Abstract
The present study proposes a computer-aided classification (CAC) system for three kidney classes, viz. normal, medical renal disease (MRD) and cyst using B-mode ultrasound images. Thirty-five B-mode kidney ultrasound images consisting of 11 normal images, 8 MRD images and 16 cyst images have been used. Regions of interest (ROIs) have been marked by the radiologist from the parenchyma region of the kidney in case of normal and MRD cases and from regions inside lesions for cyst cases. To evaluate the contribution of texture features extracted from de-speckled images for the classification task, original images have been pre-processed by eight de-speckling methods. Six categories of texture features are extracted. One-against-one multi-class support vector machine (SVM) classifier has been used for the present work. Based on overall classification accuracy (OCA), features from ROIs of original images are concatenated with the features from ROIs of pre-processed images. On the basis of OCA, few feature sets are considered for feature selection. Differential evolution feature selection (DEFS) has been used to select optimal features for the classification task. DEFS process is repeated 30 times to obtain 30 subsets. Run-length matrix features from ROIs of images pre-processed by Lee’s sigma concatenated with that of enhanced Lee method have resulted in an average accuracy (in %) and standard deviation of 86.3 ± 1.6. The results obtained in the study indicate that the performance of the proposed CAC system is promising, and it can be used by the radiologists in routine clinical practice for the classification of renal diseases.
Keywords: Ultrasound kidney images, Texture features, Feature selection, Support vector machine, Classification
Introduction
Ultrasound imaging has many advantages like real-time, inexpensive, non-radioactive and non-invasive nature and thus it has wide applications in radiology. However, the disadvantages are inter- and intra-observer variability associated with subjective interpretation, artefacts due to patient mobility and equipment limitations. These limitations restraint the clarity of subjective diagnosis. Thus, there has been a significant interest among researchers to provide possible objective aid to radiologists to assist them in decision-making.
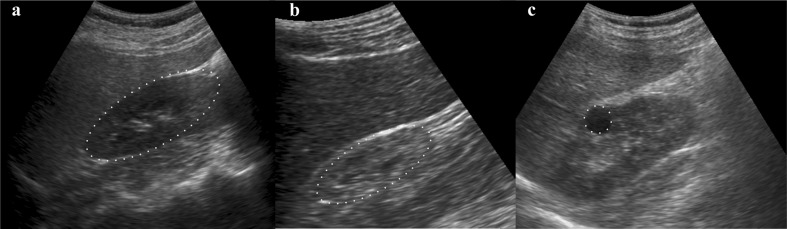
In the present study, three classes of kidney ultrasound images are considered for classification, viz. normal, medical renal disease (MRD) and cysts (Fig. 1). The bean-shaped kidney has parenchyma comprised of outer cortex and inner medulla. Renal cortex extends in between pyramids in medullary region called renal column. In case of normal kidney, cortex is hyper-echoic than liver and differentiation between cortex and medulla can be made. Disorders such as hematuria, proteinuria, pyuria, polyuria, etc. may involve diseased glomerulus, blood vessels and nephrons in renal parenchyma. When such diseases in renal parenchyma are not precisely diagnosable, they are referred as medical renal disease. In MRD, parenchyma is hyper-echoic and thus making the differentiation between cortex and medulla is very difficult. There might be reduction in kidney size. Cysts are fluid-filled regions in the kidney. They are regions with thin wall, anechoic in appearance and also exhibit posterior wall enhancement.
Fig. 1.
Sample B-mode ultrasound kidney images. a Normal, b MRD and c cyst. The dots forming oblongs in a and b are marked to highlight the kidney area as it is difficult to locate. The dots forming circle in c is marked to highlight only the cyst region as kidney area is clearly visible
Very few studies have been presented in the literature with regard to the classification of B-mode kidney ultrasound images. Bommanna Raja et al. have studied the classification of above-mentioned classes by extracting kidney region [1] and using different features [2]. In their study, the whole kidney area has been considered for extracting features. Whereas, the participating radiologist opined that the characteristic changes in MRD with respect to normal are in the parenchyma of kidney and the cysts are focal in nature. Therefore, in the present work, ROIs are obtained from the parenchyma region of kidney in case of normal and MRD classes and from within lesion in case of cyst class.
Speckle noise in B-mode ultrasound images makes visual diagnosis a difficult task, but it also contains diagnostic information which is preferred by the radiologists. Various de-speckling methods have been proposed in the literature. From the studies [3, 4], it has been noted that the de-speckling methods can be oriented towards improving either texture classification or segmentation. Hence, in the present work, few standard de-speckling methods have been utilized before feature extraction to evaluate the contribution of these methods towards improving the performance of classification.
When ultrasound waves enter the body, some are attenuated as body absorbs energy. Every substance such as muscles, fat, etc. has a property called ‘acoustic impedance’. This depends on the density of the substance and the speed of ultrasound in it. When an ultrasound wave pass from one substance to another with different acoustic impedance, two things happen to it. Part of ultrasound wave continues into the second substance by slightly bending away from its original direction referred as refraction. Part of ultrasound wave is reflected back to the probe. The amount that reflected back is directly proportional to the difference in the acoustic impedance between the substances. Reflected waves are important as they provide information for the machine to form an image. As the ultrasound wave crosses from one tissue to the next, each with different acoustic impedance, some of the wave is reflected back at each crossing. Multiple reflected waves return to the probe and machine uses this information to display an image showing different tissues. The term echogenic is used to describe the amount of echo (waves) being reflected by any structure in relation to surrounding structures. The term echo-texture describes the amount of echo being reflected by a structure with respect to its normal echo. Thus, selection of appropriate texture features while designing a CAC system plays a vital role.
Recent studies have utilized different features such as first-order statistics (FOS)-based features [2, 5, 6], grey-level co-occurrence matrices (GLCM)-based features [2, 7–9], run-length matrix (RLM) [8, 10], moment invariants [11], Laws’ texture energy measures [12, 13], wavelet-based features [14] and many more for the classification task. In the present work, gradient-based features [15] are also considered to evaluate the performance of different texture features in classifying B-mode kidney ultrasound images.
The interaction among features in performing the given classification task is another important aspect which should be considered. This can be accomplished by removing unnecessary features. Basically feature space dimensionality reduction can be approached either by transformation-based feature reduction or by feature selection. In the present work, feature selection approach has been used. Differential evolution feature selection (DEFS), being the recent method, has shown better performance with competitive methods [16].
