Abstract
Background
While choices about genetic testing are increasingly common for patients and families, and public opinion surveys suggest public interest in genomics, it is not known how adults from the general population value genetic testing for heritable conditions. We sought to understand in a US sample the relative value of the characteristics of genetic tests to identify risk of hereditary colorectal cancer, among the first genomic applications with evidence to support its translation to clinical settings.
Methods
A Web-enabled choice-format conjoint survey was conducted with adults age 50 and older from a probability-based US panel. Participants were asked to make a series of choices between two hypothetical blood tests that differed in risk of false negative test, privacy, and cost. Random parameters logit models were used to estimate preferences, the dollar value of genetic information, and intent to have genetic testing.
Results
A total of 355 individuals completed choice-format questions. Cost and privacy were more highly valued than reducing the chance of a false negative result. Most (97%, 95% Confidence Interval (CI): 95% to 99%) would have genetic testing to reduce the risk of dying from colorectal cancer in the best scenario (no false negatives, results disclosed to primary care physician). Only 41% (95% CI: 25% to 57%) would have genetic testing in the worst case (20% false negatives, results disclosed to insurance company).
Conclusions
Given the characteristics and levels included in the choice, if false negative test results are unlikely and results are shared with a primary care physician, the majority would have genetic testing. As genomic services become widely available, primary care professionals will need to be increasingly knowledgeable about genetic testing decisions.
Personalized medicine—health care targeted to the characteristics of individuals, including genetics—has developed rapidly during the last decade. Patients and members of the public say they would be tested to prevent disease in themselves or family members.1,2 Some personalized medicine applications are used to determine optimal treatments and are only relevant to individuals with specific conditions. Others are used to identify at-risk individuals who might benefit from more intensive screening or prophylactic treatment and are relevant to larger segments of the asymptomatic population.
Genetic testing used in risk assessment for hereditary colorectal cancer, specifically Lynch syndrome or hereditary nonpolyposis colorectal cancer (HNPCC), has empirical evidence for its clinical validity and utility.3,4 Evidence-based guidelines for hereditary colorectal cancer have been widely disseminated,5-8 and cost-effectiveness evaluations have been conducted.9-11 The purpose of genetic testing in this case is to identify those who would benefit from more frequent and intensive screening, or from prophylactic surgery, such as colectomy.12 While the mutations associated with hereditary colorectal cancer are not common, many people will have questions about colorectal cancer, a highly prevalent disease, and whether they or related family members would benefit from genetic testing.
Many view genetics as “a very good or good thing,” 13 but patients and the public also have significant concerns about genetic testing, including cost, accuracy, and potential for discrimination based on genetics.14-17 Individuals facing these decisions make complex tradeoffs among these factors. Beyond studies of attitudes toward testing, there is little quantitative information on how individuals at risk for health conditions or members of the general public weigh costs with benefits.18 This information is critical for health professionals who will need to be prepared for discussions about genetic testing with their patients. Without knowledge of tradeoffs, it is not possible to fully understand the value of genetic testing or the factors associated with its adoption and utilization.19 In this study, we sought to examine the relative value of specific characteristics of genetic testing for hereditary colorectal cancer in a probability-based sample of adults 50 years of age and older from the general US population, a group for whom routine colorectal cancer screening is relevant.
Methods
We used a choice-format conjoint survey to measure the value of genetic testing in a probability-based sample of adults from the US population. Choice-format conjoint, also known as choice-based conjoint (CBC), is a form of conjoint analysis. Over the past decade, this type of survey has been used increasingly to quantify preferences for characteristics of health care and policy.20-23 These surveys simulate clinical and policy decision making and provide a systematic method of eliciting tradeoffs to quantify the relative importance that individuals place on treatment characteristics or outcomes. This approach is based on the premise that medical interventions are composed of sets of characteristics (e.g., efficacy, safety) and that the relative value of a particular intervention is a function of these characteristics.24
Survey
The construction of choice alternatives used in the survey was based on seven focus groups of clinical experts, average risk community members, and patients at risk for hereditary cancer (total n=42). There were two groups of average risk community members (n=19), two groups of high risk and cancer patients (n=8), one group of genetic counselors (n=3) and two groups of physicians (n=12). The focus groups were moderated by two experienced qualitative researchers using a structured guide and were recorded using a digital audio recorder. Also, each group was observed by several of the investigators who took handwritten notes. A white board was used to record the specific characteristics and levels of these characteristics identified by focus group members as important in decisions about genetic testing. After each group, a digital photograph was taken to record the material from the white board.
Two genetic testing scenarios were presented to the groups for discussion. The questions included in the structured guide were the same for each group, but the perspective was different. For example, while patients were asked about their personal likelihood of being tested, physicians were asked how likely they thought their patients were to be tested, given a particular scenario. Each focus group included two formats: 1) an open-ended discussion of genetic testing and 2) a highly structured discussion of the specific characteristics that focus group members indicated had influenced or would influence their decisions about genetic testing.
To understand how the characteristics might influence decisions about genetic testing, the focus groups included a highly structured discussion of potential levels or categories for each characteristic (e.g., accuracy: 0%, 10%, or 20% chance of a false negative; privacy: primary care doctor, genetics health professionals, or insurance companies will receive genetic test results). During the structured discussion, we asked focus group members to identify a range of relevant levels or categories for each characteristic. For characteristics such as sensitivity, specificity, false positive, and false negative, we provided definitions in plain language and with verbal and graphical illustration appropriate for those of lower numeracy. Definitions and supporting graphics were displayed on a white board that was visible to all in the room. We then moderated a discussion of the levels and categories leading to refinement of the relevant and important levels and categories. Finally, we asked the members vote on the highest, lowest, and intermediate levels or categories that were most meaningful to decisions about genetic testing.
