Abstract
AIM: To construct a random peptide phage display library and search for peptides that specifically bind to the PreS region of hepatitis B virus (HBV).
METHODS: A phage display vector, pFuse8, based on the gene 8 product (pVIII) of M13 phage was made and used to construct a random peptide library. E.coli derived thioredoxin-PreS was purified with Thio-bond beads, and exploited as the bait protein for library screening. Five rounds of bio-panning were performed. The PreS-binding specificities of enriched phages were characterized with phage ELISA assay.
RESULTS: A phage display vector was successfully constructed as demonstrated to present a pVIII fused HBV PreS1 epitope on the phage surface with a high efficiency. A cysteine confined random peptide library was constructed containing independent clones exceeding 5±108 clone forming unit (CFU). A pool of phages showing a PreS-binding specificity was obtained after the screening against thio-PreS with an enrichment of approximately 400 times. Five phages with high PreS-binding specificities were selected and characterized. Sequences of the peptides displayed on these phages were determined.
CONCLUSION: A phage library has been constructed, with random peptides displaying as pVIII-fusion proteins. Specific PreS-binding peptides have been obtained, which may be useful for developing antivirals against HBV infection.
Keywords: Hepatitis B virus, Phage, Peptide
INTRODUCTION
The attachment to hepatocytes by hepatitis B virus (HBV) during infection has long been proposed to be a potential target for antiviral intervention. It is thought that molecules specifically binding to HBV particles may interfere with viral attachment and hence reduce or block subsequent infection[1-3]. Ideally, these molecules should mimic the structural elements of a cell surface HBV receptor. Unfortunately, although a few host proteins have been demonstrated to interact with HBV particles or viral surface antigens, such a cell surface HBV receptor remains elusive[4-8]. Consequently, our knowledge on the molecular events leading to HBV attachment to hepatocytes is very limited. One longstanding notion is the role of HBV PreS region in mediating HBV attachment to the putative receptor on hepatocytes[8-11]. Specifically, the aa 21-47 segment within the PreS1 domain is believed to be essential for this process[11]. It is noteworthy that other important functions of the PreS region have also been reported, for example, in the assembly and budding of HBV virion[12-14]. Therefore, the PreS region has become a potential target for screening antivirals against HBV infection.
Strategies for ligands discovery are usually based on procedures for assembling a large number of compounds to produce a diverse array of molecules, followed by a screening process against targets of interest. Among various technologies for ligand discovery, phage display has evolved into one of the main approaches for obtaining lead molecules[15,16]. Filamentous phage M13 has a rather simple architecture composed of five different structure proteins. The product of gene 8 (pVIII) is a major capsid protein. There are approximately 2 700 pVIII molecules all over a phage particle, while other structural proteins are located at the tail of the phage particle with only a few copies of each[17]. Thus, by fusing with pVIII, hundreds or thousands of ligands can theoretically be displayed on the surface of the phage particle, which should help strengthen its interaction with a target[15,16]. Thus, pVIII fusion protein based phage display will offer a greater chance to find mildly binding ligands.
In this study, we have constructed a structurally constrained phage display library in which random peptides are displayed as N-terminal fusions to pVIII and confined in a loop by a pair of flanking cysteine residues[15,18,19]. Recombinant thio-PreS protein was used as target in screening for the PreS-binding peptides. A pool of phages showing PreS-binding specificity was obtained with an enrichment factor of approximately 400. Phages with high PreS-binding specificities were selected and characterized. Sequences of the peptides displayed on five of PreS-binding phages were determined.
MATERIALS AND METHODS
Materials
The pCANTAB5E vector, T7 DNA sequencing kit, and HRP-conjugated M13 specific monoclonal antibody (anti-M13 mAb) are products of Amersham Pharmacia Biotech (Piscataway, USA). XL1-blue F’ E.coli strain and helper phage VCSM13 were purchased from Stratagene (La Jolla, USA). PreS1 specific mAb 125E11 has been previously described[20,21]. Maxisorp F16 module (Nunc, Denmark) was used for protein coating. ThiobondTM affinity resin and the thioredoxin specific mAb (anti-thio) were from Invitrogen (Carlsbad, USA). Recombinant enterokinase (rEK) was purchased from Novagen (Madison, USA).