The support vector machine (SVM) classifier has been extensively used for the classification of medical images [13, 14, 17–26]. One-against-one multi-class SVM classifier has been used for the present classification task.
Data Set Collection and Its Description
In the present work, data set consisting of 35 B-mode ultrasound kidney images, i.e. 11 normal, 8 MRD and 16 cyst images, collected from the Department of Radio-diagnosis, Himalayan Institute of Hospital and Trust (HIHT), Dehradun, India, during the period from January 2012 to May 2013 has been used. Each image is of size 800 × 600 pixels with 256 grey levels and 96-dpi resolution. These direct digital images, which are recorded by Siemens ACUSON X300 ultrasound machine from 35 patients, are used for analysis. Either the left or right kidney from longitudinal plane is considered. The compliance of patients and ethical committee of HIHT for making use of these images solely for the research and academics has been obtained initially.
Feature Extraction
The objective/numerical values representing image characteristics are called features. Feature extraction involves two stages as follows:
-
ROI Selection
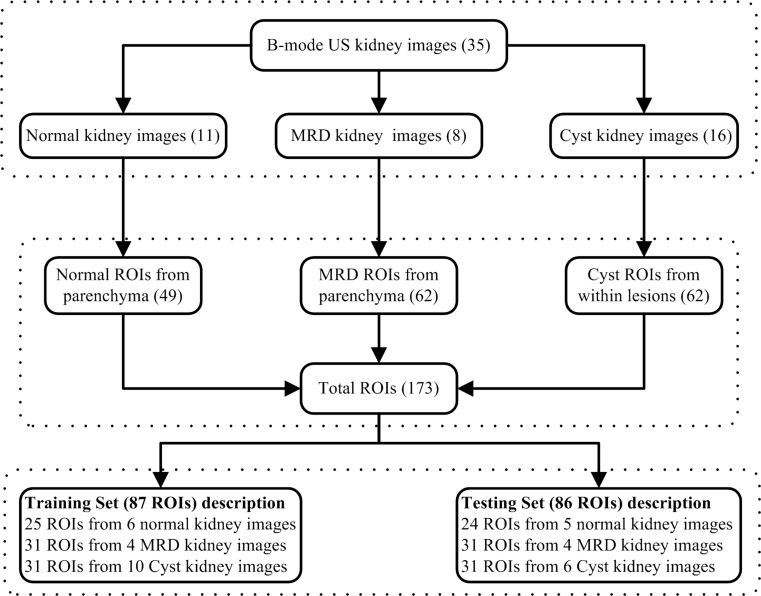
As advised by experienced participating radiologist that the distinguishing characteristics of MRD and normal are prominent in parenchyma region of kidney, ROIs are obtained from parenchyma region in case of these. In case of cysts, ROIs within lesions are selected. ROI size of 32 × 32 pixels has been used to extract maximum non-overlapping ROIs [13, 14]. A simple user-friendly MATLAB program written by the author was used by the participating radiologist to pinpoint the location where the window has to be placed, and the necessary details of it are saved. The data consisted of 49 normal ROIs from 11 normal images, 62 MRD ROIs from 8 MRD images and 62 cyst ROIs from 16 cyst images. The bifurcation of data set into training dataset and testing dataset is shown in Fig. 2.
-
Texture Features
Six categories of texture features are selected for the present study.- Gradient-based (Grad) features: 5 features—mean, variance, kurtosis, skewness and percentage of pixels with non-zero gradient [28]. In this study, 3 × 3 neighbourhood has been considered for calculating absolute gradient value.
- Moment invariant (MI) features: 7 features—a set of seven moments invariant to translation, rotation and scaling derived from the second and third normalized central moments are estimated [11].
- Grey-level co-occurrence matrix (GLCM) features: 420 features—energy, entropy, dissimilarity, contrast, inverse difference, correlation, homogeneity, autocorrelation, cluster shade, cluster prominence, maximum probability, sum of squares, sum average, sum variance, sum entropy, difference variance, difference entropy, information measures of correlation, maximum correlation coefficient, inverse difference normalized and inverse difference moment normalized [2]. Twenty-one features for five values of inter-pixel distance d, i.e., d = 1, 2, 3, 4 and 5 along four orientations θ = 0o, 45o, 90o and 135o result in 420 features.
- Run-length matrix (RLM) features: 44 features—short run emphasis, long run emphasis, grey-level non-uniformity, run-length non-uniformity, run percentage, low grey-level run emphasis, high grey-level rum emphasis, short run low grey-level emphasis, short run high grey-level emphasis, long run low grey-level emphasis and Long run high grey-level emphasis [8]. Eleven features along four orientations θ = 0o, 45o, 90o and 135o result in total 44 RLM-based features.
- Laws’ texture features: 150 features—10 first-order statistics features for 15 texture rotational invariance (TR) images result in 150 Laws’ texture features [12].
Fig. 2.
Data set description
De-speckling Methods
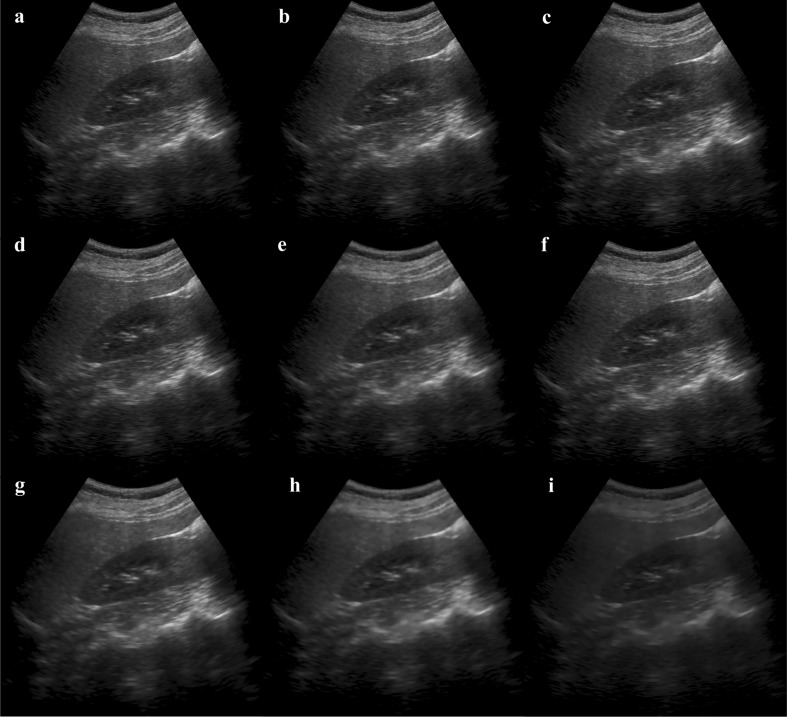
Recent review [3] on comparison of 15 de-specking methods on echocardiographic images concludes that oriented speckle reducing anisotropic diffusion (OSRAD) method is best for medical application. In [29], OSRAD has been recommended as a pre-processing step for segmentation task. In [4], speckle reducing anisotropic diffusion (SRAD) has been modified for pre-processing the images for the classification task. Modified SRAD has shown improvement with respect to SRAD. In the present work, the contribution of features from different de-speckling methods for the task of classification has been considered. Eight de-speckling methods, Lee [30], Lee’s sigma [31], enhanced Lee [32], Frost [33], Kaun [34], geometric filter [35], SRAD [36] and detail preserving anisotropic diffusion (DPAD) [37], have been considered for analysis. A sample of normal kidney image and the corresponding de-speckled images are shown in Fig. 3.