Three experienced qualitative investigators coded the verified transcripts of the recordings and the notes using a content analysis approach.25 First, the three coders reviewed the transcripts and defined a total of ten unique coding categories. Second, after the categories were clearly defined and a coding manual developed, the investigators then conducted an initial round of coding each transcript. Discrepancies in coding were resolved through discussion and the coding manual was refined. A final round of coding the transcripts was then conducted again with discrepancies being resolved through discussion.
Analysis of the transcripts generated a range of test characteristics important to decisions about genetic testing for cancer risk. While the discussions of the patient and community member groups weighed heavily in the considerations, input from all of the groups was considered in selecting the characteristics and levels to be used in the choice task. Among these, we selected specific test features based on two considerations: 1) frequency of mentions and 2) conceptual distinctness. This allowed us to construct choice alternatives that included as many relevant characteristics as possible while constraining the total number to reduce the cognitive burden of the choice task. In addition, we considered the potential of selected characteristics to provide information on the relative importance of one test characteristic compared to another. For example, while concerns about privacy are well known, no quantitative information is available on the importance of privacy compared to other characteristics, such as accuracy.
The final selection of levels and categories was based on both the focus group discussions and the current literature. For example, cost levels were based on what focus group members said they would pay for genetic testing and what was published in the literature on typical copayments for genetic tests to identify Lynch syndrome. In addition, categories or levels were selected to provide contrasts that had not been examined in previous studies and that would reveal new information about the characteristics. For example, privacy categories were based on focus group mentions of primary care doctors, genetic counselors and other genetic specialists, and insurance companies as potential test result recipients in addition to the person being tested. While previous surveys have suggested that the public values primary care physicians as sources of information about genetic testing, the relative value of involving primary care compared to other health professionals, such as genetic specialists, also valued for their knowledge, is not known.
The final choice alternatives included three genetic test characteristics: accuracy (false negative results), privacy (who other than the person being tested has access to the results), and cost (personal cost not covered by insurance).25,26 While test accuracy can be described in several ways, we selected one dimension of accuracy to include among the choice alternatives in order to examine comparisons between distinct characteristics. Among the dimensions of accuracy, we selected chance of false negative test result because our focus groups and prior studies have indicated that both patients and community members see as important having information about cancer or a predisposition for cancer so that something can be done and see as concerning having a cancer risk or a genetic mutation even when test results are normal.25,27,28
Each characteristic was associated with three of four levels or categories as shown in Table 1. One level or category was used in each choice task alternative. The fractional factorial design used to create the survey versions was generated using SAS Version 9.2. To create test profiles for the choice questions, we employed a D-optimal algorithm to construct a fractional factorial main-effects experimental design in SAS Version 9.2 resulting in 36 choice pairs.29-31 The final experimental design consisted of four survey versions, each containing 9 choice questions. Each respondent was randomly assigned to one of the four versions and the 9 choice questions were randomized in each survey version.
Table 1. Domains, characteristics, and levels for the choice tasks1.
| Domain | Characteristic | Levels |
|---|---|---|
| Risk2 | Chance that you will get colorectal cancer | 10 out of 100 (10%) 25 out of 100 (25%) 50 out of 100 (50%) |
| Accuracy | Chance of a false negative test result (the test result says people DO NOT HAVE the gene when people actually DO HAVE it) |
0 out of 10 times (0%) 1 out of 10 times (10%) 2 out of 10 times (20%) |
| Privacy | In addition to you, who else sees the test results | Your primary care doctor Your genetics health professionals Your life insurance and health insurance companies |
| Cost | Personal cost to you not covered by insurance | $250 $500 $1,000 or $1,5003 |
The domains, characteristics, and levels were developed using focus groups of high-risk clinic patients, average risk community members, and clinicians, and pretested using structured interviews of community members.
Risk of colorectal cancer given the presence of the genetic mutation
Half the participants saw $1,000, and half the participants saw $1,500
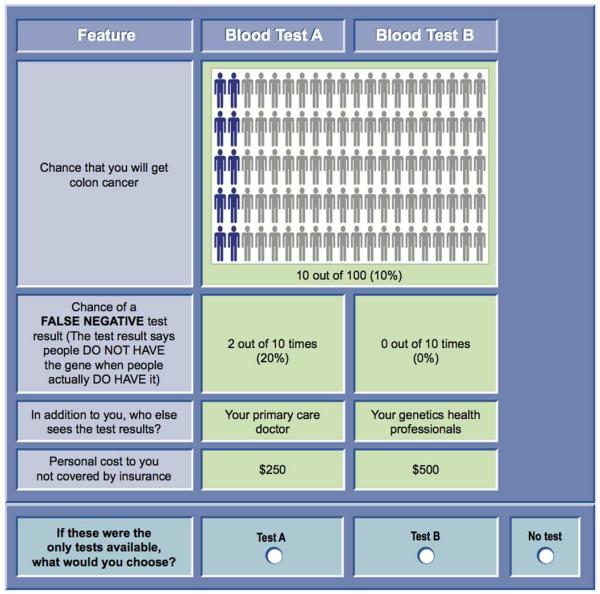
Each choice task included two hypothetical genetic test alternatives (Blood Test A, Blood Test B) and a no-test option. The “no test” option was included so we could estimate predicted test uptake. Including the “no test” option was also important in making the choice scenario more realistic to participants, as in the real-world “no test” is an option they can choose.
Each set of alternatives (Blood Test A, Blood Test B) was presented with a hypothetical level of colorectal cancer risk (i.e., 10%, 25%, 50%) (Figure 1) to provide a baseline context for each choice task. The level of was varied throughout the choice tasks in the questionnaire. In the full experimental design (36 choice questions), each of the three baseline risk levels occurred exactly 33% of the time (24/72). In addition, in each of the four versions, each baseline risk occurred 33% of the time (6/18).
Figure 1.
Choice question example.
