.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridsdale J. A., Hendzel M. J., Delcuve G. P., Davie J. R. Histone acetylation alters the capacity of the H1 histones to condense transcriptionally active/competent chromatin. J Biol Chem. 1990 Mar 25;265(9):5150–5156. [PubMed] [Google Scholar]
- Roth S. Y., Shimizu M., Johnson L., Grunstein M., Simpson R. T. Stable nucleosome positioning and complete repression by the yeast alpha 2 repressor are disrupted by amino-terminal mutations in histone H4. Genes Dev. 1992 Mar;6(3):411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Van Dyke M. W., Gregor P. D., Roeder R. G. Multiple forms of the human gene-specific transcription factor USF. I. Complete purification and identification of USF from HeLa cell nuclei. J Biol Chem. 1988 Aug 25;263(24):11985–11993. [PubMed] [Google Scholar]
- Sirito M., Lin Q., Maity T., Sawadogo M. Ubiquitous expression of the 43- and 44-kDa forms of transcription factor USF in mammalian cells. Nucleic Acids Res. 1994 Feb 11;22(3):427–433. doi: 10.1093/nar/22.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J., Hörz W. Histones, nucleosomes and transcription. Curr Opin Genet Dev. 1993 Apr;3(2):219–225. doi: 10.1016/0959-437x(93)90026-l. [DOI] [PubMed] [Google Scholar]
- Tazi J., Bird A. Alternative chromatin structure at CpG islands. Cell. 1990 Mar 23;60(6):909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- Turner B. M., Birley A. J., Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992 Apr 17;69(2):375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- Turner B. M. Decoding the nucleosome. Cell. 1993 Oct 8;75(1):5–8. [PubMed] [Google Scholar]
- Turner B. M. Histone acetylation and control of gene expression. J Cell Sci. 1991 May;99(Pt 1):13–20. doi: 10.1242/jcs.99.1.13. [DOI] [PubMed] [Google Scholar]
- Turner B. M., O'Neill L. P. Histone acetylation in chromatin and chromosomes. Semin Cell Biol. 1995 Aug;6(4):229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- Vavra K. J., Allis C. D., Gorovsky M. A. Regulation of histone acetylation in Tetrahymena macro- and micronuclei. J Biol Chem. 1982 Mar 10;257(5):2591–2598. [PubMed] [Google Scholar]
- Vettese-Dadey M., Walter P., Chen H., Juan L. J., Workman J. L. Role of the histone amino termini in facilitated binding of a transcription factor, GAL4-AH, to nucleosome cores. Mol Cell Biol. 1994 Feb;14(2):970–981. doi: 10.1128/mcb.14.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J., Chen T. A., Sterner R., Berger M., Winston F., Allfrey V. G. Affinity chromatography of mammalian and yeast nucleosomes. Two modes of binding of transcriptionally active mammalian nucleosomes to organomercurial-agarose columns, and contrasting behavior of the active nucleosomes of yeast. J Biol Chem. 1990 Apr 5;265(10):5736–5746. [PubMed] [Google Scholar]
- Wan J. S., Mann R. K., Grunstein M. Yeast histone H3 and H4 N termini function through different GAL1 regulatory elements to repress and activate transcription. Proc Natl Acad Sci U S A. 1995 Jun 6;92(12):5664–5668. doi: 10.1073/pnas.92.12.5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992 Nov;8(11):387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P. Nucleosome positioning and modification: chromatin structures that potentiate transcription. Trends Biochem Sci. 1994 Jun;19(6):240–244. doi: 10.1016/0968-0004(94)90148-1. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Kingston R. E. Nucleosome core displacement in vitro via a metastable transcription factor-nucleosome complex. Science. 1992 Dec 11;258(5089):1780–1784. doi: 10.1126/science.1465613. [DOI] [PubMed] [Google Scholar]