Fig. 3.
A sample of normal kidney image and the de-speckled images. a Original image. b Lee. c Lee’s sigma. d Enhanced Lee. e Frost. f Kaun. g Geometric. h SRAD. i DPAD
Differential Evolution Feature Selection
Feature selection is an essential step towards efficient performance of a classifier as it excludes irrelevant features while forming a subset, hence reduce calculations and time of processing. Differential evolution (DE) has its advantages over other population-based strategy in optimality and convergence speed. Rami khushaba et al. modified DE and utilized roulette wheel supplied with probabilities of relevant feature distribution for feature selection. DEFS stops when the fixed number of iterations is completed. For more details, please refer to [16].
In the present work, for obtaining optimal values to the parameters of feature selection such as desired number of features, population size and the number of iterations, an optimization study has been performed. A 3D matrix has been formed with population size of 25, 50, 75 and 100; number of iterations of 25, 50, 75 and 100; and desired number of features of 5, 10, 20, 30,…,100. For each position of this matrix, DEFS procedure is executed 30 times to obtain 30 subsets. Average accuracy and standard deviation of these 30 subsets are tabulated. The average time taken to generate a subset is also noted down. This matrix revealed that, for desired the number of features equal to 10, population size 50 and the number of iterations equal to 100 are optimal as for overall classification accuracy and computational time are concerned. Hence, these values are utilized in the DEFS procedure. DEFS is a wrapper method, and k-nearest neighbour (KNN) classifier is used in the present work to provide classification error rate as a fitness function.
Classification
Given a sample, an ability to assign it to the concerned class is the task of the classifier. After extracting and selecting appropriate features which represent the classes, the classifier has training and testing phases to go through.
SVM Classifier
SVM classifier is known for building a hyper-plane in the feature space of higher dimensions, optimum enough to separate the classes with minimal error. Herein, Gaussian radial basis function kernel is employed for performing non-linear mapping of data from input space to feature space. To prevent the domination among the data of varied ranges, min-max normalization has been used to rescale the feature values between 0 and 1 [38, 39]. For multi-class classification, one-against-one approach has been implemented in MATLAB. While the kernel parameter γ controls the curvature of the decision boundary, the soft margin constant C of SVM increase the margin with minimum error possible. To train the SVM, the optimal values are obtained by grid search in parameter space such as γ ϵ {2−5, 2−4,…, 25} and C ϵ {2−5, 2−4,…, 25} using 10-fold cross-validation on training data.
Performance Measures
Accuracy alone is not acceptable in measuring the performance of a classifier, particularly when different number of samples is there in individual classes. Sensitivity and specificity are used often in medical applications. The higher the value of these parameters, the better the classification [40].
Results
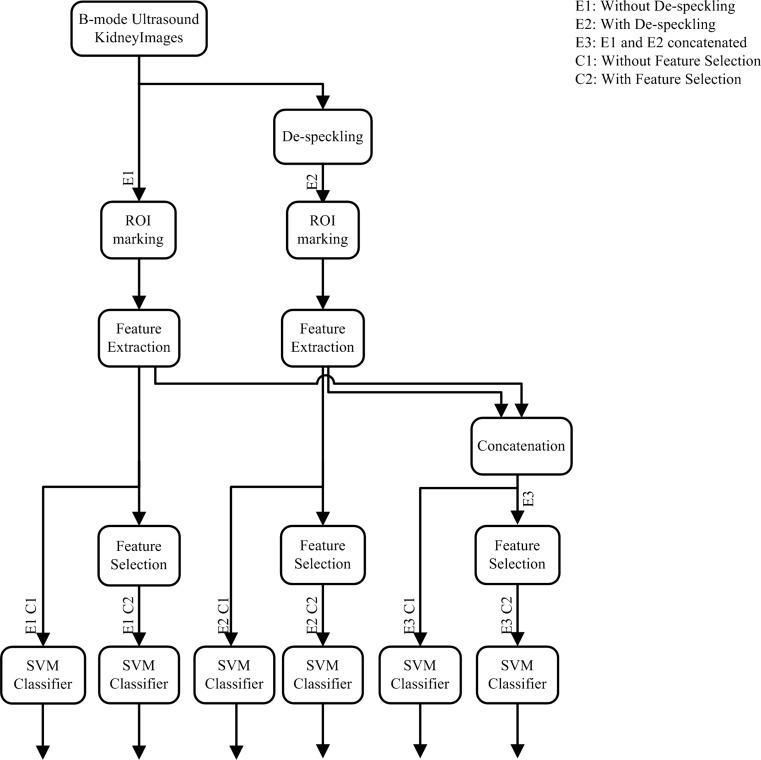
The de-speckling filter parameters and their values used in the present study are tabulated in Table 1. The block diagram representation indicating workflow of the present study is given in Fig. 4. Three experiments are conducted with two cases in each, i.e. without feature selection (C1) and with feature selection (C2). In experiment 1 (E1), ROIs are marked and extracted from original (without de-speckling) US images followed by feature extraction. E1C1 represents features from experiment 1 without feature selection; similarly E1C2 represents features from experiment 1 with feature selection. These features are used for designing separate SVM classifiers. Experiment 2 starts with de-speckling the US images; ROIs are extracted from the same locations (markings) which are saved in experiment 1. This is followed by feature extraction. E2C1 and E2C2 represent features from experiment 2 without and with feature selection, respectively. Again, classification is carried out by different SVM classifiers. In experiment 3, features from E1 and E2 are concatenated forming larger feature set. E2 has features from ROIs of images de-speckled by eight methods, giving rise to that many feature sets. The combinations considered for concatenation are mentioned in the subsection experiment 3. E3C1 represents concatenated features set and is passed through SVM classifier. E3C2 represents concatenated features from experiment 3 after feature selection and is passed through SVM classifier which is shown separately.