Construction of random peptide phage display library
pCANTAB5E was utilized as the parental vector for constructing a pVIII fusion protein display vector. The coding sequence for M13 pVIII was amplified with specific primers (pVIII-forward: 5’-TAAGGATCCGGTGGTGCT-GAGGGTGACGATCCCGC-3’, pVIII-reverse: 5’-A GGAATTCATGTACCGTAACACTG-3’) using the VCSM13 genome as template. PCR product was digested with BamHI and EcoRI, and inserted into the same sites in pCANTAB5E, replacing the original gene 3. The resulted vector was named pFuse8 (Figure 1A). To construct pFuse8-E that encodes a PreS1 epitope fused with pVIII (Figure 1A), annealed oligonucleotides (sense: 5’-CAGCAATGGCA-GGC AGCCTGAACCCTAGCTGGGATGGT GGCGGAGGATCCG-3’, antisense: 5’-AATTCGGATC-CTCCGCCACC ATCCCAGCTAGGGTTCAGGCT GCCTGCCATTGCTGGCT-3’, the epitope coding sequence is underlined) were ligated to pFuse8 digested with SfiI and BamHI.
Figure 1.
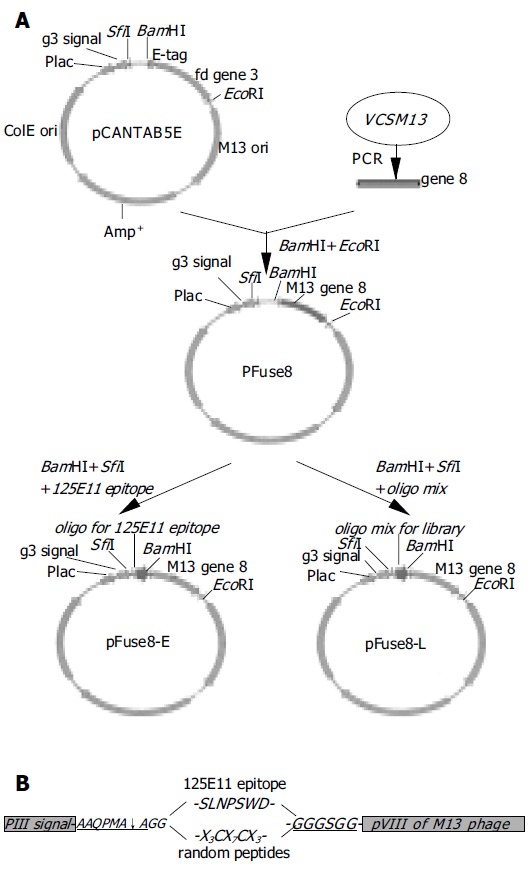
Diagram of library construction. A: a schematic representation of the steps in the construction of the phagemid encoding the pVIII fusion protein. pCANTAB5E, a pIII displaying vector; pFuse8, the vector harboring gene 8 coding sequence in replace of gene 3; pFuse8-E, the coding sequence for a specific epitope recognized by the 125E11 mAb is inserted into pFuse8; pFuse8-L, the coding sequences for random peptides are inserted into pFuse8; B: Inserted sequences in pFuse8-E and pFuse8-L after translation. SLNPSWD is the specific epitope recognized by the 125E11 mAb. In pFuse8-L, the epitope is replaced with X3CX7CX3.
For the construction of peptide library, antisense oligonu-cleotides containing random nucleotides(5’-ATTGC-GGATCCACACC(MNN)3GCA(MNN)7ACA(MNN) 3GCCTGCTGCCATTGCTGC-3’, N = A, T, C or G; M = A or C) was synthesized (Invitrogen). A complementary primer (5’-CAGCAA TGGCAGCAGGC-3’) was annealed to the 3’ end of the long oligonucleotides, followed by the fill-in reaction with the Klenow polymerase to generate the double-stranded DNA. The DNA duplex was then digested with BamHI and ligated into pFuse8 cut with SfiI and BamHI. The ligation mixture was used to transform XL1-blue F’ competent cells by electroporation. The phage library was established by co-infection of helper phage VCSM13 (MOI, 10:1) to the transformed cells as previously described[22]. Complexity of the library was estimated according to the number of independent transformed clones.
