Abstract
In this paper, we propose a new method for stitching multiple fluoroscopic images taken by a C-arm instrument. We employ an X-ray radiolucent ruler with numbered graduations while acquiring the images, and the image stitching is based on detecting and matching ruler parts in the images to the corresponding parts of a virtual ruler. To achieve this goal, we first detect the regular spaced graduations on the ruler and the numbers. After graduation labeling, for each image, we have the location and the associated number for every graduation on the ruler. Then, we initialize the panoramic X-ray image with the virtual ruler, and we “paste” each image by aligning the detected ruler part on the original image, to the corresponding part of the virtual ruler on the panoramic image. Our method is based on ruler matching but without the requirement of matching similar feature points in pairwise images, and thus, we do not necessarily require overlap between the images. We tested our method on eight different datasets of X-ray images, including long bones and a complete spine. Qualitative and quantitative experiments show that our method achieves good results.
Keywords: Radiographic image enhancement, Image fusion, Image processing, Image stitching, Number detection, Long bone, Spine
Introduction
Fluoroscopic X-ray images play a key role in a variety of surgical interventions such as long bone fracture reduction and spinal deformity corrections. In long bone fracture reduction, a mobile fluoroscopic X-ray machine called C-arm is typically used in the operating room (OR) to achieve a well-aligned reduction. However, malalignment after long bone fracture fixation has been reported to be up to 28 % [1]. The main reason is attributed to insufficient intraoperative visualization of the entire limb axis [1]. Spinal deformity correction is another type of surgery where the C-arm is used a lot. Such a surgery usually involves corrective maneuvers which aim to improve the sagittal and/or coronary profile. However, especially in longer instrumentation intraoperative estimation of the amount of correction is difficult. Mostly, anteroposterior (AP) and lateral (LAT) fluoroscopic images are used but have the disadvantage to depict only a small portion of the spine in a single C-arm image due to the limited field of view of a C-arm machine. Thus, multiple images have to be acquired and fused in the brain of the surgeon to assess spinal balance. Unfortunately, this method is prone to mistakes due to distortion and imprecise handling of the C-arm. Therefore, it would be of great help to provide accurate image fusion of the multiple small fluoroscopic images to be able to display the entire spine at once.
In literature, there exists some work for obtaining panoramic X-ray images from multiple intraoperatively acquired C-arm images. These methods can be largely classified into two categories: those that require a significant modification of the C-arm machine [2] and those that try to solve the problem by purely image registration and panorama creation techniques developed in the computer vision field [3], therefore avoiding significant modification of the hardware [4–8]. Examples of methods in the first categories include the method introduced by Wang et al. [2], where parallax-free panoramic X-ray images were generated by enabling the C-arm to rotate around its X-ray source center, relative to the patient’s table. Due to the requirement of modification of the hardware for the methods in the first category, methods in the second category are often used. For example, Yaniv and Jokowicz [4] introduced a method to obtain an X-ray panoramic image intraoperatively. Their method depends on a feature-based alignment of the detected graduations of an radiolucent X-ray ruler and they reported their evaluation results only on long bones, i.e., humerus, femur, and tibia. Although methods in this category have the advantage that they do not need a significant modification of the hardware and thus can be used for any type of C-arm, one of the main limitations common to all the methods in this category is that they require overlap between neighboring images used to generate the final stitched image. Such a constraint may lead to longer tedious image acquisition maneuvers and larger radiation exposure. In this paper, we propose a new method for stitching multiple fluoroscopic images. We employ an X-ray radiolucent ruler with numbered graduations while acquiring the images, and the image stitching is based on detecting and matching corresponding ruler parts. The pipeline of our method is shown in Fig. 1. During image acquisition, we utilize a radiolucent X-ray ruler, and we get multiple images of an anatomical structure. Then, on each image, we perform the ruler detection. Specifically, we detect the graduations on the ruler and the numbers and then all graduations are labeled with an associated number. Finally, to stitch the images together, we first create an empty stitched image with the entire virtual ruler, and then, for each original image, we “paste” it onto the stitched image by aligning the detected ruler part on that image, to the corresponding part on the virtual ruler on the stitched image.
Fig. 1.