The survey included several questions in addition to the choice tasks. Because genetic test results for colorectal cancer have consequent risk reduction recommendations, we included a separate question on choice of risk reduction strategy given hereditary colorectal cancer in the family. In this question, we asked participants to choose either colectomy to eliminate the risk, or colonoscopy to reduce the risk, of dying from colorectal cancer. If colectomy was selected, the participant was asked about the maximum that he or she would pay for a colectomy. If colonoscopy was selected, the participant was asked to identify the maximum that he or she would pay for colonoscopies over a lifetime. Several independent questions were included in the survey to measure relevant person characteristics (e.g., cancer history) that were mentioned in the focus groups, but that cannot be manipulated experimentally.
Because understanding numerical probabilities is often cognitively challenging, we used pictographs to provide graphical risk information.32 Plain language definitions and pictures were used to define concepts used in the choice tasks, such as false negative test result, genetics health professionals, colectomy, and colonoscopy.
We tested and refined the initial version of the survey using structured interviews with ten community-dwelling adults age 50 and older. We asked participants to “think aloud” as they completed the survey and then asked debriefing questions to determine whether they understood definitions and instructions and accepted the hypothetical context of the survey. Survey questions and the choice task were refined after initial feedback from the first five participants. Using the final version of the survey, we confirmed that participants saw the characteristics and levels as relevant and of concern, and were willing to accept tradeoffs among the levels of characteristics.
Based on the International Society for Pharmacoeconomics and Outcomes Research recommendations for good research practice, we used a standard algorithm to construct a fractional-factorial experimental design. This type of design is advantageous because it maximizes the statistical information obtainable from choice tasks using fewer questions than required in a full factorial design (i.e., all combinations of characteristics and levels or categories).33,34 The final survey consisted of four versions, each including nine choice tasks. Participants were randomly assigned to one of the four versions.
Each survey was designed to take approximately 20 minutes to complete. In addition to choice tasks, the survey included a written introduction to the topic of genetic testing for colorectal cancer and questions about demographics, personal and family history of cancer, experience with genetic testing, and comprehension of risk information.
The final survey was approved by the University of California at San Francisco Committee on Human Research Participation and the RTI International Office of Research Protection and Ethics (Research Triangle Park, NC). Participants were required to provide online informed consent by clicking on the statement “Yes, I agree to participate.”
Procedures
Knowledge Networks administered the Web-enabled survey to members of their online survey panel in April 2010. Using a combined random-digit dial sample from the United States landline population and an address-based sample from the United States Postal Service National Address File, the panel is a representative, probability-based sample of the United States population. Probability-based samples are preferred to non-probability based samples because of their greater representativeness of the United States population.35 Knowledge Networks panel members who do not already have Internet service are provided with Web access.
Participants were eligible for the survey if they were 50 years of age or older and a United States resident. Of the 650 members of the Knowledge Networks panel asked to participate in the survey in April 2010, 451 were determined to be eligible and completed the survey (70% participation rate). The conjoint analysis included the 355 surveys that met data quality standards needed for analytic model assumptions. Those with incomplete responses to the question on value of risk reduction with colectomy or colonoscopy (n=5) and those who selected the same genetic test alternative indiscriminately (Test A, Test B, no test) for all nine choices (n=91) were excluded from the analysis as is standard.36 These response patterns may indicate that the survey respondents were not paying attention to the choice questions. Survey pretesting demonstrated that participants similar to those in the online panel understood the instructions for the choice questions making poor comprehension of the choice task an unlikely explanation for these responses. However, because these responses inflate error in the model estimates, it is common practice to omit such participants in the analysis.
Analysis
We estimated random-parameters logit models using NLOGIT 4.0 (Econometric Software, Inc., Plainview, New York) to obtain preference parameters from the choice-format questions.37,38 This analysis yields relative preference weights for all levels of each characteristic included in the survey and estimates of the relative importance of each characteristic over the range of levels included in the survey. The observed pattern of answers to the choice questions in a conjoint survey reveals the relative importance of each characteristic to participants, the rate at which participants are willing to accept tradeoffs among characteristics and levels, and the relative value of different combinations of levels.
The final model included all of the characteristics shown in Table 1—colorectal cancer risk, accuracy, privacy, and the personal cost of the test not covered by insurance. We used effects coding in the model in order to estimate the reference levels as the sum of the rest of the preference weights for each attribute. To estimate the standard errors, we used the variance-covariance matrix. Once we obtained the standard error, we estimated the Z-score as the beta/standard error and calculated the p-value.
We obtained the participant-specific value of avoiding colorectal cancer risk by combining the specific colorectal cancer risk information shown to that participant (i.e., 10%, 25%, 50%) and that participant’s response to the questions about willingness to undergo colectomy or colonoscopy to eliminate or reduce the risk of having colorectal cancer. In the analysis, the baseline risk variable was interacted with the risk reduction level chosen in the questions about willingness to undergo colectomy or colonoscopy. This value was then used to adjust the estimate for risk of a false negative test result. This was done to provide individual-level information on the value of test accuracy, or avoiding the consequences of a false negative test.
To provide a common metric for the value of genetic test alternatives, we calculated the monetary value of genetic testing across participants as the difference in the value of a particular genetic testing alternative and the value of the no-test alternative divided by the value per dollar of cost. The overall value of genetic testing compared to no testing was calculated by setting the test characteristics (e.g., cost, privacy, accuracy) to the mean values in the experimental design. This calculation provides the mean maximum dollar amounts participants would pay for privacy (who other than the person being tested has access to genetic test results) and for test accuracy (risk of a false negative test result). The value of test accuracy was adjusted by the value of risk reduction as measured by the questions on risk reduction strategy. Thus, the value of accuracy depends partly on the perceived value of avoiding a mistake (i.e., an undetected genetic mutation).
Results
Sample Characteristics
Participant age was on average 63 years (range 50 to 96) (Table 2). More than half were white (77%), married (62%), and had at least some college education (54%). A majority had health insurance (89%) and over half had previous experience with colonoscopy (62%). Relatively few had experience with genetic testing (8%), had a personal history of cancer (14%), or had a family history of colon cancer (14%).