Table 1.
De-speckling filter parameters
| Method | Abbreviation | Filter parameters |
|---|---|---|
| Lee filter | Lee | z = 5 |
| Lee’s sigma filter | SLee | z = 5 |
| Enhanced Lee filter | EnLee | z = 5 |
| Frost filter | Frost | z = 5 |
| Kaun filter | Kaun | z = 5 |
| Geometric filter | Geo | n = 3 |
| Speckle reducing anisotropic diffusion | SRAD | s = 0.2, n = 100, z = 5 |
| Detail preserving anisotropic diffusion | DPAD | s = 0.2, n = 100, z = 5 |
z mask size, n number of iterations, s step size
Fig. 4.
CAC system for kidney ultrasound images
The classification results without feature selection are tabulated in Table 2. For feature selection, DEFS process is repeated 30 times to obtain 30 subsets [16] and the subset with best classification result has been shown in Table 3. Average accuracy and standard deviation values of 30 subsets are shown in Table 4.
Table 2.
Classification results without feature selection
| Experiment-case number | Features/de-speckle/seta | Accuracy (%) | Sensitivity (%) | Specificity (%) | Overall accuracy (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cyst | MRD | Normal | Cyst | MRD | Normal | Cyst | MRD | Normal | |||
| E1C1 | All | 93 | 86 | 83.7 | 96.7 | 87 | 54.1 | 90.9 | 85.4 | 95.1 | 81.3 |
| FOS | 100 | 77 | 77.9 | 100 | 77.4 | 50 | 100 | 78.1 | 88.7 | 77.9 | |
| Grad | 98.8 | 86 | 87.2 | 96 | 80.6 | 79.1 | 100 | 89 | 90.3 | 86 | |
| MI | 95.3 | 75.5 | 70.9 | 87 | 80 | 37.5 | 100 | 72.7 | 83.8 | 70.9 | |
| RLM | 93 | 87.2 | 84.8 | 93.5 | 80.5 | 70.8 | 92.7 | 90.9 | 90.3 | 82.5 | |
| GLCM | 93 | 82.5 | 82 | 90.3 | 83.8 | 58.3 | 94.5 | 81.8 | 91.9 | 79 | |
| Laws | 96.5 | 82 | 79 | 97 | 77 | 58 | 96.3 | 85 | 87 | 79 | |
| Grad + RLM | 97.6 | 90.6 | 90 | 93.4 | 83.9 | 91.6 | 100 | 94.4 | 90.2 | 89.5 | |
| E2C1 | All Lee | 95.3 | 82.5 | 80.2 | 96.7 | 87.1 | 45.8 | 94.5 | 80 | 93.5 | 79 |
| All Lee’s sigma | 93 | 86 | 83.7 | 96.7 | 87.1 | 54.1 | 90.9 | 85.5 | 95.1 | 81.3 | |
| All enhanced Lee | 98.8 | 82.5 | 83.7 | 96.7 | 83.8 | 62.5 | 100 | 81.8 | 91.9 | 82.5 | |
| All Frost | 90.6 | 82.5 | 84.8 | 96.7 | 80.6 | 54.1 | 87.2 | 83.6 | 96.7 | 79 | |
| All Kaun | 95.3 | 83.7 | 81.3 | 96.7 | 90.3 | 45.8 | 94.5 | 80 | 95.1 | 80.2 | |
| All geometric | 89.5 | 82.5 | 81.3 | 90.3 | 90.3 | 41.6 | 89 | 78.1 | 96.7 | 76.7 | |
| All SRAD | 90.6 | 75.5 | 73.2 | 100 | 64.5 | 37.5 | 85.4 | 81.8 | 87 | 69.7 | |
| All DPAD | 97.6 | 80.2 | 80.2 | 100 | 77.4 | 54.1 | 96.3 | 81.8 | 90.3 | 79 | |
| Grad + RLM enhanced Lee | 98.8 | 90.6 | 91.8 | 96.7 | 83.8 | 91.6 | 100 | 94.5 | 91.9 | 90.6 | |
aPlease refer result section for the description of representations used in the second column
The significance of the values in italics are the best results obtained and these values are mentioned in the description of results
Table 3.
