INTRODUCTION
Pseudohypoparathyroidism (PHP) is characterized by resistance to parathyroid hormone (PTH) in proximal renal tubules that leads to hypocalcemia and hyperphosphatemia (1). Different PHP variants have been recognized, which are all associated with loss or severely reduced expression of the alpha-subunit of the stimulatory G protein (Gsα) in this portion of the kidney thus causing impaired signal transduction of PTH and other hormones via the cAMP/PKA signaling pathway (2, 3). Gsα is encoded by GNAS located on the long arm of chromosome 20 (20q13.3), a complex imprinted locus that generates multiple sense and antisense transcripts. Through the utilization of alternative first exons and promoters, GNAS furthermore gives rise to several additional transcripts. These include the A/B transcript, which may encode an amino-terminally truncated form of Gsα (4) and non-coding antisense transcripts (AS) (5, 6), as well as transcripts encoding the extra-large Gαs variant (XLαs) and a 55-kDa neuroendocrine secretory protein (NESP55).
Patients affected by PHP type Ia (PHP1A) show resistance to several hormones that mediate their actions through G protein-coupled receptors and display various features of Albright's Hereditary Osteodystrophy (AHO), including short stature, round face, obesity, brachydactyly, ectopic ossifications, and/or various degrees of mental retardation (1). PHP1A is caused by inactivating heterozygous mutations involving one of the 13 GNAS exons or introns on the maternal allele. Subjects presenting with certain AHO features, but without hormonal resistance, obesity, and mental abnormalities, are classified as pseudopseudohypoparathyroidism (PPHP). This disorder is also caused by mutations affecting Gsα, but these are located on the paternal GNAS allele, rather than the maternal allele as in PHP1A (7).
Resistance toward PTH in the proximal renal tubules occurs also in PHP type Ib (PHPIB). These patients can furthermore show resistance towards other hormones, particularly towards TSH, and they may present with AHO features that can be indistinguishable from those observed in PHP1A (8–11). PHP1B is not caused by GNAS mutations involving the region encoding Gsα, but instead by loss-of-methylation (LOM) at GNAS exon A/B located within a differentially methylated region (DMR) (7). LOM can also be observed at additional GNAS exons, namely AS and XL, which is usually associated with a gain-of-methylation (GOM) at GNAS exon NESP.
The autosomal dominant (AD) form of PHP1B (AD-PHP1B) can be caused by maternal heterozygous deletions in STX16 (3-kb, 4.4-kb, and 24.6-kb deletions), the gene encoding syntaxin-16 located approximately 220 kb upstream of GNAS exon A/B (12). These STX16 deletions are associated with LOM affecting only GNAS exon A/B (12–14), which leads through unknown mechanisms to a reduction or loss of Gsα expression from the maternal allele. Indistinguishable LOM affecting only GNAS exon A/B occurs also with a maternal deletion comprising exon NESP and the upstream region (15).
AD-PHP1B can also be caused by maternally inherited deletions involving NESP and/or AS, which are associated with loss of all maternal GNAS methylation imprints (16, 17). Similarly broad methylation changes are also observed in most sporadic PHP1B (sporPHP1B) patients, but the molecular mechanism underlying these epigenetic changes remains unclear for the majority of these patients. In a few sporPHP1B patients the disease is caused by paternal uniparental isodisomy involving chromosome 20q (patUPD20q), which includes the GNAS locus (18–22). Such chromosomal rearrangements have not yet been described for the Japanese population. We therefore investigated 23 Japanese sporPHP1B patients, all of whom showed broad methylation changes involving all four DMRs of the GNAS locus, which led to the identification of two patients with patUPD20q. We furthermore mapped the known duplications and assessed the frequency of UPD20q based on this and previous reports, which will help guiding the evaluation and genetic counseling of sporPHP1B patients.
MATERIALS AND METHODS
Patients and healthy family members
We investigated 23 Japanese sporPHP1B cases, who had presented with PTH-resistant hypocalcemia and hyperphosphatemia, and mild AHO features in some; clinical and laboratory information of most of these patients was previously reported (23). None of the available parents, siblings, and children showed abnormalities in calcium and phosphate homeostasis. Clinical features, biochemical results, and epigenetic findings for each patient are presented in Table 1.
Table 1.