Pipeline of our method. For clarity, only two images are used in this example
Methods
Image Acquisition
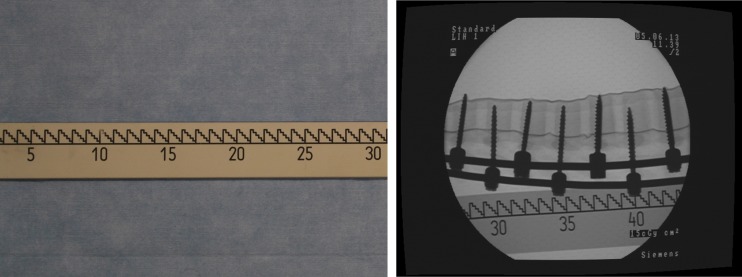
The X-ray images of this study are acquired using the Siemens SIREMOBIL IsoC3D C-arm (Siemens, Erlangen, Germany). During image acquisition, we utilize a radiolucent X-ray ruler as shown in Fig. 2 as the image stitching reference and follow the image acquisition protocol as suggested in Yaniv and Joskowicz [4] to acquire images. The ruler has a length of 125 cm and a major graduation interval as 1 cm. Moreover, numbers are present on the ruler for every 5 cm. During the acquisition, the ruler is encapsulated inside a sterilized plastic bag and is placed approximately parallel to the anatomical structure being imaged. We further require that the relative position between the ruler and the imaged anatomical structure be fixed. We also require that in each image, at least one graduation number is visible (to seed the number labeling of the graduations, as explained later).
Fig. 2.
Image acquisition. Left the radiolucent X-ray ruler. Right example X-ray image taken with the ruler
Note that our method does not rely on the pairwise stitching of successive images; therefore, we make no requirement about the overlap between the images while acquiring images.
Ruler Detection
In this subsection, we present our method for ruler detection, which is performed in two steps: graduation detection and number detection.
Graduation Detection
To detect the graduations in the image, we first detect the main line on the ruler, as in Fig. 3a. We detect lines in the image using the standard Hough Transform algorithm and detect the corners using Harris method [9]. The detected lines and corners are shown as yellow lines and circles in Fig. 3a, respectively. As we can see, due to the graduation, many corners are detected along the main line. Therefore, the main line (the red line) is identified as the line along which the points have the minimal average distance to the closest corner point.
Fig. 3.
Graduation detection. a Main line detection. b Variation of image intensity along a line slightly above the main line. c The final result of graduation detection
After the main line is identified, the image is rotated accordingly to make the main line horizontal. Thus, the rotation variation is eliminated. Then, we take a parallel line which is slightly above the main line (we use gap of 5 pixels in our implementation), as in Fig. 3b. We can see that the image intensity varies periodically along this line due to the regular ruler pattern and get local minima when we encounter the graduation. Therefore, the graduation locations are identified by nonmaximum suppression on the intensity curve over the line. Figure 3c shows the final result of graduation detection.
Number Detection
To detect the numbers in the image, we first detect all the digits from “1”,“2”,…,“0”. To this end, we employ Histogram of Oriented Gradients (HoG) feature [10] and Random Forest (RF) classifier [11].
For each digit, we collect a few (around 10) samples by manually annotating the bounding boxes on a set of training images. In addition, we create a “bg” class which represents the non-number backgrounds. We collect 1000 samples for the bg class by randomly sampling small patches in the non-number regions of the training images. Therefore, our training data contains 11 classes (10 digits + “bg”), as shown in Fig. 4a.
Fig. 4.
Number detection. a The training data for the 10 digits and a “bg” class. b Merging digits into numbers and labeling the closest graduations. c All the graduations are labeled with a corresponding number
For each training image patch, we calculate multilevel HoG as the visual feature. Specifically, each patch is divided into two levels of blocks (1 x 1 and 2 x 2), where each block is further uniformly divided into 2 x 2 cells. For each cell, the HoG feature is calculated by histogramming the gradient direction of each pixel into 18 directional bins. Then, the feature of each cell is concatenated to form the final feature vector. Therefore, finally, each image patch is represented by a multilevel HoG feature vector of (1 x 1 + 2 x 2) x 4 x 18 = 360 dimensions.
Then, for each number class, we train a “one v.s. all” binary RF classifier. For example, when we train the RF classifier of class “4”, we use the training samples of class “4” as the positive samples, and the samples of classes “1”…“3”,“5”…“0” and “bg” as negative samples. After training, we got an RF classifier for each digit. Given a new visual feature, the RF classifier will tell us the probability that this feature corresponds to an instance of the digit class which it represents.
