Abstract
Traditional plating and culturing methods used to quantify bacteria commonly require hours to days from sampling to results. We present here a simple, sensitive and rapid electrochemical method for bacteria detection in drinking water based on gold nanoparticle-enzyme complexes. The gold nanoparticles were functionalized with positively charged quaternary amine headgroups that could bind to enzymes through electrostatic interactions, resulting in inhibition of enzymatic activity. In the presence of bacteria, the nanoparticles released from the enzymes and preferentially bound to the bacteria, resulting in an increase in enzyme activity, releasing a redox-active phenol from the substrate. We employed this strategy for the electrochemical sensing of Escherichia coli and Staphylococcus aureus, resulting in a rapid detection (<1h) with high sensitivity (102 CFU·mL−1).
Introduction
Each year an estimated 47.8 million people in United States become sick resulting from contaminated food.1 As part of the newly established Food Safety Modernization Act, the Food and Drug Administration (FDA) has given special attention to the need for sanitary water.2 In developing countries, drinking water contaminated with bacteria results in millions of illnesses each year.3 Traditional plating and culturing methods used to detect bacteria require hours to days for results. Thus, a rapid and sensitive approach for the detection of bacterial contamination is needed. Advanced methods such as surface plasmon resonance (SPR),4 polymerase chain reaction (PCR)5 and enzyme linked immunosorbent assay (ELISA)6 require advanced laboratory settings which may not be pragmatic for resource limited areas. Low-cost and easy-to-use technologies have been proposed for pathogenic bacteria detection, including colorimetric,6 electroluminescence,7 immunomagnetic detection8 and electrochemical methods.9 Among these sensors, enzyme-based electrochemical sensors provide high, steady, and reproducible signal amplification while overcoming some disadvantages, such as slow sensing and high cost. Meanwhile, electrochemical detection is more easily interfaced with electronic devices/computers, making it useful for data processing and point-of-care (POC) applications.10 Enzymatic reactions combined with redox cycling or multi-enzyme labels per detection probe have been shown to achieve high sensitivity for the detection of pathogenic bacteria.11
Recent advances in nanotechnology have enabled new technologies for sensitive and rapid pathogen detection. Nanomaterials such as nanoparticles, nanorods, nanosheets and 3D-nanostructures have demonstrated significant advantages for sensing applications.12, 13 Gold nanoparticles (AuNPs) can be synthesized in a straightforward manner with high stability and biocompatibility. The size and shape of AuNPs can be adjusted for specific applications. Furthermore, AuNPs possess a suitable functionalization platform for a wide range of organic ligands, enabling a binding event with biological analytes.14, 15 Based on these unique properties, various biosensors have been built based on AuNPs for the identification of analytes such as proteins,16 cancer cells17 and bacteria.18
Cationic AuNPs can bind to enzymes by electrostatic interactions and have been shown to inhibit the activity of bound enzymes.16 Following the displacement of nanoparticle from the enzyme, the activity of the enzyme can be recovered. This displacement can be orchestrated using an analyte with preferential affinity for the nanoparticles. This nanoparticle-β-gal system has been reported to detect bacteria by colorimetric response.19 Here we employ this strategy to provide an enzyme-amplified electrochemical sensor for detection of bacteria, generating a simple device suitable for POC use. In this study, we compared four kinds of cationic AuNPs to inhibit β-galactosidase (β-gal) activity, which can catalyze the hydrolysis of β-galactosides into monosaccharides (Fig. 1). The AuNPs that demonstrated the optimal inhibition efficiency were selected for electrochemical detection. Escherichia coli (E. coli) XL1 was used as the analyte to displace the AuNPs from the AuNPs/β-gal complex. The recovered activity of the enzymes correlated with the concentration of E. coli and was able to be electrochemically quantified.
Fig. 1.

Schematic representation of bacteria detection based on gold nanoparticles/enzyme complexes.
Experimental section
Materials and apparatus
The β-galactosidase (β-gal), o-nitrophenyl-β-galactopyranoside (ONPG) and 4-aminophenyl-β-galactopyranoside (PAPG) were purchased from Sigma-Aldrich, USA. Mili-Q water with 18 MΩ cm−1 resistivity was used for all aqueous solutions. All other chemicals were purchased from Fisher Scientific, USA and used without further purification.