DNA sequencing
Phagemid pool of randomly picked-up clones from the library was prepared with SV minipreps kit (Promega). DNA sequencing was performed with T7 DNA sequencing kit according to the manufacturer’s instruction.
Preparation of PreS fusion protein
Prokaryotic expression plasmid pThioHisA-PreS was constructed for the synthesis of thioredoxin fused PreS (thio-PreS) in E.coli[23]. An affinity purification for the expression product was performed with ThiobondTM beads according to the manufacturer’s instruction.
Library screening
Totally 1011 phages were mixed with 30 µL of the thio-PreS bound ThiobondTM beads and incubated on a rotating wheel for 10 min. After washing with PBS containing 0.05% Tween 20 for four times, the beads were treated with 1 unit of rEK at 30°C for 6 h. Phages binding specifically to the PreS region were supposed to detach off the beads. Eluted phages in the collected supernatant were subject to propagation in XL1-blue F’ cells, followed by VSCM13 co-infection to produce a phage pool for next round of panning[22]. A total of five rounds of selection were performed.
Western blot
Western blot of thio-PreS was performed according to a standard method[24]. Thio-PreS coupled on resins with or without rEK treatment were boiled and separated on SDS-PAGE, and transferred to nitrocellulose filters. Anti-thio mAb (1:1000) was used as the primary antibody for the detection. Blots were developed using the ECL method with HRP-labeled rabbit anti-mouse Ig (1:2000, DaKo, USA).
Phage ELISA
Phage ELISA was carried out as described previously[22]. Purified thio-PreS proteins (1 µg/well) were immobilized on the microwells in PBS buffer overnight. Phages propagated from infected XL1-blue F’ cells were precipitated by 0.1 volume of PEG8000/NaCl (20% PEG8000, 3 mol/L NaCl), resuspended in PBS buffer and applied to thio-PreS coated wells, followed by an incubation at room temperature for 1 hour. An HRP-labeled anti-M13 mAb (1:1 000) was used for the detection of bound phages.
Virus capture assay
For capturing HBV virions, each microwell was coated with 1011 phages in carbonate buffer (pH 9.5) at 4°C overnight. After blocking with 1% BSA at 37°C for 2 h, cultured medium of HepG2.2.15[25] that contains HBV particles was added and incubated for 1 h at 37°C, followed by a washing process with PBS buffer containing 0.05% Tween 20. An HRP-conjugated anti-HBs antibody was used to detect the captured HBV particles according to the manufacturer’s protocol (Sino-American Biotech, Shanghai).
RESULTS
Construction of pVIII based phage display vector
To construct the vector pFuse8, M13 gene 8 was amplified using the genomic DNA of VCSM13 phage as template. The PCR product was inserted into pCANTAB5E to replace the coding sequence of gene 3 and the E-tag (Figure 1A). The inserted gene 8 is located downstream to a leader signal sequence of gene 3, which is used to direct the pVIII fusion protein to the surface of the phage particle. In the phagemid pFuse8-E as shown in Figure 1B, a spacer between the leader signal and gene 8 encodes an in-frame peptide AAQPMAAG-SLNPSWD-GGGSGG. The first part of the peptide (AAQPMAAG) is a recognition sequence of the pelB signal peptidase[26]. The 7-amino acids in the middle of this peptide represents an epitope within the HBV PreS1 domain that is recognized by the 125E11 mAb (unpublished data), providing a molecular marker for detection of phage display. The ensuing glycine tether is used to link the peptide to pVIII, increasing the flexibility of the peptide.
Phages were generated by co-infection of the pFuse8-E transformed XL1-blue F’ cells with helper phage VCSM13. The displayed 125E11 epitope was readily detected by phage ELISA in a dose dependent manner with the immobilized 125E11 mAb but not with an irrelevant mAb (anti-HBs) (Figure 2). As shown in Figure 2, the lac promoter employed in the vector was somewhat leaky so that pVIII-fused epitope could be easily detected even without IPTG induction[27].
Figure 2.