Table 2. Demographic and clinical characteristics.
| Characteristic | N = 355 (%)1 |
|
|---|---|---|
| Age (mean years and range) | 63 (50, 96)2 | |
|
| ||
| Gender | Male | 176 (50) |
| Female | 179 (50) | |
|
| ||
| Children | Yes | 289 (82) |
| No | 65 (18) | |
| Missing | 1 | |
|
| ||
| Race/ethnicity | White, non-Hispanic | 273 (77) |
| Black, non-Hispanic | 37 (10) | |
| Hispanic | 19 (5) | |
| Other, non-Hispanic | 14 (4) | |
| Two or more races, non- Hispanic |
12 (3) | |
|
| ||
| Educational attainment | Less than high school | 40 (11) |
| High school | 122 (34) | |
| Some college | 107 (30) | |
| Bachelors degree or higher | 86 (24) | |
|
| ||
| Marital status | Married | 218 (61) |
| Widowed | 38 (11) | |
| Divorced | 48 (14) | |
| Separated | 6 (2) | |
| Never married | 33 (9) | |
| Living with partner | 12 (3) | |
|
| ||
| Household income level | Less than $25,000 | 78 (22) |
| $25,000 to $49,999 | 98 (28) | |
| $50,000 to $74,999 | 81 (23) | |
| $75,000 to $99,999 | 47 (13) | |
| $100,000 or more | 51 (14) | |
|
| ||
| Employment status | Working – as a paid employee |
109 (31) |
| Working – self-employed | 31 (9) | |
| Not working – retired or disabled |
174 (49) | |
| Not working – other3 | 37 (11) | |
|
| ||
| Health insurance | Yes | 316 (89) |
|
| ||
| Colonoscopy ever | Yes | 219 (62) |
|
| ||
| Genetic testing ever | Yes | 29 (8) |
|
| ||
| Blood relative with colon cancer |
Yes | 49 (14) |
|
| ||
| Personal history of cancer ever | Yes | 51 (14) |
N=354 for colonoscopy, genetic testing, and cancer; N=352 for relative with colon cancer. Percent may not total to 100 due to rounding.
Range of age
Includes “not working, but looking for work,” and “not working, other”
Value of Genetic Test Characteristics
All three genetic test characteristics (i.e., cost, privacy, accuracy) were included in estimating a random-parameters logit model. Table 3 shows the logit model estimates. For each level of each characteristic, log odds (LO), also called preferences weights, are shown with their standard errors, standardized estimates (Z-scores), and p-values. Table 3 also shows the estimates of the relative importance of each characteristic relative to the other characteristics included in the model and conditional on the ranges of the levels used for the characteristics. The 95% confidence interval is shown for the relative importance weights.
Table 3. Preference weights and relative importance of characteristics.
| Characteristics | Levels | Coef1 | SE2 | Z-score3 | p | Relative Importance 4 |
95% CI5 | |
|---|---|---|---|---|---|---|---|---|
| Chance of a false negative test result (the test result says people DO NOT HAVE the gene when people actually DO HAVE it)6 |
0 out of 10 times (0%) |
0.665 | 0.084 | 7.872 | 0.000 | 1.339 | 1.021 | 1.657 |
| 1 out of 10 times (10%) |
0.009 | 0.059 | 0.149 | 0.882 | ||||
| 2 out of 10 times (20%) |
−0.674 | 0.088 | −7.647 | 0.000 | ||||
| In addition to you, who else sees the test results |
Your primary care doctor |
0.978 | 0.092 | 10.576 | 0.000 | 2.639 | 2.240 | 3.038 |
| Your genetics health professionals |
0.684 | 0.083 | 8.190 | 0.000 | ||||
| Your life insurance and health insurance companies |
−1.661 | 0.124 | −13.393 | 0.000 | ||||
| Personal cost to you not covered by insurance |
$250 | 0.496 | 0.040 | 12.251 | 0.000 | 3.302 | 2.759 | 3.845 |
| $500 | −0.165 | 0.013 | −12.251 | 0.000 | ||||
| $1,000 | −1.486 | 0.121 | −12.251 | 0.000 | ||||
| $1,500 | −2.806 | 0.229 | −12.251 | 0.000 | ||||
| Genetic testing preference7 | Test | −1.643 | 0.145 | −11.336 | 0.000 | 1.643 | 1.357 | 1.929 |
| No test | −3.286 | 0.290 | −11.336 | 0.000 | ||||
Log odds (also termed preference weights) relative to the mean effect of the characteristic, which are normalized at zero using Z-scores to clearly distinguish where the differences occur between the log odds. The marginal log odds from the random-parameters logit model can be interpreted as weights indicating the relative strength of preference for each characteristic level. With this model, the relative changes between characteristic levels are the main focus. For example, the largest non-cost improvement in genetic testing features occurs between life insurance and health insurance companies and the primary care doctor as test result recipients.
Standard error
The Z-score is the coefficient divided by the standard error. Z-scores are used to identify statistically significant differences between attribute levels. Z-score ≥1.96 corresponds to a p-value≤0.05.
Relative importance represents the weight of each characteristic (over the levels of each characteristic included in the survey), which is estimated by taking the difference in the parameter estimates between the best and worst level for each attribute.
CI=Confidence Interval
Estimate for chance of a false negative was adjusted for the value of eliminating or reducing the risk of colorectal cancer shown in each choice question (i.e., 10%, 25%, or 50%)
Comparison of no testing to the alternative of the average test
Higher log odds indicate higher preferences or values for a characteristic or levels of a characteristic. The level with the greatest value is assigned a preference weight of 10 (i.e., genetic testing results shared with primary care doctor) and the level with the lowest value ($1,500 on personal cost not covered by insurance) is assigned a preference weight of 0. Usually, the highest and lowest coefficients coincide with the same attribute and we can scale between 0 and 10 for the most important attribute. In this model, the highest and lowest coefficient occurred for different attributes and we used the same process as we would if they occurred for the same attribute. All other characteristic levels were scaled relative to these two levels. Characteristic levels that are more highly valued have higher preference weights than those that are less valued.