UV-vis absorption spectra were measured using a Synergy™ Biotek Instrument (Winooksi, VT). Inc. A battery-powered and handheld potentiostat (Palmsens BV, Netherlands) was used for electrochemical analysis. Gold electrodes were purchased from Micrux Technologies.
Synthesis of gold nanoparticles (AuNPs)
The AuNPs were synthesized and functionalized according to previously reported procedures.18, 20–22 The thiol ligands for gold nanoparticles are engineered: 1) Synthetic ligands can be fabricated onto the surface of AuNPs via the strong metal-ligand interaction between Au and S;23, 24 2) The C11-alkane part ensures stability to the nanoparticles; 3) The tetraethylene glycol moiety minimizes nonspecific protein adsorption and contributes to the biocompatibility of nanoparticles;25, 26 4) The quaternary amine provides a permanent positive surface charge for gold nanoparticles as well as the scaffold for functionality design. In this study, we utilized the above features and tuned the headgroup hydrophobicity by changing the structure of quaternary amines. Briefly, a place exchange reaction was carried out using a suspension of 1-pentanethiol-stablized gold cores (~ 2 nm) in anhydrous dichloromethane. A solution of each one of the ligands (obtained according to the reported procedures) was prepared by dissolving in dry dichloromethane and methanol mixture (v/v = 9:1). The two solutions were mixed and kept under constant stirring for 96 hours at room temperature under N2 protection. The solvent was evaporated afterwards and the residue was washed several times with n-hexanes. The nanoparticles were dispersed in Mili-Q water and dialyzed using 10,000 MWCO SnakeSkin Dialysis Tubing (Thermo Scientific, USA) for 120 hours. The concentration of the AuNP solution was measured according to the reported method by UV spectroscopy on a Molecular Devices SpectraMax M2 at 506 nm.27 The characterization results of NP1-NP4 were shown in Section 5 of the Electronic Supplementary Information.
Activity titration
Assays were conducted in sodium phosphate buffer (PB buffer, 5 mM, pH 7.4) at room temperature. Activity titrations were performed in 96-well plate by increasing the concentration of cationic AuNPs with respect to constant β-gal in 200 μL volume. The optimal ratio of β-gal and ONPG in absence of AuNPs was determined by adjusting the absorbance value to approximately 1.000. After the same amount of β-gal (60 μL, 21.4 nM) was pipetted into 96-well plate, increasing volumes of AuNPs (40 nM) were added. The volumes were then adjusted to a total of 200 μL with PB buffer and the mixture was incubated for 30 minutes. ONPG solution (30 μL, 5 mM) was then added and the absorbance kinetics were quantified at 405 nm every 20 seconds for 15 minutes.
For the activity titration using electrochemical method, the procedure was similar to the colorimetric method. Instead of ONPG, PAPG was used as the electrochemical substrate. Before the activity titration, cyclic voltammetry (CV) was performed in a control experiment where AuNPs were absent. Differential pulse voltammetry (DPV) was chosen to test the redox properties of the hydrolysis product of PAPG, 4-aminophenol. PAPG solution (30 μL, 5 mM) was added into β-gal solution (60 μL, 21.4 nM) and the volume of the solution was adjusted to 200 μL with PB buffer. The mixture was incubated for 1 hour and subsequently measured by DPV to record the CV scan. The activity titration was conducted afterwards using DPV every 1 minute for 15 minutes (condition: t equilibration: 3 seconds; E range: −0.1 V to 0.4 V; E step: 5 mV; E pulse: 50 mV; t pulse: 50 ms and scan rate: 50 mV·s−1). The optimization of experimental parameters is described in the Supplementary Information (Fig. S1). The data represented the inhibition of enzymatic activity. All experiments were performed in triplicate.
Bacterial culture
Escherichia coli (E. coli) XL1 in 30% glycerol was streaked on a Luria Bertani (LB) plate and incubated overnight at 37 °C. Then, a single colony was inoculated into LB broth (50 mL) and agitated overnight at 37 °C. Bacteria were harvested by centrifugation (7000 × g, 2 min) and washed three times with PB buffer. The bacterial concentration was directly quantified by plating as a standard method on LB agar plates, using tenfold serial dilutions of the bacteria stock solution. The serial diluted bacteria in PB buffer at various concentrations (103–107 CFU·mL−1) were used for the experiments.