Laboratory and epigenetic findings for the investigated PHP1B patients
| Serum biochemical values at diagnosis | MS-MLPA | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Code | Sex | Age at diagnosis (years) |
symptoms | Calcium | Phosphate | intact PTH |
ALP | 1,25(OH)2D | TSH | AHO features |
NESP | AS | XL | A/B | References |
| (mg/dL) | (mg/dL) | (pg/mL) | (IU/L) | (pg/mL) | (µU/mL) | ||||||||||
| (8.5–10.2) | (2.4–4.3)* | (10–65) | (115–359) | (20–60) | (0.5–5) | ||||||||||
| P3 | Male | 14 | none | 7.8 | 4..8 | 129 | 493 | N/D | 1.5 | − | + | − | ± | − | 23 |
| P7 | Male | 6 | loc, convulsion | 4.6 | 10.3 | 347 | N/D | 45.6 | 5.9 | RF | + | − | − | − | 23 |
| P8 | Male | 3 | convulsion | 5.2 | 6.8 | 720 | 681 | 22.0 | 4.8 | Ob | + | − | ± | − | 23 |
| P9 | Female | 9 | loc | 6.3 | 7.0 | 870** | 258 | N/D | 2.4 | Ob | + | − | ± | − | 23 |
| P13 | Female | 6 | convulsion | 6.0 | 6.6 | 288 | 325 | 24.1 | 2.8 | − | + | − | ± | − | 23 |
| P18 | Male | 12 | muscle cramp | 4.8 | 9.1 | 139 | 479 | 23.6 | 1.4 | − | + | − | − | − | 23 |
| P20 | Male | 11 | convulsion | 7.8 | 9.6 | 137 | 833 | 46.1 | N/D | − | + | − | − | − | 23 |
| P21 | Female | 12 | convulsion | 5.7 | 9.1 | 1600** | 451 | 31.0 | 2.3 | Ob, RF, MR | + | − | − | − | 23 |
| P23 | Female | 8 | convulsion | 6.7 | 8.2 | 134 | 579 | 41.3 | 5.9 | Ob, RF | + | − | − | − | 23 |
| P24 | Female | 35 | tetany | 5.3 | 5.2 | 96 | 237 | 24.5 | 4.4 | Ob | + | − | ± | − | 23 |
| P25 | Male | 8 | convulsion | 7.1 | 8.6 | 340 | 784 | 51.2 | 8.4 | RF | + | − | − | − | 23 |
| P27 | Female | 12 | convulsion | 5.4 | 9.5 | 360 | 711 | 59.1 | N/D | RF, SM | + | − | − | − | 23 |
| P29 | Male | 15 | convulsion | 4.8 | 7.2 | 190 | 1084 | 60.8 | 2.1 | − | + | − | − | − | 23 |
| P32 | Male | 9 | none | 6.7 | 7.4 | 360 | N/D | 49.2 | 1.6 | − | + | − | − | − | 23 |
| P40 | Female | 9 | headache | 8.3 | 5.9 | 480 | 727 | 47.3 | 2.7 | − | + | − | − | − | 23 |
| P44 | Female | 7 | convulsion | 7.6 | 9.3 | 330 | 779 | 50.7 | 5.8 | RF | + | − | − | − | |
| P46 | Male | 4 | convulsion | 7.0 | 7.5 | 300 | 1613 | N/D | 4.4 | − | + | − | − | − | |
| P52 | Male | 6 | convulsion | 6.4 | 8.5 | 349 | 726 | 60.5 | 3.3 | − | + | − | ± | − | |
| P54 | Male | 13 | convulsion | 5.7 | 9.4 | 118 | 926 | 5.9 | N/D | − | + | − | − | − | 23 |
| P55 | Male | 4 | convulsion | low | high | high | N/D | N/D | 7.2*** | − | + | − | ± | − | |
| P56 | Male | 10 | N/D | 5.2 | 6.3 | 354 | N/D | N/D | N/D | N/D | + | − | − | − | |
| P57 | Female | 5 | N/D | 7.3 | 7.6 | 473 | 918 | 44.1 | 16.5 | RF | + | − | − | − | |
| P58 | Male | 25 | loc, tetany | 7.2 | 5.2 | 457 | 263 | 13.0 | 1.5 | − | + | − | − | − | |
adult normal range;
elevated PTH was documented by a mid-PTH assay (normal range: 90–270 pg/mL), if the intact PTH assay was not available;
data at the age of 14 years during treatment with alfacalcidol; MS-MLPA, Methylation-Specific Multiplex Ligation-dependent Probe Amplification; N/D, not determined; loc, loss of consciousness; RF, round face; Ob, Obesity; MR, mental retardation; SM, short metacarpals
Molecular studies
The study was approved by the Ethics Committee of Chiba University and the Massachusetts General Hospital. Genetic analyses were performed after obtaining informed consent from the patient or parents. Genomic DNA was extracted from peripheral blood leukocytes, as described (23).
Methylation analysis
Southern blot analysis and methylation specific-polymerase chain reaction (MS-PCR) were performed, as described (23). Multiplex ligation-dependent probe amplification (MLPA) and methylation specific-MLPA (MS-MLPA) were performed using the SALSA MLPA kit ME031 GNAS (MRC-Holland, Amsterdam, The Netherland) following the manufacturer’s instructions. Analysis of the PCR products was performed on an ABI3130 genetic analyzer and using the GeneMapper Software (Applied Biosystems) at the DNA Core Facility of the Massachusetts General Hospital.
Analysis of SNPs and microsatellite markers
The PCR to search for single nucleotide polymorphism was performed with QIAGEN Taq DNA polymerase and the other reagents supplied with the same kit following the manufacture’s protocols. PCR primers are listed in Supplemental Table 4. The PCR products were purified using ExoSap-IT (Affymetrix) and sequenced at the DNA Core Facility of the Massachusetts General Hospital. Analysis of microsatellite markers across the entire chromosome 20 was performed by the Center for Human Genetic Research of the Massachusetts General Hospital.
RESULTS
Laboratory and clinical findings in our cohort of sporPHP1B patients
We investigated a total of 23 Japanese subjects with sporPHP1B; sixteen of these patients were recently described (23), while the additional patients had previously not been reported (see Table 1). Parents and available siblings of our patients have/had no mineral ion abnormalities. All 23 patients showed, when first diagnosed, hypocalcemia and hyperphosphatemia associated with a significant increase in serum PTH levels. Seven patients also had elevated TSH levels. Ten patients presented with mild AHO features. Taken together, these findings in our cohort of sporPHP1B cases were similar to those observed by us (23,24) (Table 2) and others (8,9,11,21).
Table 2.