During the testing stage, given a new image, we detect the digits using the sliding-window method. We regularly sample detection windows throughout the image.1 For each detection window, we calculate the multilevel HoG feature, and the RF classifiers for the 10 digits will give 10 scores. In this way, after all detection windows are processed, we essentially get the score map for each of the 10 digits on every location of the image. Based on these response images, we perform a thresholded nonmaximum suppression which returns the bounding boxes of the detected numbers.
After all the digits are detected, we merge the digits to numbers based on the location proximity in the image, and then, the graduation which is closest to the center of the number is associated with this number, as shown in Fig. 4b.
Finally, using the already associated graduations as seeds, all the other graduations are labeled with their corresponding numbers, as in Fig. 4c.
Image Stitching
After graduation detection and number detection, for every image, we know the location of every graduation and its corresponding number. To stitch the image, we first initialize an empty image with sufficient size. We establish a virtual ruler in this stitched image, and then, each original image is pasted into this stitched image by aligning the detected graduations in the image and the corresponding graduations on the virtual ruler in the stitched image. The alignment is calculated by the optimal similarity (rigid + scale) transformation between the corresponding graduation coordinates in the original image and the stitched image. This process is visualized in Fig. 1 bottom row.
Results
Experimental Setup
In this paper, we conduct our study using the following four structures:
Cadever Femur: a complete cadaver femur.
Lower Extremity: a complete plastic leg including both femur and tibia.
Plastic Femur: a complete plastic femur.
Spine: a complete spine phantom with deformity correction devices.
Each of the four structures are imaged in this way twice both in AP and LAT views, yielding eight datasets (image sequences) as summarized in Table 1, based on which we perform our image stitching experiments.
Table 1.
Summary of the eight datasets (image sequences) used in our study
| Structure | View direction | Number of images |
|---|---|---|
| Cadaver Femur | AP | 12 |
| Cadaver Femur | LAT | 8 |
| Lower Extremity | AP | 16 |
| Lower Extremity | LAT | 16 |
| Plastic Femur | AP | 11 |
| Plastic Femur | LAT | 9 |
| Spine | AP | 16 |
| Spine | LAT | 14 |
Because our method does not require overlap between the images, for each datasets, we perform our image stitching method on four different levels. The first level uses all the available images in the dataset which involves most overlap. The second, third, and fourth levels use every two, three, and four images, respectively. Therefore, in the higher levels, the overlap is reduced, and often in the last (fourth) level, there is no image overlap at all. In the following, we present our results using all the datasets on all levels.
Qualitative Results
The qualitative results on our datasets at different levels are shown in Fig. 5. Due to the page limit, we only show five out of the eight datasets. We observe similar results on the other datasets. Visually, we can see that our method generates satisfactory results: The bone structures and the ruler show continuous contours. Also, note that our approach generates good results on all levels, demonstrating that we do not require image overlap. The feedback from several orthopedic surgeons about our image stitching results indicates that they are accurate enough for clinical usage. Please note that the holes (white dots) seen from the stitching results of the Cadaver Femur are not caused by our method. This is because this bone was used to test screw fixation before and thus the holes were created after removing those screws from the bone.
Fig. 5.
Qualitative results of image stitching on four datasets
Quantitative Results
Distance Measurement
For quantitative evaluation, we conducted distance measurements and angle measurements. The distance measurement evaluation is conducted by measuring the physical distance on all the anatomical structures, which are regarded as the ground truth, and then comparing them with distance measurements on the stitched images. In order to get robust physical distance measurements, we have chosen different anatomical landmarks for different structures. The same landmarks are then used to get the distance measurements on the stitched images. Table 2 summarizes the results. A mean error of 1.5 ± 0.9 mm was found.
Table 2.
The ground truth and the distances measured on the stitched images for different anatomical structures, in millimeters
| Structure | View | Ground truth | Measured from stitched images | Error |
|---|---|---|---|---|
| Cadaver Femur | AP | 432 | 432 | 0 |
| Cadaver Femur | LAT | 440 | 442 | 2 |
| Lower Extremity | AP | 834 | 832 | 2 |
| Lower Extremity | LAT | 830 | 832 | 2 |
| Plastic Femur | AP | 458 | 460 | 2 |
| Plastic Femur | LAT | 380 | 382 | 2 |
| Spine | AP | 486 | 484 | 2 |
| Spine | LAT | 730 | 730 | 0 |
Angle Measurement
The angle measurement evaluation was conducted by taking the angle of anatomical landmarks depending on the acquisition orientation and the anatomical structure, as described in [12]. For all anatomic structures both in AP and LAT, except the spine, we have measured the angle (alpha) between the proximal anatomic axis (PAA) and distal anatomic axis (DAA). For the AP dataset of the lower extremity, we have measured the anatomic tibofemural angle (ATA) between the femur and the tibia; for the femur in AP position, we have measured the neck shaft angle (NSA); and for the tibia in AP position, we have measured the medial proximal tibial angle (MPTA). We have repeated the angle measurements for all levels and performed one sample t test, taking the level 1 measured angles as a value for a normal distribution for testing the null hypothesis of the measured angles of the remaining levels. The significant level was chosen as α = 0.05. Table 3 summarizes the results of the angle measurements and the t test. The results showed that no statistically significant difference was found (all p values were greater than 0.05). Thus, we can conclude that our method aligns the images correctly even when we do not have significant overlap.