Estimated preference weights for personal cost not covered by insurance and risk of false negative results were consistent with the natural ordering of these categories. That is, lower cost was valued more than higher cost and better clinical outcomes were valued more than worse clinical outcomes. For privacy, genetic testing results shared with primary care doctors and genetics health professionals (LO=0.978 and 0.684, respectively) were more highly valued than results shared with life and health insurance companies (LO=−1.661). Also, participants preferred the average test shown to no test (LO=−1.643 versus −3.286). The no-test option was preferred in only 13.6% of the choices.
The relative importance of each characteristic can be interpreted as indicating the relative value for each characteristic level compared to the mean effect of that characteristic and considering the other characteristics and levels and categories included in the model. For effect-coded characteristics (e.g., privacy) the mean effect is zero. For cost, which was modeled as linear, the mean effect is the mean level shown for cost in the survey. For the interaction of risk reduction and baseline risk on the false negative levels, the mean effect is the effect of the combined variable. Personal cost of genetic testing not covered by insurance (LO=3.302; CI=2.759; 3.845) was considered most important relative to the other characteristics. Among the clinical characteristics, privacy (LO=2.639; 95% CI=2.240; 3.038) was relatively more important than accuracy (LO=1.339; CI=1.021; 1.657).
The overall monetary value of testing relative to no testing was $622 (95% CI, $476, $778) (Table 4). We also calculated the marginal monetary value for any change in preference weight from a less valued to a more valued level of accuracy and privacy. The largest marginal value was $999 (95% CI=$815, $1193) for sharing results with primary care doctors compared to insurance companies. The value of sharing test results with genetics health professionals compared to insurance companies was somewhat less at $888 (95% CI=$719, $1,063). The value of reducing the chance of a false negative test result from 20% to 0 and from 10% to 0 was $507 (95% CI=$379, $640) and $248 (95% CI=$161, $338), respectively.
Table 4. Monetary value of test characteristics.
| Characteristic, Comparison1 | Mean2 (95% CI3) |
|---|---|
| Chance of a false negative test result | |
| 1 out of 10 times (10%) to 0 out of 10 times (0%) | $248 ($161, $338) |
| 2 out of 10 times (20%) to 0 out of 10 times (0%) | $507 ($379, $640) |
| 2 out of 10 times (20%) to 1 out of 10 times (10%) | $258 ($164, $356) |
| Who else sees the test results | |
| Genetics health professionals to primary care doctor | $111 ($18, $208) |
| Life insurance and health insurance companies to primary care doctor |
$999 ($815, $1,193) |
| Life insurance and health insurance companies to genetics health professionals |
$888 ($719, $1,063) |
| Genetic-testing preference | |
| Test vs. no test | $622 ($476, $778) |
A comparison between levels of a characteristic (e.g., 20% to 10% false negative test result)
Mean value in dollars of an improvement from one level to another of a characteristic. For example, the mean value in dollars of disclosing genetic test results to a primary care doctor rather than a genetics health professional is $111.
CI=Confidence Interval
Genetic Testing Intentions
When cost was set at $500, almost all participants (97%, 95% CI=95%, 99%) would be willing to have genetic testing done given the best scenario—results shared with the primary care doctor and no chance of a false negative test. In contrast, less than half (41%, 95% CI=25%, 57%) would be tested with results disclosed to life and health insurance companies and a 20% chance of a false negative test.
Discussion
In this study of adults in a probability-based online panel drawn from the US general population, we found strong interest in genetic testing to identify hereditary colorectal cancer risk when personal cost not covered by insurance was set at $500 and given the best test features included in the choice task—results shared with primary care physician and no chance of a false negative test result. In this situation, almost all (97%) would have testing, similar to the interest expressed in members of families newly identified with hereditary colorectal cancer.16 In contrast, when genetic information would be released to life and health insurance companies and the chance of a false negative result was 20%, interest in genetic testing fell to less than half of the sample. In analyses that included the personal cost of testing not covered by insurance, both cost and privacy were the most important factors in test decisions.
Acceptability of genetic testing is an important question studied primarily in families at risk for hereditary cancers,15,39 but is not well understood in the general population. In our study, we found that US adults would pay on average $622 in out-of-pocket costs for genetic testing. A population-based study in Canada found that fewer (27%) would pay more than $500 in out-of-pocket costs for genetic testing.40 In contrast to samples of adults from the general population in the US and in Canada, patients at low, medium, and high risk for breast cancer in Great Britain would pay up to 3,000 pounds sterling (approximately $4,850) for genetic testing, a value above the cost of testing to the National Health Plan.41
Despite public enthusiasm for the use of genomics to individualize health care,13,42 there are concerns about the privacy of genetic tests.16,43,44 The Genetics Information and Nondiscrimination Act (GINA) was enacted in 2008 to supplement existing state law on genetic discrimination, providing a minimum level of protection against health insurance and employment discrimination based on genetics.45 The findings of our population-based choice-format survey are consistent with recent public opinion studies suggesting ongoing concerns about the privacy of genetic information not fully answered by recent policy.46
One of the central findings of our work is that adults from the general population value the participation of primary care in the genetic testing process as much as they value genetics specialists (e.g., genetic counselors) as compared to disclosure of genetic test results to insurance companies. Our results quantify the relative value of primary care professionals compared to genetic specialists, information not captured in opinion surveys indicating general positive attitudes toward primary care involvement in genetic consultation.47 Because the work force of genetics professionals is limited,48 and patient and public knowledge of hereditary cancer and appropriate surveillance is incomplete,49,50 primary care has been identified as a potential resource in the delivery of genomic health services.51 Evidence that primary care professionals are not yet fully prepared to discuss the costs and benefits of genetic testing with their patients suggests an opportunity for professional development.52-55
Our results should be interpreted in light of several considerations. First, our study provides information on stated choices and not real-life decisions about genetic testing. However, our findings can be seen as indicating the perceived value of genetic testing to members of the general public, and this information is necessary for decision analyses and policy development. Second, while the sample size is small, according to published guidelines for choice-format conjoint analyses, the sample is more than sufficient to conduct the tests based on the experimental design.34 Further, the small confidence intervals associated with our estimates suggest robust results. Third, our results are not generalizable to the general US population. While our sample was recruited from a probability-based online panel of US adults, it is not representative of the entire US population possibly considering genetic testing. Fourth, as in other choice-format studies, our findings on the importance of each characteristic should be interpreted as relative to the other characteristics and levels included in the survey. While additional research will be needed to clarify the relative value of characteristics not included in this study, the rigorous methods used to develop and pretest the survey should increase confidence in the external validity of our findings and should provide a strong basis for future studies of test characteristics and other factors.