Best classification results with feature selection
| Experiment-case number | Features/de-speckle/seta | Accuracy (%) | Sensitivity (%) | Specificity (%) | Overall accuracy | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cyst | MRD | Normal | Cyst | MRD | Normal | Cyst | MRD | Normal | (%) | ||
| E1C2 | All | 96 | 90.7 | 91.8 | 93 | 90.3 | 83.3 | 98.1 | 90.9 | 95.2 | 89.5 |
| RLM | 97.7 | 89 | 87.1 | 96.8 | 80.7 | 83.2 | 98 | 94.5 | 88.6 | 88.3 | |
| GLCM | 94.1 | 87 | 90.7 | 87.1 | 83.7 | 87.5 | 98.2 | 94.6 | 88.7 | 86 | |
| Laws | 100 | 90.7 | 90.6 | 100 | 87.1 | 83.3 | 100 | 92.7 | 93.5 | 90.6 | |
| Grad + RLM | 97.6 | 91.8 | 89.5 | 93.5 | 87 | 87.5 | 100 | 94.5 | 90.3 | 89.5 | |
| E2C2 | Grad + RLM enhanced Lee | 100 | 90.6 | 90.6 | 100 | 77.4 | 95.8 | 100 | 98.1 | 88.7 | 90.6 |
| All Lee’s sigma | 100 | 90.6 | 90.6 | 100 | 83.8 | 87.5 | 100 | 94.5 | 91.9 | 90.6 | |
| All enhanced Lee | 100 | 90.6 | 90.6 | 100 | 80.6 | 91.6 | 100 | 96.3 | 90.3 | 90.6 | |
| E3C2 | RLM set 1 | 97.6 | 88.3 | 88.3 | 96.7 | 77.4 | 87.5 | 98.1 | 94.5 | 88.7 | 87.2 |
| RLM set 2 | 98.8 | 89.5 | 90.6 | 96.7 | 83.8 | 87.5 | 100 | 92.7 | 91.9 | 89.5 | |
| RLM set 3 | 98.8 | 90.6 | 91.6 | 96.7 | 87.1 | 87.5 | 100 | 92.7 | 93.5 | 90.6 | |
| RLM set 4 | 98.8 | 90.6 | 91.8 | 96.7 | 77.4 | 100 | 100 | 98.1 | 88.7 | 90.6 | |
| GLCM set 1 | 97.6 | 86 | 88.3 | 93.5 | 80.6 | 83.3 | 100 | 89 | 90.3 | 86 | |
| GLCM set 2 | 95.3 | 91.8 | 91.8 | 93.5 | 87 | 87.5 | 96.3 | 94.5 | 93.5 | 89.5 | |
| GLCM set 3 | 96.5 | 90.6 | 91.8 | 90.3 | 90.3 | 87.5 | 100 | 90.9 | 93.5 | 89.5 | |
| GLCM set 4 | 94.1 | 89.5 | 90.6 | 83.8 | 83.8 | 95.8 | 100 | 92.7 | 88.7 | 87.2 | |
| Laws set 1 | 100 | 88.3 | 88.3 | 100 | 90.3 | 70.8 | 100 | 87.2 | 95.1 | 88.3 | |
| Laws set 2 | 100 | 91.8 | 91.8 | 100 | 93.5 | 79.1 | 100 | 90.9 | 96.7 | 91.8 | |
| Laws set 3 | 100 | 94.1 | 94.1 | 100 | 90.3 | 91.6 | 100 | 96.3 | 95.1 | 94.1 | |
| Laws set 4 | 97.6 | 90.6 | 93.3 | 96.7 | 93.5 | 79.1 | 98.1 | 89 | 98.3 | 90.6 | |
| Gr + RLM set 1 | 97.6 | 87.2 | 87.2 | 93.5 | 77.4 | 87.5 | 100 | 92.7 | 87 | 86 | |
| Gr + RLM set 2 | 100 | 91.8 | 91.8 | 100 | 80.6 | 95.8 | 100 | 98.1 | 90.3 | 91.8 | |
| Gr + RLM set 3 | 97.6 | 89.5 | 91.8 | 93.5 | 80.6 | 95.8 | 100 | 94.5 | 90.3 | 89.5 | |
| Gr + RLM set 4 | 96.5 | 91.8 | 93 | 93.5 | 83.8 | 95.8 | 98.1 | 96.3 | 91.9 | 90.6 | |
| All sets 1–4 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |
aPlease refer result section for the description of representations used in the second column
The significance of the values in italics are the best results obtained and these values are mentioned in the description of results
Table 4.
Classification results from 30 subsets
| Experiment-case number | Features/de-speckle/seta | Average accuracy (%) and standard deviation |
|---|---|---|
| E1C2 | All | 84 ± 3.4 |
| RLM | 84 ± 1.7 | |
| GLCM | 82.4 ± 3.1 | |
| Laws | 83.1 ± 3.7 | |
| Grad + RLM | 84.7 ± 2.4 | |
| E2C2 | Grad + RLM enhanced Lee | 86.4 ± 2.5 |
| All Lee’s sigma | 84.5 ± 3.4 | |
| All enhanced Lee | 84.7 ± 3 | |
| E3C2 | RLM set 1 | 83.5 ± 1.5 |
| RLM set 2 | 86.3 ± 1.6 | |
| RLM set 3 | 86.2 ± 2.7 | |
| RLM set 4 | 84.6 ± 2.9 | |
| GLCM set 1 | 82.2 ± 2.2 | |
| GLCM set 2 | 85.3 ± 3 | |
| GLCM set 3 | 84.2 ± 2.7 | |
| GLCM set 4 | 83.6 ± 2.7 | |
| Laws set 1 | 84 ± 2.6 | |
| Laws set 2 | 82.4 ± 4.1 | |
| Laws set 3 | 82.2 ± 3.8 | |
| Laws set 4 | 82.8 ± 4.1 | |
| Gr + RLM set 1 | 83.9 ± 1.4 | |
| Gr + RLM set 2 | 86.2 ± 1.6 | |
| Gr + RLM set 3 | 86.2 ± 2.2 | |
| Gr + RLM set 4 | 85.9 ± 2.2 | |
| All set 1 | 85.8 ± 5.4 | |
| All set 2 | 84.7 ± 5 | |
| All set 3 | 84.9 ± 3.4 | |
| All set 4 | 86.7 ± 5.9 |
aPlease refer result section for the description of representations used in the second column
The significance of the values in italics are the best results obtained and these values are mentioned in the description of results
In Tables 2, 3 and 4, the second column, i.e. features/de-speckle/set, represents features from a particular feature’s category of which the name is mentioned. Similarly, for features from de-speckling method and sets (sets are defined in subsection experiment 3). The feature category ‘All’ represents features from all categories concatenated together and ‘Grad + RLM’ indicates that features from those two categories are concatenated. Combinatorial representations like ‘All Enhanced Lee’ denote features from all categories extracted from the ROIs of images de-speckled by enhanced Lee method. ‘All set 1-4’ means ‘All set 1’, ‘All set 2’, ‘All set 3’ and ‘All set 4’.
Experiment 1
Features from all texture features’ categories are extracted from the ROIs of original B-mode kidney ultrasound images.
Case 1
Results obtained for individual and all features together without feature selection.
Table 2 shows that the overall classification accuracy (OCA) obtained from all features is 81.3 %. Among individual feature categories, gradient (five features) has performed well with 86 % OCA value. When best two feature categories (gradient—86 % and RLM—82.5 %) are concatenated, it gave an OCA of 89.5 % showing an increase in the performance without feature selection.
Case 2
Feature categories having more than 10 features (i.e., RLM, GLCM, Laws, All, Grad + RLM) are subjected to feature selection before classification.
With feature selection (refer Table 3), the performance of all features increased from 81.3 to 89.5 %, that of RLM features increased from 82.5 to 88.3 % and that of GLCM features increased from 79 to 86 %. Laws’ features out-performed with an OCA of 90.6 %. These results show the significance of feature selection and Laws’ features.
Experiment 2
Texture features are extracted from the ROIs of de-speckled ultrasound images.
Case 1
Classification results are obtained for ROIs of images de-speckled by different methods, considering all features (refer Table 2). Lee’s sigma features resulted in OCA of 81.3 % same as that of original images, and enhanced Lee filter features has shown an increase in OCA of 82.5 %. Hence these two are considered for case 2 study.