Analyte bacteria response
From the activity titration, the AuNPs having the strongest inhibition response were selected and the corresponding optimal concentration of AuNPs/enzyme complex was determined. For bacteria detection, varying concentrations of E. coli were incubated with the complex. After adding PAPG, the enzyme activity was measured electrochemically using DPV. In this experiment, increasing concentrations of bacteria (20 μL) were added into the AuNPs/enzyme complex. Then, PAPG (30 μL, 5 mM) was added into the above solution and incubated. The enzyme activity was detected electrochemically (condition: t equilibration: 3 seconds; E range: −0.1 V to 0.4 V; E step: 5 mV; E pulse: 50 mV; t pulse: 50 ms and scan rate: 50 mV·s−1) at 16 min and 28 min.
Results and discussion
Principle of bacteria detection based on AuNPs/β-gal complex
The mechanism for bacterial detection is shown in Fig. 2. β-Galactosidase (β-gal) (pI = 4.6) has previously been used as a reporter enzyme for sensing applications.16, 19, 28 Furthermore, it has been shown that β-gal can interact with cationic gold nanoparticles (AuNPs) without denaturation.16 β-gal activity was inhibited by the binding of positively charged gold nanoparticles to negatively charged residues around β-gal active site through electrostatic interactions.28 The “turn-off” point was determined by optimizing the ratio between AuNPs and β-gal. Under this condition, the AuNPs/β-gal complex does not catalyze the hydrolysis of β-galactosides into monosaccharaides. Bacteria with surface anionic glycocalyx structures preferentially competed for the functionalized nanoparticles with the β-gal, resulting in increased enzymatic activity. The recovered β-gal catalyzed the PAPG substrate to produce an electrochemically active product that could be quantified with a low-cost device.
Fig. 2.
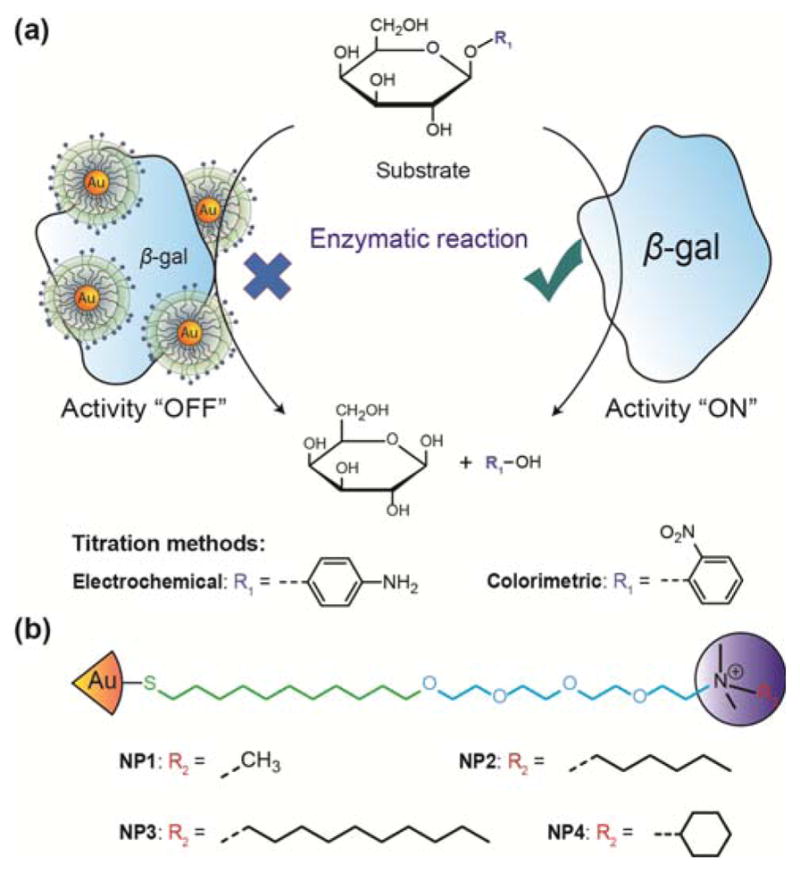
(a) Schematic illustration of β-galactosidase activity titration using gold nanoparticles. (b) The schematic structure of NP1-NP4.
Inhibition efficiencies of AuNPs
AuNPs modified with quaternary amine headgroups could reversibly bind to β-gal16, 19 efficiently compared to other nanomaterials, such as carbon nanotubes29 and graphene oxide30, 31 which have been reported as enzyme inhibitors.