Age at diagnosis and laboratory findings in our Japanese cohort of sporadic PHP1B patients in comparison to our previously reported data for Caucasian patients.
| Age at diagnosis (years) |
Calcium (mg/dl) |
Phosphate (mg/dl) |
PTH (pg/ml) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| sporPHP1B Linglart et al., 2007 |
sporPHP1B this report |
sporPHP1B Linglart et al., 2007 |
sporPHP1B this report |
sporPHP1B Linglart et al., 2007 |
sporPHP1B this report |
sporPHP1B Linglart et al., 2007 |
sporPHP1B this report |
||||
| Median | 10.0 | 9.0 | #NUM! | 5.7 | 6.4 | #NUM! | 8.0 | 7.6 | #NUM! | 402 | 335 |
| Mean | 10.0 | 10.6 | #DIV/0 | 6.0 | 6.3 | #DIV/0 | 8.2 | 7.7 | #DIV/0 | 634 | 305 |
| SD | 4.61 | 6.98 | #DIV/0 | 1.17 | 1.08 | #DIV/0 | 1.35 | 1.56 | #DIV/0 | 742.9 | 154.3 |
| SEM | 0.98 | 1.45 | #DIV/0 | 0.25 | 0.23 | #DIV/0 | 0.29 | 0.33 | #DIV/0 | 158.4 | 34.5 |
| N | 22 | 23 | 0 | 22 | 22 | 0 | 22 | 22 | 0 | 22 | 20 |
GNAS methylation status and search for deletions in GNAS and STX16
MS-MLPA of genomic DNA of all patients revealed broad GNAS methylation changes (see Table 1) that are indistinguishable from those previously reported by us (23, 24) and others (8,9,11,21). MLPA provided no evidence for an allelic loss within GNAS or STX16; the 3-kb deletion in STX16, which is the most frequent cause of AD-PHP1B (24–26), was furthermore excluded in all 23 patients by PCR analysis, as previously described (12). The child of patient P24 is reportedly healthy and showed no epigenetic change at the GNAS locus.
Analysis of SNPs and microsatellite markers at the GNAS locus
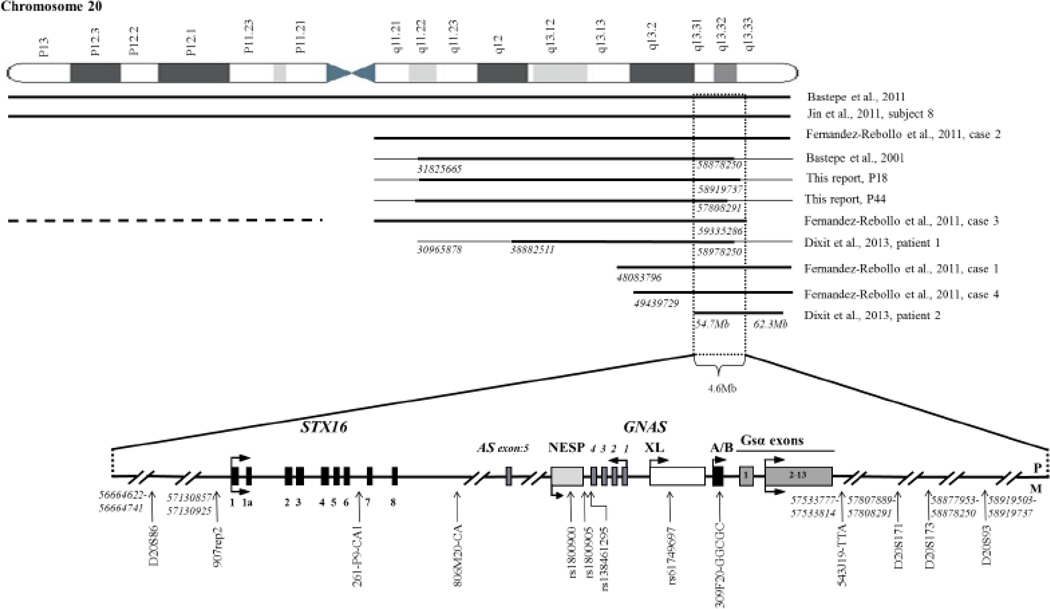
Paternal uniparental disomy of chromosome 20q (patUPD20q) and the associated methylation changes at all four GNAS DMRs had provided a molecular explanation for some of the sporPHP1B patients (18–22). Most of these cases had revealed isodisomy rather than heterodisomy, and we therefore first analyzed three frequent SNPs (rs1800900, rs1800905, and rs138461295) and the pentanucleotide repeat polymorphism at GNAS exon A/B (309F20-GGCGC) (Fig. 1 and Suppl. Table 1) to explore the possibility of patUPD20q in our cohort.
Fig. 1.
Schematic representation of the region on chromosome 20q extending from the GNAS locus to syntaxin 16 (STX16). Each horizontal thick line shows the extent of paternal uniparental isodisomy for the patients described in this report and previously described individuals. The region not conclusively defined as UPD is shown by a thin horizontal line. A horizontal broken line shows the position of the paternal heterodisomy of chromosome 20p. The two dotted vertical lines delineate the smallest region in which duplication of the paternal chromosome 20q may lead to sporadic PHP1B. Exons are indicated by boxes, introns by lines; arrows show direction of transcription. P, paternal; M, maternal. Numbers in italic were provided by hg19 (GRCh37) assembly. For the patient described in Bastepe et al., 2001, two additional microsatellite markers were analyzed (see Supplemental Table 3).
Eight patients were heterozygous for two or more of these variants, making a duplication of the paternal chromosome 20q unlikely. Eleven patients were homozygous for the four SNPs within GNAS and four patients were heterozygous for one of these variants. The latter fifteen individuals were therefore analyzed further through the analysis of six polymorphic microsatellite markers surrounding the GNAS locus (see Fig. 1 and Suppl. Table 2). Two of these individuals revealed homozygosity for all six markers raising the possibility of patUPD20q.