Case 2
Subsets obtained from feature selection by considering all features in case of Lee’s sigma have increased performance from 81.3 to 90.6 % OCA and that of enhanced Lee filter is from 82.5 to 90.6 % OCA (refer Table 3). Concatenation of gradient and RLM features from enhanced Lee filter gave an OCA of 90.6 % in cases 1 and 2.
Experiment 3
Here, all features of ROIs from original image, images de-speckled by Lee’s sigma and enhanced Lee filter are concatenated to form four sets:
Set 1: Original image features and Lee’s sigma filter features
Set 2: Lee’s sigma filter features and enhanced Lee filter features
Set 3: Original image features and enhanced Lee filter features
Set 4: Original images features, Lee’s sigma filter features and enhanced Lee filter features
From Table 3, comparing the results of experiment 1 (E1C2) and experiment 3 (E3C2), RLM features have an increased OCA from 88.3 to 90.6 % (both set 3 and set 4). GLCM features have shown better improvement in OCA from 86 to 89.5 % (both set 2 and set 3). Similarly, Laws’ features have shown an increase from 90.6 to 94.1 % OCA. For all features together, set 1, set 2, set 3 and set 4 have shown an OCA of 100 %, whereas it is 89.5 % in E1C2. These results show the significance of combining features from ROIs of original image with that of de-speckled images.
Classification Results from 30 Subsets
The features that are passed through feature selection process have been repeated 30 times to obtain 30 subsets as mentioned earlier. The average accuracies with standard deviation are tabulated in Table 4. A feature set could be considered better for classification task if the standard deviation is less and the average accuracy is more. In other words, that feature set would result with better accuracy with most of the subsets. In that respect, experiment 1 shows that RLM features from the ROIs of original images perform better with average accuracy (in %) and standard deviation (AASD) of 84 ± 1.7. RLM features in combination with gradient features from the ROIs of original images (in E1C2) and from the ROIs of images de-speckled by enhanced Lee method (in E2C2) showed an average accuracy (in %) and standard deviation of 84.7 ± 2.4 and 86.4 ± 2.5, respectively. This indicates the contribution of features from de-speckled images. From Table 4, comparing the results of E1C2 and E3C2, RLM features have shown an increased AASD from 84 ± 1.7 to 86.3 ± 1.6 (set 2). GLCM features have performed better with AASD from 82.4 ± 3.1 to 85.3 ± 3 (set 2). Similarly, Laws’ features performed better with AASD from 83.1 ± 3.7 to 84 ± 2.6 (set 1). Grad + RLM features are also on the same line with AASD from 84.7 ± 2.4 to 86.2 ± 1.6 (set 2). These results indicate the positive influence on classification by concatenating features from the ROIs of original images with that of de-speckled images in different combinations (as set 1, set 2, etc.).
Discussions
In literature, few attempts have been made towards the classification of ultrasound kidney images. In [2], authors have extracted 36 features from six categories. The process of ranking the features has been employed to reduce the number of features. The classification efficiency of the hybrid fuzzy system mentioned is 96 % for normal, 92 % for MRD and 96 % for cyst. In [41], authors have obtained 86.6 % for normal, 76.6 % for MRD and 83.3 % for cyst using Gabor wavelet features. There has been an attempt to classify the ultrasound kidney images using content descriptive power spectral features [42] and multi-scale differential features [43]. In [44], authors have used 28 multiple descriptive features with 13 highly significant for the classification. With ANN as a classifier, accuracy of 90.4 % for normal, 86.6 % for MRD and 85.7 % for cyst has been obtained.
The contribution of individual texture feature category for the classification of normal, MRD and cyst US images has been obtained. This has helped in reconsidering significant features in further processes. Concatenation of features like gradient (five features—86 % OCA) and RLM (44 features—82.5 % OCA) has shown notable performance of 89.5 % OCA (in original image) and 90.6 % OCA (in enhanced Lee image). The subsets (10 features) obtained from Grad + RLM (49 features) have two to three features of gradient. This shows the significance of gradient features in the classification of kidney images. From Table 2, FOS feature set, though 100 % accurate in classifying cyst, performed poor with MRD and normal. Gradient features showed promising improvement with accuracy of 98.8 % for cyst, 86 % for MRD and 87.2 % for normal. Grad + RLM features from images de-speckled by enhanced Lee method showed improved performance than those from original images. Accuracy has increased for cyst from 97.6 to 98.8 % and for normal from 90 to 91.8 %.
The feature selection subsets of experiment 3 that gave best results have three to six (out of 10) de-speckled image features. This indicates that features extracted from ROIs of de-speckled images have an additional contribution to make for objective classification. Thus, it is advantageous to concatenate features from ROIs of original image with those of de-speckled images. From Table 3, comparing E1C2 and E3C2, RLM features have shown improvement in accuracy for cyst from 97.7 to 98.8 %, for MRD from 89 to 90.6 % and for normal from 87.1 to 91.8 % with set 4. GLCM features have shown increased accuracy for cyst from 94.1 to 97.6 % (set 1), for MRD from 87 to 91.8 % (set 2) and for normal from 90.7 to 91.8 % (set 2 and set 3). Similarly, Laws’ features have significant improvement in accuracy for MRD from 90.7 to 94.1 % and for normal from 90.6 to 94.1 % with set 3. All features sets, i.e. all set 1, all set 2, all set 3 and all set 4, have exhibited prominent increase in accuracy for cyst from 96 to 100 %, for MRD from 90.7 to 100 % and for normal from 91.8 to 100 %. From Table 4, in E1C2, RLM features showed better performance with AASD of 84 ± 1.7. In E3C2, RLM, GLCM and Grad + RLM features performed better with set 2 compared to other sets. GLCM features of set 2 performed better (85.3 ± 3) than Laws’ features of set 1 (84 ± 2.6). But, Laws’ features consisted of less number of features and thus computations involved is less. RLM features of set 2 (88 features) performed better with AASD of 86.3 ± 1.6 with less computations involved. Thus, RLM features of set 2 are comparatively more promising with better accuracy for most of the subsets in the classification of kidney ultrasound images.