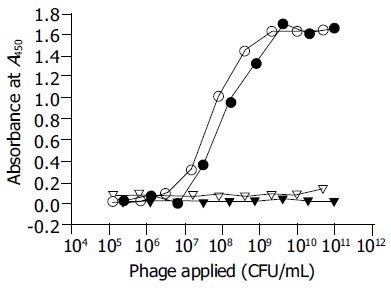
Surface display of the epitope recognized by 125E11. The SLNPSWD epitope displayed on the surface of the phage particle was detected by phage ELISA. The circles indicate the phages that bind 125E11 coated wells. Triangles represent phages captured by an irrelevant mAb (anti-HBs). The synthesis of pVIII fusion proteins with or without IPTG induction is denoted as white or black, respectively.
Construction of structure-confined random peptide phage display library
Structure-confined random peptide library has the advantage to present peptides in a fixed loop so that their conformations are relatively rigid[18,19]. In this study, random peptides fused with pVIII assume a form of X3CX7CX3, where X stands for any amino acids. Two cysteine residues frank the 7 random amino acids in the middle. Under a non-reducing environment in the periplasm of E.coli, these cysteine residues may spontaneously form a disulfide bond that constrains the conformation of the displayed peptide[18,19]. The oligonucleotides mixture coding for the random peptides was inserted into the pFuse8 vector (pFuse8-L, Figure 1A). The resulting library had the size of about 5.0 × 108 independent clones that represented a great complexity. The random property of the library was verified by DNA sequence analysis. As shown in Figure 3, in the region coding for the random peptides, all four kinds of nucleotides are equally distributed at the first two positions of the triplet codons, while T or G varies at the third position, indicating a sufficient diversity of the library.
Figure 3.

Diversity of the constructed library. In the region coding for the random peptides, four kinds of nucleotides are equally distributed at the first two loci of the triplet codons, while T or G varies at the last. Oligonucleotides for cysteine confined random peptides (5’-GCAGCAGGC-(NNS)3-TGT(NNS)7-TGC(NNS) 3-GGTGGTGGA-3’, N = A, T, C or G; S = A or C) are shown.
Screening for PreS-binding peptides
The PreS region of HBV, when synthesized alone in E.coli, is vulnerable to degradation and largely insoluble. To overcome these problems, the PreS region was fused with the C-terminus of thioredoxin (Figure 4A), which has dramatically enhanced both the stability and the solubility of the PreS region (Figure 4B). An enterokinase site located at the junction between the PreS region and the thioredoxin tag (Figure 4A) facilitates the release of the PreS region by digestion with rEK, leaving the thioredoxin tag on the affinity matrix (Figure 4C). Thus, the PreS-binding phages could be dissociated from the resin and enriched. It was expected that cleavage by rEK might decrease a nonspecific enrichment of phages binding to the thioredoxin tag. It is noteworthy that the degradation of the free PreS region should not influence the recovery of the PreS-binding phages.
Figure 4.
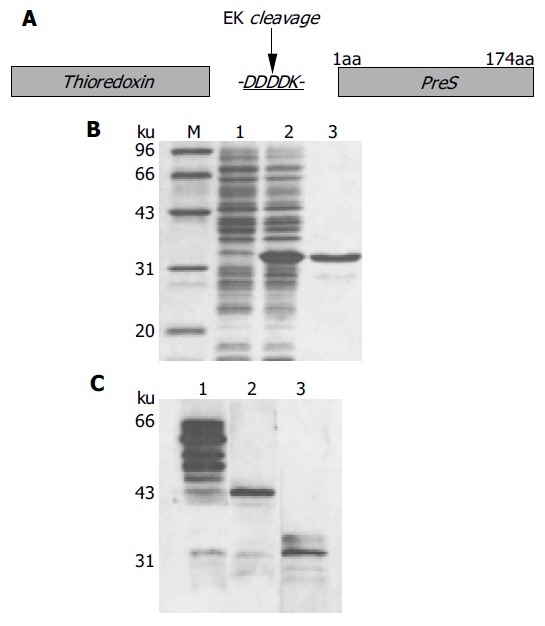
Synthesis of thio-PreS. A: a schematic representation of the thio-PreS fusion protein, DDDDK is a cleavage site of rEK; B: SDS-PAGE analysis. Lane 1, the E.coli lysate without IPTG induction; lane 2, the soluble lysate after IPTG induction, thio-PreS of 33 ku is indicated with a triangle; lane 3, the purified thio-PreS; C: Western blot with the anti-thio mAb. Lane 1, thio-PreS coupled to the ThiobondTM beads; lane 2, thio-PreS coupled beads treated in the absence of rEK; lane 3, thio-PreS coupled beads treated in the presence of rEK.