This study of adults from the general population identified out-of-pocket cost of genetic testing and the privacy of the results as the most important characteristics associated with genetic testing. However, controlling for cost, most appear willing to be tested to reduce the risk of morbidity and mortality from a heritable condition if the test results are shared with the person’s primary care physician and the chance of a false negative test result is low. As genetic testing becomes more widely available for many conditions, primary care professionals will need to be increasingly prepared for discussing decisions about genetic testing with patients, and guiding them through appropriate subspecialty consultations.
Acknowledgements
This work was supported with a grant from the National Cancer Institute (P01CA130818-1A1) Phillips, Principal Investigator, Walsh and Knight, Co-Leaders of the Preferences Project. This paper represents the perspective of the authors and does not necessarily represent the views of the Department of Veterans Affairs or the United States Government. The authors would like to thank F. Reed Johnson, Ryan Ziemiecki, Vikram Kilambi, and Lauren Donnalley from Research Triangle Institute for their assistance. All authors had full access to the data presented in this manuscript and the first and second authors take responsibility for the integrity of the data and the accuracy of the data analysis.
Financial support for this study was provided by a grant from the National Cancer Institute. The funding agreement ensured the authors’ independence in designing the study, interpreting the data, writing, and publishing the report.
Footnotes
Uri Ladabaum, MD, MS, declares a potential perceived conflict of interest with Exact Sciences Corporation (Madison, WI) and Given Imaging Ltd. (Duluth, GA)/Covidien Plc (Dublin, Ireland) for whom he has served as consultant, and Mauna Kea Technologies Inc. (Suwanee, GA), for whom he has served on an advisory board.
References
- 1.Andrykowski M, Munn R, Studts J. Interest in learning of personal genetic risk for cancer: a general population survey. Prev Med. 1996;25(5):527–536. doi: 10.1006/pmed.1996.0086. [DOI] [PubMed] [Google Scholar]
- 2.Rose A, Peters N, Shea J, Armstrong K. The association between knowledge and attitudes about genetic testing for cancer risk in the United States. J Health Commun. 2005;10(14):309–321. doi: 10.1080/10810730590950039. [DOI] [PubMed] [Google Scholar]
- 3.Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med. 2009 Jan;11(1):35–41. doi: 10.1097/GIM.0b013e31818fa2ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terdiman JP. It is time to get serious about diagnosing Lynch syndrome (hereditary nonpolyposis colorectal cancer with defective DNA mismatch repair) in the general population. Gastroenterology. 2005 Aug;129(2):741–744. doi: 10.1016/j.gastro.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 5.Engstrom P. Update: NCCN colon cancer Clinical Practice Guidelines. J Natl Compr Canc Netw. 2005 Nov;3(Suppl 1):S25–28. [PubMed] [Google Scholar]
- 6.Julie C, Tresallet C, Brouquet A, et al. Identification in daily practice of patients with Lynch syndrome (hereditary nonpolyposis colorectal cancer): revised Bethesda guidelines-based approach versus molecular screening. Am J Gastroenterol. 2008 Nov;103(11):2825–2835. doi: 10.1111/j.1572-0241.2008.02084.x. quiz 2836. [DOI] [PubMed] [Google Scholar]
- 7.Levin B, Barthel JS, Burt RW, et al. Colorectal Cancer Screening Clinical Practice Guidelines. J Natl Compr Canc Netw. 2006 Apr;4(4):384–420. doi: 10.6004/jnccn.2006.0033. [DOI] [PubMed] [Google Scholar]
- 8.American Society of Clinical Oncology policy statement update: genetic testing for cancer susceptibility. J Clin Oncol. 2003 Jun 15;21(12):2397–2406. doi: 10.1200/JCO.2003.03.189. [DOI] [PubMed] [Google Scholar]
- 9.Mvundura M, Grosse SD, Hampel H, Palomaki GE. The cost-effectiveness of genetic testing strategies for Lynch syndrome among newly diagnosed patients with colorectal cancer. Genet Med. 2010 Feb;12(2):93–104. doi: 10.1097/GIM.0b013e3181cd666c. [DOI] [PubMed] [Google Scholar]
- 10.Dinh TA, Rosner BI, Atwood JC, et al. Health benefits and cost-effectiveness of primary genetic screening for Lynch syndrome in the general population. Cancer Prev Res (Phila) 2011 Jan;4(1):9–22. doi: 10.1158/1940-6207.CAPR-10-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ladabaum U, Wang G, Terdiman J, et al. Strategies to identify the Lynch syndrome among patients with colorectal cancer: a cost-effectiveness analysis. Ann Intern Med. 2011 Jul 19;155(2):69–79. doi: 10.7326/0003-4819-155-2-201107190-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindor N, Petersen G, Hadley D, et al. Reccomendations for the care of individuals with an inherited predisposition to Lynch syndrome. JAMA. 2006;296(12):1507–1517. doi: 10.1001/jama.296.12.1507. [DOI] [PubMed] [Google Scholar]