Conclusion
A classification study of three classes of kidney ultrasound images has been performed. ROIs from parenchyma region are considered for normal and MRD images and for cysts; ROIs are taken from within lesions as they are focal in nature. Among the feature categories, gradient features and Grad + RLM features gave OCA of 86 and 89.5 %, respectively, without feature selection. The Laws’ features of ROIs from original image concatenated with those from images de-speckled by enhanced Lee method gave maximum OCA of 94.1 % after feature selection. By considering all features from the ROIs of original as well as de-speckled images followed by feature selection gave maximum OCA of 100 %. Thus, feature selection module is necessary to reduce the number of features without compromising on the OCA. RLM features extracted from the ROIs of images de-speckled by Lee’s sigma method concatenated with that of enhanced Lee method (set 2) have produced an AASD of 86.3 ± 1.6. The promising results obtained from the present study indicate the usefulness of the proposed CAC system to aid radiologists in objective classification. Thus, the study concludes with the recommendation to consider texture features from the ROIs of de-speckled images and concatenating with those of original images for elevating the performance of classification.
Acknowledgments
The authors would like to thank the reviewers for the valuable review, which led to the significant improvement in the manuscript. Author Subramanya M. B. is grateful to Dr. Jitendra Virmani for the timely suggestions and the Ministry of Human Resource and Development (MHRD), India, for the scholarship. The authors would like to acknowledge the Department of Electrical Engineering, Indian Institute of Technology, Roorkee, and Department of Radiology, Himalayan Institute of Hospital and Trust, for their support during the period of this research work.
Contributor Information
M. B. Subramanya, Phone: +91-8057646711, Email: subramanyabme@gmail.com
Vinod Kumar, Email: vinodfee@gmail.com.
Shaktidev Mukherjee, Email: mukherjee.shaktidev@gmail.com.
Manju Saini, Email: drmanjusaini@gmail.com.
References
- 1.Raja BK, Madheswaran M, Thyagarajah K. A general segmentation scheme for contouring kidney region in ultrasound kidney images using improved higher order spline interpolation. J Biol Med Sci. 2007;2(2):81–88. [Google Scholar]
- 2.Raja BK, Madheswaran M, Thyagarajah K. A hybrid fuzzy-neural system for computer-aided diagnosis of ultrasound kidney images using prominent features. J Med Syst. 2008;32(1):65–83. doi: 10.1007/s10916-007-9109-5. [DOI] [PubMed] [Google Scholar]
- 3.Finn S, Glavin M, Jones E. Echocardiographic speckle reduction comparison. IEEE Trans Ultrason Ferroelectr Freq Control. 2011;58(1):82–101. doi: 10.1109/TUFFC.2011.1776. [DOI] [PubMed] [Google Scholar]
- 4.Mittal D, Kumar V, Saxena SC, Khandelwal N, Kalra N. Enhancement of the ultrasound images by modified anisotropic diffusion method. Med Biol Eng Comput. 2010;48(12):1281–1291. doi: 10.1007/s11517-010-0650-x. [DOI] [PubMed] [Google Scholar]
- 5.Virmani J, Kumar V, Kalra N, Khandelwal N: A rapid approach for prediction of liver cirrhosis based on first order statistics. In: Proceedings of IEEE International Conference on Multimedia, Signal Processing and Communication Technologies, IMPACT-2011, 212–215, 2011
- 6.Virmani J, Kumar V, Kalra N, Khandelwal N. Neural network ensemble based CAD system for focal liver lesions from B-mode ultrasound. J Digit Imaging. 2014 doi: 10.1007/s10278-014-9685-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Virmani J, Kumar V, Kalra N, Khandelwal N: Prediction of cirrhosis based on singular value decomposition of gray level co-occurrence matrix and a neural network classifier. In: Proceedings of IEEE International Conference on Developments in E-systems Engineering, DeSe-2011, 146–151, 2011
- 8.Mittal D, Kumar V, Saxena SC, Khandelwal N, Kalra N. Neural network based focal liver lesion diagnosis using ultrasound images. Int J Comput Med Imaging Graph. 2011;35(4):315–323. doi: 10.1016/j.compmedimag.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Virmani J, Kumar V, Kalra N, Khandelwal N. SVM-based characterisation of liver cirrhosis by singular value decomposition of GLCM matrix. Int J Artif Intell Soft Comput. 2013;4(1):276–296. doi: 10.1504/IJAISC.2013.053407. [DOI] [Google Scholar]
- 10.Virmani J, Kumar V, Kalra N, Khandelwal N. PCA-SVM based CAD system for focal liver lesions from B-mode ultrasound. Def Sci J. 2013;63(5):478–486. doi: 10.14429/dsj.63.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raja B. K, Madheswaran M, and Thyagarajah K: Evaluation of tissue characteristics of kidney for diagnosis and classification using first order statistics and RTS invariants. In: Proceedings of IEEE International Conference on Signal Processing, Communications and Networking, ICSCN’07. 483–487, 2007
- 12.Virmani J, Kumar V, Kalra N, Khandelwal N: Prediction of cirrhosis from liver ultrasound B-mode images based on Laws’ masks analysis. In: Proceedings of IEEE International Conference on Image Information Processing, ICIIP-2011, 1–5, 2011
- 13.Virmani J, Kumar V, Kalra N, Khandelwal N. Characterization of primary and secondary malignant liver lesions from B-mode ultrasound. J Digit Imaging. 2013;26(6):1058–1070. doi: 10.1007/s10278-013-9578-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Virmani J, Kumar V, Kalra N, Khandelwal N. SVM-based characterization of liver ultrasound images using wavelet packet texture descriptors. J Digit Imaging. 2013;26(3):530–543. doi: 10.1007/s10278-012-9537-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szczypinski PM, Strzelecki M, Materka A, Klepaczko A. MaZda—A software package for image texture analysis. Comput Methods Prog Biomed. 2009;94(1):66–76. doi: 10.1016/j.cmpb.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Khushaba RN, Al-Ani A, Al-Jumaily A. Feature subset selection using differential evolution and a statistical repair mechanism. Exp Syst Appl. 2011;38(9):11515–11526. doi: 10.1016/j.eswa.2011.03.028. [DOI] [Google Scholar]
