Abstract
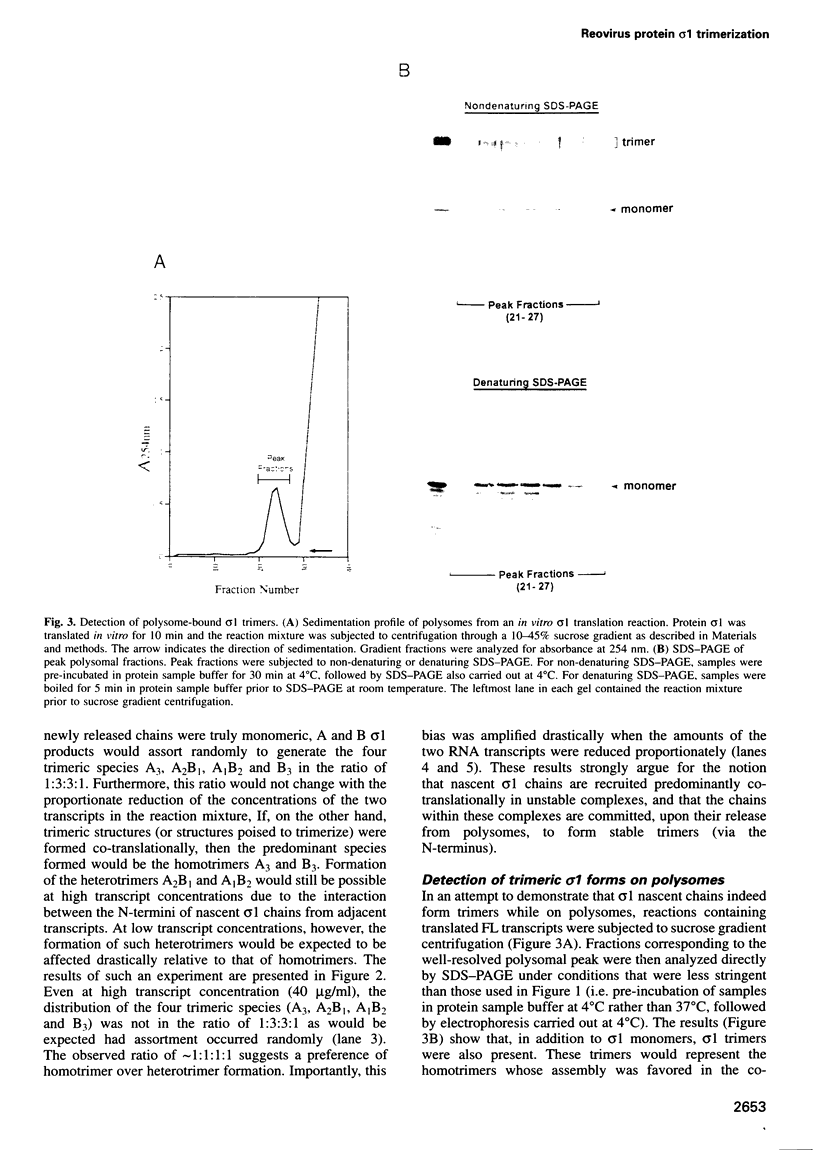
The reovirus cell attachment protein, sigma1, is a trimer with a 'lollipop' structure. Recent findings indicate that the N-terminal fibrous tail and the C-terminal globular head each possess a distinct trimerization domain. The region responsible for N-terminal trimerization (formation of a triple alpha-helical coiled-coil) is located at the N-terminal one-third of sigma1. In this study, we investigated the temporality and ATP requirement of this trimerization event in the context of sigma1 biogenesis. In vitro co-synthesis of the full-length (FL) and a C-terminally truncated (d44) sigma1 protein revealed a preference for homotrimer over heterotrimer formation, suggesting that assembly at the N-terminus occurs co-translationally. This was corroborated by the observation that polysome-associated sigma1 chains were trimeric as well as monomeric. Truncated proteins (d234 and d294) with C-terminal deletions exceeding half the length of sigma1 were found to trimerize post-translationally. This trimerization did not require ATP since it proceeded normally in the presence of apyrase. In contrast, formation of stable FL sigma1 trimers was inhibited by apyrase treatment. Collectively, our data suggest that assembly of nascent sigma1 chains at the N-terminus is intrinsically ATP independent, and occurs co-translationally when the ribosomes have traversed past the midpoint of the mRNA.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anfinsen C. B. Principles that govern the folding of protein chains. Science. 1973 Jul 20;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Banerjea A. C., Brechling K. A., Ray C. A., Erikson H., Pickup D. J., Joklik W. K. High-level synthesis of biologically active reovirus protein sigma 1 in a mammalian expression vector system. Virology. 1988 Dec;167(2):601–612. [PubMed] [Google Scholar]
- Bassel-Duby R., Jayasuriya A., Chatterjee D., Sonenberg N., Maizel J. V., Jr, Fields B. N. Sequence of reovirus haemagglutinin predicts a coiled-coil structure. 1985 May 30-Jun 5Nature. 315(6018):421–423. doi: 10.1038/315421a0. [DOI] [PubMed] [Google Scholar]
- Beckmann R. P., Mizzen L. E., Welch W. J. Interaction of Hsp 70 with newly synthesized proteins: implications for protein folding and assembly. Science. 1990 May 18;248(4957):850–854. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- Doms R. W., Lamb R. A., Rose J. K., Helenius A. Folding and assembly of viral membrane proteins. Virology. 1993 Apr;193(2):545–562. doi: 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- Doohan J. P., Samuel C. E. Biosynthesis of reovirus-specified polypeptides. Analysis of ribosome pausing during translation of reovirus S1 and S4 mRNAs in virus-infected and vector-transfected cells. J Biol Chem. 1993 Aug 25;268(24):18313–18320. [PubMed] [Google Scholar]