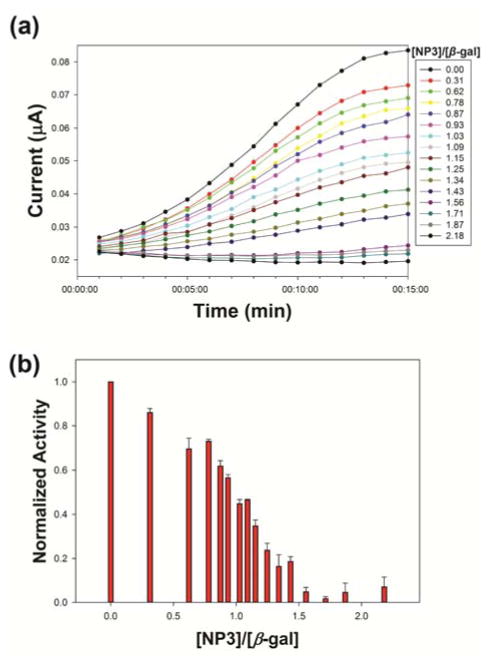
The AuNPs (NP1, NP2, NP3, and NP4) were screened using a colorimetric assay to determine the most effective ligand for detection. The results identified NP3 as most effective at reversibly inhibiting enzymatic activity (Fig. S2). To reference the colorimetric inhibition test, we also conducted an activity titration to investigate the inhibition behavior of NP3 on β-gal using PAPG as an electrochemical substrate (Fig. 3). In this study, after the hydrolysis of PAPG, the cyclic voltammetry diagram of 4-aminophenol was obtained (Fig. S1a). The oxidation peak (E: 1.35 V) was between −0.1 and 0.4 V, which was chosen as the range of measurement (E range). The activity titration was conducted with an increasing ratio of AuNPs to β-gal in the complex, using a similar procedure as the colorimetric method. The electrochemical inhibit performance of NP3 on β-gal was obtained (Fig. 3a). The results demonstrated that the inhibition ratio for NP3 and β-gal in complex was 1.43 (Fig. 3b), which was consistent with the inhibition behavior observed the colorimetric approach. NP3 has the highest headgroup hydrophobicity among the four different NPs,32, 33 resulting in the most efficient inhibition for β-gal activity. Meanwhile, the increase of headgroup hydrophobicity led to the improved inhibition performance for β-gal (Fig. S4).
Fig. 3.
(a) Electrochemical activity inhibition of β-gal after incubation with different concentrations of NP3. Inhibited activity of β-gal plotted as NP3 concentrations in PB buffer (5 mM, pH 7.4). (b) Optimal ratio of NP3/β-gal complex: 1.43. Error bars represent the standard deviation of a minimum of three replicates.
Bacteria detection with AuNPs
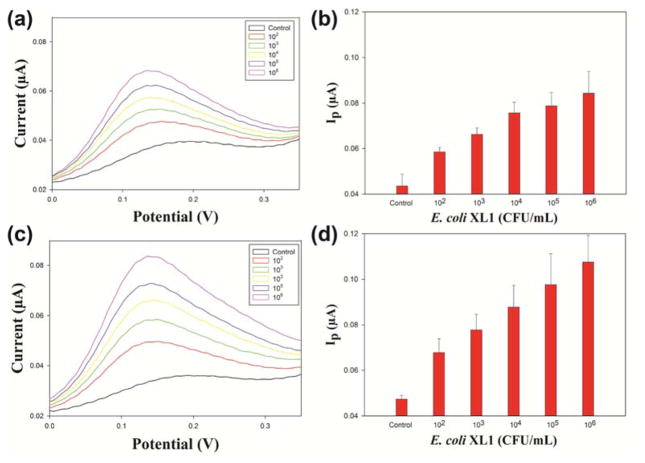
Electrochemical detection provides instant signal transduction at a low cost.34 To demonstrate that this system can be applied to an electrochemical method to detect bacteria, E. coli XL1 was chosen as model analyte. After incubating bacteria with NP3/β-gal complex, the electrochemical signal increased with the increasing concentration of E. coli (Fig. 4). Due to the stronger electrostatic interaction between the surface of bacteria and NP3, bacteria can displace the β-gal and the recovered β-gal catalyzed the electrochemical substrate, providing the electrochemical signal. Using this method we achieved detection of bacteria as low as concentration of 100 CFU·mL−1 following a 16-minute incubation (Fig. 4b). Lower levels of bacteria were not studied due to stochastic issues with the sensor volumes used (200 μL). To evaluate the performance of the sensor, accuracy and precision was calculated. The precision and accuracy of sensor operated at 16-minute incubation were calculated to be 93% and 91% respectively while the sensor operated at 28-minute incubation shows 91% precision and 77% accuracy. (For detailed calculations of accuracy and precision, please refer to Section 3 in the Electronic Supplementary Information.) As expected, longer incubation generated a stronger signal, but no significant enhancement in sensitivity (Fig. 4c, d).