Table 3.
Qualitative angular measurement
| Dataset | Angle | Level 1 | Level 2 | Level 3 | Level 4 | p-value |
|---|---|---|---|---|---|---|
| Lower extremity_AP | ATA | 0.6261 | 0.6266 | 0.6264 | 0.6269 | 0.0669 |
| Lower extremity _AP | Femur_alpha | 0.4298 | 0.6357 | 0.5951 | 0.4735 | 0.105 |
| Lower extremity_AP | Tibia_alpha | 0.1511 | 1.2323 | 1.0781 | 0.4615 | 0.0816 |
| Lower extremity_AP | NSA | 124.3893 | 123.9170 | 124.8024 | 123.9608 | 0.6294 |
| Lower extremity_AP | MPTA | 84.3098 | 84.3098 | 84.3886 | 84.3256 | 0.7863 |
| Lower extremity_LAT | Femur_alpha | 2.6016 | 3.0699 | 4.7432 | 2.7770 | 0.2688 |
| Lower extremity_LAT | Tibia_alpha | 3.3065 | 3.136 | 3.5537 | 3.4319 | 0.6414 |
| Cadaver femur _AP | Alpha | 2.2200 | 2.2605 | 2.6002 | 2.4757 | 0.151 |
| Cadaver femur _AP | NSA | 120.5407 | 120.2218 | 120.8807 | 120.5559 | 0.9551 |
| Cadaver femur_LAT | Alpha | 14.2867 | 14.2424 | 14.2613 | 14.1879 | 0.1253 |
| Plastic femur _AP | Alpha | 0.8313 | 1.6603 | 0.9938 | 1.3466 | 0.1208 |
| Plastic femur _AP | NSA | 127.9111 | 127.2723 | 128.1869 | 127.3063 | 0.3939 |
| Plastic femur _LAT | Alpha | 9.6274 | 9.5999 | 9.756 | 9.5942 | 0.8051 |
The angles are in degrees
Clinical Study
To evaluate our approach in real clinical environment, we employed our method with our clinical partner in two scoliosis correction surgeries. The X-ray images were captured by the Ziehm Vision FD Vario 3D system for intraoperative evaluation (Fig. 6, top), and the panoramic images for these two cases were constructed by our method (Fig. 6, bottom). Although errors were introduced due to parallax effect (e.g., the height of L4 in the stitched image of case 01 was shortened), our clinical partners were still satisfied with the results. The reason is simply because with the stitched images for the first time they could evaluate intraoperatively how good the surgical corrections were (i.e., the overall angular alignment of the spine column). Actually, before we introduce our technique into the operating room, such a check can only be done postoperatively with an X-ray machine, which will be too late to make any change if the surgical correction is not optimal. Now with our technique, they can get an immediate intraoperative feedback and they can change their corrections if the surgical outcome is not optimal.
Fig. 6.
Clinical study setup (top image) and image stitching results for two clinical cases (bottom two images)
Discussions and Conclusions
We have proposed a new method for stitching multiple fluoroscopic images acquired from C-arm instrument. We employ an X-ray visible ruler. During image acquisition, the relative position of the ruler with respect to the imaged structure remains unchanged, and therefore, the ruler can be used as the reference for image stitching. To this end, in each image, we first detect the main line and all graduations on the ruler. Then, we perform number detection to label and propagate the associated numbers of the graduations, and image stitching is performed accordingly. Unlike many other methods, our method does not rely on image overlapping. We conducted experiments on eight image sequences captured on different anatomical structures. Qualitative results show that our method generates satisfactory image stitching results. Distance- and angle-based quantitative measurements show that our method keeps the important geometric properties even with little or no image overlap.