- Doohan J. P., Samuel C. E. Biosynthesis of reovirus-specified polypeptides: ribosome pausing during the translation of reovirus S1 mRNA. Virology. 1992 Feb;186(2):409–425. doi: 10.1016/0042-6822(92)90006-b. [DOI] [PubMed] [Google Scholar]
- Duncan R., Horne D., Strong J. E., Leone G., Pon R. T., Yeung M. C., Lee P. W. Conformational and functional analysis of the C-terminal globular head of the reovirus cell attachment protein. Virology. 1991 Jun;182(2):810–819. doi: 10.1016/0042-6822(91)90622-i. [DOI] [PubMed] [Google Scholar]
- Duncan R., Lee P. W. Localization of two protease-sensitive regions separating distinct domains in the reovirus cell-attachment protein sigma 1. Virology. 1994 Aug 15;203(1):149–152. doi: 10.1006/viro.1994.1465. [DOI] [PubMed] [Google Scholar]
- Fedorov A. N., Baldwin T. O. Contribution of cotranslational folding to the rate of formation of native protein structure. Proc Natl Acad Sci U S A. 1995 Feb 14;92(4):1227–1231. doi: 10.1073/pnas.92.4.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J., Tang D., Leone G., Lee P. W. Binding of reovirus to receptor leads to conformational changes in viral capsid proteins that are reversible upon virus detachment. J Biol Chem. 1994 Jun 24;269(25):17043–17047. [PubMed] [Google Scholar]
- Fraser R. D., Furlong D. B., Trus B. L., Nibert M. L., Fields B. N., Steven A. C. Molecular structure of the cell-attachment protein of reovirus: correlation of computer-processed electron micrographs with sequence-based predictions. J Virol. 1990 Jun;64(6):2990–3000. doi: 10.1128/jvi.64.6.2990-3000.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J., Nimmesgern E., Ohtsuka K., Hartl F. U. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994 Jul 14;370(6485):111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- Furlong D. B., Nibert M. L., Fields B. N. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol. 1988 Jan;62(1):246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C., Welch W. J. Role of the major heat shock proteins as molecular chaperones. Annu Rev Cell Biol. 1993;9:601–634. doi: 10.1146/annurev.cb.09.110193.003125. [DOI] [PubMed] [Google Scholar]
- Hartl F. U., Hlodan R., Langer T. Molecular chaperones in protein folding: the art of avoiding sticky situations. Trends Biochem Sci. 1994 Jan;19(1):20–25. doi: 10.1016/0968-0004(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Hendrick J. P., Hartl F. U. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- Kolb V. A., Makeyev E. V., Spirin A. S. Folding of firefly luciferase during translation in a cell-free system. EMBO J. 1994 Aug 1;13(15):3631–3637. doi: 10.1002/j.1460-2075.1994.tb06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar A. A., Kommer A., Krasheninnikov I. A., Spirin A. S. Cotranslational heme binding to nascent globin chains. FEBS Lett. 1993 Jul 12;326(1-3):261–263. doi: 10.1016/0014-5793(93)81803-8. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee P. W., Hayes E. C., Joklik W. K. Protein sigma 1 is the reovirus cell attachment protein. Virology. 1981 Jan 15;108(1):156–163. doi: 10.1016/0042-6822(81)90535-3. [DOI] [PubMed] [Google Scholar]