Fig. 4.
Differential Pulse Voltammetry for the NP3/β-gal complex with increasing concentrations of E. coli (control, 1 × 102, 1 × 103, 1 × 104, 1 × 105 and 1 × 106 CFU·mL−1) following incubation for (a) 16 minutes and (c) 28 minutes. Plot of numbers of E. coli versus DPV signal after incubation for (b) 16 minutes and (d) 28 minutes. Error bars represent the standard deviation of a minimum of three replicates.
E. coli was chosen as a model strain for Gram-negative bacteria. Besides, we also successfully applied the electrochemical sensing system to Staphylococcus aureus (S. aureus) CD-48935 (Fig. S9, Fig. S10), which was used as a model strain for Gram-positive bacteria. Overall, the electrochemical sensing system in the current study has shown the capability of detecting both Gram-negative and Gram-positive model strains in drinking water.
Conclusions
In summary, we have developed a simple, sensitive and rapid electrochemical method for bacteria screening using a positively charged gold nanoparticle-β-galactosidase enzyme complex system. Using this approach in an electrochemical format, we were able to detect bacteria at concentration of 100 CFU·mL−1 within one hour. For some food or drinking water matrices with strong backgrounds, a colorimetric method can result in a misleading concentration of analyte bacteria. However, this issue can be avoided using an electrochemical approach. Our study here focuses on the detection of bacteria in water, with the future aim of applying the detection approach to multiplex systems. Additionally, electrochemical detection provides rapid quantification at a low cost and minimal instrumentation. Further study will focus on moving this gold nanoparticle-enzyme complex-based sensing system into microfluidic devices for bacteria detection in a wide range of applications resource areas.
Supplementary Material
Acknowledgments
Financial support was provided by the NIH (GM077173) and the NSF (CMMI-1025020). J.C. acknowledges the support from the Chinese Scholarship Council.
Footnotes
Electronic Supplementary Information (ESI) available. See DOI: 10.1039/b000000x/
Contributor Information
Sam R. Nugen, Email: snugen@foodsci.umass.edu.
Vincent M. Rotello, Email: rotello@chem.umass.edu.
Notes and references
- 1.Tawil N, Sacher E, Mandeville R, Meunier M. Biosens Bioelectron. 2012;37:24–29. doi: 10.1016/j.bios.2012.04.048. [DOI] [PubMed] [Google Scholar]
- 2.Jaykus L-A. ASM News. 2003;69:341–347. [Google Scholar]
- 3.Black RE. Pediatr Infect Dis J. 1993;12:751–761. doi: 10.1097/00006454-199309000-00010. [DOI] [PubMed] [Google Scholar]
- 4.Taylor AD, Ladd J, Yu Q, Chen S, Homola J, Jiang S. Biosens Bioelectron. 2006;22:752–758. doi: 10.1016/j.bios.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Bej AK, Mahbubani MH, Miller R, DiCesare JL, Haff L, Atlas RM. Mol Cell Probes. 1990;4:353–365. doi: 10.1016/0890-8508(90)90026-v. [DOI] [PubMed] [Google Scholar]
- 6.Nagata S, Tsutsumi T, Hasegawa A, Yoshida F, Ueno Y, Watanabe MF. J AOAC Int. 1997;80:408–417. [Google Scholar]
- 7.Chang J, Popp F. Biophotons. Springer; 1998. pp. 201–215. [Google Scholar]
- 8.Pyle BH, Broadaway SC, McFeters GA. Appl Environ Microbiol. 1999;65:1966–1972. doi: 10.1128/aem.65.5.1966-1972.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serra B, Morales MD, Zhang J, Reviejo AJ, Hall EH, Pingarron JM. Anal Chem. 2005;77:8115–8121. doi: 10.1021/ac051327r. [DOI] [PubMed] [Google Scholar]