- 13.Sanderson SC, Wardle J, Jarvis MJ, Humphries SE. Public interest in genetic testing for susceptibility to heart disease and cancer: a population-based survey in the UK. Prev Med. 2004 Sep;39(3):458–464. doi: 10.1016/j.ypmed.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 14.Armstrong K, Calzone K, Stopfer J, Fitzgerald G, Coyne J, Weber B. Factors associated with decisions about clinical BRCA1/2 testing. Cancer Epidemiol Biomarkers Prev. 2000 Nov;9(11):1251–1254. [PubMed] [Google Scholar]
- 15.Gritz ER, Peterson SK, Vernon SW, et al. Psychological impact of genetic testing for hereditary nonpolyposis colorectal cancer. J Clin Oncol. 2005 Mar 20;23(9):1902–1910. doi: 10.1200/JCO.2005.07.102. [DOI] [PubMed] [Google Scholar]
- 16.Hadley DW, Jenkins J, Dimond E, et al. Genetic counseling and testing in families with hereditary nonpolyposis colorectal cancer. Arch Intern Med. 2003 Mar 10;163(5):573–582. doi: 10.1001/archinte.163.5.573. [DOI] [PubMed] [Google Scholar]
- 17.Lerman C, Hughes C, Trock BJ, et al. Genetic testing in families with hereditary nonpolyposis colon cancer. Jama. 1999 May 5;281(17):1618–1622. doi: 10.1001/jama.281.17.1618. [DOI] [PubMed] [Google Scholar]
- 18.Scheuner MT, Sieverding P, Shekelle PG. Delivery of genomic medicine for common chronic adult diseases: a systematic review. Jama. 2008 Mar 19;299(11):1320–1334. doi: 10.1001/jama.299.11.1320. [DOI] [PubMed] [Google Scholar]
- 19.Phillips KA. Closing the evidence gap in the use of emerging testing technologies in clinical practice. Jama. 2008 Dec 3;300(21):2542–2544. doi: 10.1001/jama.2008.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bridges JF. Stated preference methods in health care evaluation: an emerging methodological paradigm in health economics. Appl Health Econ Health Policy. 2003;2(4):213–224. [PubMed] [Google Scholar]
- 21.Fraenkel L. Conjoint Analysis at the Individual Patient Level: Issues to Consider as We Move from a Research to a Clinical Tool. Patient. 2008 Oct 1;1(4):251–253. doi: 10.2165/1312067-200801040-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall DA, Johnson FR, Phillips KA, Marshall JK, Thabane L, Kulin NA. Measuring patient preferences for colorectal cancer screening using a choice-format survey. Value in Health. 2007 Sep-Oct;10(5):415–430. doi: 10.1111/j.1524-4733.2007.00196.x. [DOI] [PubMed] [Google Scholar]
- 23.Phillips KA, Johnson FR, Maddala T. Measuring what people value: a comparison of “attitude” and “preference” surveys. Health Serv Res. 2002 Dec;37(6):1659–1679. doi: 10.1111/1475-6773.01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green P. Design of Choice Experiments Involving Multifactor Alternatives. Journal of Consumer Research. 1974;1(2):61–68. [Google Scholar]
- 25.Walsh J, Arora M, Hosenfeld C, Ladabaum U, Kuppermann M, Knight SJ. Preferences for genetic testing to identify hereditary colorectal cancer: perspectives of high-risk patients, community members, and clinicians. J Cancer Educ. 2012 Mar;27(1):112–119. doi: 10.1007/s13187-011-0286-z. [DOI] [PubMed] [Google Scholar]
- 26.Issa AM, Tufail W, Hutchinson J, Tenorio J, Baliga MP. Assessing patient readiness for the clinical adoption of personalized medicine. Public Health Genomics. 2009;12(3):163–169. doi: 10.1159/000189629. [DOI] [PubMed] [Google Scholar]
- 27.Michie S, Weinman J, Miller J, Collins V, Halliday J, Marteau TM. Predictive genetic testing: high risk expectations in the face of low risk information. J Behav Med. 2002 Feb;25(1):33–50. doi: 10.1023/a:1013537701374. [DOI] [PubMed] [Google Scholar]
- 28.Michie S, Smith JA, Senior V, Marteau TM. Understanding why negative genetic test results sometimes fail to reassure. Am J Med Genet A. 2003 Jun 15;119A(3):340–347. doi: 10.1002/ajmg.a.20200. [DOI] [PubMed] [Google Scholar]
- 29.Johnson FR, Lancsar E, Marshall D, et al. Constructing Experimental Designs for Discrete-Choice Experiments: Report of the ISPOR Conjoint Analysis Experimental Design Good Research Practices Task Force. Value in health: the journal of the International Society for Pharmacoeconomics and Outcomes Research. 2013;16(1):3–13. doi: 10.1016/j.jval.2012.08.2223. [DOI] [PubMed] [Google Scholar]
- 30.Kuhfeld WF, Tobias RD, Garratt M. Efficient Experimental Design with Marketing Research Applications. Journal of Marketing Research. 1994;31(4):545–557. [Google Scholar]
- 31.Kuhfeld W. Marketing research methods in SAS: experimental design, choice, conjoint, and graphical techniques. SAS Institute Inc.; Cary, NC: 2010. [Google Scholar]
- 32.Hawley ST, Volk RJ, Krishnamurthy P, Jibaja-Weiss M, Vernon SW, Kneuper S. Preferences for colorectal cancer screening among racially/ethnically diverse primary care patients. Med Care. 2008 Sep;46(9 Suppl 1):S10–16. doi: 10.1097/MLR.0b013e31817d932e. [DOI] [PubMed] [Google Scholar]
- 33.Kanninen BJ. Optimal design for multinomial choice experiments. Journal of Marketing Research. 2002 May;39(2):214–227. [Google Scholar]
- 34.Marshall DA, Hauber AB, Bridges JF, et al. Assessing the quality of conjoint analysis applications in health: A pilot evaluation of the ISPOR checklist for good research practice in conjoint analysis. Value in Health. 2009 May;12(3):A31–A31. [Google Scholar]