Analysis of patients and parents through microsatellite markers across the entire chromosome 20
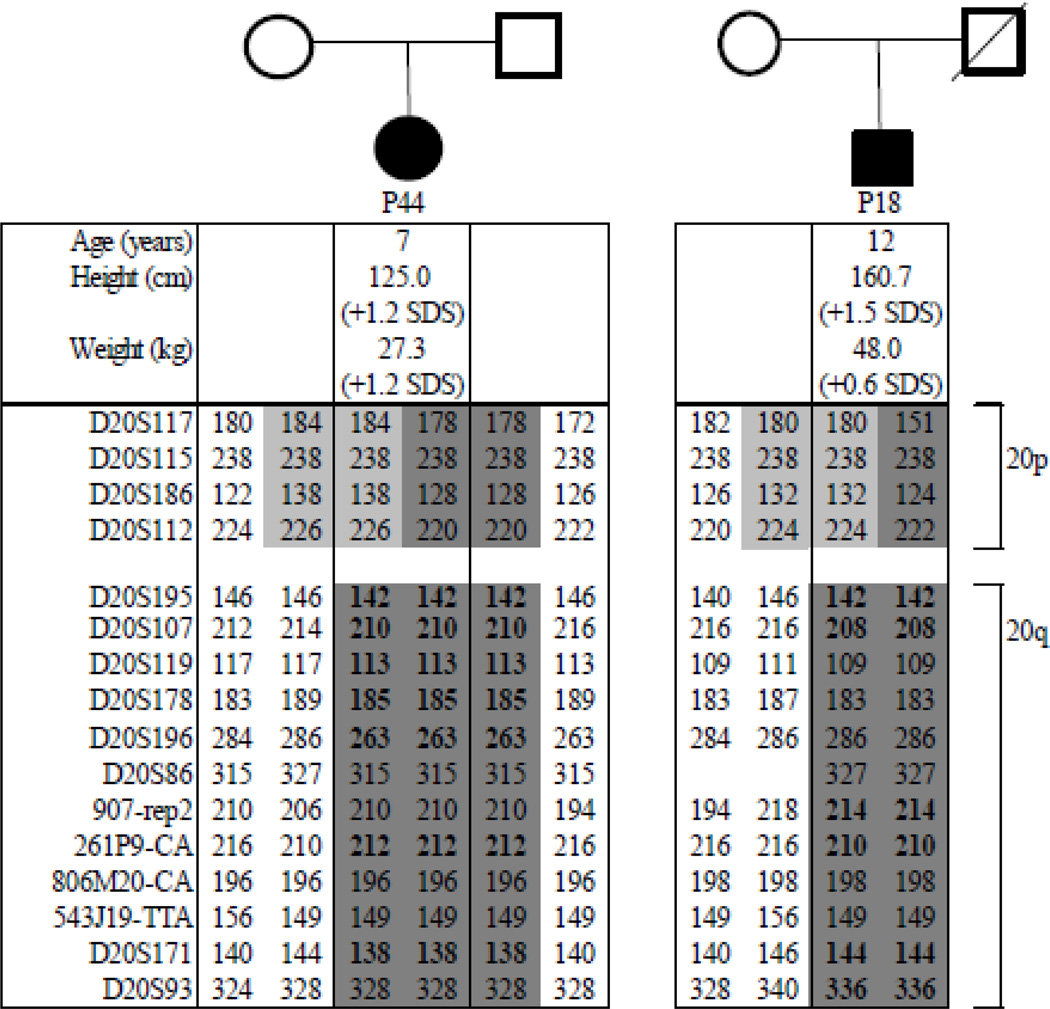
We furthermore analyzed numerous polymorphic markers across the entire chromosome 20 for patient P44 and both of her parents, as well as for patient P18 and her mother (her father is deceased). The studies revealed paternal isodisomy involving the entire long arm of chromosome 20, yet showed bi-parental inheritance for the short arm of chromosome 20. Furthermore, MLPA revealed no evidence for an allelic loss in the STX16-GNAS region leading us to the conclusion that both sporPHP1B cases are affected by patUPD20q (Fig. 2). The non-identical healthy twin brother of P44 showed no evidence for GNAS methylation abnormalities, as determined by MS-MLPA. Analysis of microsatellite markers 907-rep2, 261P9-CA, and D20S171 furthermore excluded patUPD20q; other markers were not informative (data not shown). Analysis of these and additional polymorphic markers provided no evidence for patUPD20q for the other investigated patients.
Fig. 2.
Microsatellite analysis of pedigrees P44 and P18: Affected individuals are represented by filled symbols; the parents are depicted by open symbols (circles=females; squares=male). Results for each microsatellite marker are shown below both pedigrees. Bold numbers indicate fully informative markers for the region with paternal UPD. Identical alleles have the same gray background colors.
DISCUSSION
Our Japanese sporPHP1B cases affected both sexes equally, and their ages and laboratory abnormalities at diagnosis were not significantly different from those previously reported by us (see Table 2) and others (8,9,11) for Caucasian sporPHP1B cases. Furthermore, no significant differences in clinical and laboratory findings were observed for the two patUPD20q patients presented herein and other previously reported cases with duplications involving the long arm of chromosome 20 (see Table 3). Patients with patUPD comprising the long arm of chromosome 20, the entire chromosome, and only segments of chromosome 20q showed no significant differences with regards to age at disease onset as well as levels of PTH, calcium, and phosphate at presentation. This suggests that no another imprinted gene or functionally relevant polymorphisms on chromosome 20 contributes to mineral ion homeostasis.
Table 3.