- Leone G., Duncan R., Lee P. W. Trimerization of the reovirus cell attachment protein (sigma 1) induces conformational changes in sigma 1 necessary for its cell-binding function. Virology. 1991 Oct;184(2):758–761. doi: 10.1016/0042-6822(91)90447-j. [DOI] [PubMed] [Google Scholar]
- Leone G., Duncan R., Mah D. C., Price A., Cashdollar L. W., Lee P. W. The N-terminal heptad repeat region of reovirus cell attachment protein sigma 1 is responsible for sigma 1 oligomer stability and possesses intrinsic oligomerization function. Virology. 1991 May;182(1):336–345. doi: 10.1016/0042-6822(91)90677-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G., Mah D. C., Lee P. W. The incorporation of reovirus cell attachment protein sigma 1 into virions requires the N-terminal hydrophobic tail and the adjacent heptad repeat region. Virology. 1991 May;182(1):346–350. doi: 10.1016/0042-6822(91)90678-5. [DOI] [PubMed] [Google Scholar]
- Leone G., Maybaum L., Lee P. W. The reovirus cell attachment protein possesses two independently active trimerization domains: basis of dominant negative effects. Cell. 1992 Oct 30;71(3):479–488. doi: 10.1016/0092-8674(92)90516-f. [DOI] [PubMed] [Google Scholar]
- Mah D. C., Leone G., Jankowski J. M., Lee P. W. The N-terminal quarter of reovirus cell attachment protein sigma 1 possesses intrinsic virion-anchoring function. Virology. 1990 Nov;179(1):95–103. doi: 10.1016/0042-6822(90)90278-y. [DOI] [PubMed] [Google Scholar]
- Nagata L., Masri S. A., Pon R. T., Lee P. W. Analysis of functional domains on reovirus cell attachment protein sigma 1 using cloned S1 gene deletion mutants. Virology. 1987 Sep;160(1):162–168. doi: 10.1016/0042-6822(87)90056-0. [DOI] [PubMed] [Google Scholar]
- Strong J. E., Leone G., Duncan R., Sharma R. K., Lee P. W. Biochemical and biophysical characterization of the reovirus cell attachment protein sigma 1: evidence that it is a homotrimer. Virology. 1991 Sep;184(1):23–32. doi: 10.1016/0042-6822(91)90818-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. L., Duncan R., Lee P. W. Site-directed mutagenesis of the C-terminal portion of reovirus protein sigma 1: evidence for a conformation-dependent receptor binding domain. Virology. 1992 Jan;186(1):219–227. doi: 10.1016/0042-6822(92)90076-2. [DOI] [PubMed] [Google Scholar]
- Yeung M. C., Lim D., Duncan R., Shahrabadi M. S., Cashdollar L. W., Lee P. W. The cell attachment proteins of type 1 and type 3 reovirus are differentially susceptible to trypsin and chymotrypsin. Virology. 1989 May;170(1):62–70. doi: 10.1016/0042-6822(89)90352-8. [DOI] [PubMed] [Google Scholar]