A total of five rounds of screening were performed. As shown in Table 1, the PreS-binding phages were greatly enriched as evidenced by a steadily rising enrichment factor (EF, phage eluted/phage applied). An approximately 400-fold of enrichment (EF5th/EF1st) was achieved as estimated by the titer of the phages after the screening. The pool of phages from the final round of selection bound to the thio-PreS immobilized wells specifically in a dose-dependent manner, in sharp contrast to the original pool of phages before selection (Figure 5). A much weaker noise was noticed with the thioredoxin immobilized wells serving as a control, indicating that the thioredoxin-binding phages were also selected, though they might only be a small minority.
Table 1.
Enrichment of phages binding to thio-PreS coupled beads
| Round of panning |
Thio-PreS coupled beads |
Thioredoxin coupled beads |
||||
| Phage applied | Phage eluted | EF1 | Phage applied | Phage eluted | EF | |
| 1st | 2.0 ± 1011 | 2.6 ± 104 | 1.3 ± 10-7 | 2.0 ± 1011 | 3.4 ± 104 | 1.7 ± 10-7 |
| 2nd | 1.0 ± 1011 | 1.2 ± 104 | 1.2 ± 10-7 | 1.0 ± 1011 | 9.5 ± 103 | 9.5 ± 10-8 |
| 3rd | 1.0 ± 1011 | 9.3 ± 104 | 9.3 ± 10-7 | 1.0 ± 1011 | 3.9 ± 104 | 1.2 ± 10-7 |
| 4th | 5.0 ± 1010 | 3.5 ± 105 | 7.0 ± 10-6 | 5.0 ± 1010 | 3.6 ± 104 | 7.2 ± 10-7 |
| 5th | 5.0 ± 1010 | 2.6 ± 106 | 5.1 ± 10-5 | 5.0 ± 1010 | 8.8 ± 104 | 1.8 ± 10-6 |
Enrichment factor (EF) = phage eluted/phage applied.
Figure 5.
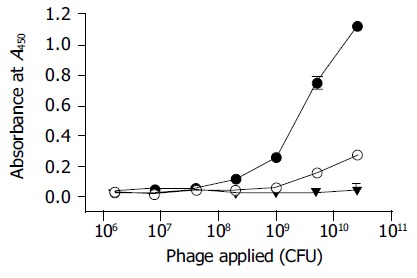
Phage ELISA of the enriched phages after the final round of screening. Black dots, the enriched phages bind to thio-PreS; Empty dots, the enriched phages bind to thioredoxin as a control; Black triangles, phages in the original library do not bind to thio-PreS.
Characterization of the PreS-binding phages
The specificity of the phages with regard to PreS-binding was further characterized by virus capture assay. When coated on microplate wells, the pool of phages from each round of selection was tested for their abilities to capture HBV virions from the cultured medium of HepG2.2.15. The binding capacity to HBV virions increased greatly after the final round of selection (Figure 6), suggesting that the PreS-binding phages were truly selected. Phages from the final round of selection were picked for a further analysis. The specificities of these phages with regard to PreS-binding were analyzed with phage ELISA assay. Phages with a strong binding capacity to thioredoxin were considered nonspecific and discarded (data not shown). The corresponding phagemids of five of the PreS-binding phages were subjected to DNA sequencing. Amino acid sequences of the potential PreS-binding peptides were deduced (Figure 7).
Figure 6.
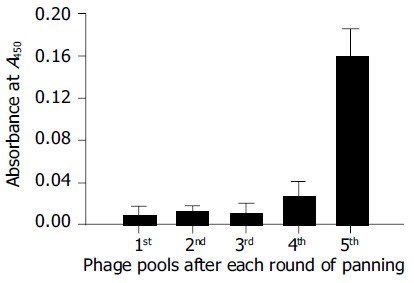
Virus capture assay. The phage pool (1011 CFU) after each round of screening shows an increasing binding capacity to HBV virions in the culture medium of HepG2.2.15.