- 35.Baker R, Blumberg SJ, Brick JM, et al. Research Synthesis: American Association of Public Opinion Research (AAPOR) Report on Online Panels. Public Opin. Q. 2010 Win74(4):711–781. [Google Scholar]
- 36.Johnson FR, Ozdemir S, Mansfield C, et al. Crohn’s disease patients’ risk-benefit preferences: serious adverse event risks versus treatment efficacy. Gastroenterology. 2007 Sep;133(3):769–779. doi: 10.1053/j.gastro.2007.04.075. [DOI] [PubMed] [Google Scholar]
- 37.Train K. Discrete choice methods with simulation. Cambridge University Press; Cambridge: 2003. [Google Scholar]
- 38.Train K, Sonnier G. Mixed logit with bounded distributions of correlated partworths. In: Scarpa R, Alberini A, editors. Applications of simulation methods in environmental and resource economics. Springer Publisher; Dordrecht, Netherlands: 2005. [Google Scholar]
- 39.Manne SL, Chung DC, Weinberg DS, et al. Knowledge and attitudes about microsatellite instability testing among high-risk individuals diagnosed with colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2007 Oct;16(10):2110–2117. doi: 10.1158/1055-9965.EPI-07-0412. [DOI] [PubMed] [Google Scholar]
- 40.Ries NM, Hyde-Lay R, Caulfield T. Willingness to pay for genetic testing: a study of attitudes in a Canadian population. Public Health Genomics. 2010;13(5):292–300. doi: 10.1159/000253120. [DOI] [PubMed] [Google Scholar]
- 41.Griffith GL, Edwards RT, Williams JM, et al. Patient preferences and National Health Service costs: a cost-consequences analysis of cancer genetic services. Fam Cancer. 2009;8(4):265–275. doi: 10.1007/s10689-008-9217-5. [DOI] [PubMed] [Google Scholar]
- 42.Graham ID, Logan DM, Hughes-Benzie R, et al. How interested is the public in genetic testing for colon cancer susceptibility? Report of a cross-sectional population survey. Cancer Prev Control. 1998 Aug;2(4):167–172. [PubMed] [Google Scholar]
- 43.Freedman AN, Wideroff L, Olson L, et al. US physicians’ attitudes toward genetic testing for cancer susceptibility. Am J Med Genet A. 2003 Jul 1;120(1):63–71. doi: 10.1002/ajmg.a.10192. [DOI] [PubMed] [Google Scholar]
- 44.Hall M, Olopade OI. Confronting genetic testing disparities: knowledge is power. Jama. 2005 Apr 13;293(14):1783–1785. doi: 10.1001/jama.293.14.1783. [DOI] [PubMed] [Google Scholar]
- 45.Payne PW, Jr., Goldstein MM, Jarawan H, Rosenbaum S. Health insurance and the Genetic Information Nondiscrimination Act of 2008: implications for public health policy and practice. Public Health Rep. 2009 Mar-Apr;124(2):328–331. doi: 10.1177/003335490912400222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haga SB, O’Daniel JM, Tindall GM, Lipkus IR, Agans R. Survey of US public attitudes toward pharmacogenetic testing. Pharmacogenomics J. 2011 Feb 15; doi: 10.1038/tpj.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller FA, Carroll JC, Wilson BJ, et al. The primary care physician role in cancer genetics: a qualitative study of patient experience. Fam Pract. 2010 Oct;27(5):563–569. doi: 10.1093/fampra/cmq035. [DOI] [PubMed] [Google Scholar]
- 48.Chou AF, Norris AI, Williamson L, Garcia K, Baysinger J, Mulvihill JJ. Quality assurance in medical and public health genetics services: a systematic review. Am J Med Genet C Semin Med Genet. 2009 Aug 15;151C(3):214–234. doi: 10.1002/ajmg.c.30219. [DOI] [PubMed] [Google Scholar]
- 49.Baer HJ, Brawarsky P, Murray MF, Haas JS. Familial risk of cancer and knowledge and use of genetic testing. J Gen Intern Med. 2010 Jul;25(7):717–724. doi: 10.1007/s11606-010-1334-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh H, Schiesser R, Anand G, Richardson PA, El-Serag HB. Underdiagnosis of Lynch syndrome involves more than family history criteria. Clin Gastroenterol Hepatol. 2010 Jun;8(6):523–529. doi: 10.1016/j.cgh.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Armstrong K, Weber B, Ubel PA, Guerra C, Schwartz JS. Interest in BRCA1/2 testing in a primary care population. Prev Med. 2002 Jun;34(6):590–595. doi: 10.1006/pmed.2002.1022. [DOI] [PubMed] [Google Scholar]
- 52.Bethea J, Qureshi N, Drury N, Guilbert P. The impact of genetic outreach education and support to primary care on practitioner’s confidence and competence in dealing with familial cancers. Community Genet. 2008;11(5):289–294. doi: 10.1159/000121400. [DOI] [PubMed] [Google Scholar]
- 53.Haga SB, Carrig MM, O’Daniel JM, et al. Genomic Risk Profiling: Attitudes and Use in Personal and Clinical Care of Primary Care Physicians Who Offer Risk Profiling. J Gen Intern Med. 2011 Feb 11; doi: 10.1007/s11606-011-1651-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schroy PC, 3rd, Barrison AF, Ling BS, Wilson S, Geller AC. Family history and colorectal cancer screening: a survey of physician knowledge and practice patterns. Am J Gastroenterol. 2002 Apr;97(4):1031–1036. doi: 10.1111/j.1572-0241.2002.05624.x. [DOI] [PubMed] [Google Scholar]
- 55.Wideroff L, Vadaparampil ST, Greene MH, Taplin S, Olson L, Freedman AN. Hereditary breast/ovarian and colorectal cancer genetics knowledge in a national sample of US physicians. J Med Genet. 2005 Oct;42(10):749–755. doi: 10.1136/jmg.2004.030296. [DOI] [PMC free article] [PubMed] [Google Scholar]