- 17.Virmani J, Kumar V, Kalra N, Khandelwal N. Prediction of liver cirrhosis based on multiresolution texture descriptors from B-mode ultrasound. Int J Converg Comput. 2013;1(1):19–37. doi: 10.1504/IJCONVC.2013.054658. [DOI] [Google Scholar]
- 18.Wan J, Zhou S: Features extraction based on wavelet packet transform for b-mode ultrasound images. In: Proceedings of IEEE International Congress on Image and Signal Processing, CISP-2010, 2:949–955, 2010
- 19.Lee C, Chen S H: Gabor wavelets and SVM classifier for liver diseases classification from CT images. In: Proceedings of IEEE International Conference on Systems, Man, and Cybernetics, 548–552, 2006
- 20.Nawaz S, Dar A H: Hepatic lesions classification by ensemble of SVMs using statistical features based on co-occurrence matrix. In: Proceedings of 4th IEEE International Conference on Emerging Technologies, ICET-2008, 21–26, 2008
- 21.Huang YL, Chen DR, Jiang YR, Kuo J, Wu HK, Moon WK. Computer-aided diagnosis using morphological features for classifying breast lesions on ultrasound. Ultrasound Obstet Gynecol. 2008;32:565–572. doi: 10.1002/uog.5205. [DOI] [PubMed] [Google Scholar]
- 22.Moayedi F, Azimifar Z, Boostani R, Katebi S: Contourlet based mammography mass classification. In: Proceedings of ICIAR 2007, LNCS 4633, 923–934, 2007
- 23.Huang YL, Wang KL, Chen DR. Diagnosis of breast tumors with ultrasonic texture analysis using support vector machines. Neural Comput & Applic. 2006;15(2):164–169. doi: 10.1007/s00521-005-0019-5. [DOI] [Google Scholar]
- 24.Reddy TK, Kumaravel N: A comparison of wavelet, curvelet and contourlet based texture classification algorithms for characterization of bone quality in dental CT. In: Proceedings of International Conference on Environmental, Biomedical and Biotechnology, IPCBEE 16:60–65, 2011
- 25.Tsiaparas N, Golemati S, Andreadis I, Stoitsis J: Multiscale geometric texture analysis of ultrasound images of carotid atherosclerosis. In: Proceedings of 10th IEEE International Conference on Information Technology and Applications in Biomedicine, ITAB-2010, 1–4, 2010
- 26.Minhas F, Sabih D, Hussain M. Automated classification of liver disorders using ultrasound images. J Med Syst. 2012;36(5):3163–3172. doi: 10.1007/s10916-011-9803-1. [DOI] [PubMed] [Google Scholar]
- 27.Contributors Wikipedia: Percentile. Available at: http://en.wikipedia.org/wiki/Percentile. Accessed: 28 September 2013
- 28.Materka A, Strzelecki M, Nski P. S: MaZda User’s Manual. 2006
- 29.Krissian K, Westin CF, Kikinis R, Vosburgh KG. Oriented speckle reducing anisotropic diffusion. IEEE Trans Image Proc. 2007;16(5):1412–1424. doi: 10.1109/TIP.2007.891803. [DOI] [PubMed] [Google Scholar]
- 30.Lee JS. Digital image enhancement and noise filtering by use of local statistics. IEEE Trans Pattern Anal Mach Intell. 1980;2(2):165–168. doi: 10.1109/TPAMI.1980.4766994. [DOI] [PubMed] [Google Scholar]
- 31.Lee JS. Digital image smoothing and the sigma filter. Comput Gr Image Process. 1983;24(2):255–269. doi: 10.1016/0734-189X(83)90047-6. [DOI] [Google Scholar]
- 32.Lopes A, Touzi R, Nezry E. Adaptive speckle filters and scene heterogeneity. IEEE Trans Geosci Remote. 1990;28(6):992–1000. doi: 10.1109/36.62623. [DOI] [Google Scholar]
- 33.Frost VS, Stiles JA, Shanmugan KS, Holtzman JC. A model for radar images and its application to adaptive digital filtering of multiplicative noise. IEEE Trans Pattern Anal Mach Intell. 1982;4(2):157–166. doi: 10.1109/TPAMI.1982.4767223. [DOI] [PubMed] [Google Scholar]
- 34.Kuan D, Sawchuk A, Strand T, Chavel P. Adaptive restoration of images with speckle. IEEE Trans Acoust Speech. 1987;35(3):373–383. doi: 10.1109/TASSP.1987.1165131. [DOI] [Google Scholar]
- 35.Crimmins TR. Geometric filter for speckle reduction. Appl Opt. 1985;24(10):1438–1443. doi: 10.1364/AO.24.001438. [DOI] [PubMed] [Google Scholar]
- 36.Yu Y, Acton ST. Speckle reducing anisotropic diffusion. IEEE Trans Image Proc. 2002;11(11):1260–1270. doi: 10.1109/TIP.2002.804276. [DOI] [PubMed] [Google Scholar]
- 37.Aja-Fernndez S, Alberola-Lpez C. On the estimation of the coefficient of variation for anisotropic diffusion speckle filtering. IEEE Trans Image Proc. 2006;15(9):2694–2701. doi: 10.1109/TIP.2006.877360. [DOI] [PubMed] [Google Scholar]
- 38.Burges CJC. A tutorial on support vector machines for pattern recognition. Data Min Knowl Disc. 1998;2(2):121–167. doi: 10.1023/A:1009715923555. [DOI] [Google Scholar]
- 39.Ben-Hur A, Weston J: A user's guide to support vector machines. Data Min Tech Life Sci, Humana Press: 223-239, 2010 [DOI] [PubMed]
- 40.Contributors Wikipedia: Sensitivity and specificity. Available at: http://en.wikipedia.org/wiki/Sensitivity_and_specificity. Accessed: 12 September 2013
- 41.Raja BK, Madheswaran M, Thyagarajah K. Texture pattern analysis of kidney tissues for disorder identification and classification using dominant Gabor wavelet. Mach Vision Appl. 2010;21(3):287–300. doi: 10.1007/s00138-008-0159-6. [DOI] [Google Scholar]
- 42.Raja BK, Madheswaran M, Thyagarajah K. Ultrasound kidney image analysis for computerized disorder identification and classification using content descriptive power spectral features. J Med Syst. 2007;31(5):307–317. doi: 10.1007/s10916-007-9068-x. [DOI] [PubMed] [Google Scholar]
- 43.Raja BK, Madheswaran M, Thyagarajah K. Quantitative and qualitative evaluation of US kidney images for disorder classification using multi-scale differential features. ICGST-BIME J. 2007;7(1):1–8. [Google Scholar]
- 44.Raja B. K, Madheswaran M, and Thyagarajah K: Analysis of ultrasound kidney images using content descriptive multiple features for disorder identification and ANN based classification. International conference on Computing: Theory and Applications, ICCTA’07 382–388, 2007