Age at diagnosis, height, and laboratory findings in patients with sporPHP1B due to different forms of patUPD20
| Serum biochemical values at diagnosis | |||||||
|---|---|---|---|---|---|---|---|
| case | Type of patUPD20q |
Sex | Age at diagnosis (years) |
Calcium | Phosphate | intact PTH | TSH |
| (mg/dL) | (mg/dL) | (pg/mL) | (µU/mL) | ||||
| (8.5–10.2) | (2.4–4.3) | (10–65) | (0.5–5) | ||||
| Bastepe et al., 2011 | entire chromosome | Female | 3.5 | 7.2 | 8.0 | 3685 | N/D |
| Jin et al., 2011 | entire chromosome | Male | 8 | 5.6 | 5.6 | 677 | 7.2 |
| Bastepe et al., 2001 | entire q arm | Male | 5 | 7.2 | 8.1 | 113 | N/D |
| Fernandez-Rebollo et al., case 2 | entire q arm | Male | 9 | 8.8 | 6.5 | 940 | 2.0 |
| this report, P18 | entire q arm | Male | 12 | 4.8 | 9.1 | 139 | 1.4 |
| this report, P44 | entire q arm | Female | 7 | 7.6 | 9.3 | 330 | 5.8 |
| Fernandez-Rebollo et al., case 3 | segmental 20p heterodisomy + 20q interstitial isodisomy | Male | 5 | 4.8 | 7.4 | 292 | 2.8 |
| Fernandez-Rebollo et al., case 1 | Segmental 20q13.13-qter | Female | 26 | 5.2 | 5.9 | 601 | N/D |
| Fernandez-Rebollo et al., case 4 | Segmental 20q13.13-qter | Male | 46 | 6.4 | 4.6 | 127 | 1.2 |
| Dixit et al., patient 1 | Segmental 20q12–q13.33 | Male | 13 | 5.0 | 9.5 | 345 | N/D |
| Dixit et al., patient 2 | Segmental 20q13.31–q13.32 | Male | 5.5* | 9.0* | 6.1* | 422* | 35.0 |
Patients without duplication of the region comprising NNAT are listed in italics.
treatment with cholecalciferol commenced
Dixit et al. had proposed a novel phenotype related to PHP1B due to patUPD20q, namely a relatively high birth weight and obesity, which was noticed during infancy and persisted until later in life (22). Moreover, macrocephaly and tall stature had been observed as possible additional changes (20). However, at the ages of 7 and 12 years, respectively, our two patUPD20q patients were only slightly above average in height and neither was obese (Fig. 2). Besides GNAS, the long arm of chromosome comprises only one other imprinted gene, namely NNAT (20q11.2–q12) encoding neuronatin. Loss of this paternally expressed gene has been implicated in obesity (35;38–41), while biparental NNAT expression as in patUPD20q does not seem to be associated with changes in weight. However, six of seven previously reported patients with patUPD20q comprising the NNAT region were taller than average, while three patients whose UPD regions do not extend to this locus were below average for height (see Table 3). This could imply that NNAT contributes to growth.
UPD is the state in which a chromosomal region or segment is inherited only from a single parent. The duplicated region may vary from segmental (interstitial or telomeric) to an entire chromosome. Several mechanisms resulting in the formation of UPD have been proposed, including monosomy rescue, trisomy rescue, gamate complementation, and post-fertilization errors (27). Clinically relevant consequences resulting from UPD include, besides trisomic mosaicism, genomic imprinting disorders and homozygosity for a recessive mutation, or a combination of both latter conditions. For example, a homozygous mutation in the adenosine deaminase was recently shown to lead to severe combined immunodeficiency because of a paternal duplication of the entire chromosome 20, which most likely caused PHP1B besides ADA-SCID (28).
UPD is a very rare event with an estimated frequency in newborns of 0.029% (29). More frequently, UPD is observed as a cause of imprinting disorders. For example, Silver-Russel-Syndrome (SRS) is caused in 5–10% of the cases by matUPD7 (30, 31), while 3–5% of patients with Angelman syndrome (AS) showed patUPD15 (32) and 20% of patients with Prader-Willi syndrome (PWS) revealed matUPD15 (33). Furthermore, patUPD6 was detected in 41% of the patients with Transient Neonatal Diabetes Mellitus (TNDM) (34) and segmental patUPD11p accounted for 20% of the cases with Beckwith-Wiedemann syndrome (BWS) (35). In addition, it appears plausible that matUPD15 causes some forms of central precocious puberty since mutations in the imprinted gene MKRN3 were found only in 15 out of 32 investigated patients (36).
Since the first description of patUPD20q as a cause of sporPHP1B (18), an additional 10 patients with this disorder have been reported (19–22), including those described in this manuscript. Most of these cases (6 out 11) revealed duplication of only one paternal long arm or the entire chromosome 20 (isodisomy), although the boundaries of the duplication were not always conclusively defined (see Fig. 1). The remaining sporPHP1B cases had smaller duplications involving chromosome 20q, including a duplication of only 7.6 Mb (20q13.31–q13.32) (22). Only one previous report had provided evidence for heterodisomy involving the short arm of chromosome 20 that was combined with interstitial isodisomy affecting chromosome 20q (20); this very infrequent cause of UPD could not be explored in the current study because parental DNA was not available for most patients.
The GNAS methylation changes for our patients with patUPD20q were indistinguishable from those of other reported cases with different extents of UPD. This indicates that a maternal segment that is no longer present within the duplicated 4.6 Mb region contains regulatory elements that allow establishment or maintenance of the normal methylation imprints at GNAS. Because of the telomeric boundaries of the duplicated region could not be conclusively defined in four cases, it remains uncertain whether the size of the genomic GNAS region that is critical for methylation could be smaller (see Fig.1).