Unlike the method proposed in [2], our method requires no modification of the fluoroscopic unit. We developed a robust method to detect the graduation and numbers on the ruler in order to achieve an accurate and robust image stitching. Furthermore, our method is based on aligning the detected ruler part on the original image with a virtual ruler on the panoramic image. Thus, our image stitching method does not require a significant overlap between two consecutive X-ray images, which is novel. In contrast, other ruler-based image stitching methods [4–6] require a significant overlap between two consecutive X-ray images for image stitching, which inevitably leads to additional radiation exposure. As demonstrated by both the qualitative and the quantitative results obtained from our evaluation experiment (Fig. 5 and Table 3), our method can generate stitched images with satisfactory accuracy even when there is no overlap between the consecutive X-ray images, which is a clear advantage over other ruler-based image stitching approaches.
It is worth to mention limitations of the present method. One limitation, which is common to all ruler-based image stitching methods [4–6], is that the present method cannot solve for the parallax effect, which is introduced by the stitching plane and the target plane not being on the same level [2]. For long bone applications, where the apparent contour plane of the bone is nearly planar and roughly parallel to the image plane, the parallax effect is not very significant, leading to accurate image stitching results, as demonstrated by both the qualitative and the quantitative results obtained from our evaluation experiment (see Fig. 5 and Table 3). However, for spine applications, where the apparent contour plane of the bone is not any more nearly planar, significant parallax effect can be observed, leading to less accurate image stitching results. Although the errors caused by the parallax effect are not significant when we stitched images of the phantom spine (see Fig. 5), such errors are observed on the stitching results of the two clinical cases. Another limitation of the present method is that our method requires a fronto-parallel mobile C-arm setup, i.e., the ruler plane must be parallel to the detector plane of mobile C-arm.
In summary, we presented and validated a novel ruler-based C-arm image stitching method. Our experimental results demonstrate that our method can generate stitched images with satisfactory accuracy even when there is no overlap between the consecutive X-ray images, which is a clear advantage over other existing ruler-based methods, leading to less image acquisition and less radiation exposure.
Footnotes
Since we already detected the main line of the ruler and we know the numbers appear only below this line, we could also reduce the search region using this information.
References
- 1.Ricci WM, Bellabarba C, Lewis R, Evanoff B, Herscovici D, Dipasquale T, Sanders R. Angular malalignment after intramedullary nailing of femoral shaft fractures. J Orthop Trauma. 2001;15(2):90–95. doi: 10.1097/00005131-200102000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, Traub J, Weidert S, Heining SM, Euler E, Navab N. Parallax-free intra-operative x-ray image stitching. Med Image Anal. 2010;14(5):674–686. doi: 10.1016/j.media.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Szeliski R. Image alignment and stitching: a tutorial. Found Trends Comput Graph. 2006;2(1):1–104. doi: 10.1561/0600000009. [DOI] [Google Scholar]
- 4.Yaniv Z, Joskowicz L. Long bone panoramas from fluoroscopic X-ray images. IEEE Trans Med Imaging. 2004;23(1):26–35. doi: 10.1109/TMI.2003.819931. [DOI] [PubMed] [Google Scholar]
- 5.Eeuwijk AHW, Lobregt S, Gerritsen FA. A novel method for digital x-ray imaging of the complete spine. Lect Notes Comput Sci. 1997;1205:519–530. doi: 10.1007/BFb0029275. [DOI] [Google Scholar]
- 6.Gooßen A, Schlüter M, Hensel M. Pralow T, Grigat R: Ruler-Based Automatic Stitching of Spatially Overlapping Radiographs. Bildverarbeitung fr die Medizin, 2008
- 7.Verdonck B, Nijlunsing R, Melman N, Geijer H. Image quality and x-ray dose for translation reconstruction overview imaging of the spine, colon and legs. Int. Congr. Ser. 2001;1230:531–537. doi: 10.1016/S0531-5131(01)00083-8. [DOI] [Google Scholar]
- 8.Qi Z, Cooperstock J: Overcoming parallax and sampling density issues in image mosaicing of non-planar scenes. Br Mach Vis Conf. 2007
- 9.Harris C, Stephens M: A combined corner and edge detector. Proc. Fourth Alvey Vision Conference, 1988
- 10.Dalal N, Triggs B: Histograms of oriented gradients for human detection. IEEE Comput Soc Conf Comput. Vis Pattern Recog, 2005
- 11.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]