- 10.Wang J. Biosens Bioelectron. 2006;21:1887–1892. doi: 10.1016/j.bios.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 11.Yang H. Curr Opin Chem Biol. 2012;16:422–428. doi: 10.1016/j.cbpa.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 12.De M, Ghosh PS, Rotello VM. Adv Mater. 2008;20:4225–4241. [Google Scholar]
- 13.Guo S, Wang E. Nano Today. 2011;6:240–264. [Google Scholar]
- 14.Saha K, Agasti SS, Kim C, Li X, Rotello VM. Chem Rev. 2012;112:2739–2779. doi: 10.1021/cr2001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Z, Le ND, Gupta A, Rotello VM. Chem Soc Rev. 2015 doi: 10.1039/C1034CS00387J. [DOI] [Google Scholar]
- 16.Miranda OR, Chen H-T, You C-C, Mortenson DE, Yang X-C, Bunz UHF, Rotello VM. J Am Chem Soc. 2010;132:5285–5289. doi: 10.1021/ja1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajaj A, Miranda OR, Kim I-B, Phillips RL, Jerry DJ, Bunz UH, Rotello VM. Proc Natl Acad Sci USA. 2009;106:10912–10916. doi: 10.1073/pnas.0900975106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips RL, Miranda OR, Mortenson DE, Subramani C, Rotello VM, Bunz UHF. Soft Matter. 2009;5:607–612. [Google Scholar]
- 19.Miranda OR, Li XN, Garcia-Gonzalez L, Zhu ZJ, Yan B, Bunz UHF, Rotello VM. J Am Chem Soc. 2011;133:9650–9653. doi: 10.1021/ja2021729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hostetler MJ, Templeton AC, Murray RW. Langmuir. 1999;15:3782–3789. [Google Scholar]
- 21.De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UHF, Rotello VM. Nat Chem. 2009;1:461–465. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brust M, Walker M, Bethell D, Schiffrin DJ, Whyman R. J Chem Soc, Chem Commun. 1994:801–802. [Google Scholar]
- 23.Pakiari A, Jamshidi Z. The Journal of Physical Chemistry A. 2010;114:9212–9221. doi: 10.1021/jp100423b. [DOI] [PubMed] [Google Scholar]
- 24.Ding Y, Jiang Z, Saha K, Kim CS, Kim ST, Landis RF, Rotello VM. Mol Ther. 2014;22:1075–1083. doi: 10.1038/mt.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You C-C, De M, Han G, Rotello VM. J Am Chem Soc. 2005;127:12873–12881. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- 26.Hong R, Fischer NO, Verma A, Goodman CM, Emrick T, Rotello VM. J Am Chem Soc. 2004;126:739–743. doi: 10.1021/ja037470o. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Atwater M, Wang J, Huo Q. Colloids Surf, B. 2007;58:3–7. doi: 10.1016/j.colsurfb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Wu LJ, Guo SS, Fu HE, Chen GN, Yang HH. Nanoscale. 2013;5:619–623. doi: 10.1039/c2nr32704j. [DOI] [PubMed] [Google Scholar]
- 29.Zhang B, Xing Y, Li Z, Zhou H, Mu Q, Yan B. Nano Lett. 2009;9:2280–2284. doi: 10.1021/nl900437n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De M, Chou SS, Dravid VP. J Am Chem Soc. 2011;133:17524–17527. doi: 10.1021/ja208427j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang XJ, Zhao CQ, Ju EG, Ren JS, Qu XG. Chem Commun. 2013;49:8611–8613. doi: 10.1039/c3cc44632h. [DOI] [PubMed] [Google Scholar]
- 32.Moyano DF, Goldsmith M, Solfiell DJ, Landesman-Milo D, Miranda OR, Peer D, Rotello VM. J Am Chem Soc. 2012;134:3965–3967. doi: 10.1021/ja2108905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saha K, Moyano DF, Rotello VM. Mater Horiz. 2014;1:102–105. doi: 10.1039/C3MH00075C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katz E, Willner I, Wang J. Electroanalysis. 2004;16:19–44. [Google Scholar]
- 35.Li X, Robinson SM, Gupta A, Saha K, Jiang Z, Moyano DF, Sahar A, Riley MA, Rotello VM. ACS Nano. 2014;8:10682–10686. doi: 10.1021/nn5042625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.