Figure 7.

Amino acid sequences of PreS specific oligopeptides. The amino acid sequences of the peptides encoded by the phagemids derived from five of the PreS-binding phages were deduced from DNA sequencing analysis. The C in bold indicates the invariable residues of cysteines.
DISCUSSION
The filamentous phage display system, whereby the expressed peptides are displayed as fusions to phage coat proteins, has been effective in the discovery of ligands[15,16]. Commonly exploited strategy is the surface display of N-terminus fusions to the minor coat protein pIII (product of phage gene 3). It represent a low-valent surface display, as only three to five copies of pIII are present at the tail of the phage particle, providing a selection platform for an interaction with a high affinity[15]. In our work, a pVIII fusion protein based vector was successfully constructed, and proved to work well in peptide displaying. Since pVIII is a major coat protein with thousands of molecules on a single phage, thus pVIII fusion protein based phage display will offer a greater chance to find mildly or weakly binding ligands. As PreS-binding peptides are concerned, the pVIII fusion protein based system may be a better choice than the pIII fusion protein based system. It is supposed that the affinity of the HBV virion to its putative receptor on hepatocytes may not be strong, because an ensuing intracellular event of dissociation may occur closely after the penetration of the viral particle during the infection.
Linear oligopeptides likely present various solution conformations that may be quite different from the structure in the native or bound forms of a protein. It has been demonstrated that the structurally active region in native protein may be mimicked by cyclic amino acid sequences of constrained peptides displayed on the phage surface[15,18,19]. A pair of flanked cysteine residues was adopted to confine the random peptides in the middle by presumably forming an intra-molecular disulfide bond that provides a more defined scaffold when folded in the periplasmic environment of E.coli. In this study, we have constructed a conformation constrained random peptide phage display library and performed the screening with this library, in the hope that potential peptides with more defined conformations may better fulfill the structural features required for PreS-binding. On the other hand, such a conformation-constrained library has its weaknesses. It is likely that during the interaction process between PreS and a potential peptide, both partners may exert their influences on each other so that the conformation of the potential peptide adjusts to better bind PreS. In such a situation, the peptide with a constrained conformation may be too rigid to accommodate itself upon binding PreS, which will probably result in a reduced binding affinity.
During the life cycle of HBV, the PreS region of the large surface antigen (LHBs) plays a crucial role in virion assembly and budding as well as in viral attachment to hepatocytes[8-12,28,13]. Given the pivotal role of the PreS region in HBV life cycle, the specific PreS-binding ligands are expected to be useful as inhibitors to block viral entry to hepatocytes, or to interfere with the process of virion assembly in HBV infected cells. Similar ideas have been pursued widely in the field of HIV research and proved to be a potential way for antiviral drug development[29,30]. Moreover, studies on the PreS-binding peptides will likely provide helpful information about the interaction between HBV virion and hepatocytes that may aid the identification of a yet-to-be-found receptor of HBV.
The binding capacity of the pool of phages to HBV virions increased greatly after the final round of selection. Thus, the thio-PreS fusion protein was proved to be an 10 effective bait protein that might mimic the conformation adopted by the PreS region in the native virions. In addition, the affinity screening for the PreS-binding phages represents a promising approach for discovering HBV binding ligands. Several peptides encoded by the phagemids of five of the PreS-binding phages were determined. No consensus sequence or conserved amino acids were found in these five peptides. Therefore, these peptides likely bind to different fragments of the PreS region. On the other hand, sequences of more PreS-binding peptides need to be determined and the binding properties of the PreS-binding peptides await further investigations, which are currently on the way.
ACKNOWLEDGMENTS
125E11 is a kind gift from Professor Zhu-Chuan Zhang.