We had previously reported 22 sporPHP1B patients, one of whom was later shown to have a duplication of the entire chromosome 20 (24, 19). Fernandez-Rebollo et al. identified patUPD20q in four out of twenty sporPHP1B patients (20), while Jin et al. observed patUPD20 in one out of seven Korean sporPHP1B cases (21). We now found two additional patUPD20q cases in our cohort of 23 Japanese sporPHP1B patients, suggesting that about 10% of all sporPHP1B cases may be caused by duplication of the paternal long arm of chromosome 20, and that all racial backgrounds are equally affected.
In comparison to the large duplicated regions identified in patUPD20 patients to date, about 20% of all patients affected by BWS revealed small duplicated segments in the 11p15.5 region that can be as small as 2.7 Mb (35, 42). Some of these patUPD11p patients were shown to be mosaic implying that postzygotic recombination events had occurred (35, 43, 44), i.e., a mechanism different from that for non-mosaic UPDs involving large chromosomal regions. It is conceivable that similarly small paternally duplicated regions comprising the GNAS locus can be a cause of some sporPHP1B cases or that mosaicism involving the chromosome 20q13 region could explain this disease variant.
In conclusion, two patients with patUPD20q were identified among 23 Japanese sporPHP1B cases. When combined with data from previous reports, these findings suggest that duplication of the paternal long arm of chromosome 20 may be a more frequent cause of sporPHP1B than initially thought and that patUPD20q should be considered in all sporPHP1B cases with broad GNAS methylation changes. Establishing patUPD20q would provide a molecular definition of their disease thus allowing appropriate genetic counseling.
Supplementary Material
Highlights.
Pseudohypoparathyroidism type Ib (PHP1B) is caused by proximal tubular resistance to parathyroid hormone.
In few sporPHP1B patients the disease is caused by paternal uniparental isodisomy involving chromosome 20q (patUPD20q).
We investigated 23 Japanese sporadic PHP1B cases to determine whether patUPD20q can be their cause of PHP1B.
PatUPD20q was confirmed for two patients.
Paternal duplication of the chromosomal region comprising the GNAS locus appears to be a relatively common cause of sporPHP1B.
ACKNOWLEDGEMENTS
We thank Drs. Kenji Fujieda, Shoko Ikema, Tomohiro Ishii, Hiroshi Mochizuki, Fumio Morohashi, Ayuko Narita, Masaya Osaki, Akira Otake, Tatsuya Shiina, Yoshihito Takahashi, Tomoyuki Takagi, Hiroyuki Yamaoka, Kazuko Yonamine, Masanori Yoshida for their help in obtaining blood samples and laboratory results. Furthermore, we are grateful for continuous support and interest of all families for participation in this study. This work was supported by the National Institutes of Health (R01 DK46718-22 to H.J.) This study was also supported in part by grants from Ministry of Health, Labor and Welfare, Japan.
Authors’ role: Study design; RT and HJ. Patients’ sample and clinical data collection: MM, MN and KK. Study conduct: RT, MM, AM, MR, KK, TT, IK, KS, KM, FN, and NS. Data analysis: RT and HJ. Data interpretation: RT and HJ. Drafting manuscript: RT. Revising manuscript: MM, AM, TT, KM, FN, NS, and HJ. Approving final version of manuscript: all authors. RT and HJ take responsibility for integrity of the data analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement: All authors state that they have no conflicts of interest.
REFERENCES
- 1.Mantovani G. Clinical review: Pseudohypoparathyroidism: diagnosis and treatment. J Clin Endocrinol Metab. 2011;96:3020–3030. doi: 10.1210/jc.2011-1048. [DOI] [PubMed] [Google Scholar]
- 2.Weinstein LS, Yu S, Warner DR, Liu J. Endocrine manifestations of stimulatory G protein alpha-subunit mutations and the role of genomic imprinting. Endocr Rev. 2001;22:675–705. doi: 10.1210/edrv.22.5.0439. [DOI] [PubMed] [Google Scholar]
- 3.Bastepe M, Jüppner H. Pseudohypoparathyroidism. New insights into an old disease. Endocrinol Metab Clin North Am. 2000;29:569–589. doi: 10.1016/s0889-8529(05)70151-1. [DOI] [PubMed] [Google Scholar]
- 4.Puzhko S, Goodyer CG, Kerachian MA, et al. Parathyroid hormone signaling via Gαs is selectively inhibited by an NH(2)-terminally truncated Gαs: implications for pseudohypoparathyroidism. J Bone Miner Res. 2011;26:2473–2485. doi: 10.1002/jbmr.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine MA. An update on the clinical and molecular characteristics of pseudohypoparathyroidism. Curr Opin Endocrinol Diabetes Obes. 2012;19:443–451. doi: 10.1097/MED.0b013e32835a255c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastepe M. The GNAS locus and pseudohypoparathyroidism. Adv Exp Med Biol. 2008;626:27–40. doi: 10.1007/978-0-387-77576-0_3. [DOI] [PubMed] [Google Scholar]
- 7.Turan S, Bastepe M. The GNAS complex locus and human diseases associated with loss-of-function mutations or epimutations within this imprinted gene. Horm Res Paediatr. 2013;80:229–241. doi: 10.1159/000355384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Nanclares GP, Fernandez-Rebollo E, Santin I, et al. Epigenetic defects of GNAS in patients with pseudohypoparathyroidism and mild features of Albright's hereditary osteodystrophy. J Clin Endocrinol Metab. 2007;92:2370–2373. doi: 10.1210/jc.2006-2287. 92:2370–2373. [DOI] [PubMed] [Google Scholar]