Footnotes
Supported by Basic Research Program from Ministry of Science and Technology of China, No. G1999054105, and special funds for Sino-France Center for Life Science and Genome Research from Chinese Academy of Sciences and Pasteur Institute in France
Co-correspondent: You-Hua Xie
Science Editor Zhu LH Language Editor Elsevier HK
References
- 1.Urban S, Gripon P. Inhibition of duck hepatitis B virus infection by a myristoylated pre-S peptide of the large viral surface protein. J Virol. 2002;76:1986–1990. doi: 10.1128/JVI.76.4.1986-1990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hong HJ, Ryu CJ, Hur H, Kim S, Oh HK, Oh MS, Park SY. In vivo neutralization of hepatitis B virus infection by an anti-preS1 humanized antibody in chimpanzees. Virology. 2004;318:134–141. doi: 10.1016/j.virol.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Park SS, Ryu CJ, Gripon P, Guguen-Guillouzo C, Hong HJ. Generation and characterization of a humanized antibody with specificity for preS2 surface antigen of hepatitis B virus. Hybridoma. 1996;15:435–441. doi: 10.1089/hyb.1996.15.435. [DOI] [PubMed] [Google Scholar]
- 4.Stefas I, Rucheton M, D'Angeac AD, Morel-Baccard C, Seigneurin JM, Zarski JP, Martin M, Cerutti M, Bossy JP, Missé D, et al. Hepatitis B virus Dane particles bind to human plasma apolipoprotein H. Hepatology. 2001;33:207–217. doi: 10.1053/jhep.2001.20531. [DOI] [PubMed] [Google Scholar]
- 5.Ryu CJ, Cho DY, Gripon P, Kim HS, Guguen-Guillouzo C, Hong HJ. An 80-kilodalton protein that binds to the pre-S1 domain of hepatitis B virus. J Virol. 2000;74:110–116. doi: 10.1128/jvi.74.1.110-116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Falco S, Ruvoletto MG, Verdoliva A, Ruvo M, Raucci A, Marino M, Senatore S, Cassani G, Alberti A, Pontisso P, et al. Cloning and expression of a novel hepatitis B virus-binding protein from HepG2 cells. J Biol Chem. 2001;276:36613–36623. doi: 10.1074/jbc.M102377200. [DOI] [PubMed] [Google Scholar]
- 7.Gong ZJ, De Meyer S, van Pelt J, Hertogs K, Depla E, Soumillion A, Fevery J, Yap SH. Transfection of a rat hepatoma cell line with a construct expressing human liver annexin V confers susceptibility to hepatitis B virus infection. Hepatology. 1999;29:576–584. doi: 10.1002/hep.510290238. [DOI] [PubMed] [Google Scholar]
- 8.De Meyer S, Gong ZJ, Suwandhi W, van Pelt J, Soumillion A, Yap SH. Organ and species specificity of hepatitis B virus (HBV) infection: a review of literature with a special reference to preferential attachment of HBV to human hepatocytes. J Viral Hepat. 1997;4:145–153. doi: 10.1046/j.1365-2893.1997.00126.x. [DOI] [PubMed] [Google Scholar]
- 9.Le Seyec J, Chouteau P, Cannie I, Guguen-Guillouzo C, Gripon P. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J Virol. 1999;73:2052–2057. doi: 10.1128/jvi.73.3.2052-2057.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pontisso P, Alberti A. The role of preS1 in the interaction of hepatitis B virus with human hepatocytes. Hepatology. 1991;14:405–406. [PubMed] [Google Scholar]
- 11.Neurath AR, Kent SB, Strick N, Parker K. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell. 1986;46:429–436. doi: 10.1016/0092-8674(86)90663-x. [DOI] [PubMed] [Google Scholar]
- 12.Bruss V. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J Virol. 1997;71:9350–9357. doi: 10.1128/jvi.71.12.9350-9357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ponsel D, Bruss V. Mapping of amino acid side chains on the surface of hepatitis B virus capsids required for envelopment and virion formation. J Virol. 2003;77:416–422. doi: 10.1128/JVI.77.1.416-422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poisson F, Severac A, Hourioux C, Goudeau A, Roingeard P. Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology. 1997;228:115–120. doi: 10.1006/viro.1996.8367. [DOI] [PubMed] [Google Scholar]
- 15.Burritt JB, Bond CW, Doss KW, Jesaitis AJ. Filamentous phage display of oligopeptide libraries. Anal Biochem. 1996;238:1–13. doi: 10.1006/abio.1996.0241. [DOI] [PubMed] [Google Scholar]
- 16.Sidhu SS. Phage display in pharmaceutical biotechnology. Curr Opin Biotechnol. 2000;11:610–616. doi: 10.1016/s0958-1669(00)00152-x. [DOI] [PubMed] [Google Scholar]
- 17.Cabilly S. The basic structure of filamentous phage and its use in the display of combinatorial peptide libraries. Mol Biotechnol. 1999;12:143–148. doi: 10.1385/MB:12:2:143. [DOI] [PubMed] [Google Scholar]
- 18.Hoess RH, Mack AJ, Walton H, Reilly TM. Identification of a structural epitope by using a peptide library displayed on filamentous bacteriophage. J Immunol. 1994;153:724–729. [PubMed] [Google Scholar]
- 19.Luzzago A, Felici F, Tramontano A, Pessi A, Cortese R. Mimicking of discontinuous epitopes by phage-displayed peptides, I. Epitope mapping of human H ferritin using a phage library of constrained peptides. Gene. 1993;128:51–57. doi: 10.1016/0378-1119(93)90152-s. [DOI] [PubMed] [Google Scholar]
- 20.Yang HL, Jin Y, Cao HT, Xu X, Li GD, Wang Y, Zhang ZC. Affinity Purification of Hepatitis B Virus Surface Antigen Containing PreS1 Region. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 1996;28:412–417. [PubMed] [Google Scholar]
- 21.Hui J, Li G, Kong Y, Wang Y. Expression and characterization of chimeric hepatitis B surface antigen particles carrying preS epitopes. J Biotechnol. 1999;72:49–59. doi: 10.1016/s0168-1656(99)00049-8. [DOI] [PubMed] [Google Scholar]
- 22.Zhang XX, Deng Q, Zhang SY, Liu J, Cai Q, Lu ZM, Wang Y. Broadly cross-reactive mimotope of hypervariable region 1 of hepatitis C virus derived from DNA shuffling and screened by phage display library. J Med Virol. 2003;71:511–517. doi: 10.1002/jmv.10521. [DOI] [PubMed] [Google Scholar]
- 23.Deng Q, Kong YY, Xie YH, Wang Y. Expression and purification of the complete PreS region of hepatitis B Virus. World J Gastroenterol. 2005;11:3060–3064. doi: 10.3748/wjg.v11.i20.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: A laboratory manual.2nd ed. Cold Spring Harbor Laboratory Press. [Google Scholar]
- 25.Sells MA, Zelent AZ, Shvartsman M, Acs G. Replicative intermediates of hepatitis B virus in HepG2 cells that produce infectious virions. J Virol. 1988;62:2836–2844. doi: 10.1128/jvi.62.8.2836-2844.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eerola R, Saviranta P, Lilja H, Pettersson K, Lövgren T, Karp M. Expression of prostate specific antigen on the surface of a filamentous phage. Biochem Biophys Res Commun. 1994;200:1346–1352. doi: 10.1006/bbrc.1994.1599. [DOI] [PubMed] [Google Scholar]
- 27.Crameri R, Jaussi R, Menz G, Blaser K. Display of expression products of cDNA libraries on phage surfaces. A versatile screening system for selective isolation of genes by specific gene-product/ligand interaction. Eur J Biochem. 1994;226:53–58. doi: 10.1111/j.1432-1033.1994.tb20025.x. [DOI] [PubMed] [Google Scholar]
- 28.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Gaubin M, Briant L, Srikantan V, Murali R, Saragovi U, Weiner D, Devaux C, Autiero M, Piatier-Tonneau D, et al. Synthetic CD4 exocyclics inhibit binding of human immunodeficiency virus type 1 envelope to CD4 and virus replication in T lymphocytes. Nat Biotechnol. 1997;15:150–154. doi: 10.1038/nbt0297-150. [DOI] [PubMed] [Google Scholar]
- 30.Choi YH, Rho WS, Kim ND, Park SJ, Shin DH, Kim JW, Im SH, Won HS, Lee CW, Chae CB, et al. Short peptides with induced beta-turn inhibit the interaction between HIV-1 gp120 and CD4. J Med Chem. 2001;44:1356–1363. doi: 10.1021/jm000403+. [DOI] [PubMed] [Google Scholar]