- 9.Mantovani G, de Sanctis L, Barbieri AM, et al. Pseudohypoparathyroidism and GNAS epigenetic defects: clinical evaluation of albright hereditary osteodystrophy and molecular analysis in 40 patients. J Clin Endocrinol Metab. 2010;95:651–658. doi: 10.1210/jc.2009-0176. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez J, Perera E, Jan de Beur S, et al. Madelung-like deformity in pseudohypoparathyroidism type 1b. J Clin Endocrinol Metab. 2011;96:E1507–E1511. doi: 10.1210/jc.2011-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez-Rebollo E, Lecumberri B, Gaztambide S, et al. Endocrine profile and phenotype-(epi)genotype correlation in Spanish patients with pseudohypoparathyroidism. J Clin Endocrinol Metab. 2013;98:E996–E1006. doi: 10.1210/jc.2012-4164. [DOI] [PubMed] [Google Scholar]
- 12.Bastepe M, Fröhlich LF, Hendy GN, et al. Autosomal dominant pseudohypoparathyroidism type Ib is associated with a heterozygous microdeletion that likely disrupts a putative imprinting control element of GNAS. J Clin Invest. 2003;112:1255–1263. doi: 10.1172/JCI19159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linglart A, Gensure RC, Olney RC, Jüppner H, Bastepe M. A novel STX16 deletion in autosomal dominant pseudohypoparathyroidism type Ib redefines the boundaries of a cis-acting imprinting control element of GNAS. Am J Hum Genet. 2005;76:804–814. doi: 10.1086/429932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elli FM, de Sanctis L, Peverelli E, et al. Autosomal Dominant Pseudohypoparathyroidism Type Ib: A Novel Inherited Deletion Ablating STX16 Causes Loss of Imprinting at the A/B DMR. J Clin Endocrinol Metab. 2014;99:E724–E728. doi: 10.1210/jc.2013-3704. [DOI] [PubMed] [Google Scholar]
- 15.Richard N, Abeguile G, Coudray N, et al. A new deletion ablating NESP55 causes loss of maternal imprint of A/B GNAS and autosomal dominant pseudohypoparathyroidism type Ib. J Clin Endocrinol Metab. 2012;97:E863–E867. doi: 10.1210/jc.2011-2804. [DOI] [PubMed] [Google Scholar]
- 16.Bastepe M, Fröhlich LF, Linglart A, et al. Deletion of the NESP55 differentially methylated region causes loss of maternal GNAS imprints and pseudohypoparathyroidism type Ib. Nat Genet. 2005;37:25–27. doi: 10.1038/ng1487. [DOI] [PubMed] [Google Scholar]
- 17.Chillambhi S, Turan S, Hwang DY, Chen HC, Jüppner H, Bastepe M. Deletion of the noncoding GNAS antisense transcript causes pseudohypoparathyroidism type Ib and biparental defects of GNAS methylation in cis. J Clin Endocrinol Metab. 2010;95:3993–4002. doi: 10.1210/jc.2009-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bastepe M, Lane AH, Jüppner H. Paternal uniparental isodisomy of chromosome 20q--and the resulting changes in GNAS1 methylation--as a plausible cause of pseudohypoparathyroidism. Am J Hum Genet. 2001;68:1283–1289. doi: 10.1086/320117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bastepe M, Altug-Teber O, Agarwal C, Oberfield SE, Bonin M, Jüppner H. Paternal uniparental isodisomy of the entire chromosome 20 as a molecular cause of pseudohypoparathyroidism type Ib (PHP-Ib) Bone. 2011;48:659–662. doi: 10.1016/j.bone.2010.10.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez-Rebollo E, Lecumberri B, Garin I, et al. New mechanisms involved in paternal 20q disomy associated with pseudohypoparathyroidism. Eur J Endocrinol. 2010;163:953–962. doi: 10.1530/EJE-10-0435. [DOI] [PubMed] [Google Scholar]
- 21.Jin HY, Lee BH, Choi JH, et al. Clinical characterization and identification of two novel mutations of the GNAS gene in patients with pseudohypoparathyroidism and pseudopseudohypoparathyroidism. Clin Endocrinol (Oxf) 2011;75:207–213. doi: 10.1111/j.1365-2265.2011.04026.x. [DOI] [PubMed] [Google Scholar]
- 22.Dixit A, Chandler KE, Lever M, et al. Pseudohypoparathyroidism type 1b due to paternal uniparental disomy of chromosome 20q. J Clin Endocrinol Metab. 2013;98:E103–E108. doi: 10.1210/jc.2012-2639. [DOI] [PubMed] [Google Scholar]
- 23.Kinoshita K, Minagawa M, Takatani T, Takatani R, Ohashi M, Kohno Y. Establishment of diagnosis by bisulfite-treated methylation-specific PCR method and analysis of clinical characteristics of pseudohypoparathyroidism type 1b. Endocr J. 2011;58:879–887. doi: 10.1507/endocrj.k10e-364. [DOI] [PubMed] [Google Scholar]
- 24.Linglart A, Bastepe M, Jüppner H. Similar clinical and laboratory findings in patients with symptomatic autosomal dominant and sporadic pseudohypoparathyroidism type Ib despite different epigenetic changes at the GNAS locus. Clin Endocrinol (Oxf) 2007;67:822–831. doi: 10.1111/j.1365-2265.2007.02969.x. [DOI] [PubMed] [Google Scholar]
- 25.Turan S, Ignatius J, Moilanen JS, et al. De novo STX16 deletions: an infrequent cause of pseudohypoparathyroidism type Ib that should be excluded in sporadic cases. J Clin Endocrinol Metab. 2012;97:E2314–E2319. doi: 10.1210/jc.2012-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mantovani G, Bondioni S, Linglart A, et al. Genetic analysis and evaluation of resistance to thyrotropin and growth hormone-releasing hormone in pseudohypoparathyroidism type Ib. J Clin Endocrinol Metab. 2007;92:3738–3742. doi: 10.1210/jc.2007-0869. [DOI] [PubMed] [Google Scholar]
- 27.Yamazawa K, Ogata T, Ferguson-Smith AC. Uniparental disomy and human disease: an overview. Am J Med Genet C Semin Med Genet. 2010;154C:329–334. doi: 10.1002/ajmg.c.30270. [DOI] [PubMed] [Google Scholar]
- 28.Geelen J, Pfundt R, Meijer J, et al. Severe phenotype of severe combined immunodeficiency caused by adenosine deaminase deficiency in a patient with a homozygous mutation due to uniparental disomy. J Allergy Clin Immunol. 2013;132:222–223. doi: 10.1016/j.jaci.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Robinson WP. Mechanisms leading to uniparental disomy and their clinical consequences. Bioessays. 2000;22:452–459. doi: 10.1002/(SICI)1521-1878(200005)22:5<452::AID-BIES7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 30.Kotzot D, Schmitt S, Bernasconi F, et al. Uniparental disomy 7 in Silver-Russell syndrome and primordial growth retardation. Hum Mol Genet. 1995;4:583–587. doi: 10.1093/hmg/4.4.583. [DOI] [PubMed] [Google Scholar]
- 31.Eggermann T, Wollmann HA, Kuner R, et al. Molecular studies in 37 Silver-Russell syndrome patients: frequency and etiology of uniparental disomy. Hum Genet. 1997;100:415–419. doi: 10.1007/s004390050526. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y, Lev-Lehman E, Bressler J, Tsai TF, Beaudet AL. Genetics of Angelman syndrome. Am J Hum Genet. 1999;65:1–6. doi: 10.1086/302473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mascari MJ, Gottlieb W, Rogan PK, et al. The frequency of uniparental disomy in Prader-Willi syndrome. Implications for molecular diagnosis. N Engl J Med. 1992;326:1599–1607. doi: 10.1056/NEJM199206113262404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Docherty LE, Kabwama S, Lehmann A, et al. Clinical presentation of 6q24 transient neonatal diabetes mellitus (6q24 TNDM) and genotype-phenotype correlation in an international cohort of patients. Diabetologia. 2013;56:758–762. doi: 10.1007/s00125-013-2832-1. [DOI] [PubMed] [Google Scholar]
- 35.Cooper WN, Curley R, Macdonald F, Maher ER. Mitotic recombination and uniparental disomy in Beckwith-Wiedemann syndrome. Genomics. 2007;89:613–617. doi: 10.1016/j.ygeno.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Abreu AP, Dauber A, Macedo DB, et al. Central precocious puberty caused by mutations in the imprinted gene MKRN3. N Engl J Med. 2013;368:2467–2475. doi: 10.1056/NEJMoa1302160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dou D, Joseph R. 1996 Cloning of human neuronatin gene and its localization to chromosome-20q 11.2–12: the deduced protein is a novel "proteolipid'. Brain Res. 1996;723:8–22. doi: 10.1016/0006-8993(96)00167-9. [DOI] [PubMed] [Google Scholar]
- 38.Chu K, Tsai MJ. Neuronatin, a downstream target of BETA2/NeuroD1 in the pancreas, is involved in glucose-mediated insulin secretion. Diabetes. 2005;54:1064–1073. doi: 10.2337/diabetes.54.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mzhavia N, Yu S, Ikeda S, Chu TT, Goldberg I, Dansky HM. Neuronatin: a new inflammation gene expressed on the aortic endothelium of diabetic mice. Diabetes. 2008;57:2774–2783. doi: 10.2337/db07-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gburcik V, Cleasby ME, Timmons JA. Loss of neuronatin promotes "browning" of primary mouse adipocytes while reducing Glut1-mediated glucose disposal. Am J Physiol Endocrinol Metab. 2013;304:E885–E894. doi: 10.1152/ajpendo.00463.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vrang N, Meyre D, Froguel P, et al. The imprinted gene neuronatin is regulated by metabolic status and associated with obesity. Obesity (Silver Spring) 2010;18:1289–1296. doi: 10.1038/oby.2009.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slatter RE, Elliott M, Welham K, et al. Mosaic uniparental disomy in Beckwith-Wiedemann syndrome. J Med Genet. 1994;31:749–753. doi: 10.1136/jmg.31.10.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Catchpoole D, Lam WW, Valler D, et al. Epigenetic modification and uniparental inheritance of H19 in Beckwith-Wiedemann syndrome. J Med Genet. 1997;34:353–359. doi: 10.1136/jmg.34.5.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henry I, Puech A, Riesewijk A, et al. Somatic mosaicism for partial paternal isodisomy in Wiedemann-Beckwith syndrome: a post-fertilization event. Eur J Hum Genet. 1993;1:19–29. doi: 10.1159/000472384. [DOI] [PubMed] [Google Scholar]
- 45.Bastepe M, Pincus JE, Sugimoto T, et al. Positional dissociation between the genetic mutation responsible for pseudohypoparathyroidism type Ib and the associated methylation defect at exon A/B: evidence for a long-range regulatory element within the imprinted GNAS1 locus. Hum Mol Genet. 2001;1:1231–1241. doi: 10.1093/hmg/10.12.1231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.